Abstract
Rad51 is a conserved protein essential for recombinational repair of double-stranded DNA breaks (DSBs) in somatic cells and during meiosis in germ cells. Yeast Rad51 mutants are viable but show meiosis defects. In the mouse, RAD51 deletions cause early embryonic death, suggesting that in higher eukaryotes Rad51 is required for viability. Here we report the identification of SpnA as the Drosophila Rad51 gene, whose sequence among the five known Drosophila Rad51-like genes is most closely related to the Rad51 homologs of human and yeast. DmRad51/spnA null mutants are viable but oogenesis is disrupted by the activation of a meiotic recombination checkpoint. We show that the meiotic phenotypes result from an inability to effectively repair DSBs. Our study further demonstrates that in Drosophila the Rad51-dependent homologous recombination pathway is not essential for DNA repair in the soma, unless exposed to DNA damaging agents. We therefore propose that under normal conditions a second, Rad51-independent, repair pathway prevents the lethal effects of DNA damage.
Keywords: Drosophila/DSB repair/meiosis/Rad51/spindle-A
Introduction
Chromosomal integrity is essential for proper embryonic and postembryonic development, prolonged survival and successful reproduction. Highly conserved repair mechanisms exist in all organisms, from bacteria to mammals, to recognize and repair DNA damage (Sancar, 1996; Wood, 1996; Sekelsky et al., 2000; Wood et al., 2001). The repair of double-stranded DNA breaks (DSBs) is a necessary mechanism for recombining parental genomes during meiosis and is used as a defense mechanism after DNA damage caused by irradiation or chemical agents. The presence of DNA damage activates a cell cycle checkpoint (Melo and Toczyski, 2002). This allows time for the cell to correct the damage so as not to propagate the defect or affect normal cellular functions. In Saccharomyces cerevisiae mutations in the same genes show increased sensitivity to ionizing radiation and meiotic phenotypes, such as chromosome non-disjunction and/or rearrangements (Symington, 2002). This suggested a functional relationship between the mechanisms of mitotic DNA repair and meiotic recombination.
Genetic studies in S.cerevisiae led to the discovery of the Rad52 epistasis group of DSB repair genes (Symington, 2002). A core protein in this pathway is Rad51, which is related to the bacterial RecA protein. Rad51 has DNA-dependent ATPase activity and catalyzes strand exchange between homologous DNA molecules. Rad51 and Rad51-related proteins are found from yeast to humans (Shinohara and Ogawa, 1999; Thacker, 1999). In yeast, the Rad51 null mutant is viable but shows sporulation defects (Shinohara et al., 1992). The mouse Rad51 knockout is embryonic lethal (Lim and Hasty, 1996; Tsuzuki et al., 1996), thus the role of Rad51 in mouse meiosis could not be studied. In both mouse and yeast, a meiosis-specific Rad51-related gene, Dmc1, has been identified and shown to be required for chromosome synapsis and strand exchange during prophase of meiosis I (Bishop et al., 1992; Pittman et al., 1998).
In contrast to yeast, Drosophila members of the Rad52 epistasis group were not identified on the basis of meiosis defects or mutagen sensitivity. Rather, mutations in the Drosophila RAD51-related gene, spindle-B (spnB), and the Rad54 homolog, okra, were discovered as maternal-effect mutants with altered patterning of the eggshell, the so-called spindle phenotype (Morris and Lehmann, 1999). It was shown that this phenotype, observed in spnB and okra mutants, was due to reduction in the levels of the morphogen Gurken, a TGFα-like protein that controls both dorso–ventral patterning of the egg and antero–posterior polarity of the embryo (Ghabrial et al., 1998). Schüpbach and colleagues suggested that the activation of a meiotic checkpoint, which resulted in defective Gurken translation, was the result of a failure to repair DNA breaks in mutants for okra, spnB and spindle-D (spnD), another Rad51-related protein (Ghabrial and Schüpbach, 1999; Abdu et al., 2003). Accordingly, the spindle phenotype was suppressed by mutants for the Spo11 homolog, mei-W68, which are defective in double-stranded break formation and thus are unable to activate the checkpoint (Ghabrial and Schüpbach, 1999). Spn mutants were also suppressed in combination with mutants of known transducers of cell cycle checkpoints, such as Drosophila mei-41, an ATR/ATM phosphatidylinositol 3-kinase-like protein, and the Drosophila homolog of Chk2 kinase, chk2/mnk/loki (Ghabrial and Schüpbach, 1999; Abdu et al., 2002). A target for the meiotic checkpoint in Drosophila is the ATP-dependent helicase Vasa (Styhler et al., 1998; Tomancak et al., 1998), which is phosphorylated upon checkpoint activation and may regulate Gurken translation (Ghabrial and Schüpbach, 1999). Sequence analysis indicates that there are at least five Drosophila genes that show significant homology to yeast and human Rad51 (this report). It remained unclear whether these genes have distinct functions in DSB repair and whether the activation of the meiotic checkpoint was a consequence of the failure to repair DSBs. Furthermore, while mutations in spnB showed meiotic defects, they did not affect DNA repair in somatic cells (Ghabrial et al., 1998), raising the possibility that in Drosophila distinct sets of Rad51-like genes may control DSB repair either in the germline or in the soma.
Here we report the identification of spnA as the Drosophila Rad51 gene, whose sequence among the five known Drosophila Rad51-like genes is most closely related to the Rad51 homologs of human and yeast. We show that spnA mutants exhibit the spindle eggshell phenotype. In spnA oocytes synapse of homologous chromosomes is correctly initiated during meiosis but its resolution is delayed and unrepaired double-stranded breaks persist longer than in wild type causing the activation of a meiotic recombination checkpoint. spnA null mutants are viable but show sensitivity to irradiation, suggesting that SpnA acts in the soma but that other repair mechanisms compensate in the absence of SpnA. Analysis of the expression pattern of the five known Drosophila Rad51 homologs together with the analysis of the mutant phenotype of three of these genes suggest that the Drosophila Rad51 genes act in concert during oogenesis and that only a subset of them are used for repair in the soma.
Results
spindle-A encodes a Drosophila Rad51 ortholog
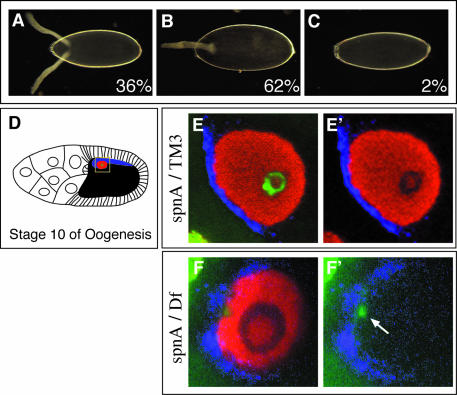
We identified 22 new alleles of spindle-A by mutagenesis screening (see Supplementary data available at The EMBO Journal Online). spnA was originally identified as a maternal-effect lethal mutation affecting egg patterning (Tearle and Nusslein-Volhard, 1987). All new spnA alleles are viable in trans to the original spnA alleles (spnA003, spnA050 and spnA057) and in trans to a deficiency for the region (see below), suggesting that they are not affecting a function essential for viability. Like the original spnA alleles, the new alleles show ∼100% maternal-effect embryonic lethality. All mutant lines produce a spectrum of eggshell ventralization phenotypes similar to those described for known mutations affecting the EGFR signaling pathway, ranging from fused dorsal appendages (Figure 1B) to complete ventralization (Figure 1C; Gonzalez-Reyes et al., 1997). For most spindle-class mutants, the eggshell phenotype has been attributed to a defect in RNA and protein localization or protein synthesis of the EGFR ligand Gurken (Grk) (Gonzalez-Reyes et al., 1997). In spnA mutants, the level and distribution of Grk protein is disrupted (Figure 1F and F′; Gonzalez-Reyes et al., 1997). In one example, instead of the normal crescent of Grk protein along the dorsal-anterior side of the wild-type oocyte nucleus facing the somatic follicle cells (Figure 1E and E′), Grk protein distribution is less coherent and only found in a few spots along the mutant oocyte nucleus. Another common feature shared among the spindle-class mutants is a disruption in oocyte nuclear morphology (Gonzalez-Reyes et al., 1997; Ghabrial et al., 1998). Mature wild-type oocytes contain highly compact chromatin called the karyosome (Figure 1E and E′). In spnA mutant oocytes, as well as in other spindle-class mutants, the DNA is less organized and diffuse. In contrast to the wild type, where DNA is found as a condensed sphere in the center of the nucleus, DNA in spnA mutant oocytes clusters along the periphery of the nucleus adjacent to the nuclear membrane (Figure 1F and F′).
Fig. 1. spnA mutant phenotypes. (A–C) Eggshell phenotype. (A) Wild-type with two anteriorly located dorsal appendages. (B) Mild ventralization with a single fused-dorsal appendage. (C) Completely ventralized. Percentages represent the phenotypic proportions observed per total eggs laid (n = 374) from spnA155–52 germline clones. (D–F′) Gurken expression and karyosome phenotype. (D) Diagram of a wild-type Stage 10 egg chamber. The oocyte nucleus is indicated in red, Gurken protein in blue, karyosome in green. (E and E′) A confocal section of a germinal vesicle from a spnA093A heterozygote Stage 10 egg chamber. Staining for the centrosomal protein, CP190 (red; Whitfield et al., 1995), shows the nucleoplasm. (F and F′) Germinal vesicle from a spnA093A/Df(3R)X3F Stage 10 egg chamber. (E′) is identical to (E) minus the green channel. (F′) is identical to (F) minus the red channel. The yellow box in (D) represents the area captured in (E) and (F). [(A–C) dark-field photographs using 20× objective, anterior to the left; (E and F) confocal images using a 40× objective and 4× zoom, dorsal view with anterior to the left].
We mapped spnA to the cytological region 99D01–99E01 (Supplementary figure 2). The Berkley Drosophila Genome Project predicted a Rad51-like gene (CG7948) to reside within this region of the genome. CG7948 shows strong sequence similarity to the yeast and mammalian Rad51 gene (see below). Since two other spindle-class genes, spnB and okra, encode members of the Drosophila Rad51 family and Rad54, respectively (Ghabrial et al., 1998), CG7948 was a likely candidate for spnA. Sequence analysis of spnA alleles revealed unique missense mutations within CG7948 (Table I). Thus, spindle-A encodes a Drosophila Rad51-like gene.
Table I. DmRAD51 allele characterization and comparison.
| SpnA alleles | Drosophila Rad51 | Saccharomyces cerevisiae Rad51a | Escherichia coli RecAa |
|---|---|---|---|
| 091B | L38F | L99 | |
| 050A | L57Q | L118 | |
| 093A | Q70Stop | K131 | |
| 148-15 | G112D | G173 | G54 |
| 104-7 | E115V | E176 | P57 |
| 050 | S118F | S179 | R60 |
| 087A | E125L | E186 | P67 |
| 009A | |||
| 032B | R127H | R188 | S69 |
| 119A | A138T | A199 | I80 |
| 003 | G149D | G210 | |
| 084A | G150S | G211 | R85 |
| 068A | E151L | E212 | E86 |
| 001A | R167C | R228 | Y103 K106 |
| 095A | R190C | R251 | G122 |
| 057 | A205V | A266 | A133 |
| 010A | V218M | V279 | V143 |
| 061A | D219N | D280 | D144 |
| 155-52 | L235P | L296 | I159 |
| 009D | L249F | L310 | A174 |
| 091C | A253V | A314 | A179 |
| 128-53 | V260M | V321 | L189 |
| 048B | Q265Stop | Q326 | Q194 |
| 055A | G284N | G346 | G211 |
| 116C | R295W | R357 | R222 |
Tubingen screen (original) alleles are in bold italic; 3R maternal-effect screen alleles are in italic; DmRad51 allele screen alleles are in roman.
aAmino acid comparison based on alignment with ScRad51 and RecA (McKee et al., 1996).
Drosophila Rad51/SpnA is not necessary for viability
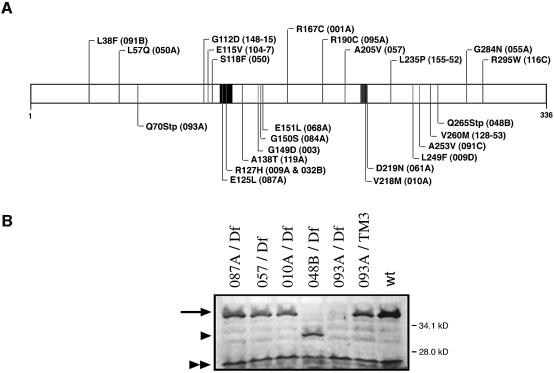
The molecular characterization of all 25 spnA alleles revealed 22 missense mutations (spnA009A and spnA032B contained the same mutation) and two stop codon mutations. Each missense mutation affects an amino acid conserved from yeast to human Rad51 (Figure 2A; Table I). Western analysis using an antibody that was raised against the entire DmRad51 protein revealed that spnA093A, which has an early stop codon at amino acid 70, produces no detectable protein and classifies as a protein null. The other nonsense allele spnA048B introduces a late stop at amino acid 265 and produces a truncated protein (Figure 2B). We also analyzed three missense alleles, spnA087A and spnA010A, which have missense mutations in the Walker-A box and Walker-B box, respectively, and spnA057, which has a change within the DmRad51 core domain. Western analysis revealed that the missense alleles produce stable proteins that are of similar size to the wild-type protein (Figure 2B). Since all alleles of spnA including the null allele spnA093A were viable in trans to the deficiency, Df(3R)X3F, we conclude that SpnA function is required for oogenesis but is not essential for normal cell viability.
Fig. 2. Molecular characterization of the spnA alleles and DmRad51 protein expression analysis of mutant ovary extracts. (A) Map of the DmRad51 mutations. Amino acid substitution and allele name are given (in parentheses). (B) Western blots of extracts prepared from wild-type and hemizygote mutant ovaries. Walker-A and -B box motifs are indicated by black rectangles. Both the wild-type (wt; Oregon R) and heterozygote line, spnA093A/TM3, show expression of a 36.6 kDa protein (arrow). Band shift (arrowhead) in spnA048B/Df(3R)X3F ovary extracts confirms the specificity of the antibody. The lower running band (double arrowhead) is a non-specific cross-reactive protein.
Drosophila has five Rad51 family members
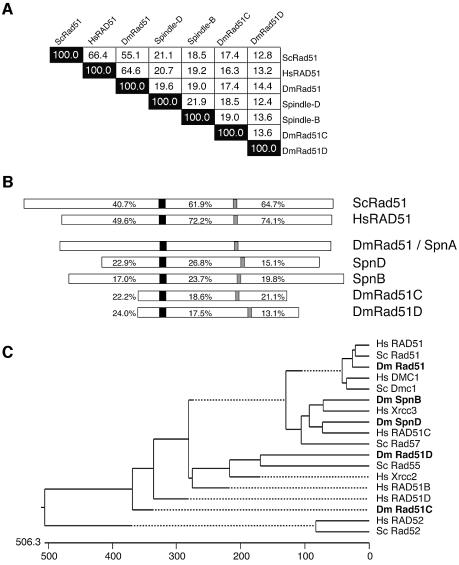
DmRad51/spnA is predicted to encode a 336 amino acid protein. It shows 55.1% identity to S.cerevisiae Rad51 over the entire protein (Figure 3A; McKee et al., 1996). Even higher conservation is observed in sequence alignment with the mouse and human Rad51 (64.6% identical and ∼80% similar) (Figure 3A; McKee et al., 1996; Sekelsky et al., 2000). There were four members of the Drosophila Rad51 family identified from the Genome Project, rad51-like (CG7948; spnA), spnB (CG3325), rad51C (CG2412) and rad51D (CG6318) (Sekelsky et al., 2000) and a Rad51C-like protein (CG31069), which was shown to be encoded by spindle-D (Figure 3A–C; Abdu et al., 2003). Alignment comparison of all five Drosophila Rad51 family members reveals that SpnA protein is the most similar to yeast and human Rad51 (Figure 3A and C). This similarity is not only restricted to the RecA core domain (72.2% identical to HsRAD51) but extends to the N-terminus (49.6%) and C-terminus (74.1%) (Figure 3B). Phylogenetic analysis shows that the other Drosophila members, SpnB, SpnD, Rad51C and Rad51D, are more similar to the Rad51 accessory proteins HsXRCC3, HsRAD51C, HsRAD51D and ScRad55 (or HsXRCC2), respectively (Figure 3C; Sekelsky et al., 2000; Abdu et al., 2003). Based on genome annotation and sequence similarity, we conclude that Spindle-A is the structural homolog of the yeast and mammalian Rad51 protein.
Fig. 3. Sequence comparison of Drosophila Rad51 family members with those of yeast and human. A multiple sequence alignment, using complete protein sequences from known S.cerevisiae, human and Drosophila Rad51 family members, was assembled by the CLUSTAL V method (Higgins and Sharp, 1989; Higgins et al., 1992), which is included in the MegAlign module of the Lasergene sequence analysis software suite (DNASTAR, Inc.), and utilizing PAM250 residue weighting. (A) Summary of alignment results. Numbers represent percent identity between proteins. (B) Each protein is presented as a diagram in actual relative size and aligned with respect to the conserved Walker-A box motif (black). The Walker-B box is indicated by a gray rectangle. The percentages represent the level of identity, within either the N-termini, the central core domains containing both Walker boxes, and the C-termini, with respect to the Drosophila Rad51 gene. (C) Phylogenetic analysis of eukaryotic Rad51 family members. ScRad52 and HsRad52 represent divergent members of the DNA repair proteins and are shown for comparison.
spnA is expressed in both the germline and the soma
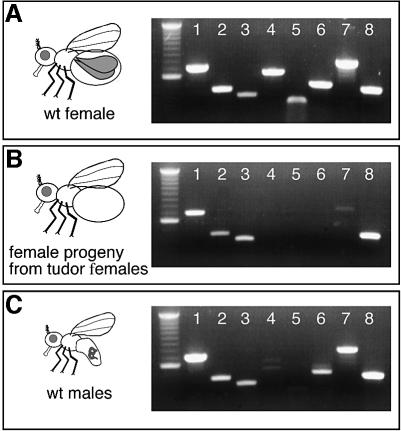
Germline specific expression and the sterility of DMC1-deficient mice defined DMC1 as a meiosis-specific component of the homologous recombination complex (Habu et al., 1996; Pittman et al., 1998). However, genome-wide search failed to identify a clear Dmc1 homolog in Drosophila. Considering that a Dmc1-like gene would be expressed exclusively in the germline, we examined whether DmRad51/SpnA or any of the other Drosophila Rad51 family members are meiosis specific by determining germline and soma gene expression of the five Drosophila Rad51 genes by RT–PCR (Figure 4A and B). Rad51 gene expression profiles from wild-type females, flies from tudor mutant females, which lack germline (Boswell and Mahowald, 1985), and to males, which fail to undergo meiotic recombination, were compared. For each Rad51 gene, primers were designed to specifically amplify a fragment of the corresponding transcript. As a control for germline-specific expression, we also analyzed oskar and nanos RNA by RT–PCR (Ephrussi et al., 1991; Wang and Lehmann, 1991). spnA, spnB and rad51C RNA are expressed in both males and females (Figure 4A and C). Furthermore, their expression is not limited to the female germline (Figure 4B). Interestingly, rad51D and spnD RNA appear to be expressed almost exclusively in the germline of adult females. Thus, Drosophila may have two Rad51 family members that are specifically involved in meiotic recombination and functionally equivalent to Dmc1.
Fig. 4. Gene expression profiles of Drosophila Rad51 family members from wild-type females, wild-type males, and germline-depleted females. RT–PCR was performed with total RNA from: (A) wild-type females (spnA/TM3); (B) female progeny of tud/Df mutant females, which lack germline tissue; and (C) wild-type males. 1, dmRad51 (687 bp); 2, spnB (436 bp); 3, dmRad51C (380 bp); 4, dmRad51D (620 bp); 5, spnD (302 bp); 6, nanos (533 bp); 7, oskar (760 bp); and 8, rp49 (430 bp). nanos and oskar represented germline controls. The residual oskar RNA expression in the germline-depleted flies may relate to its reported expression in the nervous system (Dubnau et al., 2003). The first lane in each gel is a 100 bp DNA ladder (Roche).
spnA mutants are sensitive to ionizing radiation
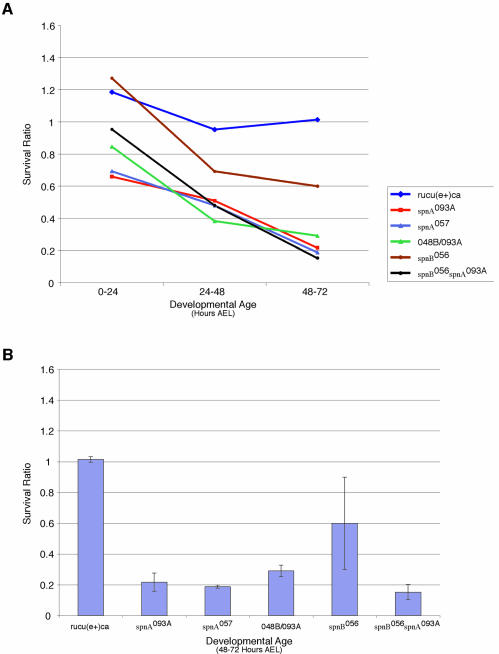
Since spnA and spnB are expressed in the soma as well as the germline, we wished to investigate a possible somatic function for these genes. To determine if these proteins function in the mitotically active cells of the soma, spnA, spnB and spnB spnA doubly mutant embryos and larvae were exposed to 20 Gy of ionizing radiation (X-rays) and examined for survival. The progeny of heterozygous parents were irradiated at either 0–24 h, 24–48 h or 48–72 h after egg laying (AEL). The survival of the irradiated progeny was compared to that of their unirradiated siblings and a survival ratio was established (see Materials and methods). At the irradiation dosage chosen, there was no apparent difference in the survival of irradiated and unirradiated control flies (survival ratio close to 1) for all irradiation times, indicating an insensitivity to this dose of irradiation (Figure 5A and B). At the same dose, spnA, spnB and spnB spnA doubly mutant embryos and larvae show an age-dependent sensitivity to ionizing radiation (Figure 5A and B). When irradiated during embryogenesis (0–24 h AEL), there was little difference in survival between mutant and control. However, during later larval stages (48–72 h AEL) there was a significant difference between the survival of spnA mutant larvae and their heterozygous control siblings (Figure 5A). This increase in sensitivity to irradiation with age is likely due to maternal proteins present in the developing embryos; as the animals get older less maternal protein is present due to degradation. Heterozygous control progeny experience the same degradation of maternal product but survive due to their ability to synthesize necessary gene product de novo. While spnA mutants show a striking sensitivity to irradiation at late third instar (48–72 h AEL), spnB mutants show only a modest sensitivity to ionizing radiation (Figure 5B). This is in contrast to the insensitivity observed when spnB mutants were exposed to MMS (Ghabrial et al., 1998). spnB spnA double mutants do not show a synergistic sensitivity to ionizing radiation (Figure 5A and B), rather, they behave similar to spnA alone, suggesting that the two genes are part of the same non-redundant pathway. Most importantly, our results show that SpnA does indeed play a role in the soma to protect against chromosomal damage inflicted by DNA damaging agents.
Fig. 5. X-ray sensitivity of spnA, spnB and spnBspnA mutants by developmental age. (A) Survival ratios of 0–24 h AEL embryos, 24–48 h AEL larvae, and 48–72 h AEL larvae after 20 Gy of ionizing radiation. (B) Survival ratios of the 48–72 h AEL third instar larval age. The average sensitivity of four independent experiments is reported with standard deviation.
dmRad51/spindle-A mutants are defective in female meiosis
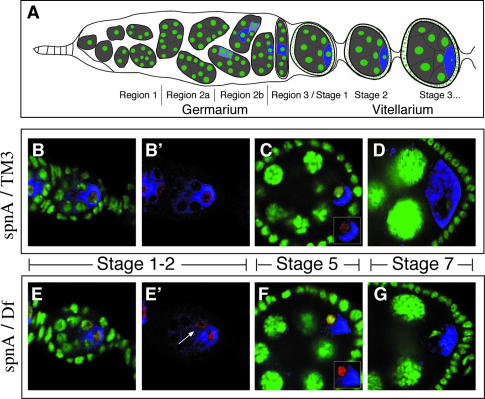
To better characterize the role of SpnA during oogenesis, we analyzed in detail the meiotic defect of the spnA null allele. We asked whether meiotic chromosome synapsis is affected in the mutants, whether DNA breaks occur during meiosis and can be repaired in the mutant, and finally whether spnA mutants indeed cause activation of a meiotic checkpoint. In Drosophila, each germline stem cell, at the anterior tip of each ovariole, divides asymmetrically to produce a new stem cell and a differentiating cystoblast (Figure 6A; Spradling, 1993). The cystoblast undergoes four rounds of mitotic division with incomplete cytokinesis to generate a cyst of 16 cells. The cells within a cyst remain interconnected by cytoplasmic bridges called ring canals. The initiation of meiosis is indicated by the appearance of the synaptonemal complex (SC), which assembles in region 2a of the germarium in the four cells of the cyst that form first and thus contain either three- or four-ring canals. The two four-ring canal cells will become the pro-oocytes as defined by the persistence of the SC and their accumulation of oocyte specific markers in region 2b (Page and Hawley, 2001). By stage 1 of oogenesis, the SC, as observed by immunostaining for the Drosophila SC component C(3)G, and oocyte markers are restricted to a single cell, the future oocyte. As the egg matures, C(3)G begins to lose association with chromatin and the SC is no longer observed.
Fig. 6. Synaptonemal complex formation and resolution during wild-type and spnA oogenesis. (A) Diagram of the anterior of a Drosophila ovary. Germline cells are in gray. The DNA is in green and blue represents expression of the oocyte-specific factor, Orb (Christerson and McKearin, 1994), with light blue representing early low levels and darker blue representing full, localized expression within the oocyte. (B–D) Confocal images of different oogenesis stages from a wild-type (spnA093A/TM3) ovary. (E–G) Confocal images of similar oogenesis stages from a spnA [spnA093A/Df(3R)X3F] ovary. DNA is stained with OliGreen (green). Oocytes are marked by immunostaining for Orb (blue). SC is made visible by anti-C(3)G immunofluorescence (red). (B′) is the same as (B) minus the green channel. (E′) is the same as (E) minus the green channel, with an arrow indicating the losing pro-oocyte. Insets shows oocyte nucleus minus the green channel. All images are single confocal sections.
We followed the distribution of C(3)G in spnA mutant germline cysts. As in the wild type, C(3)G expression is first detected in region 2A (data not shown). However, C(3)G restriction to the oocyte and its dissolution from the chromatin are delayed. At stage 1 of oogenesis, while the SC is always restricted to just the oocyte in wild type, it persists from time to time in both pro-oocytes in spnA mutants, similar to what was observed previously. (Figure 6B, B′, E and E′; Huynh and St Johnston, 2000). Furthermore, when C(3)G staining decreases in the maturing stage 5 oocyte, it remains in the mutant (Figure 6C and F). By stage 7, the staining is no longer detected in the oocyte of either wild-type or spnA mutant egg chambers (Figure 6D and G). These results suggest that synapse formation is appropriately initiated in spnA mutants but the failure to repair broken DNA causes a delay in the resolution of synapsis, first in the cyst that will not become the oocyte and subsequently in the oocyte as it progresses through meiosis.
DSBs are not processed efficiently in spnA oocytes
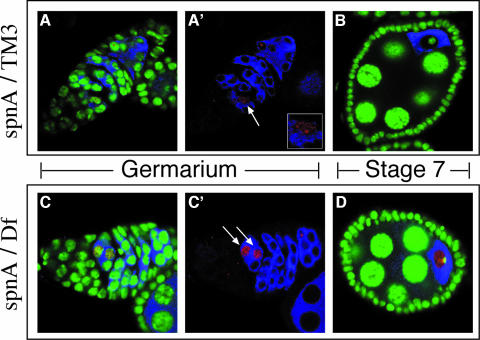
The meiotic phenotype of spnA suggests that there may be a delay in proper meiotic chromosome dynamics due to the failure to repair DSBs. To visualize DSBs cytologically, we used an antibody that recognizes γ-H2AX, a phospho-epitope of the human histone H2A variant, H2AX, which becomes phosphorylated upon DSB formation (Redon et al., 2002). The phospho-epitope is conserved in Drosophila histone variant HIS2AV and becomes phosphorylated in the event of DSBs, whether induced exogenously or during meiosis (Madigan et al., 2002; Jang et al., 2003). During wild-type meiosis, we observed few γ-HIS2AV foci, presumably due to the rapid repair of DSBs and the formation of viable recombination intermediates. When observed, γ-HIS2AV foci were found in only one cell in region 2a of the germarium (Figure 7A and A′). This early appearance of γ-HIS2AV, before the restriction of other oocyte markers to a single cell, suggests that the regulation of DSB formation and persistence may be a critical event in oocyte specification. γ-HIS2AV foci were not observed from region 2B onwards (Figure 7B). Thus, DSBs are rapidly processed and recombination intermediates are formed concomitant with oocyte specification. In contrast, spnA mutant germaria show a more robust HIS2AV activation in one or two cells of a growing cyst in region 2a (Figure 7C and C′), suggesting that in spnA DSBs form at the normal time but their resolution is delayed. Furthermore, γ-HIS2AV localization is more extensive along the DNA rather than in distinct foci as observed in the wild type, possibly due to the accumulation of unresolved breaks along the chromosomes. HIS2AV activation persists in the oocyte nucleus through later stages of oogenesis suggesting a failure to properly repair DNA breaks (Figure 7D).
Fig. 7. Persistence of γ-HIS2AV staining suggests a defect in the repair of meiotically induced DSBs. (A and B) Confocal images of different oogenesis stages from a wild-type (spnA093A/TM3) ovary. (C and D) Confocal images of similar oogenesis stages from a spnA [spnA093A/Df(3R)X3F] ovary. DNA is stained with OliGreen® (green). Early cysts and the oocytes are marked by immunostaining for Orb protein (blue). (A and A′) γ-HIS2AV speckles the DNA in one cell (A′, red, arrow), presumably the oocyte. A 2× zoom of that nucleus is provided in the A′ inset. (C and C′) γ-HIS2AV observed in the germarium of spnA ovaries. (C′, arrows) Two cells of a common cyst (D) γ-HIS2AV in spnA mutants beyond the germarium in the vitellogenic stages of oogenesis. All images are single confocal sections.
The spnA oogenesis phenotypes are linked to defects in DSB repair
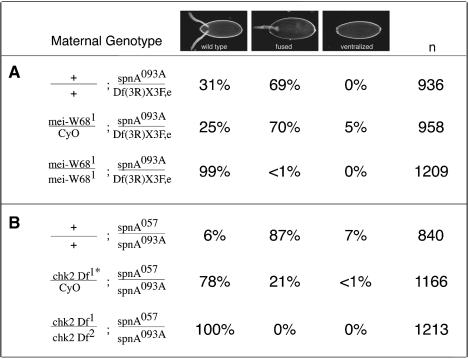
If all the defects observed in spnA mutants were due to the activation of a checkpoint upon failure to repair DSBs, one would predict that mutations, which prevented break formation in the first place, would suppress the spnA phenotype. This rationale was suggested by results in yeast where mutations in spo11 suppressed the meiotic sporulation defects of dmc1 mutations (Roeder, 1997; Bishop et al., 1999). Subsequently, it was shown that the eggshell phenotype of two Drosophila Rad51 family members, spnB and spnD, as well as the Rad54 homolog, okra, was suppressed in the absence of mei-W68, the Drosophila homolog of Spo11 (Ghabrial et al., 1998). We therefore generated double mutants between mei-W68 and spnA and examined their ability to produce properly patterned eggs. Control females that were efficient at producing DSBs during meiosis [mei-W68/+; spnA093A/Df(3R)X3F] but were defective in SpnA function, produced progeny with spindle eggshells (Table IIA). In contrast, in flies defective in DSB production and SpnA function [mei-W68/mei-W68; spnA093A/Df(3R)X3F], the spindle phenotype was rarely observed (<1%). Furthermore, the oocyte nuclear morphology and Gurken protein localization and distribution appeared normal in the double mutants (data not shown). mei-W68 also suppressed the embryonic lethality associated with loss of maternal SpnA. In this situation, embryos from doubly mutant females survived to adulthood with a frequency similar to that observed in mei-W68 progeny alone (data not shown). The fact that all phenotypes associated with spnA mutants are suppressed by mei-W68 suggests that it is indeed the role of SpnA in repair of meiotic-induced DSBs that is essential for normal oogenesis and survival.
Table II. Suppression of spnA eggshell phenotypes by mei-W68 and chk2.
*See Materials and methods for description of chk2 mutant lines.
The spnA oogenesis phenotype results from the activation of a cell cycle checkpoint
Our data show that DSBs are readily detectable by γ-HIS2AV staining and persist during oogenesis in spnA mutants. Unrepaired DSBs or unresolved recombination intermediates lead to the activation of an ATM/ATR-dependent cell cycle checkpoint in mitosis and meiosis, which often causes delays in cell cycle progression in order to repair DNA damage (Roeder and Bailis, 2000). To test whether unrepaired DSBs or unresolved recombination intermediates in spnA mutants trigger a cell cycle checkpoint that leads to defects in oocyte development, we wished to inactivate the checkpoint response.
Two genes have been implicated in checkpoint function, the Drosophila ATR homolog Mei-41 and the Chk2 homolog DmChk2/Mnk/Loki (Sibon et al., 1999; Xu et al., 2001). Since Drosophila mei-41 mutants also show a defect in meiotic recombination (Baker and Carpenter, 1972; Carpenter, 1979), it is difficult to assess the exact step in meiosis that is affected in this mutant. We therefore focused on the checkpoint protein, DmChk2. On its own, chk2 mutants do not appear to have a meiotic phenotype (Masrouha et al., 2003). As detailed in Table IIB, females doubly mutant for chk2 and spnA produced progeny with wild-type egg shape (100 versus 6%), even a reduction in the copy number of chk2 (chk2/+, spnA057/spnA093A) partially suppresses the spindle phenotype (22 versus 94%). In addition, the karyosome appears normal in the oocytes from females doubly mutant for chk2 and spnA (data not shown), suggesting that the abnormal nuclear morphology observed in spnA mutant oocytes is not the result of fragmented DNA, since the DNA breaks should persist in these double mutants. In contrast to the mei-W68;spnA doubles, deletion of chk2 did not suppress the maternal-effect embryonic lethality of spnA. Thus, we conclude that the spnA phenotype results from the activation of a Chk2-dependent meiotic checkpoint.
Discussion
Screens in Drosophila have recovered many mutations that cause disruption to normal meiotic chromosome behavior (Baker and Carpenter, 1972; Sandler, 1974; Sekelsky et al., 1999; McKim et al., 2002). They were identified based on the ability to recognize abnormal events, such as chromosome loss, non-disjunction or a change in recombination frequency. Mutagen sensitivity screens, similar to those performed in yeast, have also been conducted in Drosophila to identify genes necessary for DNA repair (Henderson, 1999). As would be expected, some of these mutagen-sensitive mutants showed meiotic defects as well (Baker et al., 1976; Baker et al., 1978; Green, 1981). Interestingly, none of the Rad52 epistasis genes of Drosophila were recovered from these types of screens. Instead, due to downstream effects on D/V patterning through the activation of a meiotic checkpoint, the spindle oogenesis phenotype has proven to be an effective assay by which to uncover these genes. Thus far, four members of the Rad52 epistasis group in Drosophila have been found through this approach, (this study; Ghabrial et al., 1998; Abdu et al., 2003).
In Drosophila, there are five members of the Rad51 family. Our analysis confirms that Spindle-A is the structural and functional homolog of the yeast and mammalian Rad51 protein. Biochemical analysis of in vitro purified DmRad51 has shown that it has strand exchange capabilities (Alexiadis and Kadonaga, 2002). The other Rad51 paralogs show greater sequence homology to Rad51 accessory proteins, which have been shown to promote Rad51 foci formation on DNA. Here we show that both rad51D and spnD, in the adult, are expressed specifically in the germline. Therefore, we suggest that they are Rad51 accessory proteins involved in meiotic recombination, compensating for a lack of a Drosophila Dmc1 homolog. Initial studies on spnB revealed a striking similarity to Dmc1, namely its importance in meiotic recombination and its resistance to the effects of MMS (Ghabrial et al., 1998). However, we show that spnB RNA is expressed in the soma as well as the germline. Moreover, we present evidence that spnB mutant larvae are less tolerant than their wild-type siblings to the DNA damaging effects of ionizing radiation. Based on its sequence homology to XRCC3, it is possible that SpnB functions as a necessary partner for DmRad51 during meiotic recombination and takes on a supporting role in Rad51 stabilization during DSB repair of the soma (Liu et al., 1998; Brenneman et al., 2002).
SpnA is not essential for viability
Analysis of Rad51 function in vertebrate development has been difficult due to the early embryonic lethality of RAD51–/– mice. Vertebrate cell culture studies have suggested an essential role of RAD51 in the repair of breaks generated during DNA replication (Sonoda et al., 1998), thus providing some explanation for the embryonic lethality in mice. Here we show that Drosophila Rad51 null animals can survive to adulthood. Therefore, the requirement for Rad51 in the repair of DNA breaks occurring during DNA replication may not be conserved. However, other possibilities exist. First, maternal Rad51 may persist to repair DSBs occurring throughout embryogenesis. However, we show that female flies doubly mutant for mei-W68 and spnA produce embryos that survive to adulthood, suggesting that neither maternal nor zygotic DmRad51 function are essential for viability. Another possibility is that the Rad51 genes may have partially overlapping, redundant functions. However, neither of the other family members shows strong homology to Rad51. Additionally, flies doubly mutant for the spnA and its closest relative, spnB, are viable (Gonzalez-Reyes et al., 1997) and the next closest paralog, spnD, is expressed specifically in the germline, though we only tested adult animals. An alternative explanation, and the one we favor, is the existence of an alternative repair pathway that can compensate in the event of homologous recombination failure. Homologous recombination has been considered the major DNA repair pathway in Drosophila (Engels et al., 1990; Kurkulos et al., 1994; Nassif et al., 1994). Recent evidence in Drosophila has shown that when the homologous recombination pathway is compromised, the error-prone non-homologous end joining (NHEJ) pathway can compensate and prevent a lethal outcome (Adams et al., 2003). Therefore, in Drosophila, we would predict that an efficient cooperation must exist between the homologous recombination and NHEJ pathways to prevent the lethal effects of DNA DSBs, presumably with homologous recombination being the primary choice and NHEJ playing a backup role.
SpnA is necessary for oogenesis
During meiotic recombination, crossing over between homologous chromosomes guarantees their proper segregation. Defects in the proper formation of recombination intermediates result in the activation of a pachytene, or meiotic recombination, checkpoint (Roeder and Bailis, 2000). In mice, if defects in chromosomal synapsis or meiotic recombination persist, the result is the activation of the pachytene checkpoint and removal of the arrested germ cells most probably by apoptosis (Odorisio et al., 1998; Pittman et al., 1998; Baudat et al., 2000; Romanienko and Camerini-Otero, 2000). In our study, we show that a meiotic recombination checkpoint is activated in response to a loss of SpnA function. spnA mutant females do not show an appreciable defect in egg deposition, suggesting that the apoptotic pathway is not activated in response to the meiotic recombination checkpoint. Moreover, the p53 protein, a strong inducer of apoptosis during the mitotic cell cycle, has been shown not to be involved in the Drosophila meiotic recombination checkpoint (Abdu et al., 2002). Instead, as our data indicate, the unsuccessful processing of meiotic-induced DSBs results in a Chk2-dependent delay of the meiotic cell cycle. Concomitant with this delay, we observe a defect in the EGFR/TGFα signaling pathway, which results in the production of eggs with dorsal/ventral patterning defects. Thus, our results show a coupling between progression through the meiotic cell cycle and oocyte patterning and development. The ATP-dependent helicase Vasa has been implicated in mediating at least two aspects of meiotic checkpoint activation, Gurken translation and karyosome formation. It remains unclear if Vasa is directly activated by the checkpoint transducer kinase Chk2/Mnk and how defects in DSB repair lead to checkpoint activation. The spindle eggshell phenotype has proven to be an efficient assay to identify genes that lead to the activation of the meiotic checkpoint, making Drosophila an excellent genetic system to identify additional components that regulate the interplay between DNA repair, cell cycle progression and cell differentiation during meiosis and possibly, as our studies suggest, also mitosis.
Materials and methods
Fly stocks
mei-W681 was obtained from the Bloomington Drosophila Stock Center. chk2 mutants, kindly provided by B.Suter, were used as a transallelic combination of two deficiency lines that uncover the chk2 gene locus: w; Df(2L)pr2b, P[w+, barren+]/CyO and w; Df(2L)be408, P[w+, CG107278+]/CyO (Masrouha et al., 2003). tudor mutant adults were the progeny of tudwc8 bw/Df(2R)PurP133 c px sp females. The genetic markers used are described in Lindsley and Zimm (1992). Flies were raised on standard cornmeal–molasses medium at 25°C. Embryos were collected on agar–apple juice plates.
Eggshell/chorion preparation
Embryos produced from germline clones of the mutagenized lines were washed once in PBS before being placed into a drop of 100% Hoyer’s Mounting media (Ashburner, 1989). Darkfield images were adapted using Adobe Photoshop® 7.0 software.
Allele sequencing
Genomic DNA from flies transheterozygous for each allele of spnA and a deletion of the dmRAD51 gene [Df(3R)X3F] and from control starting strains (Line FRT 161-48 for mutants from the 3R maternal screen and ru cu[e+]ca for mutants from the spnA allele screen) was prepared as described (Ashburner, 1989). Multiple independent PCRs, following standard manufacturer’s protocol, were used to amplify the 1.35 kb genomic region of DmRAD51 (CG7948). Sequencing in both the forward and reverse direction, as described in Supplementary figure 2, was performed on an ABI Prism 3700 machine (Rockefeller University DNA Sequencing Resource Center). SeqMan II (DNASTAR, Inc.) and EditView (Applied Biosystems) were used for analysis of sequencing data.
Western blot analyses
Ovarian extracts were prepared as previously described (Gavis and Lehmann, 1994). Polyclonal anti-DmRad51 was generated using the full-length recombinant Rad51 protein containing a His6 tag at the C-terminus as antigen. The protein was purified from IPTG-induced Escherichia coli BL21 cells containing a Rad51/pET30b expression vector by metal chelation chromatography (Novagen), and used to immunize rabbits. The anti-Rad51 antibody was used at a dilution of 1:1000 (PBST + 2.5% dry milk). Horseradish peroxidase-conjugated goat anti-rabbit was used at 1:5000 (PBST + 2.5% dry milk) and signal was obtained using the LumiGLO®? Chemiluminescent Substrate System (KPL).
Mutagen sensitivity
Virgin heterozygote males and females of the specified alleles were mated in yeasted vials for 2 days. The progeny of these vials served as untreated controls. After 2 days, the parents were transferred to newly yeasted vials every day for 4 days. The embryos from these vials were then subjected to 20 Gy of ionizing radiation, using a Torrex 150D X-ray irradiator cabinet, either 0–24, 24–48 or 48–72 h AEL. Upon eclosion, the number of homozygous or transheterozygous mutant progeny was compared to the heterozygous progeny (N = number of mutant/number of wild-type [hets]) for both the untreated and treated populations. These numbers were then compared to establish the level of DNA damage sensitivity (Ntreated/Nuntreated = X). If X equaled ∼1, then there was no sensitivity. If X < 1, then the mutant embryos exhibited sensitivity to the ionizing radiation.
Ovary fixation and immunofluorescence
Antibody staining was done as described (de Cuevas et al., 1996). DNA staining was performed by incubating the samples for 30 min at room temperature in PBS + 0.1% Tween-20 containing OliGreen® (1:5000; Molecular Probes) and 5 µg/ml RNase A followed by rinsing three times and washing in PBST for 30 min.
All antibodies were diluted in PBS + 0.1% Tween-20 + 0.2% BSA and used at the following dilutions: mouse monoclonal anti-Gurken antibodies ID12 and IF12 at 1:5. (Gift from T.Schupbach), mouse monoclonal anti-Orb antibodies 4H8 and 6H4 at 1:5 (Hybriboma Bank), guinea pig anti-C3G serum at 1:500 (kind gift from R.S.Hawley), rabbit serum for polyclonal anti-CP190 antibody Rb188 at 1:500 (gift from W.Whitfield), commercial rabbit anti-phoshoH2A.X (Ser139) at 1:100 (Upstate Biotechnologies, Lake Placid, NY). Secondary antibodies Alexa-488-conjugated anti-mouse IgG (Molecular Probes), Alexa-488-conjugated anti-rabbit IgG (Molecular Probes), Cy3-conjugated anti-guinea pig IgG (Jackson Immunoresearch), Cy5-conjugated anti-rabbit IgG (Jackson Immunoresearch) and Cy3-conjugated anti-mouse IgG (Jackson Immunoresearch) were each used at 1:500. Images were collected on a Leica DM RBE confocal microscope equipped with a Leica PL APO 40×/1.25NA oil objective using the Leica TCS NT program. Images were adapted using Adobe Photoshop® 7.0 software.
RT–PCRs
Total RNA was obtained from 10–15 whole adult animals using Trizol Reagent® (Invitrogen) and following the manufacturer’s protocol. RNA samples were treated using DNA-free™ (Ambion). RT–PCR was performed using SuperScript™ One-Step RT–PCR with Platinum® Taq (Invitrogen). Control experiments, using Platinum® Taq minus RT, were performed to confirm the absence of contaminating genomic DNA. No signal was ever obtained from the RNA preparation. The primers are listed in Supplementary data S4.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank R.S.Hawley, T.Schüpbach, W.Whitfield and the Developmental Studies Hybridoma Bank for providing antibodies. We thank the Bloomington Stock Center for providing fly stocks. We are grateful to B.Suter for providing the chk2 fly stocks prior to publication. We thank T.Schüpbach and U.Abdu for sharing unpublished data and results prior to their publication. We are indebted to C.Yohn for organizing the 3R maternal-effect screen, as well as to the other members of the screen team: Z.Forbes, J.Morris, C.Navarro, M.Samuels, J.Stein and H.Zinszner. We also thank the members of the Lehmann laboratory for critically reading the manuscript. Finally, we thank B.McKee and J.Sekelsky for their comments and suggestions. S.Yoo generated the Rad51 antibody in the laboratory of B.McKee. E.S.-V. was supported by a grant to R.L. (RO1 HD41900) from the NIH. S.Y. was supported by a grant to B.McKee (RO1 GM40489) from the NIH. R.L. is an HHMI investigator.
References
- Abdu U., Brodsky,M. and Schüpbach,T. (2002) Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr. Biol., 12, 1645–1651. [DOI] [PubMed] [Google Scholar]
- Abdu U., Gonzalez-Reyes,A., Ghabrial,A. and Schüpbach,T. (2003) The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics, 165, 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M.D., McVey,M. and Sekelsky,J.J. (2003) Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science, 299, 265–267. [DOI] [PubMed] [Google Scholar]
- Alexiadis V. and Kadonaga,J.T. (2002) Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev., 16, 2767–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. (1989) Drosophila. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Baker B.S. and Carpenter,A.T. (1972) Genetic analysis of sex chromosomal meiotic mutants in Drosophilia melanogaster. Genetics, 71, 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S., Boyd,J.B., Carpenter,A.T., Green,M.M., Nguyen,T.D., Ripoll,P. and Smith,P.D. (1976) Genetic controls of meiotic recombination and somatic DNA metabolism in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 73, 4140–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.S., Carpenter,A.T. and Ripoll,P. (1978) The utilization during mitotic cell division of loci controlling meiotic recombination and disjunction in Drosophila melanogaster. Genetics, 90, 531–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Manova,K., Yuen,J.P., Jasin,M. and Keeney,S. (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell, 6, 989–998. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Nikolski,Y., Oshiro,J., Chon,J., Shinohara,M. and Chen,X. (1999) High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells, 4, 425–444. [DOI] [PubMed] [Google Scholar]
- Boswell R.E. and Mahowald,A.P. (1985) tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell, 43, 97–104. [DOI] [PubMed] [Google Scholar]
- Brenneman M.A., Wagener,B.M., Miller,C.A., Allen,C. and Nickoloff,J.A. (2002) XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell, 10, 387–395. [DOI] [PubMed] [Google Scholar]
- Carpenter A.T. (1979) Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma, 75, 259–292. [DOI] [PubMed] [Google Scholar]
- Christerson L.B. and McKearin,D.M. (1994) orb is required for anteroposterior and dorsoventral patterning during Drosophila oogenesis. Genes Dev., 8, 614–628. [DOI] [PubMed] [Google Scholar]
- de Cuevas M., Lee,J.K. and Spradling,A.C. (1996) α-spectrin is required for germline cell division and differentiation in the Drosophila ovary. Development, 122, 3959–3968. [DOI] [PubMed] [Google Scholar]
- Dubnau J. et al. (2003) The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol., 13, 286–296. [DOI] [PubMed] [Google Scholar]
- Engels W.R., Johnson-Schlitz,D.M., Eggleston,W.B. and Sved,J. (1990) High-frequency P element loss in Drosophila is homolog dependent. Cell, 62, 515–525. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson,L.K. and Lehmann,R. (1991) Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell, 66, 37–50. [DOI] [PubMed] [Google Scholar]
- Gavis E.R. and Lehmann,R. (1994) Translational regulation of nanos by RNA localization. Nature, 369, 315–318. [DOI] [PubMed] [Google Scholar]
- Ghabrial A. and Schüpbach,T. (1999) Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol., 1, 354–357. [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Ray,R.P. and Schupbach,T. (1998) okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev., 12, 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A., Elliott,H. and St Johnston,D. (1997) Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development, 124, 4927–4937. [DOI] [PubMed] [Google Scholar]
- Green M.M. (1981) mus(3)312D1, a mutagen sensitive mutant with profound effects on female meiosis in Drosophila melanogaster. Chromosoma, 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Habu T., Taki,T., West,A., Nishimune,Y. and Morita,T. (1996) The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res., 24, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson D.S. (1999) DNA repair defects and other (mus)takes in Drosophila melanogaster. Methods, 18, 377–400. [DOI] [PubMed] [Google Scholar]
- Higgins D.G. and Sharp,P.M. (1989) Fast and sensitive multiple alignments on a microcomputer. Comput. Appl. Biosci., 5, 151–153. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Bleasby,A.J. and Fuchs,R. (1992) CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci., 8, 189–191. [DOI] [PubMed] [Google Scholar]
- Huynh J.R. and St Johnston,D. (2000) The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development, 127, 2785–2794. [DOI] [PubMed] [Google Scholar]
- Jang J.K., Sherizen,D.E., Bhagat,R., Manheim,E.A. and McKim,K.S. (2003) Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci., 116, 3069–3077. [DOI] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg,J.M., Roy,D. and Mount,S.M. (1994) P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics, 136, 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D.L. and Zimm,G.G. (1992) The Genome of Drosophila melanogaster. Academic Press, San Diego, CA. [Google Scholar]
- Liu N. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- Madigan J.P., Chotkowski,H.L. and Glaser,R.L. (2002) DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res., 30, 3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrouha N., Yang,L., Hijal,S., Larochelle,S. and Suter,B. (2003) The Drosophila chk2 gene loci is essential for embryonic DNA double-strand-break checkpoints induced in S phase or G2. Genetics, 163, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B.D., Ren,X. and Hong,C. (1996) A recA-like gene in Drosophila melanogaster that is expressed at high levels in female but not male meiotic tissues. Chromosoma, 104, 479–488. [DOI] [PubMed] [Google Scholar]
- McKim K.S., Jang,J.K. and Manheim,E.A. (2002) Meiotic recombination and chromosome segregation in Drosophila females. Annu. Rev. Genet., 36, 205–232. [DOI] [PubMed] [Google Scholar]
- Melo J. and Toczyski,D. (2002) A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol., 14, 237–245. [DOI] [PubMed] [Google Scholar]
- Morris J. and Lehmann,R. (1999) Drosophila oogenesis: versatile spn doctors. Curr. Biol., 9, R55–R58. [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney,J., Pal,S., Engels,W.R. and Gloor,G.B. (1994) Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol., 14, 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorisio T., Rodriguez,T.A., Evans,E.P., Clarke,A.R. and Burgoyne,P.S. (1998) The meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nat. Genet., 18, 257–261. [DOI] [PubMed] [Google Scholar]
- Page S.L. and Hawley,R.S. (2001) c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev., 15, 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman D.L., Cobb,J., Schimenti,K.J., Wilson,L.A., Cooper,D.M., Brignull,E., Handel,M.A. and Schimenti,J.C. (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell, 1, 697–705. [DOI] [PubMed] [Google Scholar]
- Redon C., Pilch,D., Rogakou,E., Sedelnikova,O., Newrock,K. and Bonner,W. (2002) Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev., 12, 162–169. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev., 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. and Bailis,J.M. (2000) The pachytene checkpoint. Trends Genet., 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J. and Camerini-Otero,R.D. (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell, 6, 975–987. [DOI] [PubMed] [Google Scholar]
- Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- Sandler L. (1974) Some observations on the study of the genetic control of meiosis in Drosophila melanogaster. Genetics, 78, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J.J. et al. (1999) Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics, 152, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J.J., Brodsky,M.H. and Burtis,K.C. (2000) DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol., 150, F31–F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A. and Ogawa,T. (1999) Rad51/RecA protein families and the associated proteins in eukaryotes. Mutat. Res., 435, 13–21. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein (published erratum appears in Cell, 1992, 71, following 180). Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Sibon O.C., Laurencon,A., Hawley,R. and Theurkauf,W.E. (1999) The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol., 9, 302–312. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C. (1993) Developmental genetics of oogenesis. In Bate,M. and Martinez-Arias,A. (eds), The Development of Drosophila melanogaster. Vol. I. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1–70. [Google Scholar]
- Styhler S., Nakamura,A., Swan,A., Suter,B. and Lasko,P. (1998) vasa is required for GURKEN accumulation in the oocyte and is involved in oocyte differentiation and germline cyst development. Development, 125, 1569–1578. [DOI] [PubMed] [Google Scholar]
- Symington L.S. (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev., 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tearle R. and Nusslein-Volhard,C. (1987) Tubingen mutants and stock list. Drosophila Inform. Serv., 66, 209–226. [Google Scholar]
- Thacker J. (1999) A surfeit of RAD51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Guichet,A., Zavorszky,P. and Ephrussi,A. (1998) Oocyte polarity depends on regulation of gurken by Vasa. Development, 125, 1723–1732. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. and Lehmann,R. (1991) Nanos is the localized posterior determinant in Drosophila. Cell, 66, 637–647. [DOI] [PubMed] [Google Scholar]
- Whitfield W.G., Chaplin,M.A., Oegema,K., Parry,H. and Glover,D.M. (1995) The 190 kDa centrosome-associated protein of Drosophila melanogaster contains four zinc finger motifs and binds to specific sites on polytene chromosomes. J. Cell Sci., 108, 3377–3387. [DOI] [PubMed] [Google Scholar]
- Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- Wood R.D., Mitchell,M., Sgouros,J. and Lindahl,T. (2001) Human DNA repair genes. Science, 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- Xu J., Xin,S. and Du,W. (2001) Drosophila Chk2 is required for DNA damage-mediated cell cycle arrest and apoptosis. FEBS Lett., 508, 394–398. [DOI] [PubMed] [Google Scholar]