Abstract
The co-presence of histoincompatible fetal and maternal cells is a characteristic of human placental inflammation. Villitis of unknown etiology (VUE), a destructive inflammatory lesion of villous placenta, is characterized by participation of Hofbauer cells (placental macrophages) and maternal T cells. In contrast to acute chorioamnionitis of infection-related origin, the fundamental immunopathology of VUE is unknown. This study was performed to investigate the placental transcriptome of VUE and to determine whether VUE is associated with systemic maternal and/or fetal inflammatory response(s). Comparison of the transcriptome between term placentas without and with VUE revealed differential expression of 206 genes associated with pathways related to immune response. The mRNA expression of a subset of chemokines and their receptors (CXCL9, CXCL10, CXCL11, CXCL13, CCL4, CCL5, CXCR3, CCR5) was higher in VUE placentas than in normal placentas (p < 0.05). Analysis of blood cell mRNA showed a higher expression of CXCL9 and CXCL13 in the mother, and CXCL11 and CXCL13 in the fetus of VUE cases (p < 0.05). The median concentrations of CXCL9, CXCL10, and CXCL11 in maternal and fetal plasma were higher in VUE (p < 0.05). Comparison of preterm cases without and with acute chorioamnionitis revealed elevated CXCL9, CXCL10, CXCL11, and CXCL13 concentrations in fetal plasma (p < 0.05), but not in maternal plasma with chorioamnionitis. We report for the first time the placental transcriptome of VUE. A systemic derangement of CXC chemokines in maternal and fetal circulation distinguishes VUE from acute chorioamnionitis. We propose that VUE be a unique state combining maternal allograft rejection and maternal antifetal graft-vs-host disease mechanisms.
Keywords: human, inflammation, chemokines, graft versus host disease, transplantation
Introduction
The human placenta represents the anatomical and functional feto-maternal interface. When a robust inflammatory reaction during pregnancy is mounted (i.e., microbial-induced acute chorioamnionitis), both maternal and fetal inflammatory responses can be observed. One of the histologic manifestations of such an inflammatory response is infiltration of both maternal and fetal neutrophils in the placenta (1). However, this localized inflammatory lesion (acute chorioamnionitis) is associated with systemic elevation of pro-inflammatory cytokines in both maternal and fetal circulation. A clinical complex akin to sepsis characterized by an elevated fetal plasma IL-6 concentration has been used to define the fetal inflammatory response syndrome (2–4).
Villitis of unknown etiology (VUE)4 is another major placental inflammatory lesion, which is quite distinct from acute chorioamnionitis. VUE, an enigmatic and destructive inflammatory lesion, involves the villous placenta and is characterized by infiltration of predominantly CD8+ maternal T cells into the chorionic villi (5,6). VUE is a relatively common lesion found in 5–15% of term placentas, and it is associated with intrauterine fetal growth restriction, fetal death, and a wide range of perinatal morbidity (7–9). While the immunologic mechanisms implicated in VUE appear to be analogous to graft rejection by the mother (10,11), its fundamental pathology remains to be determined. Recent studies have shown that in addition to the maternal T cells, resident placental macrophages (Hofbauer cells) of fetal origin are key participants in this inflammatory process (6,12).
A unique feature of VUE is the interaction of leukocytes from two distinct hosts (mother and fetus), suggesting that both hosts are engaged in either the local or the systemic inflammatory response. While the nature of the local and systemic inflammatory responses of acute chorioamnionitis are relatively well-documented (2–4), those of VUE have not been fully addressed. Genome-wide expression analysis is a powerful tool, which often provides comprehensive information about the transcriptome of a particular pathologic process (13). The present study was designed to investigate the changes in the transcriptome of placentas affected by VUE and thereby to determine key immunopathological alterations associated with this lesion.
Materials and Methods
Sample Collection
Placental tissues, maternal blood, and cord blood samples were retrieved from the Bank of Biological Materials of the Perinatology Research Branch of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health. Patients included women who delivered after spontaneous labor at term without (TIL; n=22) and with VUE (n=22). VUE included in this study was defined as multifocal lymphohistiocytic infiltration in more than five villi on multiple slides, encompassing multifocal low-grade villitis and patchy or diffuse high-grade villitis based on histologic criteria previously defined (1) (Fig. 1). From each group, 16 pairs of placental tissues in RNAlater tissue collection (Applied Biosystems), 17 pairs of maternal blood in PAXgene blood RNA system (PreAnalytiX/BD Biosciences), 17 pairs of fetal blood from the umbilical vein in PAXgene blood RNA system, 19 pairs of maternal plasma, and 19 pairs of fetal plasma were available for analysis. For purposes of comparison, both maternal and fetal blood samples from women with preterm labor and delivery without (PTL; n=20) and with acute chorioamnionitis (PTLI; n=20) were also used. All patients provided written informed consent, and the collection and use of the samples were approved by the Institutional Review Boards of the participating institutions.
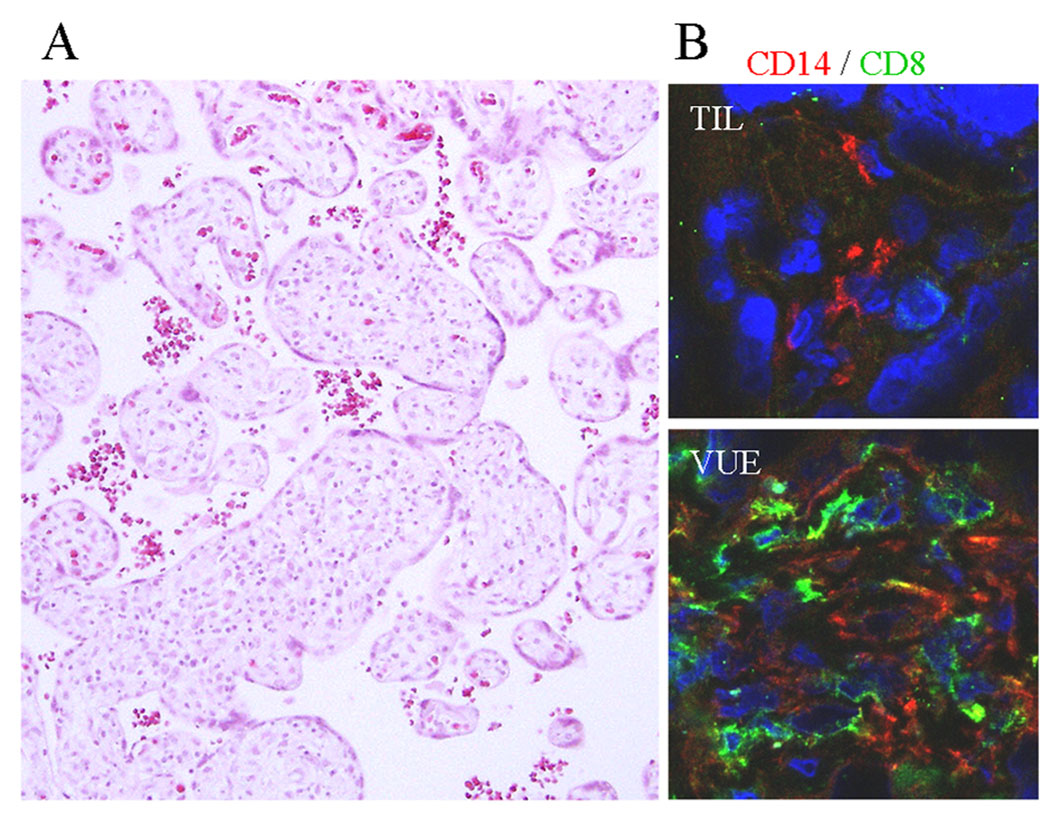
Figure 1.
Microscopic findings of VUE. A, VUE is characterized by patchy lymphocytic infiltrates in the affected chorionic villi. B, Hofbauer cells (placental tissue macrophages) in the chorionic villi of a term control placenta (TIL) are immunopositive for CD14. Chorionic villi in the placenta with VUE show infiltration of CD8+ T cells in addition to CD14+ Hofbauer cells. (CD14, red; CD8, green).
Microarray analysis
For microarray analysis, 10 total RNA samples from each group (TIL vs VUE) were used. Isolation of total RNA from placental villous tissues in RNAlater tissue collection was performed using TRIzol (Invitrogen) followed by purification of RNA using an RNeasy Mini kit (Qiagen). Synthesis of cDNA was performed using the Affymetrix GeneChip expression 3′ amplification one-cycle cDNA synthesis kit using 1 µg of total RNA. After purification of double-stranded cDNA using the Affymetrix GeneChip sample clean-up module, the cDNA product was used for the synthesis of biotin-labeled cRNA using the GeneChip IVT labeling kit (Affymetrix). Labeled cDNAs were hybridized to an Affymetrix GeneChip human genome U133 Plus 2.0 array platform with 47,650 annotated probe sets targeting 19,886 unique genes. Arrays were scanned with the GeneChip scanner 3000 (Affymetrix).
The gene expression data were preprocessed using the RMA (robust multiarray average) algorithm (14) to obtain a background corrected and normalized intensity level for each probe set of each gene represented on the array. Data was further log2 transformed and analyzed using a linear model strategy in which the gene expression levels were fitted as a function of the inflammation severity index (ISI). The ISI was defined according to the histologic grade of VUE; ISI 1, 2, and 3 were multifocal low-grade villitis, patchy high-grade villitis, and diffuse high-grade villitis, respectively. ISI 0 was defined as no evidence of villitis (TIL group). This modeling strategy was chosen over a classical ANOVA approach because we were interested in identifying genes with a rather monotonic behavior (i.e., their expression levels are significantly correlated with the ISI). A p value was obtained for each probe set, measuring the significance of the coefficient for the ISI variable in the linear model. The limma package (15) of Bioconductor [www.bioconductor.org] was used for model fitting and p value calculations for each probe set. The p values for all probe sets of the same gene were combined using a Fisher approach, in which a global p value for each gene is obtained, giving the probability of observing a set of p values as low as or lower than those obtained for all different probe sets of a given gene. The resulting p values were corrected across the genes to account for multiple testing using the false discovery rate (FDR) algorithm (16). A gene was labeled as significant if the FDR p values for the gene were < 0.05 and the ISI coefficient was more extreme than 0.25, which corresponds to a 2-fold change in expression (increase or decrease) between the group with severe inflammation and the control group. Differentially expressed genes were further analyzed to identify enriched Gene Ontology terms, such as molecular functions and biological processes using the GOstats package in R.2.7.0 (17). Moreover, pathway analysis was performed using an over-representation approach on the Kyoto Encyclopedia of Genes and Genomes (KEGG) human signaling and metabolic pathways database (www.genome.jp/kegg/). For the signaling pathways available in KEGG, the signaling pathway impact analysis (SPIA) algorithm (18,19) was conducted, which makes full use of the pathways topology and the directed gene-gene interactions in assessing pathway significance.
Real-time quantitative RT-PCR (qRT-PCR)
Isolation of total RNA from placental villous tissue in RNAlater tissue collection was performed using TRIzol. RNA was isolated from maternal and fetal blood in PAXgene blood RNA system using the PAXgene blood RNA kit (Qiagen). DNase-treated total RNA was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) and oligo(dT) primers. qRT-PCR analyses were performed with TaqMan gene expression assays (CXCL9, Hs00171065_m1; CXCL10, Hs00171042_m1; CXCL11, Hs00171138_m1; CXCL13, Hs00757930_m1; CCL4, Hs99999148_m1; CCL5, Hs00174575_m1; CXCR3, Hs00171041_m1; CXCR5, Hs00173527_m1; CCR5, Hs00152917_m1; Applied Biosystems) using an ABI 7500 Fast real-time PCR system. The human ribosomal protein, large, P0 (RPLP0; Applied Biosystems) was used for normalization.
ELISA
Plasma was collected by centrifugation of blood obtained in EDTA tubes (BD Vacutainer; BD Diagnostics) and stored at −80°C until use. The plasma concentrations of CXCL9, CXCL10, CXCL11, and CXCL13 chemokines were measured by specific ELISA (R&D Systems) according to the manufacturer’s instructions.
Immunofluorescent staining
Immunofluorescent staining was performed using Abs to CXCL9 (R&D Systems), CXCL10 (R&D Systems), CXCL11 (R&D Systems), CXCL13 (Proteintech Group), CCL4 (R&D Systems), CCL5 (R&D Systems), CXCR3 (R&D Systems), CXCR5 (Novus Biologicals), CCR5 (BD Pharmingen), CD14 (Abcam), and CD8 (monoclonal, Dako; polyclonal, Abcam). Five-micrometer-thick frozen tissue sections were fixed with 4% (w/v) paraformaldehyde for 30 min at 4°C and permeabilized with 0.25% Triton X-100 or with an equal volume of acetone and methanol for 5 min at 4°C. To quench nonspecific binding, sections were incubated with 5% (w/v) BSA in PBS for 30 min at room temperature. Sections were incubated with a primary Ab in 1% (w/v) BSA in PBS for 1 h, followed by incubation with Alexa 568 donkey anti-goat IgG or Alexa 594 donkey anti-rabbit IgG (Invitrogen) in 1% (w/v) BSA for 30 min. For double staining, sections were subsequently incubated with a second primary Ab in 1% (w/v) BSA for 1 h, before incubation with Alexa 488 goat anti-mouse IgG (Invitrogen) in 1% (w/v) BSA in PBS for 30 min. The slides were mounted in ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). The stained sections were examined using a Leica TCS SP5 spectral confocal system (Leica Microsystems).
Statistical analysis
The median mRNA expression and the median concentration for each chemokine were compared between two groups using Mann-Whitney U tests. SPSS version 12.0 was employed for statistical analysis. A p value of <0.05 was considered statistically significant. The methods employed for microarray analysis have been described above in the appropriate section.
Results
Placental transcriptome of VUE: enrichment of immune response among differentially expressed genes
For the comparison of placental transcriptome, linear modeling was used to correlate gene expression levels with the severity of inflammation in 10 TIL (control) and 10 VUE placentas. The complete data set is available in a MIAME (Minimal Information About a Microarray Experiment)-compliant format in ArrayExpress (www.ebi.ac.uk/microarray-as/ae/) database (entry ID: E-TABM-577). The results of gene expression profiling are shown in Fig. 2. The “volcano plot” (Fig. 2A) shows the gene significance (−log10 of p value) as a function of the magnitude of expression changes with the ISI. Differential regulation was found in 206 genes out of 19,886 annotated genes on the array (FDR < 0.05, fold change of > 2), and most of them (93%) were increased in expression as a function of the severity of the inflammatory process (Table I and Fig. 2, B and C). In particular, several chemokines, MHC class I molecules (HLA-B, -C, -G), and MHC class II molecules (HLA-DM, -DO, -DP, -DQ, -DR) were up-regulated in the placentas with VUE compared with those of TIL cases.
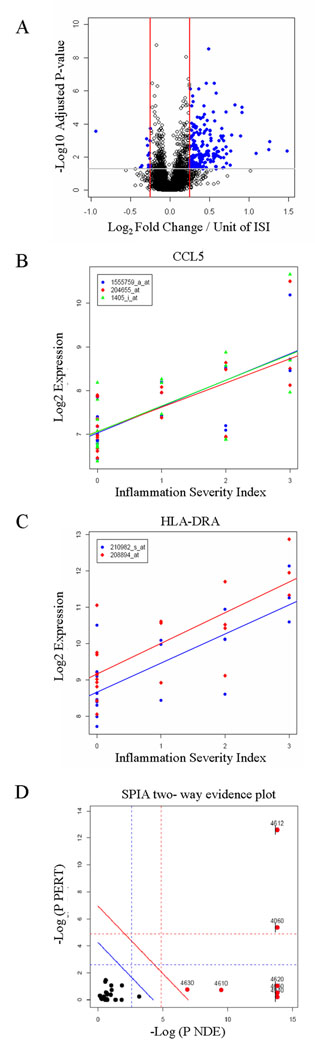
Figure 2.
Transcriptome analysis of VUE placentas using microarray. A, A volcano plot showing the relationship between the FDR-corrected p values of the genes and their fold changes (log2 of). Blue dots outside the two red lines and above the gray line have fold changes > 2 and FDR-corrected p values < 0.05. Positive values on the x-axis indicate genes whose expression increased with inflammation severity, while negative values indicate genes down-regulated with inflammation severity. A gene located at 0.25 on the x-axis increases in average 4 × 0.25 = 1 U of log2 fold change (2-fold) from the group without disease (ISI = 0) to the most severe inflammation group (ISI = 3). B and C, The expression levels of the Affymetrix probe sets for the CCL5 (B) and HLA-DRA (C) genes were plotted according to inflammation severity. Gene expression increased along with inflammation severity. D, The list of significant genes and their log-fold changes were analyzed using the SPIA algorithm to identify signaling pathways for which there is high enrichment evidence (high x-axis) and high perturbation evidence (high y-axis). Each KEGG signaling pathway analyzed is shown as a bullet point. Pathways above the oblique red line are significant at 5% after Bonferroni correction based on the combined evidence, while those above the oblique blue line are significant at 5% after FDR correction. The vertical and horizontal thresholds represent the same corrections for each type of evidence. Ag processing and presentation (KEGG ID: 4612) and cytokine-cytokine receptor interaction (KEGG ID: 4060) are the most relevant according to the signaling perturbation-based evidence (y-axis).
Table I.
Top 30 differentially expressed genes in VUE
| Gene | False Discovery Rate | Fold Change |
|---|---|---|
| STAT1 | 1.02E-15 | 2.24 |
| HLA-C | 6.14E-14 | 2.44 |
| LST1 | 2.18E-12 | 1.95 |
| FAM26F | 4.01E-12 | 3.33 |
| SOD2 | 2.93E-09 | 2.75 |
| HLA-DQB1 | 3.54E-07 | 2.58 |
| SLAMF7 | 3.54E-07 | 3.21 |
| LCP2 | 7.55E-07 | 1.70 |
| PTPRC | 7.76E-07 | 2.12 |
| CCL5 | 1.41E-06 | 3.36 |
| CLEC7A | 2.05E-06 | 1.81 |
| IL23A | 4.57E-06 | 1.83 |
| HLA-DRA | 6.90E-06 | 5.54 |
| IL21R | 8.03E-06 | 2.44 |
| LILRB1 | 8.03E-06 | 1.77 |
| ANKRD22 | 9.64E-06 | 6.65 |
| HLA-DPB1 | 1.85E-05 | 3.12 |
| HLA-DPA1 | 1.91E-05 | 4.36 |
| GBP5 | 1.98E-05 | 6.67 |
| GBP1 | 2.55E-05 | 2.47 |
| CD86 | 3.86E-05 | 2.06 |
| RAC2 | 3.86E-05 | 2.37 |
| CD74 | 4.22E-05 | 2.18 |
| LCK | 4.71E-05 | 2.24 |
| RTN1 | 6.11E-05 | 2.64 |
| FCGR3B | 6.44E-05 | 3.25 |
| ARHGAP9 | 9.22E-05 | 1.77 |
| PLAUR | 9.22E-05 | 1.76 |
| TNFAIP6 | 0.00011 | 2.46 |
| TFEC | 0.00011 | 1.87 |
Gene Ontology analysis of the significantly differentially regulated genes revealed enrichment of 78 biological processes, most of which were related to immune responses (Table II). Pathway analysis identified 10 pathways significantly associated with VUE. They included cell adhesion molecules, Ag processing and presentation, cytokine-cytokine receptor interaction, and TCR signaling pathways (Table III). The impact evidence captured by SPIA from signaling perturbations suggested that the Ag processing and presentation (KEGG ID: 4612) and the cytokine-cytokine receptor interaction (KEGG ID: 4060) were the most relevant in this study (Fig. 2D).
Table II.
Top 20 biological processes enriched in VUE
| Biologic Process | pFDR | No. Enriched Genes /No. Total Genes |
|---|---|---|
| Immune response | 8.10E-51 | 61/369 |
| Ag processing and presentation of peptide or polysaccharide antigen via MHC class II |
1.43E-13 | 10/15 |
| Inflammatory response | 3.01E-11 | 25/290 |
| Chemotaxis | 7.18E-11 | 18/139 |
| Locomotory behavior | 2.76E-09 | 18/174 |
| Cellular defense response | 1.77E-08 | 12/71 |
| Lymphocyte-mediated immunity | 1.79E-08 | 12/72 |
| Response to virus | 3.68E-08 | 12/78 |
| Regulation of immune response | 6.87E-08 | 12/83 |
| Adaptive immune response based on somatic recombination of immune receptors built from Ig superfamily domains |
6.87E-08 | 10/51 |
| Leukocyte activation | 7.51E-08 | 15/150 |
| Regulation of cell activation | 1.01E-07 | 12/87 |
| Positive regulation of response to stimulus | 7.45E-07 | 12/104 |
| Immune effector process | 7.58E-07 | 10/67 |
| Activation of immune response | 6.55E-06 | 9/63 |
| Positive regulation of leukocyte activation | 2.92E-05 | 8/56 |
| Innate immune response | 6.68E-05 | 10/107 |
| Ag processing and presentation | 7.43E-05 | 6/31 |
| Positive regulation of immune system process | 0.00028 | 7/58 |
| Death | 0.00029 | 27/796 |
Table III.
List of pathways enriched in VUE
| Pathway | KEGG ID |
pFDR | No. Enriched Genes /No. Total Genes |
|---|---|---|---|
| Type I diabetes mellitus | 4940 | 1.95E-18 | 18/41 |
| Cell adhesion molecules | 4514 | 4.66E-12 | 21/129 |
| Ag processing and presentation | 4612 | 2.15E-11 | 17/84 |
| NK cell-mediated cytotoxicity | 4650 | 1.64E-07 | 16/129 |
| Hematopoietic cell lineage | 4640 | 3.32E-06 | 12/87 |
| Cytokine-cytokine receptor interaction |
4060 | 4.07E-06 | 20/257 |
| TLR signaling pathway | 4620 | 1.38E-05 | 12/102 |
| T cell receptor signaling pathway | 4660 | 0.00021 | 10/93 |
| Alzheimer's disease | 5010 | 0.01984 | 4/28 |
| Complement and coagulation cascades |
4610 | 0.02248 | 6/69 |
Differential expression of chemokines and chemokine receptors in the placentas with VUE
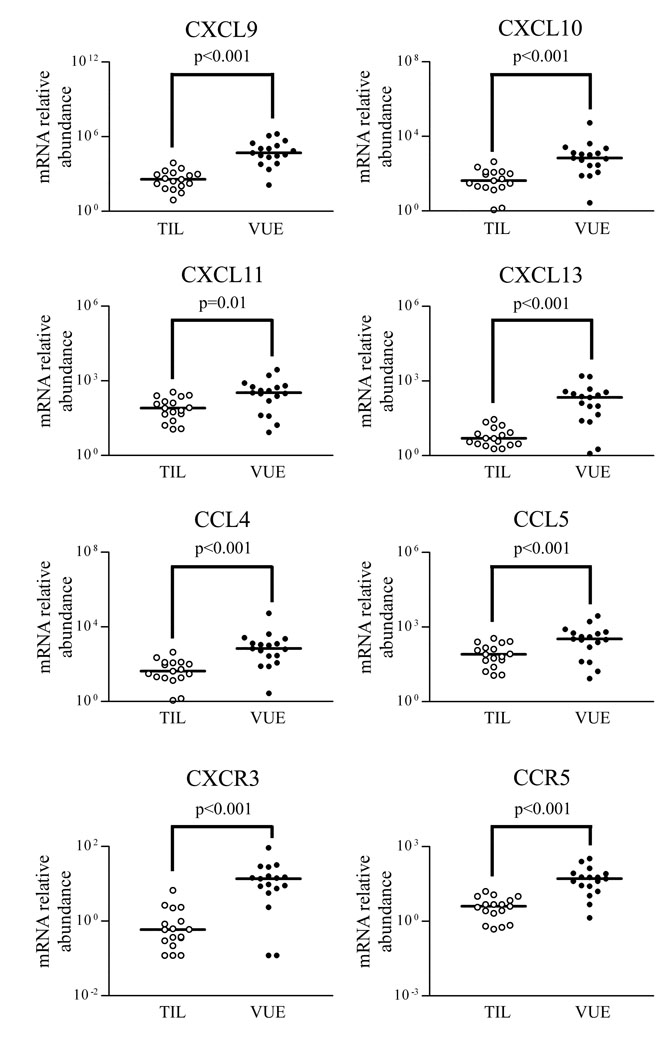
For validation of the microarray results, mRNA expression of selected chemokines (CXCL9, CXCL10, CXCL11, CXCL13, CCL4, CCL5) was further analyzed by qRT-PCR. Expression of these chemokines was significantly higher in placentas with VUE than in those without it (all p < 0.05), and mRNA expression of the corresponding receptors of the chemokines CXCR3 and CCR5 was also found to be highly expressed in VUE (p < 0.001; Fig. 3), except for CXCR5 mRNA, which was not detected in any case.
Figure 3.
Changes in mRNA expression of a subset of chemokines and receptors in VUE placentas. mRNA expressions of CXCL9, CXCL10, CXCL11, CXCL13, CCL4, and CCL5 are higher in VUE placentas compared with those in term control placentas. mRNA of CXCR3 and CCR5 is also expressed at higher levels in VUE placentas than in term control placentas.
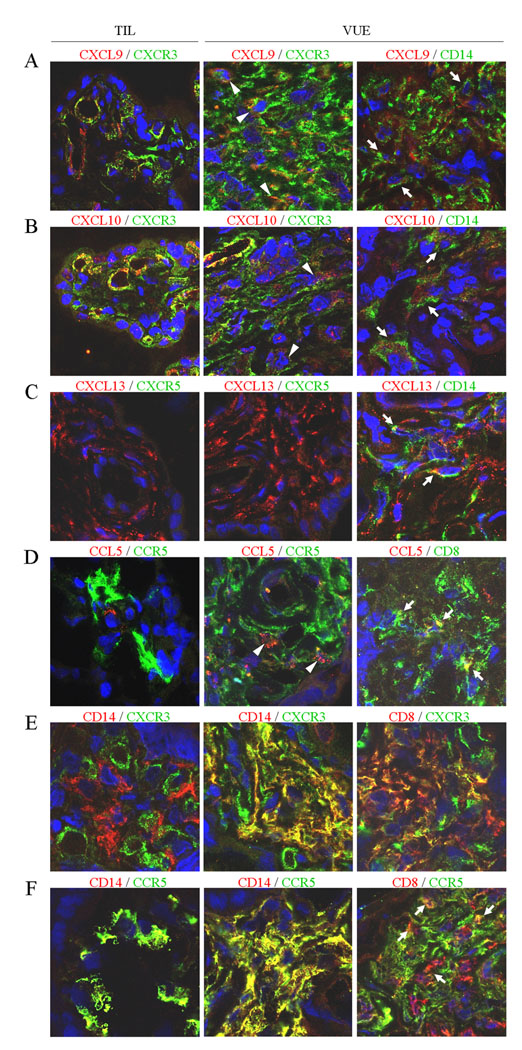
Immunofluorescent staining revealed CXCL9, CXCL10, CXCL11, and CXCL13 immunoreactivity in Hofbauer cells (placental macrophages), stromal cells, and endothelial cells to a variable extent in VUE placentas. The intensity of the staining was higher in placentas with VUE than in controls. An increase of CCL4 and CCL5 immunoreactivity was detected in Hofbauer cells and stromal cells. CD8+ T cells and CD14+ Hofbauer cells in VUE foci were positive for CXCR3 and CCR5. In control placentas, Hofbauer cells were positive for CCR5 but negative for CXCR3. CXCR5 was not detected in either VUE placentas or control placentas (Fig. 4).
Figure 4.
Immunofluorescent staining for selected chemokines and their receptors in VUE placentas. Immunofluorescent signals for corresponding chemokines, receptors, and CD Ags are indicated in green (Alexa 488) and red (Alexa 568 or Alexa 594). A–D, Increased immunoreactivity of CXCR9, CXCR10, CXCR13, and CCL5 in VUE placentas. CXCR3+ and CCR5+ leukocytes infiltrate the chorionic villi, expressing their chemokine ligands in VUE placentas. CXCR5 (C) is not found in either VUE placentas or normal term (TIL) placentas. Immunoreactivity of CXCL9 (A), CXCL10 (B), and CXCL13 (C) is localized in CD14+ Hofbauer cells (arrows) and endothelial cells of VUE placentas. CCL5 (D) is positive in some CD14+ Hofbauer cells (not shown) and CD8+ T cells (arrows). Some infiltrating leukocytes were doubly positive for both a chemokine and its receptor (arrowheads). E and F, CD14+ Hofbauer cells in TIL placentas are negative for CXCR3 (E) while being immunoreactive for CCR5 (F). Infiltrating CD8+ T cells and CD14+ Hofbauer cells in VUE foci are positive for CXCR3 and CCR5. The endothelial cells of villous capillaries are also positive for CXCR3.
Changes in chemokine mRNAs and proteins in maternal and fetal circulation
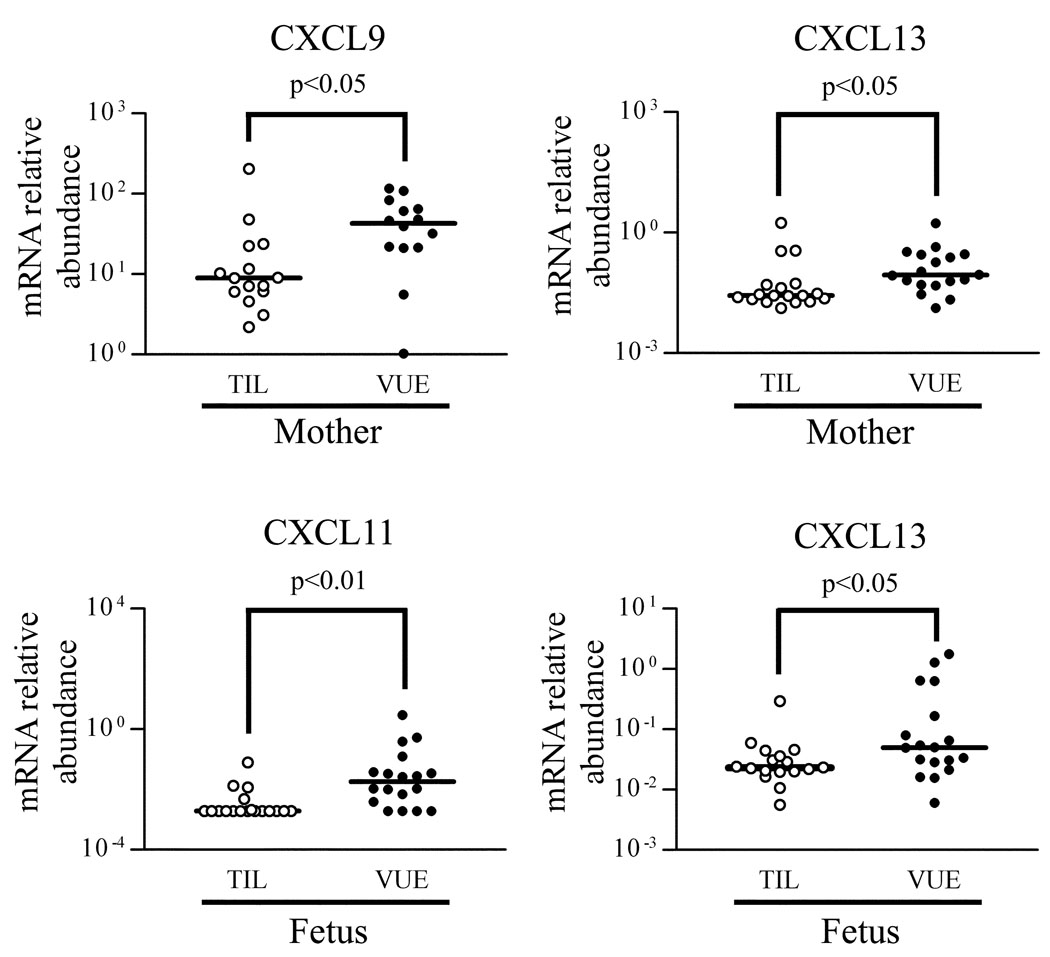
Based on the data from the analysis of the placentas, mRNA expression of chemokines (CXCL9, CXCL10, CXCL11, CXCL13, CCL4, CCL5) and their receptors (CXCR3, CXCR5, CCR5) were further analyzed by qRT-PCR using blood total RNA samples to determine whether VUE is associated with a systemic immune response. The analysis revealed a significantly higher expression of CXCL9 and CXCL13 mRNA in maternal blood of VUE cases (p < 0.05), whereas CXCL11 and CXCL13 mRNA expression was higher in fetal blood in VUE than in controls (p < 0.05; Fig. 5). Differences in mRNA expression of CXCR3, CXCR5, and CCR5 were not observed in either maternal or fetal blood between VUE and controls.
Figure 5.
Differential mRNA expression of chemokines and receptors in maternal and fetal blood. Maternal blood shows higher mRNA expression of CXCL9 and CXCL13 in VUE cases than in control cases (TIL). CXCL11 and CXCL13 mRNA expression is higher in fetal blood of VUE cases than in control cases.
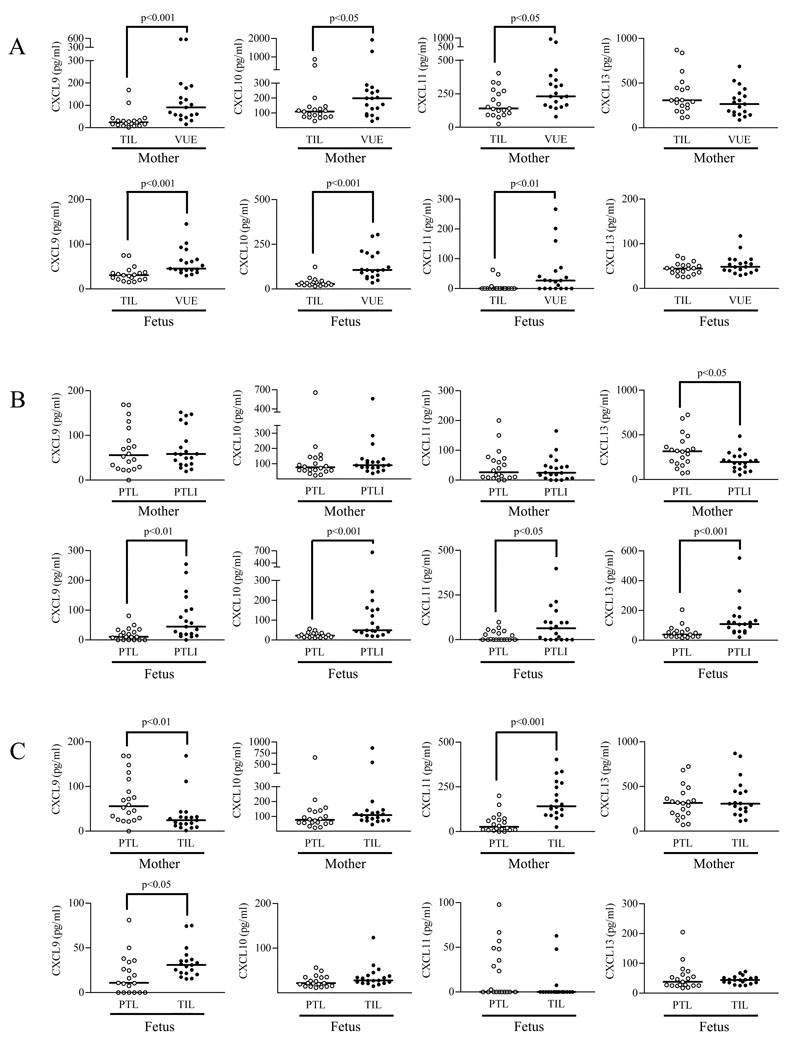
Maternal and fetal plasma concentrations of selected chemokines (CXCL9, CXCL10, CXCL11, CXCL13) were also measured by specific immunoassays. The median concentrations of CXCL9 (maternal, median (range), 24.4 (1.5–168.9) pg/ml in control vs 90.7 (16.0–574.9) pg/ml in VUE cases, p < 0.001; fetal, median (range), 31.0 (15.4–75.2) pg/ml in control vs 45.5 (29.7–145.6) pg/ml in VUE cases, p < 0.001), CXCL10 (maternal, median (range), 108.5 (45.7–868.2) pg/ml in control vs 198.3 (45.0–1928.0) pg/ml in VUE cases, p < 0.05; fetal, median (range), 27.8 (15.1–123.7) pg/ml in control vs 105.9 (35.6–304.9) pg/ml in VUE cases, p < 0.001), and CXCL11 (maternal, median (range), 140.9 (25.4–403.4) pg/ml in control vs 230.9 (79.0–933.5) pg/ml in VUE cases, p < 0.05; fetal, median (range), 0 (0–62.9) pg/ml in control vs 26.5 (0–266.7) pg/ml in VUE cases, p < 0.01) were higher in VUE cases than in control, respectively. On the other hand, the median concentration of CXCL13 was not different (maternal, median (range), 307.6 (112.1–870.5) pg/ml in control vs 265.4 (86.9–687) pg/ml in VUE cases, p > 0.05; fetal, median (range), 44.7 (26.9–99.1) pg/ml in control vs 48.7 (30.3–121.1) pg/ml in VUE cases, p > 0.05; Fig. 6A).
Figure 6.
Chemokine concentrations in maternal and fetal plasma of VUE and acute chorioamnionitis. A, The median concentrations of chemokines CXCL9, CXCL10, and CXCL11 in both maternal and fetal plasma are higher in VUE cases than in term control cases (TIL), respectively. On the other hand, the median concentration of CXCL13 is not different between VUE and control. B, The median fetal plasma concentrations of CXCL9, CXCL10, CXCL11, and CXCL13 are also higher in PTLI cases than in PTL cases. In contrast, maternal plasma concentrations of CXCL9, CXCL10, and CXCL11 are not different between PTL and PTLI cases. CXCL13 concentration is lower in PTLI cases than in PTL cases. C, Comparison between PTL and TIL cases showed that maternal plasma CXCL9 concentration is higher in PTL cases than in TIL cases, whereas CXCL11 concentration is lower in PTL than in TIL cases.
Since acute chorioamnionitis associated with microbial invasion of the amniotic cavity represents another common type of placental inflammatory lesions with both maternal and fetal responses (1–4), we measured maternal and fetal plasma concentrations of the same chemokines in cases with preterm labor and delivery without (PTL) and with acute chorioamnionitis (PTLI). These experiments were undertaken to determine whether changes in fetal and maternal plasma CXC chemokine concentrations are distinct in VUE and acute chorioamnionitis. The median fetal plasma concentrations of CXCL9 (median (range), 10.9 (0–81.2) pg/ml in PTL cases vs 44.6 (0–254.6) pg/ml in PTLI cases; p < 0.01), CXCL10 (median (range), 21.9 (12.0–56.3) pg/ml in PTL cases vs 49.1 (19.9–663.4) pg/ml in PTLI cases; p < 0.001), CXCL11 (median (range), 0 (0–97.9) pg/ml in PTL cases vs 63.5 (0–397.9) pg/ml in PTLI cases; p < 0.05), and CXCL13 (median (range), 38.4 (17.9–205.3) pg/ml in PTL cases vs 108.4 (21.2–552.1) pg/ml in PTLI cases; p < 0.001) were higher in PTLI cases than in PTL cases. Interestingly, however, no differences were found in the maternal plasma concentrations of these chemokines between PTL and PTLI cases. Furthermore, CXCL13 concentration was lower in PTLI than in PTL cases (median (range), 316 (71.4–724.9) pg/ml vs 195.5 (52.7–485.3) pg/ml, respectively, p < 0.05; Fig. 6B). Additional comparisons showed a higher CXCL9 median concentration in maternal plasma of PTL cases than in TIL cases (p < 0.01), while CXCL11 concentration was lower in PTL than in TIL cases (p < 0.001). CXCL9 median concentration in fetal plasma of PTL cases was lower than that of TIL cases (p < 0.05; Fig. 6C).
Discussion
We report, for the first time, the changes in global placental gene expression patterns in VUE. This study also uncovers another novel immunopathologic feature of VUE: the presence of a systemic CXC chemokine response both in the mother and fetus. VUE has been regarded as a placental lesion analogous to allograft rejection by the mother (host) against fetal Ags (graft) (10,11). However, the evidence supporting a fetal systemic inflammatory response strongly suggests that VUE has a component similar to graft-vs-host disease (GVHD) by maternal lymphocytes (graft) to fetal placental tissue (host) (6).
The transcriptome profiles of the placentas with VUE essentially reflect the presence of an inflammatory response involving T lymphocytes and APCs in the chorionic villi. The gene expression profile in this lesion is quite similar to that reported in other organs in the setting of either transplantation rejection or GVHD (20–25). Gene Ontology analysis indicates that VUE, allograft rejection, and GVHD share enrichment of genes involved in Ag presentation, leukocyte migration, T cell activation, and induction by IFN-γ. Up-regulation of the expression of MHC class II molecules in the placentas with VUE is consistent with previous observations in which increased expression of MHC class II Ags in villous trophoblasts and macrophages in and around areas of VUE was demonstrated (26,27). In a murine model of hepatic GVHD, mRNA for MHC class II molecules and for genes related to peptide processing for MHC molecules were up-regulated in the liver 1 wk after experimental allogeneic bone marrow transplantation (24). In acute GVHD, HLA-DR protein was expressed by human keratinocytes. This is interesting because HLA-DR is not found in normal skin or after regression of GVHD (28). A mouse renal allograft rejection model also demonstrated increased mRNA expression of MHC class II molecules in the kidney as a major feature of INF-γ-dependent, rejection-induced transcripts (29). Indeed, the expression levels of MHC class II molecules in human organ allograft show a gradient in the order of rejection, nonrejection, and normal organ before transplantation, showing the critical nature of MHC class II molecules for successful transplantation (22,30). Interestingly, HLA-DR expression in Hofbauer cells increases as a function of gestational age (31,32). Enhanced potential for Ag presentation by Hofbauer cells with increased MHC class II expression at term may be an explanation of why VUE occurs mostly at term (1).
The ligands for CXCR3 (CXCL9, CXCL10, CXCL11) and for CCR5 (CCL4, CCL5) are among the most common chemokines expressed during the course of transplantation rejection (33–35) and GVHD (25,36,37). Increased expression of these chemokines and their receptors in the placentas with VUE strongly suggests the presence of the signals required for leukocyte migration akin to those observed in transplantation rejection and GVHD. Hofbauer cells and endothelial cells in the villi of cases with VUE showed increased expression of selective chemokines, suggesting an interaction with CXCR3+ and CCR5+ T cells, which infiltrate chorionic villi. Transplant rejection and GVHD also show increased expression of chemokines in the parenchymal tissue, including renal tubules and epidermis, in addition to leukocyte infiltration (33,38). A homeostatic chemokine, CXCL13, which is not commonly found in either transplantation rejection or GVHD, except for a few examples of allograft rejection associated with B cell infiltration (39,40), was overexpressed in VUE, although B cells were minimally present in VUE (26,41). Similarly, we could not detect the expression of CXCR5, the primary CXCL13 receptor, in placentas with VUE. However, CXCL13 in VUE may be involved in the chemotaxis of activated T cells, since CXCL13 is also a potential ligand for CXCR3 in addition to IFN-γ-induced CXC chemokines without the ELR motif (42). CXC chemokines are known to regulate angiogenesis by acting on their receptors on endothelial cells. Chemokine ligands for CXCR2 and CXCR4 (CXCL1, CXCL6, CXCL8) drive proangiogenic signals, while CXCL9, CXCL10, CXCL11, and CXCL13 are antiangiogenic (43–46). This is interesting because VUE is commonly associated with obliterative changes of villous vessels (obliterative fetal vasculopathy) (47). We propose that such a finding is associated with an increase in antiangiogenic chemokine expression in the placenta.
The most meaningful and novel observation in the present study is the demonstration of a systemic derangement in chemokine concentrations in both maternal and fetal circulation associated with VUE. The expression pattern of each chemokine mRNA indicates that circulating chemokines could be produced by the placenta or peripheral leukocytes. Chemokine up-regulation in systemic circulation could also be a result rather than a cause of VUE. A few studies have described changes in systemic chemokine concentrations either in allograft rejection or in GVHD. Circulating CXCL11 was elevated in patients with coronary artery disease, which developed in a transplanted heart (48). The serum concentrations of CXCL10 and CCL5 have been reported to be increased in patients with cutaneous GVHD (38). Interestingly, VUE-associated changes in the mother are distinct from those observed in acute chorioamnionitis in which systemic CXC chemokine concentrations were not changed (CXCL9, CXCL10, CXCL11) or even decreased (CXCL13), while those in the fetus were similar to those of acute chorioamnionitis. This stark difference in maternal plasma chemokine concentrations and the similarity in fetal plasma concentrations between VUE and acute chorioamnionitis strongly suggest that the immune responses mounted by the mother are more finely tuned and specific depending on the etiology of the inflammatory process. Another incidental but intriguing finding was the differences in maternal plasma concentrations of CXCL9 and CXCL11 between PTL and TIL patients. Whether those differences are related to gestational age or to the pathologic vs physiologic nature of labor needs further study. Nevertheless, this is the first evidence showing that the maternal systemic chemokine profile varies between preterm labor and spontaneous labor at term.
Pregnancy has been likened to a semiallograft. However, the fetus is not a simple allograft because it may also be a host, as demonstrated by the development of microchimerism (maternal cells in fetal circulation) (49). VUE is a unique fetal response associated with maternal T cell infiltration in the placenta. The overall findings reported herein indicate that VUE is akin to placental GVHD in the fetal compartment and that increased circulating antiangiogenic chemokines, in addition to inflammatory destruction of the placenta, may have a causal link in the development of fetal growth restriction or fetal death. Systemic involvement of the skin or the gastrointestinal tract is a typical clinical presentation of GVHD (50). From this perspective, VUE could be an atypical form of GVHD with its full-blown histologic lesion confined to the placenta. Tissue damage due to underlying diseases or previous treatment is a prerequisite for the development of GVHD (51,52). This prerequisite is met during pregnancy because there is ongoing damage to the villous tree as the pregnancy progresses (53). Restricted tissue damage in the placental villous tissue, exposure of villous tissue to maternal circulation isolated from fetal circulation, and the absence of lymphatics in the placenta may be plausible explanations for the confinement of VUE to the placenta. Secondary lymphoid organs are important for T cell activation, but they are not an absolute requirement for allorecognition (54). Fetal placental macrophages (Hofbauer cells) seem to play a key role in establishing chemokine-chemokine receptor interaction.
In conclusion, we propose that VUE is a disorder characterized by distinct up-regulation of local and systemic CXC chemokines both in the mother and the fetus. Such a distinct pattern of chemokine up-regulation clearly differs from that observed in acute chorioamnionitis due to microbial invasion of the amniotic cavity. Therefore, we propose that placental VUE be considered a unique and genuine pathologic link between maternal allograft transplantation rejection and fetal GVHD. VUE is the only example in human biology where two hosts in intimate contact deploy a bidirectional inflammatory response resulting in a unique form of tissue destruction.
Footnotes
This work was supported by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services.
Abbreviations used in this paper: VUE, villitis of unknown etiology; GVHD, graft-vs-host disease; FDR, false discovery rate; ISI, inflammation severity index; KEGG, Kyoto Encyclopedia of Genes and Genomes; PTL, preterm labor and delivery without acute chorioamnionitis; PTLI, preterm labor and delivery with acute chorioamnionitis; qRT-PCR, quantitative RT-PCR; SPIA, signaling pathway impact analysis; TIL, term in labor.
Reference List
- 1.Kraus FT, Redline RW, Gersell DJ, Nelson DM, Diche JM. Inflammation and infection. In: Kraus FT, Redline RW, Gersell DJ, Nelson DM, Diche JM, editors. Placental Pathology Atlas of Non Tumor Pathology 3. 1st ed. Washington, DC: AFIP; 2004. pp. 75–116. [Google Scholar]
- 2.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am. J. Obstet. Gynecol. 1998;179:194–202. doi: 10.1016/s0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 3.Murtha AP, Greig PC, Jimmerson CE, Roitman-Johnson B, Allen J, Herbert WN. Maternal serum interleukin-6 concentrations in patients with preterm premature rupture of membranes and evidence of infection. Am. J. Obstet. Gynecol. 1996;175:966–969. doi: 10.1016/s0002-9378(96)80033-0. [DOI] [PubMed] [Google Scholar]
- 4.Shimoya K, Matsuzaki N, Taniguchi T, Okada T, Saji F, Murata Y. Interleukin-8 level in maternal serum as a marker for screening of histological chorioamnionitis at term. Int. J. Gynaecol. Obstet. 1997;57:153–159. doi: 10.1016/s0020-7292(97)02891-9. [DOI] [PubMed] [Google Scholar]
- 5.Brito H, Juliano P, Altemani C, Altemani A. Is the immunohistochemical study of the inflammatory infiltrate helpful in distinguishing villitis of unknown etiology from non-specific infection villitis? Placenta. 2005;26:839–841. doi: 10.1016/j.placenta.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Kim JS, Romero R, Kim MR, Kim YM, Friel L, Espinoza J, Kim CJ. Involvement of Hofbauer cells and maternal T cells in villitis of unknown aetiology. Histopathology. 2008;52:457–464. doi: 10.1111/j.1365-2559.2008.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becroft DM, Thompson JM, Mitchell EA. Placental villitis of unknown origin: epidemiologic associations. Am. J. Obstet. Gynecol. 2005;192:264–271. doi: 10.1016/j.ajog.2004.06.062. [DOI] [PubMed] [Google Scholar]
- 8.Redline RW. Villitis of unknown etiology: noninfectious chronic villitis in the placenta. Hum. Pathol. 2007;38:1439–1446. doi: 10.1016/j.humpath.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 9.Boog G. Chronic villitis of unknown etiology. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008;136:9–15. doi: 10.1016/j.ejogrb.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 10.Labarrere CA, Faulk WP. Maternal cells in chorionic villi from placentae of normal and abnormal human pregnancies. Am. J. Reprod. Immunol. 1995;33:54–59. doi: 10.1111/j.1600-0897.1995.tb01138.x. [DOI] [PubMed] [Google Scholar]
- 11.Redline RW, Patterson P. Villitis of unknown etiology is associated with major infiltration of fetal tissue by maternal inflammatory cells. Am. J. Pathol. 1993;143:473–479. [PMC free article] [PubMed] [Google Scholar]
- 12.Myerson D, Parkin RK, Benirschke K, Tschetter CN, Hyde SR. The pathogenesis of villitis of unknown etiology: analysis with a new conjoint immunohistochemistry-in situ hybridization procedure to identify specific maternal and fetal cells. Pediatr. Dev. Pathol. 2006;9:257–265. doi: 10.2350/08-05-0103.1. [DOI] [PubMed] [Google Scholar]
- 13.Weniger M, Engelmann JC, Schultz J. Genome Expression Pathway Analysis Tool--analysis and visualization of microarray gene expression data under genomic, proteomic and metabolic context. BMC Bioinformatics. 2007;8:179. doi: 10.1186/1471-2105-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. Limma: linear models for microarray data. New York, NY: Springer; 2005. [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 17.Gentleman R. Using GO for statistical analyses. In: Antoch J, editor. Compstat 2004 Proceedings in Computational Statistics. Prague: Springer; 2004. pp. 171–180. [Google Scholar]
- 18.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, Kim CJ, Kusanovic JP, Romero R. A Novel Signaling Pathway Impact Analysis (SPIA) Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akalin E, Hendrix RC, Polavarapu RG, Pearson TC, Neylan JF, Larsen CP, Lakkis FG. Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation. 2001;72:948–953. doi: 10.1097/00007890-200109150-00034. [DOI] [PubMed] [Google Scholar]
- 21.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, Pauly DF, Baron H, Wohlgemuth JG, Woodward RN, Klingler TM, Walther D, Lal PG, Rosenberg S, Hunt S. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am. J. Transplant. 2006;6:150–160. doi: 10.1111/j.1600-6143.2005.01175.x. [DOI] [PubMed] [Google Scholar]
- 22.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, Chismar JD, Horvath S, Mondala T, Gilmartin T, Cook DJ, Kay SA, Walker JR, Salomon DR. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am. J. Transplant. 2004;4:1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimino VJ, Lande JD, Berryman TR, King RA, Hertz MI. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am. J. Respir. Crit. Care Med. 2003;168:1237–1242. doi: 10.1164/rccm.200305-644OC. [DOI] [PubMed] [Google Scholar]
- 24.Ichiba T, Teshima T, Kuick R, Misek DE, Liu C, Takada Y, Maeda Y, Reddy P, Williams DL, Hanash SM, Ferrara JL. Early changes in gene expression profiles of hepatic GVHD uncovered by oligonucleotide microarrays. Blood. 2003;102:763–771. doi: 10.1182/blood-2002-09-2748. [DOI] [PubMed] [Google Scholar]
- 25.Sugerman PB, Faber SB, Willis LM, Petrovic A, Murphy GF, Pappo J, Silberstein D, van den Brink MR. Kinetics of gene expression in murine cutaneous graft-versus-host disease. Am. J. Pathol. 2004;164:2189–2202. doi: 10.1016/S0002-9440(10)63776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labarrere CA, McIntyre JA, Faulk WP. Immunohistologic evidence that villitis in human normal term placentas is an immunologic lesion. Am. J. Obstet. Gynecol. 1990;162:515–522. doi: 10.1016/0002-9378(90)90421-3. [DOI] [PubMed] [Google Scholar]
- 27.Labarrere CA, Faulk WP. MHC class II reactivity of human villous trophoblast in chronic inflammation of unestablished etiology. Transplantation. 1990;50:812–816. doi: 10.1097/00007890-199011000-00014. [DOI] [PubMed] [Google Scholar]
- 28.Favre A, Cerri A, Bacigalupo A, Lanino E, Berti E, Grossi CE. Immunohistochemical study of skin lesions in acute and chronic graft versus host disease following bone marrow transplantation. Am. J. Surg. Pathol. 1997;21:23–34. doi: 10.1097/00000478-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 29.Famulski KS, Einecke G, Reeve J, Ramassar V, Allanach K, Mueller T, Hidalgo LG, Zhu LF, Halloran PF. Changes in the transcriptome in allograft rejection: IFN-gamma-induced transcripts in mouse kidney allografts. Am. J. Transplant. 2006;6:1342–1354. doi: 10.1111/j.1600-6143.2006.01337.x. [DOI] [PubMed] [Google Scholar]
- 30.Hubscher SG, Adams DH, Elias E. Changes in the expression of major histocompatibility complex class II antigens in liver allograft rejection. J. Pathol. 1990;162:165–171. doi: 10.1002/path.1711620210. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein J, Braverman M, Salafia C, Buckley P. The phenotype of human placental macrophages and its variation with gestational age. Am. J. Pathol. 1988;133:648–659. [PMC free article] [PubMed] [Google Scholar]
- 32.Sutton L, Gadd M, Mason DY, Redman CW. Cells bearing class II MHC antigens in the human placenta and amniochorion. Immunology. 1986;58:23–29. [PMC free article] [PubMed] [Google Scholar]
- 33.Panzer U, Reinking RR, Steinmetz OM, Zahner G, Sudbeck U, Fehr S, Pfalzer B, Schneider A, Thaiss F, Mack M, Conrad S, Huland H, Helmchen U, Stahl RA. CXCR3 and CCR5 positive T-cell recruitment in acute human renal allograft rejection. Transplantation. 2004;78:1341–1350. doi: 10.1097/01.tp.0000140483.59664.64. [DOI] [PubMed] [Google Scholar]
- 34.Tan J, Zhou G. Chemokine receptors and transplantation. Cell. Mol. Immunol. 2005;2:343–349. [PubMed] [Google Scholar]
- 35.Zhao DX, Hu Y, Miller GG, Luster AD, Mitchell RN, Libby P. Differential expression of the IFN-gamma-inducible CXCR3-binding chemokines, IFN-inducible protein 10, monokine induced by IFN, and IFN-inducible T cell alpha chemoattractant in human cardiac allografts: association with cardiac allograft vasculopathy and acute rejection. J. Immunol. 2002;169:1556–1560. doi: 10.4049/jimmunol.169.3.1556. [DOI] [PubMed] [Google Scholar]
- 36.Wysocki CA, Burkett SB, Panoskaltsis-Mortari A, Kirby SL, Luster AD, McKinnon K, Blazar BR, Serody JS. Differential roles for CCR5 expression on donor T cells during graft-versus-host disease based on pretransplant conditioning. J. Immunol. 2004;173:845–854. doi: 10.4049/jimmunol.173.2.845. [DOI] [PubMed] [Google Scholar]
- 37.Wysocki CA, Panoskaltsis-Mortari A, Blazar BR, Serody JS. Leukocyte migration and graft-versus-host disease. Blood. 2005;105:4191–4199. doi: 10.1182/blood-2004-12-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piper KP, Horlock C, Curnow SJ, Arrazi J, Nicholls S, Mahendra P, Craddock C, Moss PA. CXCL10-CXCR3 interactions play an important role in the pathogenesis of acute graft-versus-host disease in the skin following allogeneic stem-cell transplantation. Blood. 2007;110:3827–3832. doi: 10.1182/blood-2006-12-061408. [DOI] [PubMed] [Google Scholar]
- 39.Di Carlo E, D'Antuono T, Contento S, Di Nicola M, Ballone E, Sorrentino C. Quilty effect has the features of lymphoid neogenesis and shares CXCL13-CXCR5 pathway with recurrent acute cardiac rejections. Am. J. Transplant. 2007;7:201–210. doi: 10.1111/j.1600-6143.2006.01584.x. [DOI] [PubMed] [Google Scholar]
- 40.Steinmetz OM, Panzer U, Kneissler U, Harendza S, Lipp M, Helmchen U, Stahl RA. BCA-1/CXCL13 expression is associated with CXCR5-positive B-cell cluster formation in acute renal transplant rejection. Kidney Int. 2005;67:1616–1621. doi: 10.1111/j.1523-1755.2005.00244.x. [DOI] [PubMed] [Google Scholar]
- 41.Kapur P, Rakheja D, Gomez AM, Sheffield J, Sanchez P, Rogers BB. Characterization of inflammation in syphilitic villitis and in villitis of unknown etiology. Pediatr. Dev. Pathol. 2004;7:453–458. doi: 10.1007/s10024-004-2124-3. [DOI] [PubMed] [Google Scholar]
- 42.Jenh CH, Cox MA, Hipkin W, Lu T, Pugliese-Sivo C, Gonsiorek W, Chou CC, Narula SK, Zavodny PJ. Human B cell-attracting chemokine 1 (BCA-1; CXCL13) is an agonist for the human CXCR3 receptor. Cytokine. 2001;15:113–121. doi: 10.1006/cyto.2001.0923. [DOI] [PubMed] [Google Scholar]
- 43.Addison CL, Daniel TO, Burdick MD, Liu H, Ehlert JE, Xue YY, Buechi L, Walz A, Richmond A, Strieter RM. The CXC chemokine receptor 2, CXCR2, is the putative receptor for ELR+ CXC chemokine-induced angiogenic activity. J. Immunol. 2000;165:5269–5277. doi: 10.4049/jimmunol.165.9.5269. [DOI] [PubMed] [Google Scholar]
- 44.Romagnani P, Lasagni L, Annunziato F, Serio M, Romagnani S. CXC chemokines: the regulatory link between inflammation and angiogenesis. Trends Immunol. 2004;25:201–209. doi: 10.1016/j.it.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Romagnani P, Annunziato F, Lasagni L, Lazzeri E, Beltrame C, Francalanci M, Uguccioni M, Galli G, Cosmi L, Maurenzig L, Baggiolini M, Maggi E, Romagnani S, Serio M. Cell cycle-dependent expression of CXC chemokine receptor 3 by endothelial cells mediates angiostatic activity. J. Clin. Invest. 2001;107:53–63. doi: 10.1172/JCI9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spinetti G, Camarda G, Bernardini G, Romano Di Peppe S, Capogrossi MC, Napolitano M. The chemokine CXCL13 (BCA-1) inhibits FGF-2 effects on endothelial cells. Biochem. Biophys. Res. Commun. 2001;289:19–24. doi: 10.1006/bbrc.2001.5924. [DOI] [PubMed] [Google Scholar]
- 47.Redline RW, Ariel I, Baergen RN, Desa DJ, Kraus FT, Roberts DJ, Sander CM. Fetal vascular obstructive lesions: nosology and reproducibility of placental reaction patterns. Pediatr. Dev. Pathol. 2004;7:443–452. doi: 10.1007/s10024-004-2020-x. [DOI] [PubMed] [Google Scholar]
- 48.Kao J, Kobashigawa J, Fishbein MC, MacLellan WR, Burdick MD, Belperio JA, Strieter RM. Elevated serum levels of the CXCR3 chemokine ITAC are associated with the development of transplant coronary artery disease. Circulation. 2003;107:1958–1961. doi: 10.1161/01.CIR.0000069270.16498.75. [DOI] [PubMed] [Google Scholar]
- 49.Hall JM, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Detection of maternal cells in human umbilical cord blood using fluorescence in situ hybridization. Blood. 1995;86:2829–2832. [PubMed] [Google Scholar]
- 50.Shlomchik WD. Graft-versus-host disease. Nat. Rev. Immunol. 2007;7:340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 51.Hill GR, Ferrara JL. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 2000;95:2754–2759. [PubMed] [Google Scholar]
- 52.Xun CQ, Thompson JS, Jennings CD, Brown SA, Widmer MB. Effect of total body irradiation, busulfan-cyclophosphamide, or cyclophosphamide conditioning on inflammatory cytokine release and development of acute and chronic graft-versus-host disease in H-2-incompatible transplanted SCID mice. Blood. 1994;83:2360–2367. [PubMed] [Google Scholar]
- 53.Watson AL, Burton GJ. A microscopical study of wound repair in the human placenta. Microsc. Res. Tech. 1998;42:351–368. doi: 10.1002/(SICI)1097-0029(19980901)42:5<351::AID-JEMT6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 54.Zhou P, Hwang KW, Palucki D, Kim O, Newell KA, Fu YX, Alegre ML. Secondary lymphoid organs are important but not absolutely required for allograft responses. Am. J. Transplant. 2003;3:259–266. doi: 10.1034/j.1600-6143.2003.00067.x. [DOI] [PubMed] [Google Scholar]