Abstract
Breast cancer metastasis suppressor 1 (BRMS1) is a predominantly nuclear protein that differentially regulates expression of multiple genes, leading to suppression of metastasis without blocking orthotopic tumor growth in multiple human and murine cancer cells of diverse origins. We hypothesized that miR-146 may be involved in the ability of BRMS1 to supress metastasis because miR-146 expression is altered by BRMS1 and because BRMS1 and miR-146 are both associated with decreased signaling through the nuclear factor-κB pathway. BRMS1 significantly up-regulates miR-146a by 6- to 60-fold in metastatic MDA-MB-231 and MDA-MB-435 cells, respectively, and miR-146b by 40-fold in MDA-MB-435 as measured by real-time quantitative reverse transcription-PCR. Transduction of miR-146a or miR-146b into MDA-MB-231 down-regulated expression of epidermal growth factor receptor, inhibited invasion and migration in vitro, and suppressed experimental lung metastasis by 69% and 84%, respectively (mean ± SE: empty vector = 39 ± 6, miR-146a = 12 ± 1, miR-146b = 6 ± 1). These results further support the recent notion that modulating the levels of miR-146a or miR-146b could have a therapeutic potential to suppress breast cancer metastasis.
Introduction
Metastasis suppressors regulate diverse pathways to block metastasis without preventing orthotopic tumor growth (1). Breast cancer metastasis suppressor 1 (BRMS1) affects multiple steps in the metastatic cascade leading to ∼90% metastasis suppression in xenograft models of breast carcinoma, melanoma, and ovarian carcinoma (2, 3). Mechanistically, BRMS1 regulates expression of multiple genes linked to metastasis, including osteopontin (OPN), urokinase-type plasminogen activator (uPA), epidermal growth factor receptor (EGFR), fascin, and connexins (reviewed in ref. 1). Transcriptional regulation by BRMS1 has been proposed to occur through interaction with large SIN3:HDAC chromatin remodeling complexes based on multiple protein-protein interaction studies (3, 4). A second, although not mutually exclusive, mechanism involves the negative regulation of nuclear factor-κB (NF-κB) through direct interaction with RelA/p65 and inhibition of IκBα phosphorylation (5, 6). Regardless of the specific mechanism by which BRMS1 regulates transcription, multiple changes in gene and protein expression have been verified by microarray and proteomic analyses when BRMS1 was reexpressed in cells (7–9).
We had hypothesized that BRMS1 could exert its antimetastatic action by differentially regulating expression of microRNA (miRNA or miR). These are transcribed genes processed to single-stranded regulatory RNA of ∼22 nucleotides (10). Mature miR represses protein expression primarily through base pairing of a seed region with the 3′ untranslated region (UTR) of the target mRNA leading to inhibition of translation and/or mRNA degradation. The hypothesis is supported by microarrays showing multiple changes in miR expression on ectopic expression of BRMS1.7
Because miR-146 plays a role in regulating NF-κB (11, 12) and because miR-146 is more abundantly expressed in BRMS1-expressing cells, we chose this miR to directly test whether miR-146 could be a downstream mediator of metastasis suppression by BRMS1. miR-146a and miR-146b are distinct genes encoded on chromosomes 5q33 and 10q24, respectively. The mature products have similar targets because they differ only by two nucleotides near the 3′ end. Recently, miR-146a and miR-146b were shown to inhibit both migration and invasion, suggesting that they play a role in metastasis (12). In this report, we show that BRMS1 up-regulates miR-146a in two breast cancer cell lines, MDA-MB-231 and MDA-MB-435, and miR-146b in MDA-MB-435, but not MDA-MB-231. miR-146a and miR-146b both reduce EGFR expression and suppress metastasis. These results add to the growing list of miR genes that regulate metastasis and provide a potential mechanism for how BRMS1 suppresses metastasis by altering the expression profile of metastasis-associated miR.
Materials and Methods
Cell lines and cell culture
MDA-MB-231, MDA-MB-435, MDA-MB-436, and T47D cell lines were cultured in a mixture (1:1, v/v) of DMEM and Ham's F12 medium (Invitrogen) supplemented with 2 mmol/L l-glutamine (Invitrogen), 0.02 mmol/L nonessential amino acids (Mediatech), and 5% fetal bovine serum (Atlanta Biologicals). The immortalized, nontumorigenic breast epithelial cell line MCF10A was cultured in a mixture (1:1, v/v) of DMEM and Ham's F12 medium supplemented with 5% horse serum, 2 mmol/L l-glutamine, 0.02 mmol/L nonessential amino acids, 10 ng/mL EGF, 0.5 μg/mL hydrocortisone, 0.1 μg/mL cholera toxin, and 10 μg/mL insulin (Sigma). MCF7 was cultured in MEM with l-glutamine and Earle's salts (Invitrogen) supplemented with 10% fetal bovine serum, 0.1 mmol/L nonessential amino acids, 10 μg/mL insulin, and 1 mmol/L sodium pyruvate. Neither antibiotics nor antimycotics were used. All cell lines were confirmed negative for Mycoplasma spp. infection using a PCR-based test (TaKaRa). Transduction of cells with BRMS1 or miR-146 was performed as previously described (2, 3, 12).
Quantitative real-time reverse transcription-PCR
miRNA expression was determined by collecting total RNA from 70% to 90% confluent cell cultures. Medium was aspirated, and cells were rinsed in ice-cold calcium- and magnesium-free Dulbecco's PBS. Cells were lysed with Qiazol (Qiagen) and RNA was separated using the miRNeasy system for purifying total RNA (Qiagen). MiScript primers specific for mature miRNA were hs-miR-146a_1 and hs-miR-146b_1 (Qiagen). All samples were normalized to small nRNA U6 and fold changes were calculated as previously described (13).
Western blotting
Total protein lysates were prepared as previously described (12) and probed for EGFR expression (rabbit polyclonal; Santa Cruz Biotechnology, Inc.). Relative expression level was determined by densitometry normalized to the expression of β-actin (mouse monoclonal; Santa Cruz Biotechnology).
Metastasis assays
Cells at 80% to 90% confluence were detached using 0.2 mmol/L EDTA in Ca+2-free, Mg+2-free, and NaHC03-free HBSS. Viable cells were counted using a hemacytometer and resuspended at a final concentration of 2.5 × 106/mL in ice-cold HBSS. Female athymic mice (3−4 wk; Harlan Sprague Dawley) were injected with 0.2 mL cell suspension into the lateral tail vein. Mice were necropsied 10 wk after inoculation following anesthesia with ketamine:xylazine and euthanasia by cervical dislocation. All organs were observed for the presence of macroscopic metastases. Lungs were removed and fixed in a mixture of Bouin's fixative and neutral buffered formalin (1:5, v/v). Mice were maintained under the guidelines of the NIH and the University of Alabama at Birmingham Institutional Animal Care and Use Committee. Food and water were provided ad libitum.
Statistical analyses
The number of lung metastases was compared for miR-146a–transduced or miR-146b–transduced cell lines to the vector only–transduced line. A Kruskal-Wallis ANOVA of ranks procedure was used with Dunn's post hoc test. Calculations were performed using SigmaStat statistical analysis software (SPSS, Inc.). Statistical significance was defined as a probability P ≤ 0.05.
Results
BRMS1 up-regulates miR-146
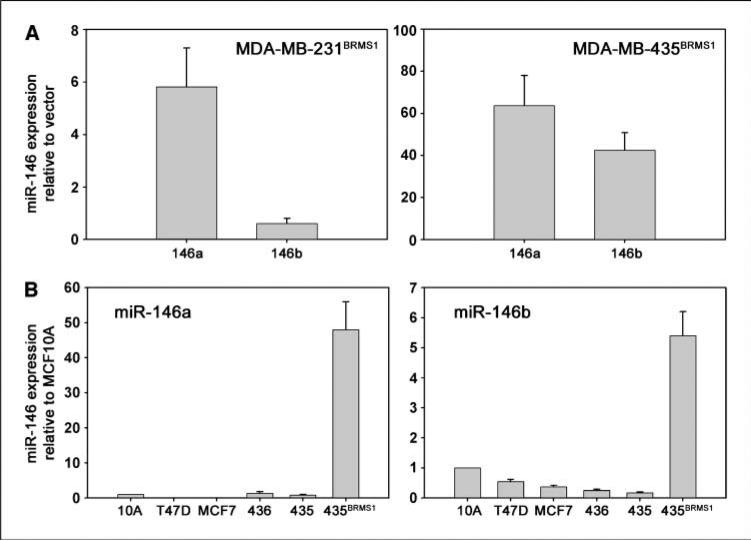
BRMS1 has already been shown to regulate the expression of multiple genes involved in metastasis (1). Microarrays comparing the expression profile of miR between breast cancer cell lines with and without BRMS1 (at near physiologic expression levels) showed alteration of the expression of multiple miR genes that already have been implicated in the process of invasion and metastasis.7 We chose to focus studies with miR-146 because it was reported to be involved with regulation of NF-κB. miR-146a was significantly up-regulated by introduction of BRMS1 into the MDA-MB-231 and MDA-MB-435 breast cancer cell lines (Fig. 1A). Additionally, miR-146b was significantly up-regulated by BRMS1 in MDA-MB-435, but not MDA-MB-231 (Fig. 1A).
Figure 1.
BRMS1 up-regulates miR-146. A, ectopic expression of BRMS1 in MDA-MB-231 or MDA-MB-435 breast cancer cell lines significantly enhanced the expression of miR-146a as shown by real-time RT-PCR (∼6-fold and ∼60-fold, respectively). miR-146b expression was also increased in MDA-MB-435 (∼40-fold). B, endogenous expression levels of miR-146a and miR-146b in multiple breast-derived cell lines generally show decreased levels in tumorigenic but weakly/nonmetastatic cell lines (MCF7, T47D, and MDA-MB-436) compared with the immortalized, nontumorigenic breast epithelial cell line (MCF10A) and a further decrease was found in metastatic MDA-MB-435. Please note scale differences.
To assess whether miR-146a or miR-146b levels were altered in other breast cancer cell lines, a panel of breast-derived cell lines was analyzed to measure relative levels of miR-146. Basal expression levels generally decreased for both miR-146a and miR-146b in tumorigenic but weakly/nonmetastatic cell lines MCF7, T47D, and MDA-MB-436 compared with the immortalized but not tumorigenic breast epithelial MCF10A and a further decrease was found in metastatic MDA-MB-435 (Fig. 1B).
To address whether the miR-146 changes were specific to BRMS1, cells were transfected with small interfering RNA targeting BRMS1. With a 70% to 90% reduction in BRMS1 expression, miR-146a and miR-146b were reduced by 20% to 50% at 48 hours (data not shown). The magnitude of the changes varied between experiments, but the trends were comparable, perhaps explained by miR stability and shared processing machinery (10).
miR-146a and miR-146b down-regulate EGFR
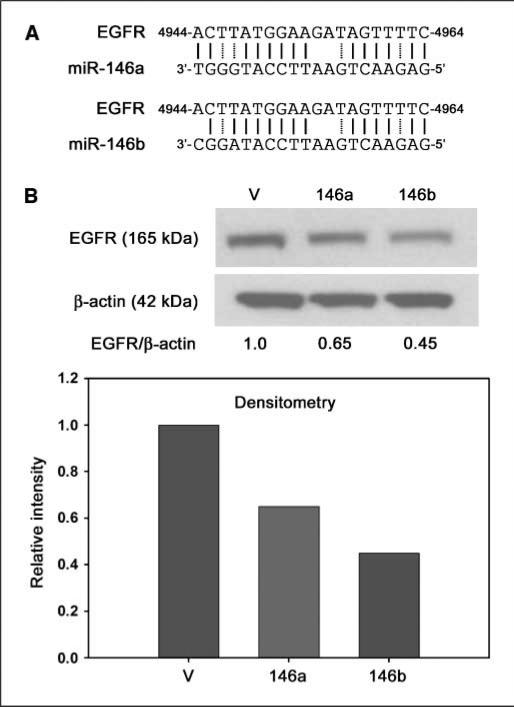
Recently, BRMS1 was shown to directly down-regulate transcription of EGFR (13). However, it was recently discovered that EGFR is also a predicted target of miR-146a and miR-146b, in addition to nine other human miR, based on predicted base pairing using miRBase targets (Sanger database; ref. 14). The predicted target sequence is in the 3′ UTR at position 4944 to 4964 of EGFR (numbering based on NG_007726; Fig. 2A). Because miR-146 was significantly up-regulated by BRMS1 and because EGFR is a predicted target of miR-146, we tested whether ectopic expression of either miR-146a or miR-146b could alter EGFR protein expression. A ∼50% reduction in protein was shown by immunoblotting and densitometry in the MDA-MB-231 cell line on transduction with miR-146a or miR-146b (Fig. 2B). It is important to note that there is undetectable basal BRMS1 expression in MDA-MB-231 by real-time quantitative reverse transcription-PCR (RT-PCR) or immunoblot.
Figure 2.
EGFR protein expression is decreased by miR-146. A, EGFR is a predicted target of miR-146a and miR-146b. The EGFR target sequence is located in the 3′ UTR (numbering is based on accession number NG_007726). Solid lines indicate binding sites and dashed lines represent low-affinity T-G matches. B, ectopic expression of miR-146a or miR-146b in MDA-MB-231 resulted in a ∼50% reduction in EGFR protein levels as measured by immunoblot and densitometry.
miR-146a and miR-146b suppress metastasis
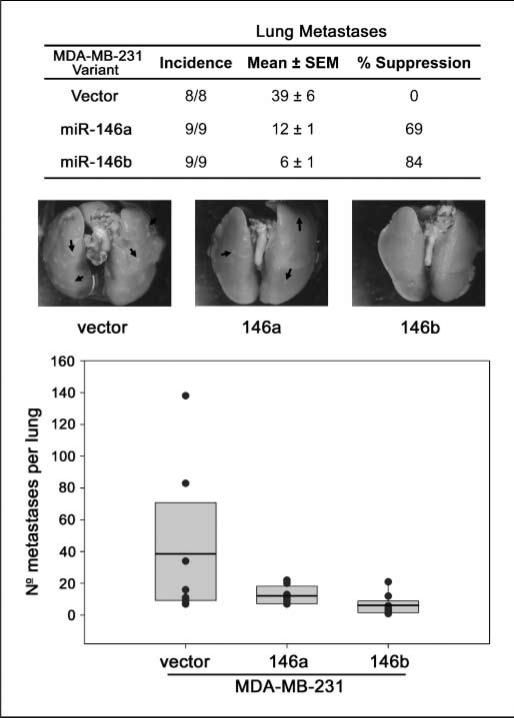
Recently, miR-146a and miR-146b were shown to inhibit migration and invasion in vitro (12). To determine whether either miR could also suppress metastasis, an experimental metastasis assay was performed. MDA-MB-231 cells transduced with miR-146a or miR-146b suppressed pulmonary metastasis by 69% and 84%, respectively (Fig. 3).
Figure 3.
Metastasis is suppressed by miR-146. MDA-MB-231 cells expressing human miR-146a or miR-146b or vector only were injected into the lateral tail vein of athymic mice and the lungs were analyzed for macroscopic metastases. The data are shown graphically with black dots representing the number of pulmonary metastases from each mouse, the box represents the 10th and 90th percentile, and the black line is the mean for each group. The table lists the incidence and the mean number of pulmonary metastases. Representative images for each group are pictured with arrows highlighting some of the individual lung metastases.
Discussion
There is a growing list of miR genes that play specific roles in suppressing or enhancing metastasis. We hypothesized that BRMS1, a metastasis suppressor that regulates the expression of multiple genes, might directly or indirectly regulate expression of miR genes. We found a decrease in several prometastatic miR and an increase in metastasis-suppressive miR on ectopic expression of BRMS1.7 Of the many miR expression changes, we focused on miR-146 because it is involved with regulation of the NF-κB signaling pathway with which BRMS1 was already implicated and because miR-146a and miR-146b have recently been shown to inhibit migration and invasion (12).
Ectopic BRMS1 expression significantly increased miR-146 expression. Moreover, transduction of miR-146a or miR-146b into MDA-MB-231 cells resulted in suppression of metastasis by 69% to 84%. Although there have already been multiple mechanisms associated with BRMS1 suppression of metastasis, results presented in this study, for the first time, suggest that up-regulation of miR-146, whether direct or indirect, contributes to the ability of BRMS1 to suppress metastasis.
Precisely how a BRMS1-miR-146 axis could regulate metastasis is more difficult to determine. However, several possibilities can be deduced from the literature, noting that miR-146a and miR-146b are distinct genes encoded on different chromosomes. The data presented here emphasize that, despite their similarities, both miRs are independently regulated. Recently, a G/C polymorphism was identified in the pre-miR-146a sequence that reduced mature miR-146a levels (15). In addition, the transcription factor PLZF down-regulated miR-146a (16). It is likely, then, that miR-146a and miR-146b are regulated by different mechanisms depending on cell type and the environment in which cells find themselves. In fact, the difference noted between the MDA-MB-231 and MDA-MB-435 cell lines is not completely surprising because the lines are unrelated. In addition, NF-κB is constitutively active in MDA-MB-231 but not in MDA-MB-435 (17). Because NF-κB plays a role in regulating miR-146a and because BRMS1 regulates NF-κB (5, 6), there exists a potential connection that may relate to differences between the regulation of miR-146a and miR-146b in these different cell lines.
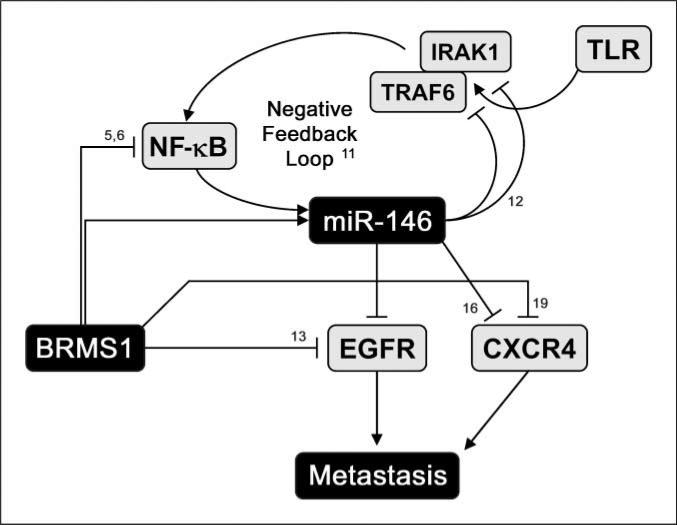
Several targets have been predicted and identified for miR-146a and miR-146b, including IRAK1 (11, 12), TRAF6 (11, 12), interleukin (IL)-8 (12), IL-6 (12), CXCR4 (16), and matrix metalloproteinase-9 (12). EGFR is an additional predicted target (14) and we recently showed that BRMS1 reexpression reduced EGFR in MDA-MB-231 cells by ∼50% (13). Although EGFR transcription is directly down-regulated by BRMS1, miR-146 could be a redundant mechanism and a working model is proposed (Fig. 4). This possibility is supported by complete obliteration of EGFR expression in BRMS1-expressing MDA-MB-435 cells (3, 13) in which both miR-146a and miR-146b were up-regulated.
Figure 4.
Working model for BRMS1-miR-146 axes in metastasis suppression. BRMS1 directly down-regulates transcription of EGFR (13), a predicted target of miR-146. Ectopic expression of miR-146a or miR-146b in cells that do not express BRMS1 has ∼50% lower EGFR. BRMS1 may regulate CXCR4 directly or via miR-146. Both EGFR and CXCR4 correlate with increased metastasis. miR-146 up-regulation by BRMS1 could be direct or indirect (by affecting NF-κB). However, the connection through NF-κB is paradoxical with a negative feedback loop described for NF-κB and miR-146a in response to inflammatory stimulation (11).
The organotropism of metastasis is mediated, in part, by differential expression of chemokine receptors and ligands on tumor cells and in various tissues. For example, breast carcinoma cells frequently express high levels of CXCR4 and frequently colonize lung and bone, which express abundant stromal cell–derived factor-1 (SDF-1), the preferred ligand for CXCR4 (18). Interfering with CXCR4:SDF-1 interactions reduces metastatic potential (19). Although regulation of CXCR4 via a BRMS1-miR-146 axis is not directly tested here, we have observed a decrease in CXCR4 expression by microarray analysis with MDA-MB-231 and MDA-MB-435 cells following BRMS1 reexpression (data not shown). This is consistent with recent reports showing decreased CXCR4 in cells expressing miR-146a (16) or BRMS1 (20).
The biological observations notwithstanding, our findings are paradoxical with regard to NF-κB. Taganov and colleagues (11) report that NF-κB induces miR-146a following induction by lipopolysaccharide, with a corresponding induction of TRAF6 and IRAK1, the latter two that form a negative feedback loop (Fig. 4). However, prior reports show direct BRMS1-RelA binding leading to decreased NF-κB signaling (6). Based on the latter, BRMS1 should reduce miR-146 expression. We observe the opposite. The most likely explanations relate to the aforementioned cell type–specific regulatory mechanisms, including as-yet-unknown cofactors that influence transcription.
These data reinforce the growing consensus that metastasis requires coordinated expression of multiple genes. Suppression of metastasis by BRMS1 may be explained by decreased EGFR, OPN, and/or CXCR4, but we now show that miR-146 also contributes to BRMS1-mediated metastasis suppression. Likewise, the myriad downstream targets of BRMS1 can also be explained because of relative promiscuity of miRNA in targeting gene expression (10).
We also emphasize that the experiments reported here did not use superphysiologic expression of miR to achieve an effect on metastasis in contrast to many previously published reports showing functions of microRNA in cancer cells. This point is key for several reasons. Primarily, off-target effects are more likely if the expression is too high. Likewise, if miR or miR mimetics are to be translated into clinical practice, the efficacy needs to be achievable at reasonable concentrations. miR-146a and miR-146b were elevated 10- to 30-fold in transduced MDA-MB-231 (12), which is consistent with the 6- to 60-fold induction observed in BRMS1-expressing cells. Based on our findings, miR-146a or miR-146b are promising targets for antimeta-static therapy based on the findings presented here.
Acknowledgments
Grant support: USPHS grants CA87728 (D.R. Welch) and F32CA113037 (D.R. Hurst); predoctoral fellowship from the U.S. Army Medical Research and Materiel Command W81-XWH-08-1-0786 (M.D. Edmonds); postdoctoral fellowship from Susan G. Komen for the Cure PDF1122006 (K.S. Vaidya); and National Foundation for Cancer Research, Center for Metastasis Research (D.R. Welch).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank Dr. Janet Price (University of Texas M. D. Anderson Cancer Center, Houston, TX) for providing the MDA-MB-231 and MDA-MB-435 cell lines as well as Drs. Fred Miller and Herbert Soule (Karmanos Cancer Institute, Detroit, MI) for providing MCF10A and related variants.
Footnotes
M.D. Edmonds and D.R. Welch, in preparation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Vaidya KS, Welch DR. Metastasis suppressors and their roles in breast carcinoma. J Mammary Gland Biol Neoplasia. 2007;12:175–90. doi: 10.1007/s10911-007-9049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phadke PA, Vaidya KS, Nash KT, Hurst DR, Welch DR. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol. 2008;172:809–17. doi: 10.2353/ajpath.2008.070772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurst DR, Xie Y, Vaidya KS, et al. Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–44. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meehan WJ, Samant RS, Hopper JE, et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–9. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- 5.Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-κB activity. Cancer Res. 2005;65:3586–95. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol. 2006;26:8683–96. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champine PJ, Michaelson J, Weimer B, Welch DR, DeWald DB. Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exp Metastasis. 2007;24:551–65. doi: 10.1007/s10585-007-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cicek M, Samant RS, Kinter M, Welch DR, Casey G. Identification of metastasis-associated proteins through protein analysis of metastatic MDA-MB-435 and metastasis-suppressed BRMS1 transfected-MDA-MB-435 cells. Clin Exp Metastasis. 2004;21:149–57. doi: 10.1023/b:clin.0000024729.19084.f0. [DOI] [PubMed] [Google Scholar]
- 9.Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006–18. doi: 10.1021/pr0703167. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-κB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;42:5643–7. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaidya KS, Harihar S, Stafford LJ, et al. Breast cancer metastasis suppressor-1 differentially modulates growth factor signaling. J Biol Chem. 2008;283:28354–60. doi: 10.1074/jbc.M710068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la CA. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labbaye C, Spinello I, Quaranta MT, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 17.Coppe JP, Kauser K, Campisi J, Beausejour CM. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem. 2006;281:29568–74. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- 18.Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768:952–63. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SY, Lee CH, Midura BV, et al. Inhibition of the CXCR4/CXCL12 chemokine pathway reduces the development of murine pulmonary metastases. Clin Exp Metastasis. 2008;25:201–11. doi: 10.1007/s10585-007-9133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, Zhang B, Lin Y, Yang Y, Liu X, Lu F. Breast cancer metastasis suppressor 1 inhibits SDF-1α-induced migration of non-small cell lung cancer by decreasing CXCR4 expression. Cancer Lett. 2008;269:46–56. doi: 10.1016/j.canlet.2008.04.016. [DOI] [PubMed] [Google Scholar]