Abstract
The post-transcriptional role of Mss51p in mitochondrial gene expression is of great interest since MSS51 mutations suppress the respiratory defect caused by shy1 mutations. SHY1 is a Saccharomyces cerevisiae homolog of human SURF1, which when mutated causes a cytochrome oxidase assembly defect. We found that MSS51 is required for expression of the mitochondrial reporter gene ARG8m when it is inserted at the COX1 locus, but not when it is at COX2 or COX3. Unlike the COX1 mRNA-specific translational activator PET309, MSS51 has at least two targets in COX1 mRNA. MSS51 acts in the untranslated regions of the COX1 mRNA, since it was required to synthesize Arg8p when ARG8m completely replaced the COX1 codons. MSS51 also acts on a target specified by the COX1 coding region, since it was required to translate either COX1 or COX1:: ARG8m coding sequences from an ectopic COX2 locus. Mss51p was found to interact physically with newly synthesized Cox1p, suggesting that it could coordinate Cox1p synthesis with insertion into the inner membrane or cytochrome oxidase assembly.
Keywords: cytochrome c oxidase/mitochondrial DNA/translation/untranslated regions
Introduction
The inability to assemble fully functional cytochrome c oxidase underlies several encephalomyopathic disorders in humans, caused in most cases by recessive autosomal mutations (Shoubridge, 2001). Genetic studies of cytochrome oxidase deficiencies in the yeast Saccharomyces cerevisiae provide a useful tool for both identifying relevant human genes and helping to elucidate their mechanisms of action (Shoubridge, 2001; Barrientos et al., 2002a; Steinmetz et al., 2002). For example, human SURF1 mutations cause the cytochrome oxidase deficiency associated with Leigh’s syndrome (Tiranti et al., 1998; Zhu et al., 1998), apparently by interfering with efficient assembly of the complex (Tiranti et al., 1999). Mutations in the homologous yeast gene SHY1 similarly reduce cytochrome oxidase activity (Mashkevich et al., 1997) by causing a partial assembly defect (Nijtmans et al., 2001), and reduced pulse labeling of Cox1p in vivo (Barrientos et al., 2002b). Interestingly, missense mutations in the yeast nuclear gene MSS51 can compensate for the loss of Shy1p (Barrientos et al., 2002b). This finding suggests that Shy1p may have particular roles in the assembly and/or synthesis of cytochrome oxidase subunit I (Cox1p), since mss51 null mutations fail to accumulate Cox1p (Decoster et al., 1990).
Cox1p, the largest subunit of the enzyme, is coded in the mitochondrial DNA (mtDNA), synthesized on mitochondrial ribosomes and inserted into the inner membrane from the inside. Cox1p spans the mitochondrial inner membrane 12 times and is an integral part of the catalytic core of the enzyme (Tsukihara et al., 1996). Insertion of Cox1p appears to be an early step in assembly of nuclear and mitochondrially encoded subunits of the complex (Lemaire et al., 1998; Nijtmans et al., 1998). In addition to genes encoding the enzyme subunits, biogenesis of yeast cytochrome oxidase requires a large set of nuclear-encoded genes whose products function in RNA processing, translation and assembly (Tzagoloff and Dieckmann, 1990; Grivell et al., 1999). Several of these function chiefly in the expression of COX1 (Kloeckener-Gruissem et al., 1987; Pel et al., 1992).
Among the nuclear genes required for cytochrome c oxidase activity are those that encode mRNA-specific translational activators for the COX1, COX2 and COX3 mRNAs (Fox, 1996). The translational activator proteins functionally interact with the 5′-untranslated leaders (5′-UTLs) of their target mRNAs. They are bound to the inner surface of the inner membrane (Fox, 1996; Manthey et al., 1998; Green-Willms et al., 2001), and are likely to play a role in localizing organellar translation (Sanchirico et al., 1998). Interestingly, the COX1 mRNA-specific activator protein Pet309p interacts with the COX2 and COX3 mRNA-specific activator proteins, suggesting that together they may co-localize Cox1p, Cox2p and Cox3p synthesis, promoting efficient assembly of the enzyme core (Naithani et al., 2003).
The nuclear gene MSS51 previously has been suggested to encode another mRNA-specific translational activator for the COX1 mRNA (Decoster et al., 1990; Siep et al., 2000). MSS51 was first identified as a gene required for the processing of introns in the COX1 pre-mRNA (Faye and Simon, 1983). However, mss51 mutations did not affect the level of COX1 mRNA in strains lacking mitochondrial introns, but prevented Cox1p pulse-labeling and accumulation, suggesting a role in translation (Decoster et al., 1990; Siep et al., 2000). While Mss51p is associated with the mitochondrial inner membrane (Siep et al., 2000), its mechanism of action remains unclear.
We have studied here the role of Mss51p in COX1 expression and its targets of action using mitochondrial gene rearrangements and the mitochondrial reporter gene ARG8m (Steele et al., 1996). This reporter encodes a matrix-localized biosynthetic enzyme normally specified by the nuclear gene ARG8, and allows us to look directly at mitochondrial translation by scoring growth of yeast in the absence of arginine and accumulation of the reporter protein. Our results confirm that Mss51p is specifically required for COX1 mRNA translation, but its mechanism is clearly distinct from that of Pet309p and the other known mRNA-specific translational activators.
Results
MSS51 is required for expression of the ARG8m mitochondrial reporter inserted at COX1
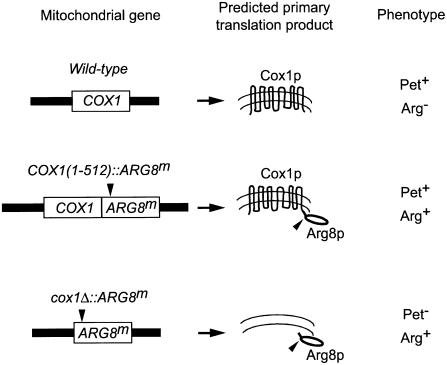
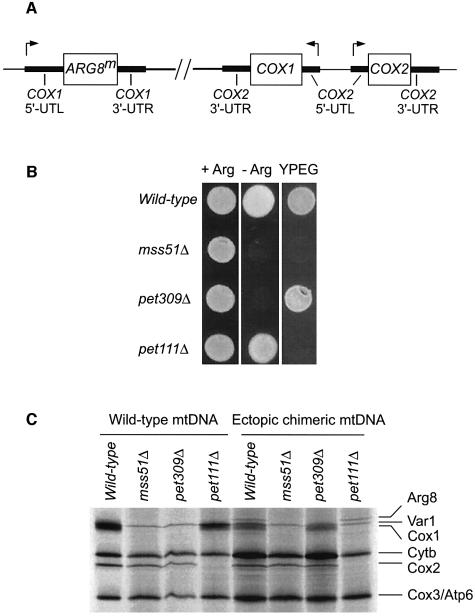
mss51 mutations strongly reduce incorporation of radioactive label into Cox1p in vivo without affecting the level of intronless COX1 mRNA (Decoster et al., 1990; Siep et al., 2000). These data suggest a defect in translation, but cannot rule out the possibility that Cox1p becomes highly unstable in the absence of Mss51p. Indeed, the fact that MSS51 mutations could suppress the assembly defect of shy1 mutants (Barrientos et al., 2002b) suggested that Mss51p could be involved in assembly. We therefore reinvestigated the role of MSS51 in COX1 expression by inserting the mitochondrial reporter gene ARG8m (Steele et al., 1996) at the end of the 512 codon intronless form of the COX1 coding sequence [COX1(1–512)::ARG8m] (Figure 1).
Fig. 1. ARG8m reporter constructs at COX1. The mitochondrial genes, predicted translation products, and arginine and respiratory phenotypes of strains bearing wild-type nuclear genomes are indicated. The open boxes represent the COX1 and ARG8m structural genes. The COX1 5′-UTL and 3′-UTR are indicated as solid boxes. The black triangles indicate the processing site for the pre-Arg8p matrix-targeting signal. In the COX1(1–512)::ARG8m construct, the ARG8m gene was fused in-frame to the complete, 512 COX1 codons, such that the COX1 stop codon was replaced by the initial methionine of the ARG8m gene. In the cox1Δ::ARG8m construct, the ARG8m gene replaced the COX1 codons, and was precisely fused to the COX1 5′-UTL and 3′-UTR.
The ARG8m product is a soluble biosynthetic enzyme normally located in the matrix and whose activity is unaffected by mss51 mutations. Since the ARG8m sequence used here specifies the cleavage site for removal of the matrix-targeting signal of pre-Arg8p, accumulation of processed Arg8p should not be affected by the instability of Cox1p fused upstream of it. We have previously shown that an unstable variant of Cox2p fused upstream of Arg8p did not affect Arg8p stability (Bonnefoy et al., 2001). Thus Arg8p provides a readout of COX1 gene expression at the level of translation.
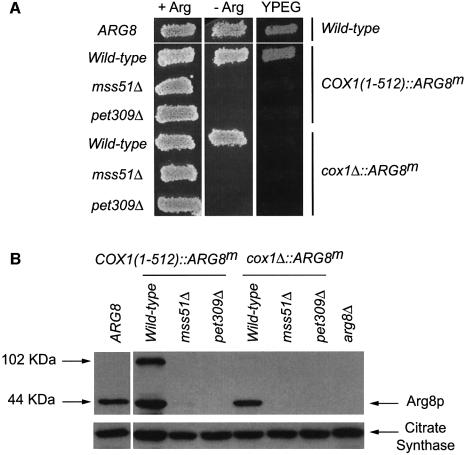
The COX1(1–512)::ARG8m gene was inserted into ρ+ mtDNA and the respiratory and Arg phenotypes of the resulting strain were examined (Figure 2A). In a wild-type nuclear background, this chimeric mitochondrial gene supported normal growth on both non-fermentable carbon sources and medium lacking arginine. However, an mss51Δ, COX1(1–512)::ARG8m strain could not grow on either a non-fermentable carbon sources or a medium lacking arginine, confirming that MSS51 is required for synthesis of Cox1p (Figure 2A). As expected, a pet309Δ, COX1(1–512)::ARG8m strain was also both Pet– and Arg– (Figure 2A). Similar results were obtained with an intron-bearing COX1(1–512)::ARG8m construct derived from strain D273-10B (data not shown).
Fig. 2. MSS51 is required for translation of ARG8m when it is inserted at COX1. (A) Growth phenotypes. Relevant nuclear and mitochondrial genotypes are indicated on the left and right side of the panel, respectively. Yeast cells were grown on YPD and replica plated to glucose minimal medium containing (+Arg) or lacking (–Arg) arginine, and complete non-fermentable medium containing ethanol and glycerol (YPEG) as indicated. Cells carrying the nuclear-encoded ARG8 gene were used for arginine growth comparison (ARG8). Cells were grown for 2 days at 30°C. (B) Steady-state accumulation of the reporter Arg8p. A 25µg aliquot of total mitochondrial proteins was separated by 12.5% SDS–PAGE, and the western blot was probed with anti-Arg8p antibody. The membrane was stripped and reprobed with antibody against citrate synthase as a loading control. An arg8 null mutant (arg8Δ) was used as a negative control. Strain details are given in the Supplementary data.
We also created strains in which ARG8m completely replaced the COX1 coding sequence (cox1Δ::ARG8m) (Figure 1). In a wild-type nuclear background, the cox1Δ:: ARG8m mitochondrial gene supported Arg+ growth (Figure 2A). Both the mss51Δ, cox1Δ::ARG8m strain and the pet309Δ, cox1Δ::ARG8m strain were Arg–, indicating that COX1 5′- and 3′-untranslated regions (UTRs) cause translation to be dependent upon Pet309p, as expected, but also on Mss51p. Pet309p accumulation was not prevented by an mss51Δ mutation (data not shown), suggesting, but not demonstrating, that the two proteins may function independently. mss51Δ mutations specifically prevented expression of ARG8m inserted at COX1. ARG8m expression was not affected by mss51Δ when it was inserted in place of the COX2 or COX3 coding sequences (Supplementary figure 1S, available at The EMBO Journal Online).
The results of western blot analysis of mitochondrial protein extracts confirmed our conclusions based on growth phenotypes (Figure 2B). Mitochondria bearing the intronless COX1(1–512)::ARG8m gene contained processed 44 kDa Arg8p corresponding to the wild-type protein, and a 102 kDa species corresponding in size to the unprocessed Cox1p–preArg8p fusion protein. Mitochon dria from the cox1Δ::ARG8m strain contained a band corresponding to the mature 44 kDa Arg8p. In addition, a band with a size close to 102 kDa was present, although its origin is uncertain. Levels of Arg8p were dramatically reduced in mss51Δ and pet309Δ mitochondria containing either the COX1(1–512)::ARG8m or cox1Δ::ARG8m reporter genes. (Overexposure revealed the presence of low levels of the 44 kDa species in cox1Δ::ARG8m mitochondria.) Similar results were obtained when the intron-containing version of the COX1(1–512)::ARG8m gene was analyzed (data not shown). As expected, the levels of ARG8m mRNA in the mss51Δ mutants bearing the intronless COX1(1–512)::ARG8m gene or the cox1Δ:: ARG8m gene were equivalent to those in otherwise wild-type strains (data not shown).
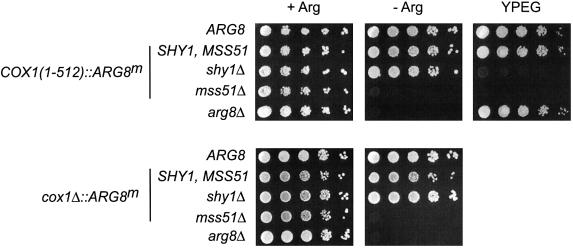
We also investigated the effect of a shy1Δ mutation on expression of the COX1(1–512)::ARG8m and cox1Δ:: ARG8m reporters. The shy1Δ mutation should not block synthesis of the Cox1p–preArg8p reporter protein if Shy1p is only required for assembly. Consistent with this expectation, the shy1Δ, COX1(1–512)::ARG8m strain exhibited an Arg+ phenotype, but failed to grow robustly on non-fermentable carbon sources (Figure 3). This result confirmed that the COX1(1–512)::ARG8m reporter produces an Arg+ phenotype if translated, even if the Cox1p moiety fails to assemble. Similarly, the shy1Δ mutation had no effect on the Arg+ phenotype of a cox1Δ::ARG8m strain (Figure 3). Western analysis confirmed that shy1Δ had no effect on accumulation of the Arg8p reporter protein encoded by COX1(1–512)::ARG8m or cox1Δ:: ARG8m (unpublished results).
Fig. 3. The shy1Δ mutation does not affect the expression of ARG8m in the COX1(1–512)::ARG8m and cox1Δ::ARG8m mitochondrial genes. Ten-fold serial dilutions of cells bearing the COX1(1–512)::ARG8m or cox1Δ::ARG8m mitochondrial alleles were spotted on glucose minimal medium containing (+Arg) or lacking (–Arg) arginine, or complete ethanol/glycerol medium (YPEG), and incubated for 3 days at 30°C. In addition, a strain that expressed the nuclear-encoded ARG8 gene (ARG8) and an arg8 null mutant (arg8Δ) were included. Strain details are given in the Supplementary data.
Taken together, these data demonstrate that MSS51 is required for the synthesis of Arg8p when ARG8m is at the COX1 locus. Moreover, the fact that the expression of the reporter in the strain carrying the cox1Δ::ARG8m gene is dramatically reduced in the mss51Δ mutant indicates that targets of MSS51 action must be located in the UTRs of the COX1 mRNA, since this reporter gene lacks the coding region of COX1 yet requires MSS51 for expression.
MSS51 function is not bypassed by placing the COX3 mRNA 5′-UTL on the COX1 mRNA
Respiratory deficiency caused by the absence of the previously characterized mRNA-specific translational activators, such as PET309, can be phenotypically suppressed by mitochondrial DNA rearrangements that fuse the affected mitochondrial coding sequence to a mitochondrial mRNA 5′-UTL containing the target of another activator (Fox, 1996). Thus, pet309Δ mutations are suppressed in strains containing chimeric COX1 mRNAs lacking introns that bear a 5′-UTL from either the COB or COX3 mRNA (Manthey, 1995; Manthey and McEwen, 1995). If the only function of MSS51 were to work together with PET309 to provide translational activation through the COX1 mRNA 5′-UTL, then mss51 mutations should be bypassed in strains containing the same chimeric mRNAs.
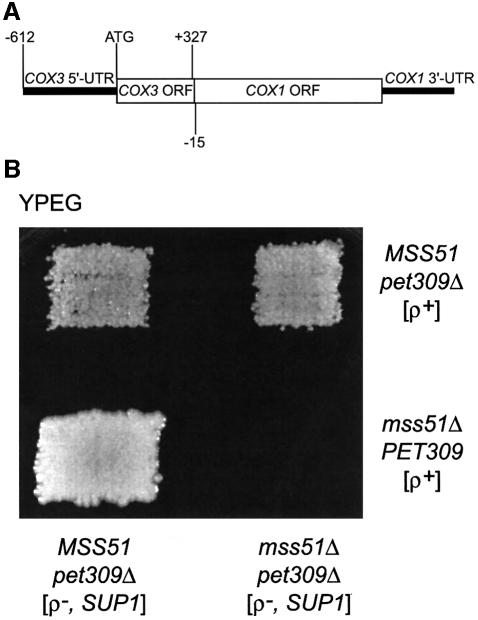
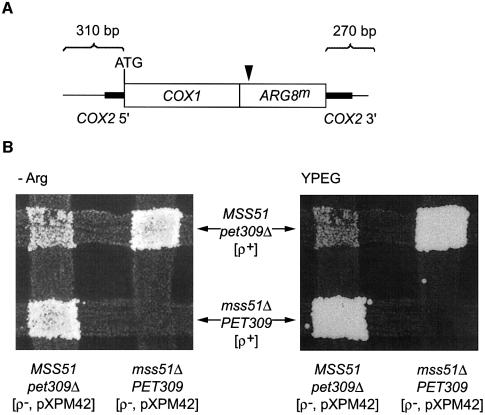
We tested the ability of a mtDNA rearrangement, previously selected to bypass pet309Δ mutations, to bypass an mss51 deletion by generating mss51Δ cells containing both wild-type ρ+ mtDNA and a ρ– mtDNA bearing the chimeric COX1 gene in a heteroplasmic state. The mitochondrial pet309Δ bypass suppressor SUP1 encodes a chimeric mRNA with the COX3 mRNA 5′-UTL and part of the COX3 coding sequence fused to the COX1 coding sequence and COX1 mRNA 3′-UTR (Manthey, 1995) (Figure 4A). We constructed an mss51Δ, pet309Δ strain carrying the SUP1, ρ– genome and mated it to an mss51Δ, PET309 strain bearing wild-type ρ+ mtDNA to generate mss51Δ homozygous cells heteroplasmic for the mtDNAs. These cells failed to respire, indicating that the presence of the COX3 5′ leader on the chimeric mRNA did not allow COX1 expression in the absence of Mss51p (Figure 4B). MSS51 cells homozygous for pet309Δ were respiratory competent as expected (Manthey, 1995). Thus, MSS51 does not simply work in conjunction with PET309 to activate translation through the mRNA 5′-UTL.
Fig. 4. MSS51 function cannot be bypassed by a pet309Δ suppressor. (A) Representation of the mtDNA rearrangement termed SUP1 (Manthey, 1995) that allows COX1 expression in a pet309Δ background. SUP1 arose from a recombination event between the COX3 locus and the COX1 5′-UTL sequence. The resulting chimeric gene has the entire COX3 5′-UTL sequence and the first 327 nucleotides of the COX3 coding region fused to 15 nucleotides of the COX1 5′-UTR and the rest of COX1. (B) Haploid cells containing a ρ– mtDNA bearing the SUP1 gene were patched in vertical stripes. Their relevant nuclear genotypes are indicated. Cells containing wild-type, ρ+ mitochondria were patched on horizontal stripes. Their relevant nuclear genotypes are as indicated. The stripes were cross-printed on YPD medium and allowed to mate. The plate was then printed on non-fermentable YPEG medium and incubated for 5 days at 30°C. Strain details are given in the Supplementary data.
MSS51 function is not bypassed by replacing both the COX1 mRNA 5′-UTL and 3′-UTR with those of the COX2 mRNA
If MSS51 acted only through the COX1 mRNA 3′-UTR or both the 5′-UTL and the 3′-UTR of the COX1 mRNA, then replacement of both UTRs could bypass the requirement for MSS51. We therefore asked whether the COX1 coding sequence could be expressed in the absence of Mss51p from a chimeric mRNA in which both the COX1 mRNA 5′-UTL and 3′-UTR were replaced by the 5′-UTL and 3′-UTR derived from the COX2 mRNA. This construct contains both the COX2 promoter, 54 bp upstream of COX1, and the COX2 dodecamer sequence specifying the 3′ end of the mRNA, 75 bp downstream of COX1 (Dieckmann and Staples, 1994; Mireau et al., 2003). We integrated this chimeric gene ectopically into ρ+ cox1Δ:: ARG8m mtDNA at a site 295 bp upstream of COX2 (Figure 5A), where we know that inserted sequences do not prevent respiratory function (Thorsness and Fox, 1993).
Fig. 5. MSS51 is required to express the COX1 coding sequence when it is fused to the COX2 5′-UTL and 3′-UTR at an ectopic locus. (A) Schematic representation of mtDNA bearing the ectopic chimeric COX1 gene. The COX1 structural gene is flanked by 73 and 119 bp encoding the COX2 5′-UTL and 3′-UTR, respectively (Mireau et al., 2003). This construct was inserted into ρ+ mtDNA, 295 bp upstream of the COX2 gene. The endogenous COX1 gene was replaced with cox1Δ::ARG8m (Materials and methods). Open boxes indicate COX1, COX2 and ARG8m structural genes. The COX1 and COX2 5′-UTLs and 3′-UTRs are indicated as thick lines. The arrows indicate the direction of transcription for each gene. (B) Growth phenotypes of strains carrying the ectopic chimeric COX1 gene. Relevant nuclear genotypes are as indicated. Cells grown on liquid YPD were spotted on glucose minimal medium containing (+Arg) or lacking (–Arg) arginine and on non-fermentable ethanol/glycerol medium (YPEG), and incubated for 2 days at 30°C. The pet111Δ mutation removes the COX2 mRNA-specific translational activator (Poutre and Fox, 1987; Mulero and Fox, 1993). (C) Pulse-labeling of mitochondrial translation products. Cells were labeled with [35S]methionine for 30 min in the presence of cycloheximide, and proteins were analyzed as described in Materials and methods. Cytochrome c oxidase subunit 1, Cox1; subunit 2, Cox2; subunit 3, Cox3; cytochrome b, Cytb; subunit 6 of ATPase, Atp6; ARG8m gene product, Arg8; and the ribosomal protein, Var1. Strain details are given in the Supplementary data.
A strain bearing this mtDNA and a wild-type nuclear genome was respiratory competent, albeit with a less than wild-type growth rate on non-fermentable medium, indicating functional expression of the chimeric COX1 gene (Figure 5B). It was also Arg+ due to expression of cox1Δ::ARG8m. As expected, a strain bearing this mtDNA and a pet309Δ mutation could not express the cox1Δ::ARG8m reporter, but was respiratory competent, indicating that the chimeric COX1 mRNA could be translated in the absence of Pet309p. In contrast, a strain bearing this mtDNA and an mss51Δ mutation exhibited tight non-respiratory as well as Arg– phenotypes (Figure 5B). Therefore, replacement of both the 5′-UTL and 3′-UTR of the COX1 mRNA with those of the COX2 mRNA failed to bypass the requirement for Mss51p in COX1 expression. The failure to respire reflects a block in COX1 expression, since mss51 mutations do not affect expression of other mitochondrial genes (Decoster et al., 1990; Siep et al., 2000) (Supplementary figure 1S).)
We conclude from these data that Mss51p must act on a target or targets encoded within the COX1 coding sequence to allow functional expression of COX1, in addition to and independently of its role, demonstrated above, in working through the COX1 mRNA UTRs to promote translation of the reporter.
Mss51p action on a target specified by the COX1 coding sequence promotes translation
While the experiments described above demonstrate the role of Mss51p in promoting translation of reporter mRNAs bearing COX1 mRNA UTRs, it remained possible that Mss51p could play a different role in promoting expression of the COX1 coding sequence itself when this coding sequence is flanked by COX2 UTRs. For example, Mss51p could interact both with COX1 mRNA UTRs to promote translation, and with nascent Cox1p itself to stabilize the protein during cytochrome oxidase assembly.
To investigate the action of Mss51p on the COX1 coding sequence, we first examined pulse-labeled mitochondrial translation products from strains bearing the ectopic COX1 gene (Figure 5A), in comparison with strains with wild-type mtDNA. In strains with wild-type mtDNA, mss51Δ and pet309Δ mutations completely prevented labeling of Cox1p, while a pet111Δ mutation, which removes the COX2 mRNA-specific translational activator, only reduced Cox1p labeling while eliminating Cox2p, as previously observed (Poutre and Fox, 1987) (Figure 5C). In a strain bearing the ectopic COX1 chimeric gene and a wild-type nuclear genome, Cox1p labeling was reduced relative to a true wild-type, but was easily detectable. As expected, translation of the chimeric mRNA copied from this ectopic gene was not affected by the pet309Δ mutation but was eliminated by the pet111Δ mutation (Figure 5C). Labeling of Cox1p translated from the chimeric mRNA was reduced to undetectable levels by the mss51Δ mutation, suggesting that Mss51p acts at the level of COX1 codon translation, even if the COX1 codons are flanked by the COX2 5′-UTL and 3′-UTR (Figure 5C).
To confirm that Mss51p is required for translation of the chimeric COX1 mRNA bearing the COX2 5′-UTL and 3′-UTR, we fused the ARG8m reporter to its COX1 coding sequence and examined Arg8p expression. For reasons noted above, Arg8p expression from this chimeric gene provides a readout of COX1 gene expression at the level of translation that is independent of the fate of the encoded Cox1p.
We created strains containing only this construct (pXPM42, Figure 6A) as their mtDNA by mitochondrial transformation of a ρ0, and introduced either mss51Δ or pet309Δ nuclear mutations. These strains were mated with strains carrying wild-type ρ+ mtDNA and either mss51Δ or pet309Δ mutations (Figure 6B). The resulting diploids, containing both pXPM42 ρ– and wild-type ρ+ mtDNAs heteroplasmically, were then tested for their ability to grow on Arg selective medium containing glucose, and on non-fermentable medium. As expected, homozygous pet309Δ diploid cells were both Arg+ and competent for respiratory growth, albeit at less than wild-type strength (Figure 6B, upper left patch), demonstrating that this chimeric COX1(1–512)::ARG8m mRNA could be translated in the absence of Pet309p. In contrast, homozygous mss51Δ diploid cells exhibited Arg– and non-respiratory phenotypes (Figure 5B, lower right patch). The Arg– phenotype of these mss51Δ cells confirms that translation of this chimeric mRNA depends on the action of Mss51p upon a target or targets embedded in the COX1 codons. This activity must be distinct from the action of Mss51p on the UTRs of the COX1 mRNA to promote translation.
Fig. 6. Mss51p acts through the COX1 coding sequence to promote COX1 translation. (A) Schematic representation of the chimeric COX1 gene present in the plasmid pXPM42. The COX1(1–512)::ARG8m gene (represented as open boxes) was fused to 310 bp of the COX2 5′-UTL and 270 bp of the COX2 3′-UTR. The 54 bp from the COX2 promoter and 75 bp from the COX2 mRNA 3′ end are represented by thick lines. The black triangle indicates the position of the pre-Arg8p targeting signal cleavage site. (B) Expression of the chimeric COX1(1–512)::ARG8m gene in mss51Δ and pet309Δ mutants. Synthetic ρ–, haploid strains carrying pXPM42 DNA were patched in vertical stripes on YPD. Cells containing wild-type ρ+ mitochondria were patched on YPD on horizontal stripes. Their relevant genotypes are as indicated. The stripes were cross-printed on YPD and allowed to mate overnight before being replicated to glucose minimal medium lacking arginine (–Arg) and complete non-fermentable ethanol/glycerol medium (YPEG). These plates were incubated for 5 days at 30°C to reveal the phenotype of the resulting heteroplasmic diploid strains. Strain details are given in the Supplementary data.
Mss51p interacts with newly synthesized Cox1p
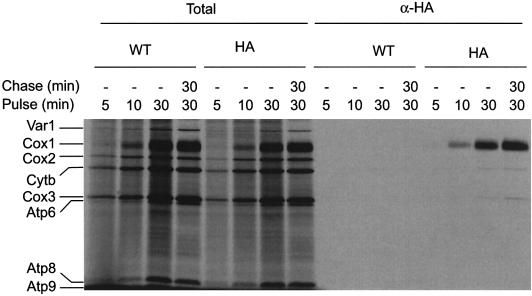
We next asked whether newly synthesized Cox1p itself could be a target of Mss51p. To explore this, we attached triple hemagglutinin (HA) epitopes to the C-terminus of Mss51p for co-immune precipitation experiments. The respiratory competence of Mss51p-HA and the untagged Mss51p cells was comparable, indicating that Mss51p-HA was functional. Translation in mitochondria isolated from strains bearing either the MSS51-HA or the MSS51 gene was carried out in the presence of [35S]methionine for 5–30 min, and then the mitochondria were solubilized with digitonin. Interaction with newly synthesized Cox1p was assessed by immunoprecipitation using an HA-specific antibody, and the immunoprecipitate was analyzed by SDS–PAGE. Radiolabeled Cox1p was detectable in both the total mitochondrial samples and in the immunoprecipitate from the MSS51-HA mitochondria after the 5 min pulse, and increased in amount during the course of the experiment (Figure 7). The labeled Cox1p was stable during a 30 min chase, as was its interaction with Mss51p-HA. The newly synthesized Cox1p was not immunoprecipitated from mitochondria containing untagged Mss51p, demonstrating specificity of the reaction. The immunoprecipitates from both extracts contained trace amounts of cytochrome b and Cox3p, although these signals were slightly stronger in the presence of Mss51p-HA. Interaction of newly synthesized Cox1p with Mss51p-HA was also observed when immunoprecipitation was performed in mitochondria solubilized with Triton X-100 (data not shown).
Fig. 7. Mss51p interacts with newly synthezised Cox1p. Translation in mitochondria isolated from MSS51 (WT) or MSS51-HA (HA) strains was performed at 25°C in the presence of [35S]methionine. Samples were removed at the indicated labeling times (pulse). A fourth sample was pulse-labeled for 30 min and then chased by addition of 10 mM cold methionine followed by incubation at 25°C for 30 min (chase). Mitochondria were washed and solubilized, and the solubilized products were immunoprecipitated with anti-HA antibody (α-HA). Translation products were analyzed by SDS–PAGE and autoradiography. The total aliquots represent 10% of the aliquots used for immunoprecipiation. Abbreviations are as in Figure 5C; ATPase subunit 8, Atp8; subunit 9, Atp9. Strain details are given in the Supplementary data.
Discussion
We have examined the role of the nuclear gene product Mss51p in translation of the mitochondrial COX1 mRNA, and have found that it is remarkably different from those of previously described mRNA-specific translational activators. The COX1, COX2 and COX3 mitochondrially coded mRNAs are translated only when their previously described mRNA-specific activators are present and can functionally interact with the target mRNA 5′-UTLs (Fox, 1996). Interaction of these activators with each other is likely to promote correct localization of translation on the inner membrane (Sanchirico et al., 1998; Naithani et al., 2003). The requirement for any one of these three translational activators can be bypassed by replacing its target 5′-UTL with the 5′-UTL of an mRNA encoding another mitochondrial membrane protein (Costanzo and Fox, 1986; Mulero and Fox, 1993; Manthey and McEwen, 1995). Our evidence shows that nuclear-encoded Mss51p has a novel role in controlling the mitochondrial synthesis of Cox1p. Unlike mutations affecting other mRNA-specific translational activators, including the COX1 mRNA-specific activator Pet309p, mss51 mutations have never been suppressed by mitochondrial gene rearrangements that allow other translational activators to work on the COX1 mRNA (Decoster et al., 1990) (our unpublished results). Here, we have confirmed that a chimeric COX3–COX1 mRNA that is translated in the absence of Pet309p (Manthey, 1995; Manthey and McEwen, 1995) cannot be expressed in the absence of Mss51p. Thus the role of Mss51p in allowing Cox1p accumulation is mechanistically distinct from that of Pet309p and the other mRNA-specific translational activators described to date. Indeed, it seemed possible that the apparent lack of Cox1p synthesis in mss51 mutants could be due to extremely rapid degradation of pulse-labeled protein.
To re-examine the role of Mss51p in Cox1p accumulation, we employed the synthetic mitochondrial reporter gene ARG8m (Steele et al., 1996). Since ARG8m expression was not blocked by mss51 mutations when it was inserted into the COX2 and COX3 loci, it is clear that neither its coding sequence nor its protein product, Arg8p, is intrinsically affected by Mss51p. ARG8m expression depends upon MSS51 function when ARG8m is substituted for the COX1 coding sequence at the COX1 locus. This clearly demonstrates that MSS51 is required for translation of the reporter mRNA bearing COX1 5′- and 3′-UTRs in the absence of COX1 codons. ARG8m expression was also MSS51 dependent when the reporter sequence was fused to the end of the COX1 coding sequence. These observations, taken together with the inability of a 5′-UTL substitution to bypass the requirement for MSS51, demonstrate that Mss51p has a specific yet novel role in promoting translation of the COX1 mRNA, that must involve at least one target outside the mRNA 5′-UTL. Our results cannot distinguish possible roles for the COX1 5′-UTL, 3′-UTR or both as the targets of Mss51p action, nor whether it acts directly on the mRNA.
Surprisingly, in addition to being required for translation of mRNAs bearing COX1 mRNA UTRs, Mss51p was also required for translation of chimeric mRNAs bearing the COX1 coding sequence, even when no COX1 untranslated sequences were present. We inserted into an ectopic site in mtDNA the COX1 coding sequence, flanked by the COX2 promoter and 5′-UTL, and by the COX2 3′-UTR. As expected, Cox1p encoded by this chimeric gene was both synthesized and functionally expressed at the level of respiratory growth in a pet309Δ strain, since the COX2 mRNA-specific activator Pet111p could substitute for Pet309p. However, Cox1p encoded by this chimeric gene could not be synthesized in the absence of Mss51p. Since Mss51p is not required for expression of either COX2 or ARG8m coding sequences at the COX2 locus, these data show that Mss51p must have a target of action specified by the COX1 coding sequence. Moreover, the requirement for Mss51p for expression of COX1 codons is at the translational level, since the expression from the ectopic COX2 locus of the reporter ARG8m fused to the COX1 coding sequence also exhibited a strong requirement for MSS51.
Our findings suggest that Mss51p has a role in translation elongation during Cox1p synthesis. It was observed previously that the negative effect of the mss51-3 mutation, which lowers the levels of wild-type Mss51p, is enhanced when combined with the paromomycin resistance mutation PR454 in mtDNA (Decoster et al., 1990; Siep et al., 2000). This supports a role for Mss51p during elongation since PR454 affects a conserved region of the mitochondrial 15S rRNA (Li et al., 1982), reducing the ability of paromomycin to interfere with the decoding process at the ribosomal A-site (Fourmy et al., 1998).
How might Mss51p function through both the UTRs and coding sequence of the COX1 mRNA to allow its translation? Mss51p could act together with Pet309p through the COX1 mRNA 5′-UTL to activate translation. However, Mss51p may have an entirely different role that depends upon the COX1 mRNA 3′-UTR or both the 5′-UTL and 3′-UTR. In any event, another target of Mss51p action maps within the COX1 coding sequence, suggesting that it could govern translation elongation through a regulatory element within the mRNA coding sequence or nascent polypeptide.
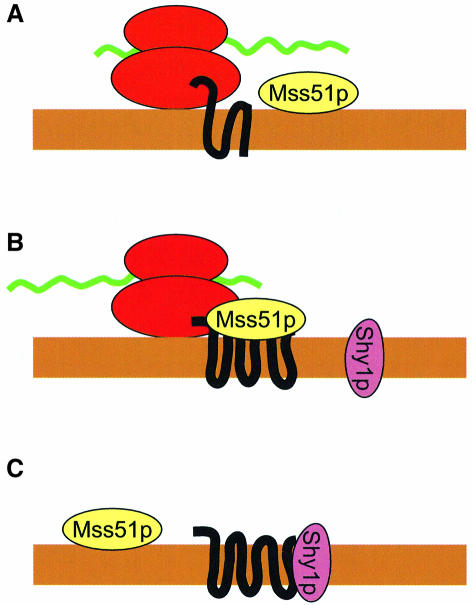
We previously have detected antagonistic translational regulatory elements embedded in the COX2 mRNA coding sequence (Bonnefoy et al., 2001; Williams and Fox, 2003). In that case, a positive element embedded in the mRNA sequence specifying the leader peptide of yeast pre-Cox2p is required to allow efficient translation through three downstream negative elements in the mRNA coding sequence. Mss51p could recognize analogous signals in the COX1 mRNA coding sequence, at the level of RNA. On the other hand, we found that Mss51p interacts with newly synthesized Cox1p, and it is tempting to speculate that Cox1p itself may be the target that Mss51p recognizes to promote translation. In one possible model, the nascent Cox1p polypeptide could inhibit completion of translation by the elongating ribosome. While we have not established this, regulation of translation by nascent chains is a well established phenomenon (Lovett and Rogers, 1996; Tenson and Ehrenberg, 2002). In this model, if Mss51p is available to bind to Cox1p, then translation could proceed to completion. The binding of Mss51p to Cox1p could then promote productive interactions with Shy1p and possibly other assembly proteins (Figure 8). Release of Mss51p at this stage of the assembly pathway would allow it to interact with another nascent Cox1p. By mechanisms such as this, Mss51p could tightly coordinate Cox1p synthesis with downstream events leading to cytochrome oxidase assembly.
Fig. 8. A model for regulation of Cox1p synthesis based on its physical interaction with Mss51p. This model is based on the possibility that the target specified by the COX1 coding sequence, through which Mss51p promotes Cox1p synthesis, is Cox1p itself. We propose the following. (A) Translation elongation of the COX1 mRNA (green line) may be arrested by interaction of the nascent Cox1p polypeptide (black, thick line) with the ribosome. (B) Interaction of Mss51p with nascent Cox1p allows completed translation. (C) Mss51p ‘hands off’ newly synthesized Cox1p to Shy1p, and possibly other proteins, for assembly. Mss51p is then released and available to interact with another Cox1p nascent polypeptide as shown in (A). The mitochondrial inner membrane is shown as a brown bar. For simplicity, the action of Mss51p through the COX1 mRNA UTRs to promote translation was not included in this model, since the target could involve the COX1 5′-UTR, the 3′-UTR or both.
Regulation of translation by feedback controls from membrane insertion is an established phenomenon. Recently, evidence has been presented that completion of Escherichia coli secM translation elongation is modulated by a nascent polypeptide sequence that interacts with the ribosome exit tunnel, blocking further translation if membrane insertion is stalled (in this case, regulating translation of the neighboring secA coding sequence) (Nakatogawa and Ito, 2002). Feedback translational control systems monitoring protein complex assembly have also been described for flagella in bacteria (Aldridge and Hughes, 2002) and for photosystem complexes in chloroplasts (Choquet et al., 2001). One can easily imagine that the presence of unassembled Cox1p in the mitochondrial inner membrane could be deleterious, and that mechanisms to prevent its synthesis in the absence of assembly may have evolved.
Mitochondrial genetic systems have diverged dramatically during their evolution from an apparent common ancestor (Gray et al., 1999). The extent to which common regulatory mechanisms may have been conserved remains unclear. The SURF1/SHY1 cytochrome oxidase assembly function appears to be conserved from yeast to humans. Proteins highly similar to S.cerevisiae Mss51p, and likely to be orthologous, are encoded in the genomes of both Neurospora crassa and Schizosaccharomyces pombe (accession Nos EAA28045 and T50191, respectively), as well as closely related budding yeasts (Cliften et al., 2003; Kellis et al., 2003). No clear homologs of Mss51p are present in currently available animal genomes; however, highly diverged orthologs may be present in animals.
Materials and methods
Strains, media and genetic methods
Saccharomyces cerevisiae strains used in this study are listed in Supplementary table IS. Standard genetic methods and media recipes were as previously described (Rose et al., 1988; Fox et al., 1991). Complete fermentable media were YPD or YPRaf (containing 2% glucose or 2% raffinose). Non-fermentable medium was YPEG (3% glycerol, 3% ethanol) or 2% lactate (Diekert et al., 2001). Minimal medium was synthetic complete (0.67% yeast nitrogen base, 2% glucose) lacking arginine. The mss51Δ::LEU2 and shy1Δ::URA3 deletion constructs were obtained by PCR. The pet309::URA3 disruption cassette was obtained from Manthey and McEwen (1995). Mss51p was tagged at its C-terminus with an HA3 epitope by transformation of DAU1 with a PCR product (Schneider et al., 1995).
Plasmids pXPM18, pXPM19, pXPM34i and pXPM42 (see below) were transformed into the ρ0 strain NAB69 by high-velocity microprojectile bombardment (Bonnefoy and Fox, 2001). Trans formants were selected by their ability to rescue arginine growth when mated with an arg8m-1 mutant (Bonnefoy and Fox, 2000), or by the ability of the resulting diploids to respire when mated with a strain carrying a cox1D369N mutation, L45 (Meunier et al., 1993).
Transformants with plasmids pXPM18 and pXPM19 (see below) were mated with XPM46 (containing intronless mtDNA; Labouesse, 1990) or NB40-36a (intron-bearing mtDNA derived from D273-10B mitochondria). Cytoductants were selected for their ability to grow on media lacking arginine. The mitochondrial transformant of plasmid pXPM34i (see below) was mated by cytoduction with XPM13a, which carries the cox2-62 mutation (Dunstan et al., 1997) and the cox1Δ::ARG8m construct, and respiring recombinants were selected. In all cases, correct integration of the different constructs into the mtDNA was confirmed by PCR and DNA sequencing.
Construction of chimeric mitochondrial genes
All plasmids were generated by the fusion PCR technique (Ho et al., 1989) using Pfu polymerase (Stratagene) or Taq polymerase (Invitrogen). Total DNA from the strain NB40-36a or plasmid pHD6 (Green-Willms et al., 2001) was used to amplify COX1 and ARG8m sequences, respectively. For the COX1(1–512)::ARG8m construct, the ARG8m codons specifying the precursor Arg8p were fused in-frame to the last exon (453 bp) of the COX1 coding sequence, and upstream 986 bp of the COX1 3′-UTR. This product was ligated into the PstI–XhoI sites of pBluescript to generate plasmid pXPM18. This plasmid was used to integrate the COX1(1–512)::ARG8m construct into the intronic COX1 locus derived from D273-10B mitochondria or the intronless COX1 locus derived from CK520 mitochondria (Labouesse, 1990). The cox1Δ::ARG8m construct was obtained as a precise fusion of the ARG8m gene specifying the precursor Arg8p downstream 790 bp of the COX1 5′-UTL and upstream 986 bp from the COX1 3′-UTR to create plasmid pXPM19 (Figure 1).
Intronless mtDNA of strain XPM46, derived from CK520, and the plasmid pJM2 (Mulero and Fox, 1993) were used as templates for PCRs to amplify COX1 or COX2 sequences, respectively. Using the QuikChange mutagenesis kit (Statagene), a SnaBI site was introduced in pJM2 at position –295 relative to the COX2 start codon to create plasmid pXPM50. The chimeric COX1 gene, with 73 and 119 bp of the COX2 5′-UTL and 3′-UTR (Mireau et al., 2003), respectively, was amplified by fusion PCR. The primers used introduced EcoRV sites at the ends of the PCR product. This amplification product was digested with EcoRV and ligated into the SnaBI site of pXPM50 to create plasmid pXPM34i. In this construct, the COX1 chimeric gene was integrated in the opposite orientation to COX2 (Figure 5).
Plasmid pXPM42 was obtained by a precise fusion of the COX1(1–512)::ARG8m codons to 310 and 271 bp of the COX2 5′-UTL and 3′-UTR, respectively (Figure 7). DNA from the strain XPM78a or from pJM2 was used to amplify COX1(1–512)::ARG8m or COX2 sequences respectively.
Analysis of mitochondrial proteins
Yeast cells were grown in complete raffinose medium until late log phase. Cells were disrupted by vortexing with glass beads, and crude mitochondria were obtained as described by Diekert et al. (2001), except that protease inhibitor cocktail (Sigma) was added instead of phenylmethylsulfonyl fluoride (PMSF). Proteins were separated by SDS–PAGE on a 12.5% gel (Laemmli, 1970), and western blots were probed with an anti-Arg8p antibody (Steele et al., 1996) or an anti-citrate synthase antibody (provided by G.Schatz). Secondary goat anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad) was detected with the ECL kit (Amersham Pharmacia Biotech).
In vivo pulse-labeling of cells with [35S]methionine was performed as previously described (Bonnefoy et al., 2001). After pulse-labeling, cells were chilled on ice in the presence of 10 mM cold methionine. Cells were disrupted by vortexing with glass beads and mitochondria were prepared according to Diekert et al. (2001). The radiolabeled proteins were separated on a 15% polyacrylamide gel and analyzed with a Storm 840 Phosphoimager (Molecular Dynamics, Sunnyvale, CA).
Translation in isolated mitochondria (3 mg protein/ml) from MSS51-HA or MSS51 strains in the presence of [35S]methionine was performed as previously described (Westermann et al., 2001). After translation, mitochondria were washed with 0.6 M sorbitol, 20 mM HEPES pH 7.4, and lysed with a buffer containing 1% digitonin, 100 mM NaCl and 20 mM Tris pH 7.4. Immunoprecipitation of labeled mitochondrial products with an anti-HA antibody (clone 3F10, Roche) was performed as previously described (Herrmann et al., 2001).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.M.Herrmann, S.Funes and W.Neupert for useful discussions and advice on the experiment of Figure 7. This work was supported by The Pew Charitable Trusts Fellowship (to X.P.-M), and by a National Institutes of Health research grant GM-29362 (to T.D.F.) and training grant GM07617 (to S.A.B.).
References
- Aldridge P. and Hughes,K.T. (2002) Regulation of flagellar assembly. Curr. Opin. Microbiol., 5, 160–165. [DOI] [PubMed] [Google Scholar]
- Barrientos A., Barros,M.H., Valnot,I., Rotig,A., Rustin,P. and Tzagoloff,A. (2002a) Cytochrome oxidase in health and disease. Gene, 286, 53–63. [DOI] [PubMed] [Google Scholar]
- Barrientos A., Korr,D. and Tzagoloff,A. (2002b) Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J., 21, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy N. and Fox,T.D. (2000) In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol. Gen. Genet., 262, 1036–1046. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N. and Fox,T.D. (2001) Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol., 65, 381–396. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Bsat,N. and Fox,T.D. (2001) Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol. Cell. Biol., 21, 2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Wostrikoff,K., Rimbault,B., Zito,F., Girard-Bascou,J., Drapier,D. and Wollman,F.A. (2001) Assembly-controlled regulation of chloroplast gene translation. Biochem. Soc. Trans, 29, 421–426. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam,P., Desikan,A., Fulton,L., Fulton,B., Majors,J., Waterston,R., Cohen,B.A. and Johnston,M. (2003) Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science, 301, 71–76. [DOI] [PubMed] [Google Scholar]
- Costanzo M.C. and Fox,T.D. (1986) Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol. Cell. Biol., 6, 3694–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decoster E., Simon,M., Hatat,D. and Faye,G. (1990) The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol. Gen. Genet., 224, 111–118. [DOI] [PubMed] [Google Scholar]
- Dieckmann C.L. and Staples,R.R. (1994) Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int. Rev. Cytol., 152, 145–181. [DOI] [PubMed] [Google Scholar]
- Diekert K., de Kroon,A.I., Kispal,G. and Lill,R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol., 65, 37–51. [DOI] [PubMed] [Google Scholar]
- Dunstan H.M., Green-Willms,N.S. and Fox,T.D. (1997) In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′-untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics, 147, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G. and Simon,M. (1983) Analysis of a yeast nuclear gene involved in the maturation of mitochondrial pre-messenger RNA of the cytochrome oxidase subunit I. Cell, 32, 77–87. [DOI] [PubMed] [Google Scholar]
- Fourmy D., Yoshizawa,S. and Puglisi,J.D. (1998) Paromomycin binding induces a local conformational change in the A-site of 16S rRNA. J. Mol. Biol., 277, 333–345. [DOI] [PubMed] [Google Scholar]
- Fox T.D. (1996) Genetics of mitochondrial translation. In Hershey,J.W.B., Matthews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 733–758. [Google Scholar]
- Fox T.D., Folley,L.S., Mulero,J.J., McMullin,T.W., Thorsness,P.E., Hedin,L.O. and Costanzo,M.C. (1991) Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol., 194, 149–165. [DOI] [PubMed] [Google Scholar]
- Gray M.W., Burger,G. and Lang,B.F. (1999) Mitochondrial evolution. Science, 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Green-Willms N.S., Butler,C.A., Dunstan,H.M. and Fox,T.D. (2001) Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem., 276, 6392–6397. [DOI] [PubMed] [Google Scholar]
- Grivell L.A., Artal-Sanz,M., Hakkaart,G., de Jong,L., Nijtmans,L.G., van Oosterum,K., Siep,M. and van der Spek,H. (1999) Mitochondrial assembly in yeast. FEBS Lett., 452, 57–60. [DOI] [PubMed] [Google Scholar]
- Herrmann J.M., Westermann,B. and Neupert,W. (2001) Analysis of protein–protein interactions in mitochondria by coimmuno precipitation and chemical cross-linking. Methods Cell Biol., 65, 217–230. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson,N., Endrizzi,M., Birren,B. and Lander,E.S. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature, 423, 241–254. [DOI] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B., McEwen,J.E. and Poyton,R.O. (1987) Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae: multiple trans-acting nuclear genes exert specific effects on the expression of each of the cytochrome c oxidase subunits encoded on mitochondrial DNA. Curr. Genet., 12, 311–322. [DOI] [PubMed] [Google Scholar]
- Labouesse M. (1990) The yeast mitochondrial leucyl-tRNA synthetase is a splicing factor for the excision of several group I introns. Mol. Gen. Genet., 224, 209–221. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lemaire C., Robineau,S. and Netter,P. (1998) Molecular and biochemical analysis of Saccharomyces cerevisiae cox1 mutants. Curr. Genet., 34, 138–145. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff,A., Underbrink-Lyon,K. and Martin,N.C. (1982) Identification of the paromomycin-resistance mutation in the 15S rRNA gene of yeast mitochondria. J. Biol. Chem., 257, 5921–5928. [PubMed] [Google Scholar]
- Lovett P.S. and Rogers,E.J. (1996) Ribosome regulation by the nascent peptide. Microbiol. Rev., 60, 366–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey G.M. (1995) A study of the nuclear gene, PET309 and its roles in regulating the expression of the mitochondrial COX1 gene in Saccharomyces cerevisiae. PhD Thesis, Microbiology and Molecular Genetics, University of California, Los Angeles, p. 160. [Google Scholar]
- Manthey G.M. and McEwen,J.E. (1995) The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J., 14, 4031–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey G.M., Przybyla-Zawislak,B.D. and McEwen,J.E. (1998) The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem., 255, 156–161. [DOI] [PubMed] [Google Scholar]
- Mashkevich G., Repetto,B., Glerum,D.M., Jin,C. and Tzagoloff,A. (1997) SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J. Biol. Chem., 272, 14356–14364. [DOI] [PubMed] [Google Scholar]
- Meunier B., Lemarre,P. and Colson,A.-M. (1993) Genetic screening in Saccharomyces cerevisiae for large numbers of mitochondrial point mutations which affect structure and function of catalytic subunits of cytochrome-c oxidase. Eur. J. Biochem., 213, 129–135. [DOI] [PubMed] [Google Scholar]
- Mireau H., Arnal,H. and Fox,T.D. (2003) Expression of Barstar as a selectable marker within yeast mitochondria. Mol. Gen. Genomics, 270, 1–8. [DOI] [PubMed] [Google Scholar]
- Mulero J.J. and Fox,T.D. (1993) PET111 acts in the 5′-leader of the Saccharomyces cerevisiae mitochondrial COX2 mRNA to promote its translation. Genetics, 133, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S., Saracco,S.A., Butler,C.A. and Fox,T.D. (2003) Interactions among COX1, COX2 and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell, 14, 324–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H. and Ito,K. (2002) The ribosomal exit tunnel functions as a discriminating gate. Cell, 108, 629–636. [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., Taanman,J.W., Muijsers,A.O., Speijer,D. and Van den Bogert,C. (1998) Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem., 254, 389–394. [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., Artal Sanz,M., Bucko,M., Farhoud,M.H., Feenstra,M., Hakkaart,G.A., Zeviani,M. and Grivell,L.A. (2001) Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett., 498, 46–51. [DOI] [PubMed] [Google Scholar]
- Pel H.J., Tzagoloff,A. and Grivell,L.A. (1992) The identification of 18 nuclear genes required for the expression of the yeast mitochondrial gene encoding cytochrome c oxidase subunit I. Curr. Genet., 21, 139–146. [DOI] [PubMed] [Google Scholar]
- Poutre C.G. and Fox,T.D. (1987) PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics, 115, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Hieter,P. (1988) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanchirico M.E., Fox,T.D. and Mason,T.L. (1998) Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J., 17, 5796–5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B.L., Seufert,W., Steiner,B., Yang,Q.H. and Futcher,A.B. (1995) Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast, 11, 1265–1274. [DOI] [PubMed] [Google Scholar]
- Shoubridge E.A. (2001) Cytochrome c oxidase deficiency. Am. J. Med. Genet., 106, 46–52. [DOI] [PubMed] [Google Scholar]
- Siep M., van Oosterum,K., Neufeglise,H., van der Spek,H. and Grivell,L.A. (2000) Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr. Genet., 37, 213–220. [DOI] [PubMed] [Google Scholar]
- Steele D.F., Butler,C.A. and Fox,T.D. (1996) Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl Acad. Sci. USA, 93, 5253–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L.M. et al. (2002) Systematic screen for human disease genes in yeast. Nat. Genet., 31, 400–404. [DOI] [PubMed] [Google Scholar]
- Tenson T. and Ehrenberg,M. (2002) Regulatory nascent peptides in the ribosomal tunnel. Cell, 108, 591–594. [DOI] [PubMed] [Google Scholar]
- Thorsness P.E. and Fox,T.D. (1993) Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics, 134, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V. et al. (1998) Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet., 63, 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V., Galimberti,C., Nijtmans,L., Bovolenta,S., Perini,M.P. and Zeviani,M. (1999) Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet., 8, 2533–2540. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama,H., Yamashita,E., Tomizaki,T., Yamaguchi,H., Shinzawa-Itoh,K., Nakashima,R., Yaono,R. and Yoshikawa,S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science, 272, 1136–1144. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A. and Dieckmann,C.L. (1990) PET genes of Saccharomyces cerevisiae. Microbiol. Rev., 54, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., Herrmann,J.M. and Neupert,W. (2001) Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol., 65, 429–438. [DOI] [PubMed] [Google Scholar]
- Williams E.H. and Fox,T.D. (2003) Antagonistic signals within the COX2 mRNA coding sequence control its translation in Saccharomyces cerevisiae mitochondria. RNA, 9, 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. et al. (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet., 20, 337–343. [DOI] [PubMed] [Google Scholar]