Abstract
Clinical studies evaluating the mRNA expression level of the BRMS1 metastasis suppressor in the progression of breast cancer have not been consistent. The purpose of this study was to characterize endogenous BRMS1 mRNA and protein in a model of the progression of breast cancer. BRMS1 protein expression was evaluated in the genetically related MCF10 cell lines representing ‘normal’ breast epithelial cells (MCF10A), pre-malignant breast disease (MCF10AT), comedo ductal carcinoma in situ (MCF10DCIS.com), and metastatic carcinoma (MCF10CAa.1 and MCF10CAd.1α) with two antibodies that recognize distinct epitopes in the BRMS1 protein. Nuclear expression of the characteristic ∼35 kDa BRMS1 protein was detected in all cell lines. Because BRMS1 was expressed in the metastatic MCF10 variants, the BRMS1 exons were sequenced to scan for possible genetic mutations. BRMS1 was wild-type with the exception of a synonymous T/C transition in exon 7. However, alternatively spliced variants were detected by RT-PCR. Two variants, BRMS1.v2 and BRMS1.v4 were only detected in the MCF10A and AT cell lines, while BRMS1 and BRMS1.v3 were detected in all lines. These results demonstrate that expression of the characteristic ∼35 kDa BRMS1 protein is not sufficient to prevent metastasis. The differential expression of alternative splice variants suggests caution should be taken when evaluating BRMS1 mRNA in clinical samples.
Keywords: Breast cancer progression, Co-immunoprecipitation, Metastasis, Splice variant, Synonymous transition
Introduction
The discovery of metastasis suppressor genes has led to functional data demonstrating that these genes play diverse roles in the suppression of metastasis [1, 2]. Breast cancer metastasis suppressor 1 (BRMS1) has been shown to affect multiple steps in the metastatic cascade leading to ∼90% suppression in xenograft model systems of breast carcinoma, melanoma, and ovarian carcinoma [3-6]. Microarray and proteomic analyses have shown multiple changes in gene and protein expression when BRMS1 was introduced [7-9]. Further mechanistic studies have demonstrated that BRMS1 regulates the expression of multiple genes that have been linked to metastasis including osteopontin (OPN), urokinase-type plasminogen activator (uPA), epidermal growth factor receptor (EGFR), and connexins [10-13]. All of these studies have been performed by re-expression of BRMS1 into cells that have little to no detectable levels of endogenous BRMS1.
Clinically, loss of BRMS1 protein has been correlated with reduced disease-free survival when patient samples were stratified by loss of estrogen or progesterone receptor (ER, PR) or expression of HER2 [14]. Consistent with the protein data, BRMS1 mRNA correlated with PR and loss of HER2 and loss of BRMS1 mRNA correlated with poor prognosis [15]. However, mRNA studies have not been completely consistent as some groups found no correlation with breast cancer markers and BRMS1 mRNA expression was associated with shorter disease-free survival [16]. The current study was undertaken to evaluate the expression level of BRMS1 in isogenic cell lines representing normal breast epithelium, pre-malignancy, ductal carcinoma in situ (DCIS), and metastasis. We demonstrate that although BRMS1 is present and localized to the nucleus, its expression is not sufficient to prevent metastasis, suggesting that other protein interactors are important for BRMS1 mediated metastasis suppression.
Materials and methods
Cell lines and cell culture
The MCF10 breast cancer progression model system was a kind gift from Drs. Fred Miller and Herbert Soule (Karmanos Cancer Institute, Detroit, MI). The immortalized breast epithelial cell line MCF10A, pre-malignant MCF10AT, comedo ductal carcinoma in situ MCF10DCIS.com, and two metastatic clones MCF10CAa.1 and MCF10CAd.1α were cultured in a mixture (1:1, v/v) of Dulbecco’s modified Eagle’s medium and Ham’s F12 medium supplemented with 5% horse serum, 2 mM l-glutamine (Invitrogen), and 0.02 mM non-essential amino acids (Mediatech, Herndon, VA) without antibiotics or antimycotics. Additional supplements for the MCF10A and MCF10AT cell lines were 10 ng/ml EGF, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxins, and 10 μg/ml insulin (Sigma). All cultures were confirmed negative for Mycoplasma spp. infection using a PCR-based test (TaKaRa, Shiga, Japan). Cells were maintained on 100 mm Corning tissue culture dishes at 37°C with 5% CO2 in a humidified atmosphere. When cultures reached 80-90% confluence they were passaged using a solution of 0.05% Trypsin/EDTA (1×) (Invitrogen).
Metastasis assays
Spontaneous and experimental metastasis assays with the MCF10CA clones were performed as described previously [3, 4], with the exception that 2 × 105 cells were injected i.v. into the lateral tail vein. For the MCF10A and MCF10AT cell lines, 2 × 106 cells were injected into the mammary fat pad. Ten mice per experimental group were used. Orthotopic tumor size was evaluated weekly by taking orthogonal measurements and plotted by the mean tumor diameter (MTD = {(diameterx) (diametery)}0.5). All animals were maintained under the guidelines of the National Institutes of Health and the University of Alabama at Birmingham Institutional Animal Care and Use Committee. Food and water were provided ad libitum.
Antibodies, co-immunoprecipitation, and western blotting
Nuclear and cytoplasmic fractions were obtained according to the manufacturer’s protocol (NE-PER kit, Thermo Fisher Scientific). Co-immunoprecipitation and western blotting was performed as previously described [13]. Monoclonal BRMS1 antibodies 1a5.7 and 3a1.21 were described previously [13, 14, 17]. Other antibodies used in this study were purchased as indicated: anti-ARID4A clone LY11 (Upstate Biotechnology, Lake Placid, NY) and anti-lamin A/C (Cell Signaling Technology, Danvers, MA).
PCR amplification of genomic DNA
Genomic DNA was collected using Trizol (Invitrogen) according to manufacturer’s protocol. Primers to the individual exons were used for PCR (see Fig. 3b) with GoTaq (Promega). The products were visualized on a 2% agarose gel and sequenced.
Fig. 3.
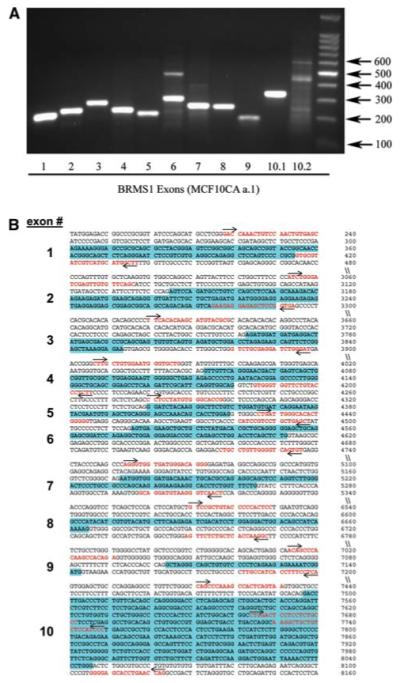
Genomic sequence of BRMS1 in cell lines reveals no mutations. a Genomic DNA was collected from the genetically related MCF10A-derived cell lines. PCR products were visualized on a 2% agarose gel. Results for MCF10CAa.1 are shown as a representative gel. The sequences were found to be wild-type except for a synonymous T/C transition in exon 7 that was identified in all MCF10A derived cell lines tested. b Primer map showing the primers in red font with an arrow indicating forward and reverse. The exons are highlighted in blue
Splice variant identification
Total RNA was collected using Trizol (Invitrogen) and RT-PCR was performed using the following primers: forward primers, 5′-ATGCCTGTCCAGCCTCCAAG-3′, 5′-AGA GAGTGAGGAGGAGCG-3′, and 5′-GACAGAGTCAGA AGAGGA-3′; reverse primers, 5′-TCAAGGTCCATCC GATTTTCT-3′, 5′-TCAGCTCCACGGCCACAG-3′, and 5′-TCACGTCTGACTCAGAGT-3′. The primers were designed based on sequences of BRMS1 splice variants found in the AceView genes database at NCBI. The linear range for each primer pair was determined independently. RT-PCR products were visualized on a 1% agarose gel and were sequence verified.
Results
Characterization of metastasis in the MCF10 model
The MCF10 breast cancer progression model system includes the immortalized breast epithelial cell line MCF10A, the pre-malignant MCF10AT, comedo ductal carcinoma in situ MCF10DCIS.com, and two metastatic clones MCF10CAa.1 and MCF10CAd.1α. The metastatic potential of mammalian cells may change under slightly different culture conditions [18]. Therefore, we characterized the ability of the two MCF10CA clones to metastasize following orthotopic mammary fat pad and i.v. tail vein injections. Both clones were able to metastasize following both injection routes (Fig. 1). Metastases were relatively small following i.v. injection but resulted in a large number for the d.1α clone (62 ± 3) compared to clone a.1 (5 ± 0.4). Very large metastases were seen following orthotopic mammary fat pad injection and resulted in approximately the same number for both clones (6 ± 4 and 5 ± 3). However, the growth rate for clone a.1 was more rapid than clone d.1α (Fig. 1d). Orthotopic mammary fat pad injection of MCF10A and MCF10AT resulted in no palpable tumors.
Fig. 1.
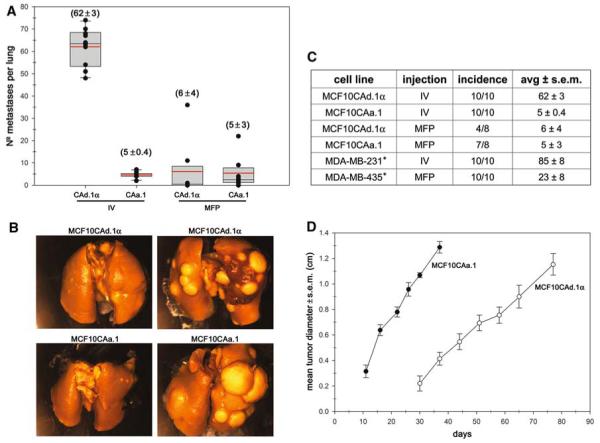
MCF10CA cell lines are metastatic. Two different clones, MCF10d.1α and a.1, were injected into the tail vein (IV) or mammary fat pad (MFP) of athymic mice. a The number of lung metastases are shown for both injection methods and cell lines. The black dots represent the number of lung metastases for each mouse, the boxes represent the 25th and 75th percentiles, and the red line is the mean (also shown in parentheses). b Representative images of the lungs. c The table shows the incidence of lung metastasis and the mean. *Two model systems routinely used are included for comparison [13]. d In vivo tumor growth curves for the MFP injected cells
BRMS1 protein is expressed in the MCF10 model
The expression of BRMS1 mRNA was previously described in the MCF10A normal immortalized cell line [3]. However, at that time, no BRMS1 antibody was available. We have generated two monoclonal BRMS1 antibodies, 3a1.21 and 1a5.7, that recognize the N- and C-terminus, respectively (Fig. 2a) [13, 14, 17]. These antibodies were used to probe for BRMS1 in the nuclear and cytoplasmic fractions. The characteristic 35 kDa BRMS1 protein was detected in the nuclear fraction of all five cell lines by the 1a5.7 BRMS1 antibody although it was reduced in MCF10CAd.1α (Fig. 2b). A lower 25 kDa protein was also detected by this antibody (but not the 3a1.21 antibody) in both the cytoplasmic and nuclear fractions and was significantly greater in the MCF10A and MCF10AT cell lines compared to the others.
Fig. 2.
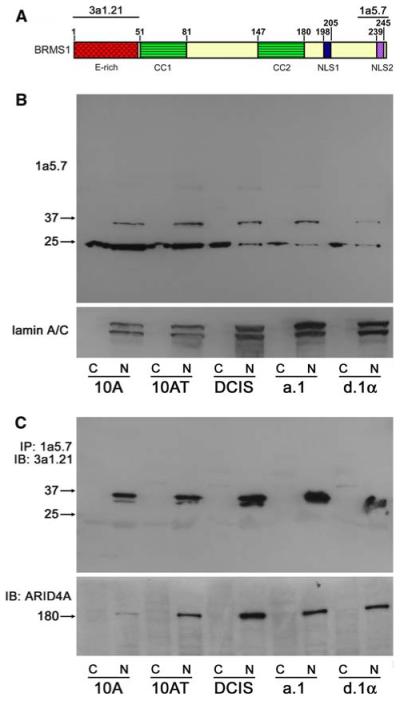
Expression of endogenous BRMS1. a Domain structure of BRMS1 showing the epitopes for the two monoclonal BRMS1 antibodies, 3a1.21 and 1a5.7. Abbreviations: E-rich, glutamate rich region; CC, coiled-coil domain; NLS, nuclear localization sequence. b The nuclear (N) and cytoplasmic (C) fractions were probed for BRMS1 expression. A lower ∼25 kDa band was detected by the 1a5.7 BRMS1 Ab. c ARID4A is associated with endogenous BRMS1 as shown by co-immunoprecipitation of BRMS1. The ∼25 kDa band was not detected with the 3a1.21 BRMS1 Ab
One of the direct protein-protein interactions of BRMS1 is with the AT-rich interactive domain 4A (ARID4A; previously known as retinoblastoma binding protein 1, RBBP1) [13]. Co-immunoprecipitation of ARID4A by 1a5.7 demonstrated that ARID4A could interact with endogenous BRMS1 in these cell lines (Fig. 2c) confirming previous studies with exogenously expressed BRMS1 in breast cancer cell lines. No significant differences in the levels of co-immunoprecipitated ARID4A were identified between the cell lines. An apparent BRMS1 doublet was also detected (Fig. 2c). It is not yet clear if this represents post-translational protein modifications and/or if this affects how BRMS1 is interacting with protein complexes.
Genomic sequence of BRMS1 is not mutated
Because the BRMS1 gene is located at 11q13.1-q13.2 [3] and karyotypic alterations frequently occur in this region during the later stages of breast cancer [19], we tested for possible genetic mutations. PCR was performed on genomic DNA with primers spanning known intron-exon splice sites and 2% agarose gel electrophoresis (Fig. 3). Sequencing of the exons revealed wild-type sequences except for a synonymous T/C transition in exon 7 that was identified in all cell lines. This data corroborates previous data showing no major genomic alterations by CGH in this region [14].
Splice variants are differentially expressed
The AceView genes database at NCBI (www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) [20] contains multiple splice variants of BRMS1. These sequences are derived from multiple cDNA databases. To test the possibility that splice variants of BRMS1 were expressed in the MCF10 breast cancer progression model, we designed multiple primers that could distinguish variant expression and performed RT-PCR followed by 1% agarose gel electrophoresis (Fig. 4a). Visualized bands were sequence verified. Three variants of BRMS1 were detected in addition to wildtype (BRMS1.v1), denoted BRMS1.v2, BRMS1.v3 and BRMS1.v4 [GenBank:NM_015399, NM_001024957, and NM_001024958, respectively; BRMS1v.4 does not currently have an NCBI accession number but is listed as AceView variant E]. Wild-type BRMS1.v1 has 10 exons spanning 741 nucleotides. BRMS1.v2 contains an alternative splice site in exon 10, BRMS1.v3 has an alternative splice site in exon 5, lacks exons 6-9, and alternatively splicesexon 10, and BRMS1.v4 lacks exon 9 (Fig. 4b). BRMS1.v1 and BRMS1.v3 were expressed in all cell lines, however, BRMS1.v2 and BRMS1.v4 were only detected in MCF10A and MCF10AT (Fig. 4a). The putative protein sequences were aligned using the ClustalW 1.8 multiple sequence alignment from the Human Genome Sequence Center at the Baylor College of Medicine (Fig. 4c). Protein for these variants in these cell lines has not yet been detected.
Fig. 4.
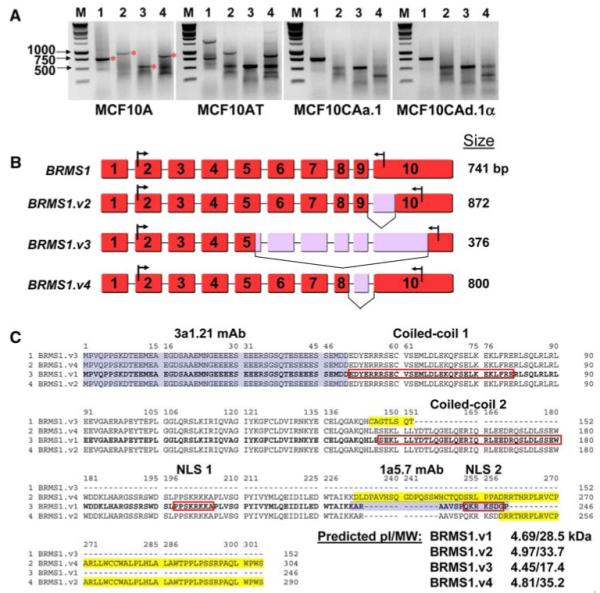
Identification of BRMS1 splice variants. a Total RNA was collected from the genetically related cell lines MCF10A, AT, CAa.1, and CAd.1α. Primers to each of the variants for RT-PCR are listed in the materials and methods. The products were visualized on a 1% agarose gel. M is marker; 1, 2, 3, and 4 correspond to BRMS1.v1, 2, 3, and 4. The bands with a red star were sequence verified. b Exon map of the splice variants. BRMS1.v1 (NM_015399) was the first reported and encodes the most abundant transcript, BRMS1.v2 (NM_001024957) uses an alternative splice site in exon 10, BRMS1.v3 (NM_001024958) uses an alternative splice site in exon 5, lacks exons 6-9 and alternatively splices exon 10, and BRMS1.v4 (AceView variant E) lacks exon 9. Numbering is based on NC_000011. The arrows in exon 10 show the stop codon and the location of the reverse primer. c Putative protein sequence alignment of the splice variants. The four variants of BRMS1 were aligned using the ClustalW 1.8 multiple sequence alignment from the Human Genome Sequence Center at the Baylor College of Medicine. BRMS1.v1 is shown in bold font; red boxes indicate coiled-coil domains and nuclear localization sequences; yellow highlights indicate differences from BRMS1.v1. Blue highlights indicate Ab recognition sites. ExPASy proteomics server was used for pI/MW prediction
Discussion
Clinical correlations of BRMS1 mRNA levels and cancer progression, development of metastasis, and/or survival have not been completely consistent thus far. In breast cancer, two studies showed inverse correlations of BRMS1 expression with metastasis [15, 21]; however, one study showed an inverse correlation with disease free survival [16] and another showed no correlation of BRMS1 with breast cancer progression [22]. Interpretation of those studies is not completely unambiguous since contaminating stromal tissues expressing BRMS1 may have skewed the findings, a point emphasized by detection of BRMS1 in ‘normal’ (immortalized but not tumorigenic) MCF10A mammary epithelial cells. To date, only one study has been published with BRMS1 protein in clinical samples [14]. Expression of BRMS1 correlated with disease free survival of breast cancer patients when stratified with ER+, PR+, and HER2-. Another preliminary study demonstrated an inverse correlation of BRMS1 protein with the metastatic potential of melanoma [23]. The goal of this study was to characterize endogenous BRMS1 using a model system of breast cancer progression. Notably, MCF10A cells are ER-negative, MCF10AT are ER-positive, MCF10DCIS.com are ER-negative, and the ER status of the MCF10CA clones are ambiguous [24, 25].
The MCF10 isogenic model has been instrumental for several studies of genes involved in breast cancer progression [26-29]. In order to validate the metastatic ability of the MCF10CA clones, we have performed metastasis assays using both orthotopic (‘spontaneous’) and i.v. (‘experimental’) metastasis xenograft models. Both clones were metastatic in both assays. Although the expression of BRMS1 did not prevent metastasis, we cannot exclude the possibility that BRMS1 inhibited metastasis. Maintaining a stable knockdown may be required to test this hypothesis but has proven difficult thus far. However, current studies with BRMS1 RNAi are being conducted to determine if knockdown of BRMS1 leads to a more aggressive metastatic phenotype.
One possibility for the inability of BRMS1 to prevent metastasis was that it could be mutated. Chromosome 11q is a region of karyotypic alteration occurring in the progression of breast cancer [19]. We therefore sequenced the exons of BRMS1 and found wild-type sequence with the exception of a synonymous T/C transition in exon 7. Recently, a synonymous single-nucleotide polymorphism has been linked to altered function in the multidrug resistance 1 (MDR1) gene [30]. However, we currently do not have any data to suggest that this transition has any functional relevance to BRMS1.
Many biological functions have been described for exogenously expressed BRMS1 that are associated with the ability to suppress metastasis. The majority of these functional roles have been linked to changes in the expression of genes that regulate particular molecular pathways. One of the mechanisms for BRMS1 regulation of gene transcription has been through a large SIN3:HDAC chromatin remodeling complex that includes a direct interaction between BRMS1 and ARID4A [13, 31]. In this study, we have validated that interaction endogenously for the first time. Previous studies suggested, however, that the direct interaction was not sufficient to suppress metastasis and the overall composition of the SIN3:HDAC complex may be important for regulation of specific genes. Although BRMS1 still interacts with key core components in this system, the entirety of the complex(es) has not yet been determined. Likewise, it is not yet known which of the myriad complexes are responsible for metastasis suppression. Our working hypothesis is that BRMS1-associated proteins are altered in MCF10 cells. As a result, the functionality of BRMS1 is hindered in some way. Understanding exactly how BRMS1 is functioning will require a detailed understanding of the dynamic composition of these protein complexes.
Consistent with reports of BRMS1 associating with chromatin remodeling complexes, BRMS1 is predominantly localized to the nucleus. The characteristic 35 kDa protein was demonstrated to be localized in the nuclear fractions, however, a second smaller protein at 25 kDa was found in both the cytoplasmic and nuclear fractions. We are currently characterizing this band to determine if it is a processed form of BRMS1 or a splice variant. Because it was identified with the 1a5.7 BRMS1 antibody, which recognizes the C-terminus of BRMS1, and not the 3a1.21 antibody, which recognizes the N-terminus, this band does not correlate with any of the splice variants identified by RT-PCR in this study. We are careful, however, not to exclude the possibility of other splice variants that have not yet been characterized. Alternatively, this band could be the result of cross reactivity of the antibody with another protein, although we have performed Blast searches of the epitope and found no matching sequences.
These studies highlight the fact that depending on the PCR primers or antibody used for a particular study may lead to significantly different results. For example, in theory, the primers used to identify BRMS1.v3 should also amplify other BRMS1 variants, but they do not. The reasons that they do not amplify other variants have not been systematically studied here. We believe that the most likely explanations are because BRMS1.v3 is the smallest variant (i.e., a preferred product) and because deletions of intervening introns and exons could significantly impact mRNA stability. It is apparent that the 3′ primers coupled with 5′ primers in exon 2 produce preferred products (i.e., BRMS1 variants), but also products of different sizes, perhaps different variants or PCR artifacts. The alternative products (i.e., those not of the predicted size) have not yet been sequenced and characterized. Therefore, interpretation of BRMS1 expression levels in clinical samples should be performed carefully and critically to understand correlations between study groups.
In summary, the data presented here raise important questions regarding interpretation of our previously published data and data from other laboratories, particularly studies using clinical material. Importantly, mRNA and protein levels for BRMS1 are not necessarily correlative. Additionally, depending upon the probes used in situ hybridization studies cannot distinguish between the alternative BRMS1 variants. And, equally important from a mechanistic point of view, the data open new directions related to how BRMS1 (and possibly BRMS1 variants or family members) interacts with other proteins in chromatin remodeling complexes to affect metastasis suppression.
Acknowledgments
This work was supported by United States Public Health Service Grants CA87728 (to DRW) and F32CA113037 (to DRH) and a grant from the National Foundation for Cancer Research Center for Metastasis Research (to DRW). We thank Drs. Fred Miller and Herbert Soule (Karmanos Cancer Institute) for generously providing the MCF10 cell line series. We also thank Alka Mehta and Blake Moore for technical assistance and Dr. Monica Richert and Alexandra Silveira for critical reading of the manuscript.
Abbreviations
- BRMS1
Breast cancer metastasis suppressor 1
- ARID4A
AT-Rich interactive domain 4A
- ER
Estrogen receptor
- PR
Progesterone receptor
Contributor Information
Douglas R. Hurst, Department of Pathology, University of Alabama at Birmingham, 1670 University Blvd., room VH-G019A, Birmingham, AL 35294-0019, USA
Yi Xie, Department of Pathology, University of Alabama at Birmingham, 1670 University Blvd., room VH-G019A, Birmingham, AL 35294-0019, USA.
Mick D. Edmonds, Department of Pathology, University of Alabama at Birmingham, 1670 University Blvd., room VH-G019A, Birmingham, AL 35294-0019, USA
Danny R. Welch, Departments of Pathology, Cell Biology, Pharmacology/Toxicology & Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, AL, USA
References
- 1.Vaidya KS, Welch DR. Metastasis suppressors and their roles in breast carcinoma. J Mamm Gland Biol Neopl. 2007;12:175–190. doi: 10.1007/s10911-007-9049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinker-Schaeffer CW, O’Keefe JP, Welch DR, Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms and clinical application. Clin Cancer Res. 2006;12:3382–3389. doi: 10.1158/1078-0432.CCR-06-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- 4.Samant RS, Seraj MJ, Saunders MM, Sakamaki T, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon LR, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2001;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- 5.Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer. 2006;16:522–531. doi: 10.1111/j.1525-1438.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- 7.Champine PJ, Michaelson J, Weimer B, Welch DR, DeWald DB. Microarray analysis reveals potential mechanisms of BRMS1-mediated metastasis suppression. Clin Exp Metastasis. 2007;24:551–565. doi: 10.1007/s10585-007-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cicek M, Samant RS, Kinter M, Welch DR, Casey G. Identification of metastasis-associated proteins through protein analysis of metastatic MDA-MB-435 and metastasis-suppressed BRMS1 transfected-MDA-MB-435 cells. Clin Exp Metastasis. 2004;21:149–157. doi: 10.1023/b:clin.0000024729.19084.f0. [DOI] [PubMed] [Google Scholar]
- 9.Rivera J, Megias D, Bravo J. Proteomics-based strategy to delineate the molecular mechanisms of the metastasis suppressor gene BRMS1. J Proteome Res. 2007;6:4006–4018. doi: 10.1021/pr0703167. [DOI] [PubMed] [Google Scholar]
- 10.Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-κB activity. Cancer Res. 2005;65:3586–3595. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- 11.Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR, Shevde LA. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders MM, Seraj MJ, Li ZY, Zhou ZY, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–1767. [PubMed] [Google Scholar]
- 13.Hurst DR, Xie Y, Vaidya KS, Mehta A, Moore BP, Accavitti-Loper MA, Samant RS, Saxena R, Silveira AC, Welch DR. Alterations of breast cancer metastasis suppressor 1:at rich interactive domain 4a interaction modify gene expression but still suppress metastasis in human breast cancer cells. J Biol Chem. 2008;283:7438–7444. doi: 10.1074/jbc.M709446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri T, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost AR, Welch DR, Casey G. Loss of BRMS1 protein expression predicts reduced disease-free survival in hormone receptor negative and HER2 positive subsets of breast cancer. Clin Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Yamashita H, Toyama T, Yamamoto Y, Kawasoe T, Iwase H. Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res. 2006;12:6410–6414. doi: 10.1158/1078-0432.CCR-06-1347. [DOI] [PubMed] [Google Scholar]
- 16.Lombardi G, Di Cristofano C, Capodanno A, Iorio MC, Aretini P, Isola P, Tancredi M, Collecchi P, Naccarato AG, Porta RP, Bevilacqua G, Caligo MA. High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int J Cancer. 2007;120:1169–1178. doi: 10.1002/ijc.22379. [DOI] [PubMed] [Google Scholar]
- 17.Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA, Shevde LA, Hopper JE, Xie Y, Welch DR, Samant RS. Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Commun. 2006;348:1429–1435. doi: 10.1016/j.bbrc.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch DR. Technical considerations for studying cancer metastasis in vivo. Clin Exp Metastasis. 1997;15:272–306. doi: 10.1023/a:1018477516367. [DOI] [PubMed] [Google Scholar]
- 19.Welch DR, Wei LL. Genetic and epigenetic regulation of human breast cancer progression and metastasis. Endocr Relat Cancer. 1998;5:155–197. [Google Scholar]
- 20.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12–S14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol. 2005;131:191–198. doi: 10.1007/s00432-004-0629-9. [DOI] [PubMed] [Google Scholar]
- 22.Kelly LM, Buggy Y, Hill A, O’Donovan N, Duggan C, McDermott EW, O’Higgins NJ, Young L, Duffy MJ. Expression of the breast cancer metastasis suppressor gene, BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumour Biol. 2005;26:213–216. doi: 10.1159/000086955. [DOI] [PubMed] [Google Scholar]
- 23.Phadke PA, Ashby-Richardson H, Shepard-Barry A, Naber SP, Welch DR. Loss of BRMS1 expression is strongly associated with metastatic potential of malignant melanoma. Lab Invest. 2008;88:99A. [Google Scholar]
- 24.Miller FR. Xenograft models of premalignant breast disease. J Mamm Gland Biol Neopl. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 25.Strickland LB, Dawson PJ, Santner SJ, Miller FR. Progression of premalignant MCF10AT generates heterogeneous malignant variants with characteristic histologic types and immunohistochemical markers. Breast Cancer Res Treat. 2000;64:235–240. doi: 10.1023/a:1026562720218. [DOI] [PubMed] [Google Scholar]
- 26.Tang BW, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM. TGF-β switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest. 2003;112:1116–1124. doi: 10.1172/JCI18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian F, Byfield SD, Parks WT, Stuelten CH, Nemani D, Zhang YE, Roberts AB. Smad-binding defective mutant of transforming growth factor β type I receptor enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 2004;64:4523–4530. doi: 10.1158/0008-5472.CAN-04-0030. [DOI] [PubMed] [Google Scholar]
- 28.Smukste I, Stockwell BR. Restoring functions of tumor suppressors with small molecules. Cancer Cell. 2003;4:419–420. doi: 10.1016/s1535-6108(03)00307-6. [DOI] [PubMed] [Google Scholar]
- 29.Peng X, Wood S, Bratescu L, Shilkaitis A, Christov K. Retinoids suppress premalignant MCF10AT but not malignant MCF10CA1a breast epithelial cells in vivo. Role of retinoic acid receptor beta2 expression. Cancer Lett. 2005;222:153–163. doi: 10.1016/j.canlet.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 31.Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]


