Abstract
We combine Gal4/UAS, FLP/FRT and fluorescent reporters to generate cell clones that provide spatial, temporal, and genetic information about the origins of individual cells in Drosophila. We name this combination the Gal4 Technique for Real-time and Clonal Expression (G-TRACE). The approach should allow for screening and the identification of real-time and lineage-traced expression patterns on a genomic scale.
The yeast transcriptional activator Gal4 has been extensively utilized as a genetic reporter in Drosophila1. To date, thousands of transgenic lines have been generated that exhibit distinct Gal4 expression patterns during development and are available for analysis2,3. In this report, we describe a Drosophila screening tool, which we name the Gal4 Technique for Real-time and Clonal Expression (G-TRACE) system, developed by the UCLA Undergraduate Research Consortium in Functional Genomics4,5. This system reveals real-time Gal4 expression, similar to traditional Gal4 analysis, but also marks cell lineages derived from Gal4-expressing cells (Fig. 1A and Supplementary Fig. 1). The key feature is that the initiation of reporter expression is Gal4-dependent whereas maintenance of reporter expression is not. Importantly, G-TRACE makes use of fluorescent protein reporters for both real-time (RFP) and lineage-based analysis (EGFP), thereby increasing screening throughput (Fig. 1A) since there is no requirement for antibody staining.
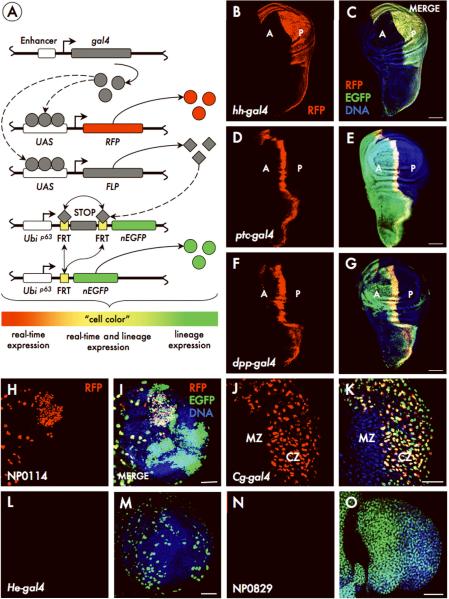
Figure. 1. The G-TRACE analysis system.
A) Schematic depicting molecular mechanisms underlying the G-TRACE system. Gal4 activates the expression of RFP and FLP recombinase. Cells expressing FLP then excise the FRT-flanked STOP cassette separating the Ubi-p63E promoter and nEGFP open reading frame. This initiates expression of EGFP, which is heritably maintained in all daughter cells. The “cell color” gradient represents the apparent color of cells subsequent to the initiation of Gal4 expression. (B-O) G-TRACE in hh-gal4 (B, C), ptc-gal4 (D, E), dpp-gal4 (F, G), NP0114 (H, I), Cg-gal4 (J, K), He-gal4 (L, M) and NP0829 (N, O) reporter lines, showing current RFP expression alone (B, D, F, H, J, L, N) and merged with lineage-traced EGFP expression (C, E, G, I, K, M, O). Images show the wing imaginal disc (B-G), the brain (H, I, L, M) or the lymph gland (J, K, N, O), all at the late third instar. A, anterior wing compartment; P, posterior wing compartment. CZ, cortical zone; MZ, medullary zone; see Supplementary Fig. 4). Scale: 50 μm.
To initiate lineage tracing, Gal4 mediates the expression of FLP recombinase that, in turn, removes an FRT-flanked transcriptional termination cassette inserted between Ubiquitin-p63E promoter fragment and the EGFP open reading frame. Thereafter, the Ubi-p63E promoter maintains EGFP expression perpetually in all subsequent daughter cells, independent of Gal4 activity. These “FLP-out” constructs have previously been utilized to assess biological function including lineage information6-10, however G-TRACE is the first system to utilize the Ubi-p63E promoter and a direct EGFP reporter.
To demonstrate the usefulness and efficacy of the G-TRACE system, we evaluated Gal4 lines with well-defined real-time expression patterns, but in which expression patterns at earlier stages are not known (Fig. 1B-G and Supplementary Fig. 2). At the third instar larval stage, hedgehog-gal4 (hh-gal4) mediates RFP expression throughout the posterior (P) compartment (Fig. 1B). Examination of the cognate lineage-traced EGFP pattern reveals uniform expression limited to the P compartment (Fig. 1C), confirming prior genetic evidence. Both patched-gal4 (ptc-gal4) and decapentaplegic-gal4 (dpp-gal4), which respond to Hedgehog signaling, express RFP in a stripe of anterior (A) compartment cells along the A/P compartment boundary (Fig. 1D and 1F). However, despite similar real-time expression at this developmental stage, EGFP expression in ptc-gal4 marks the entire A compartment (Fig. 1E) but only a portion of the A compartment in dpp-gal4 (Fig. 1G), indicating differential regulation of these genes during wing development.
The G-TRACE system works well across all tissues, including the brain and lymph gland (Supplemental Figs. 3 and 4); examples of real-time expression and lineage tracing for the Gal4 enhancer-trap (Gal4-ET) line NP0114 within the brain (Fig. 1H, I), and Collagen-gal4 (Cg-gal4) within the lymph gland (Fig. J-K) are shown. The method was also useful for identifying Gal4 lines with expression patterns restricted to early development (i.e. showing EGFP expression, but no RFP expression at later developmental stages); examples are shown for Hemese-gal4 (He-gal4), a putative blood-specific marker11, in the late third instar brain (Fig. 1L-M), and for the Gal4-ET line NP0829 in the lymph gland at the same stage (Fig. 1N-O).
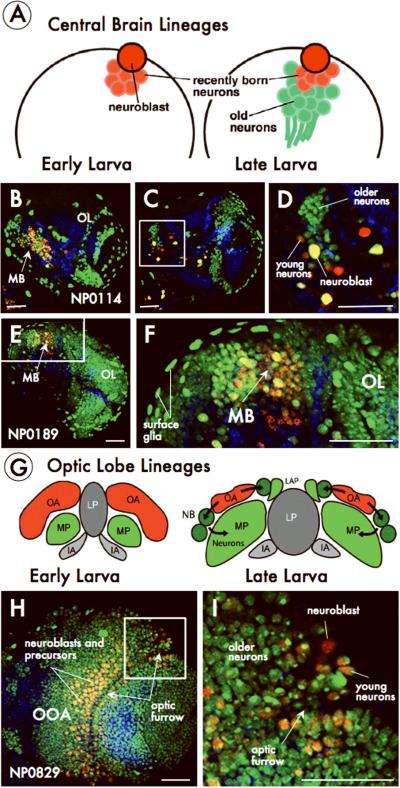
We found many lines with expression patterns within distinct brain lineages. For example, in Gal4-ET lines NP0114 (Fig. 2B-D) and NP0189 (Fig. 2E-F), current expression (third-instar) is restricted to the central brain whereas a substantial number of lineage-traced populations are found in the optic lobes and surface glia. In contrast to these lines, the Gal4-ET line NP0829 exhibits widespread current and lineage expression in the optic lobe (Fig. 2H-I). A summary of Gal4 expression patterns in four different tissues and representative examples for each Gal4 line screened during the course of this work is available in Supplementary Tables 1-4 and Supplementary Data; larger images can also be viewed online at (www.mcdb.ucla.edu/Research/Banerjee/URCFG/).
Figure. 2. G-TRACE reveals novel cell lineage markers.
A) Schematic of lineages from larval central brain neuroblasts. (B-F) G-TRACE in the Gal4-enhancer trap lines NP0114 (B-D) and NP0189 (E, F). D and F are close-ups of the boxed regions in C and E respectively. MB, mushroom body; OL, optic lobe G) Schematic depicting optic lobe development. Cross-section of a brain hemisphere at early and late stages showing the outer (OA; red) and inner (IA; gray) anlagen of the optic lobe. Neuroblasts (NB; green) at the lateral edge form neurons of the medullary primordium (MP) while medial edge neuroblasts form neurons of the laminar primordium (LAP). The inner optic anlage (IA) forms the lobula primordium (LP). (H, I) G-TRACE in the Gal4-enhancer trap line NP0829. OOA, outer optic anlage. I is a close-up of the boxed region in H. Scale: 50μm.
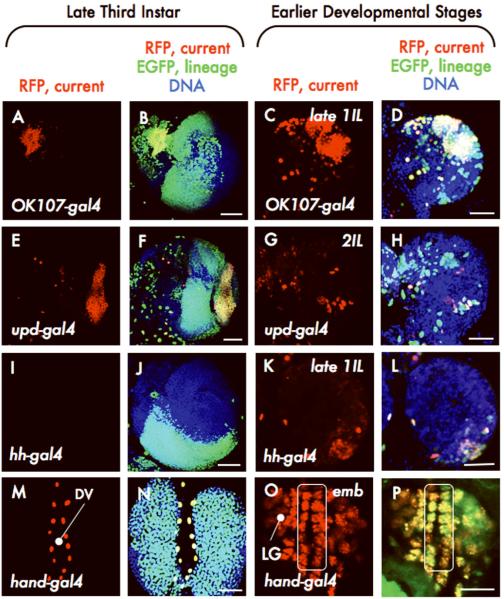
To better understand how late stage patterns arise, a subset of Gal4 lines was selected for analysis at earlier stages (Fig. 3 and Supplementary Fig. 5); in each case, we identified the earliest time point at which RFP was expressed in the tissue or area of interest. In the late third instar, real-time expression of OK107-gal4 is restricted to the mushroom body of the central brain, however a large fraction of the optic lobe expresses EGFP (Fig. 3A-B). Consistent with this pattern, OK107-gal4 is expressed by a large number of cells in the region of the developing optic lobe by the late first instar (Fig. 3C-D). unpaired-gal4 (upd-gal4) drives RFP expression in the optic lamina at the late third instar12 (Fig. 3E), while EGFP expression occurs throughout the optic lobe (Fig. 3F). The earliest expression of upd-gal4 in the developing optic lobe was during the early stages of the second instar (Fig. 3G-H). The hh-gal4 line lacks real-time expression in the late third-instar brain but generates a large cluster of EGFP-expressing cells in the posterior ventral portion of the optic lobe (Fig. 3I-J). We determined that hh-gal4 was expressed in the early first instar in a small cluster of cells located ventrally within the posterior region of the optic lobe (Fig. 3K-L). In the lymph gland, hand-gal4 expression is absent at the late third instar, however the vast majority of lymph gland cells express EGFP at this stage (Fig. 3M-N). Using G-TRACE, hand-gal4 was found to be expressed in the lymph gland in late stage embryos (Fig. 3O-P), consistent with reports of hand gene13 expression in the embryonic cardiogenic mesoderm.
Figure. 3. Stage dependence of G-TRACE patterns.
G-TRACE on OK107-gal4 (A-D), upd-gal4 (E-H), hh-gal4 (I-L), and hand-gal4 (M-P; DV, dorsal vessel; LG, lymph gland; line surrounds cardioblasts of the DV) at late and early developmental stages, as indicated. 1IL, first instar larvae; 2IL, second instar larvae; emb, embryo. Scale: (C-D, G-H, K-L, 25μm; O-P, 10μm; others, 50μm.
As exemplified in the preceding analyses, the G-TRACE system places Gal4 expression patterns in a developmental context by combining real-time expression analysis with a cell-autonomous FLP-out marking system, which is particularly valuable in providing information about gene expression, such as transient expression patterns, that may be limited or inaccessible using other reporter systems (e.g., standard Gal4 expression analysis or analysis of spatially random heat shock-induced clones). Temporal analysis (Fig. 3) can distinguish between broad EGFP expression due to small numbers of precursors that proliferate extensively (i.e. actual cell clones) and Gal4 expression among numerous post-mitotic or slowly proliferating cells at later stages. We note that for any line, a subset of Gal4-expressing cells may fail to initiate EGFP expression due to a “threshold effect”. We find that the efficiency of generating EGFP-marked cell clones correlates primarily with driver strength, i.e., the level and duration of Gal4 expression. Strong drivers, such as hh-gal4 and ubiquitin-gal4 (Supplementary Fig. 6), initiate lineage tracing in nearly all Gal4-expressing cells whereas weaker drivers, such as eyeless-gal4, cause unmarked clones to appear at high frequency (not shown). Drivers that express Gal4 in a gradient, such as hand-gal4, reproducibly generate both marked and unmarked lineages (Supplementary Fig. 6). Thus, reliable Gal4-based lineage patterns as defined by the G-TRACE system are those that are highly reproducible among multiple samples.
In summary, the G-TRACE system is very useful for the identification and characterization of Gal4 lines with early and/or transient expression patterns and in describing the development of tissues with complex lineages. Furthermore, G-TRACE has the potential to be valuable in studying stem cells and how they contribute to the growth and maintenance of tissues, as well as in studying phenomena such as cell death, senescence, and dedifferentiation. Future analyses may benefit from modified G-TRACE systems with different reporters (eg. membrane-associated GFP) or in which Gal4 activity can be modulated during development, such as with Gal80ts.
METHODS
Drosophila lines
The uncharacterized Gal4 enhancer trap lines screened in this study were obtained from the GETDB, a subdivision of the Drosophila Genetic Resource Center, Kyoto Institute of Technology, Japan. The following lines were obtained from the Bloomington Drosophila Stock Center: hh-lacZ, ptc-gal4, ptc-lacZ, dpp-gal4, dpp-lacZ, and OK107-gal4. Other drivers used in this study: Antp-gal4 (S. Cohen, Temasek Life Sciences Laboratory, Singapore), Cg-gal4 (C. Dearlolf, Massachusetts General Hospital, Boston), dome-gal4 (S. Noselli, Centre National de la Recherche Scientifique, Nice, France), Dot-gal4 (D. Kimbrell, University of California, Davis), gcm-gal4 (L. Zipursky, University of California, Los Angeles), hand-gal4 (Z. Han, University of Michigan), hh-gal4 (T. Kornberg, University of California, San Francisco), upd-gal4 (Y.H. Sun, Academia Sinica, Taiwan).
Molecular cloning and the generation of the G-TRACE system
Approximately 2 kb of DNA immediately upstream of the Ubiquitin-p63E (CG11624) open reading frame was PCR amplified as an EcoRI/KpnI fragment from a previously cloned promoter region (kindly provided by P. O'Farrell, University of California, San Francisco) and cloned into the pCRII vector (Invitrogen). Primer sequences are available in Supplementary Table 5. This EcoRI/KpnI fragment was then subcloned into the Drosophila transformation vector pStinger (Drosophila Genomics Resource Center), which contains a multiple cloning site upstream of the nuclear EGFP reading frame. A “STOP cassette”, containing a transcriptional termination sequence flanked by an inverted pair of FRT recombination sites, was isolated as a KpnI fragment from the construct D237 (kindly provided by G. Struhl, Columbia University) and subcloned into the KpnI site that lies in between the Ubi-p63E promoter and the EGFP reading frame. Transgenic flies were generated using the pUbi-63E-STOP-Stinger vector and a commercial transformation service (BestGene, Inc.). A triple-recombinant chromosome was generated using UAS-RedStinger14 (w1118; P{UAS-RedStinger}4/CyO, Bloomington Drosophila Stock Center), UAS-FLP (y1 w*; P{UAS-FLP1.D}JD1, Bloomington Drosophila Stock Center) and pUbi-63E-STOP-Stinger insert 9F6 and was balanced over CyO.
Analysis of Gal4 expression patterns
The Gal4-expressing lines were individually crossed on standard fly food at 25°C. Wandering third instar larvae were dissected in 1XPBS and tissues were fixed in 4% formaldehyde in 1XPBS for 30 min, followed by three washes (10 min each) with 1XPBS/0.4% Triton-X-100 (Fisher) detergent (PBST). During the second 1XPBST wash, To-Pro3 iodide (Invitrogen) was added to stain nuclei. Tissues were mounted on slides in VECTASHIELD (Vector Laboratories) and imaged using a Zeiss Apotome or a BioRad Radiance 2000 confocal microscope with standard techniques.
Supplementary Material
ACKNOWLEDGMENTS
KTN, EKi, NEL, EKu, ANP, DS, PT, MD, KY, AC participated as undergraduate research students in the UCLA Undergraduate Research Consortium in Functional Genomics, which is supported by a Howard Hughes Medical Institute Professor's Award to UB. CJE was supported by NIH/UCLA Vascular Biology Training Grant T32HL69766. JMO and GBC are HHMI instructors supported by the HHMI Professor's award. This work was supported by NIH grants R01EY008152 and R01HL067395 to UB. We thank G. Struhl (Columbia University) and P. O'Farrell (University of California, San Francisco) for providing DNA reagents.
Abbreviations
- G-TRACE
Gal4 Technique for Real-time Clonal Expression
- Gal4-ET
Gal4 enhancer trap
Footnotes
Editorial Summaries AOP: A Gal4-based system in Drosophila reports on gene expression at a given developmental stage combined with lineage information on expression at earlier developmental stages.
Issue: A Gal4-based system in Drosophila reports on gene expression at a given developmental stage combined with lineage information on expression at earlier developmental stages.
REFERENCES
- 1.Elliott DA, Brand AH. Methods Mol Biol. 2008;420:79–95. doi: 10.1007/978-1-59745-583-1_5. [DOI] [PubMed] [Google Scholar]
- 2.Pfeiffer BD, et al. Proc Natl Acad Sci U S A. 2008;105(28):9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi S, et al. Genesis. 2002;34(1-2):58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, et al. PLoS Biol. 2005;3(2):e59. doi: 10.1371/journal.pbio.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Call GB, et al. Genetics. 2007;177(2):689–697. doi: 10.1534/genetics.107.077735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pignoni F, Zipursky SL. Development. 1997;124(2):271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, et al. Development. 1997;124(4):761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 8.Struhl G, Basler K. Cell. 1993;72(4):527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 9.Weigmann K, Cohen SM. Development. 1999;126(17):3823–3830. doi: 10.1242/dev.126.17.3823. [DOI] [PubMed] [Google Scholar]
- 10.Jung SH, et al. Development. 2005;132(11):2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- 11.Kurucz E, et al. Proc Natl Acad Sci U S A. 2003;100(5):2622–2627. doi: 10.1073/pnas.0436940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasugi T, et al. Development. 2008;135(8):1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- 13.Han Z, Olson EN. Development. 2005;132(15):3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- 14.Barolo S, Castro B, Posakony JW. Biotechniques. 2004;36(3):436–440. 442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.