Summary
ErbB2, a metastasis-promoting oncoprotein, is overexpressed in ~25% of invasive/metastatic breast cancers, but in 50–60% of non-invasive ductal carcinomas in situ (DCIS). It has been puzzling how a subset of ErbB2-overexpressing DCIS develops into invasive breast cancer (IBC). We found that co-overexpression of 14-3-3ζ in ErbB2-overexpressing DCIS conferred a higher risk of progression to IBC. ErbB2 and 14-3-3ζ overexpression, respectively, increased cell migration and decreased cell adhesion, two prerequisites of tumor cell invasion. 14-3-3ζ overexpression reduced cell adhesion by activating the TGFβ/Smads pathway that led to ZFHX1B/SIP-1 upregulation, E-cadherin loss, and epithelial-mesenchymal transition (EMT). Importantly, patients whose breast tumors overexpressed both ErbB2 and 14-3-3ζ had higher rates of metastatic recurrence and death than those whose tumors overexpressed only one.
Introduction
ErbB2 overexpression is found in approximately 25% of invasive breast cancers (IBC) and is strongly associated with poor patient survival (Slamon et al., 1989). Overexpression of ErbB2 has been demonstrated to promote breast cancer invasion and metastasis (Yu and Hung, 2000). However, ErbB2 is overexpressed in 50-60% of ductal carcinomas in situ (DCIS) in general and 60-70% of high-grade DCIS (Nofech-Mozes et al., 2005). DCIS, a precursor of IBC, consists of clonal proliferation of malignant cells within the lumen of mammary ducts, with no evidence of invasion through the basement membrane into the surrounding stroma (Burstein et al., 2004). The apparent paradox that ErbB2, the well-known metastasis-promoting oncoprotein, is more frequently overexpressed in non-invasive DCIS than in IBC has been puzzling.
This stimulated debate about whether ErbB2 overexpression alone is sufficient to promote progression from non-invasive DCIS to IBC. The limited number of studies that have used patient follow-up data on invasive recurrence of primary DCIS have yielded ambiguous results. Some studies indicated that ErbB2-overexpressing DCIS had an increased risk of invasive recurrence (Provenzano et al., 2003), while others suggested the opposite (Perin et al., 1996; Ringberg et al., 2001). Interestingly, studies using three-dimensional (3D) culture of mammary epithelial cells (MECs) showed that ErbB2 activation in preformed, growth-arrested, mammary acini led to disruption of the well-organized acinar structure that shared several properties with DCIS in vivo, including uncontrolled cell proliferation, luminal filling, and no invasion (Muthuswamy et al., 2001). Moreover, transgenic mice expressing neu (rat homologue of human ErbB2) under its endogenous promoter developed DCIS-like mammary tumors after a long latency with rare metastasis (Andrechek et al., 2003).
These indicate that ErbB2 activation/overexpression may be involved in DCIS formation and that ErbB2 overexpression alone is not sufficient to drive invasion/metastasis. It was suggested that greater ErbB2 activity or additional genetic/epigenetic events (“second hits”) are needed for MECs to gain invasive capability and for a subset of ErbB2-overexpressing DCIS to transition into IBC (Muthuswamy et al., 2001). However, it remained unclear as to what the “second hits” are.
The transition from a normal cell to a malignant cell is a multistep process, and at least six hallmark alterations in cell physiology collectively drive the malignant progression (Hanahan and Weinberg, 2000). 14-3-3 is a family of evolutionally conserved proteins that can bind to many target proteins involved in each of these cancer hallmark alterations (Tzivion et al., 2006; Wilker and Yaffe, 2004). It is conceivable that deregulation of 14-3-3 may contribute to cancer development. Generally, 14-3-3 proteins are divided into two subgroups: 14-3-3σ is a tumor suppressor, whereas the other 14-3-3 isoforms may have oncogenic functions. Increased 14-3-3ζ expression was observed in several tumor types and in the early stages of breast diseases such as DCIS (Danes et al., 2008). This raised the interesting possibility that 14-3-3ζ overexpression might contribute to DCIS progression to IBC.
The epithelial-mesenchymal transition (EMT) is a process during which epithelial cells convert to a mesenchymal cell phenotype after losing cell polarity, disassembling cell-cell adhesion machinery, and subsequently acquiring cell motility (Guarino, 2007). EMT promotes tumor invasion and metastasis by facilitating escape of tumor cells from the original rigid constraints of the surrounding tissue architecture (Guarino, 2007). The EMT-mediated increase in invasion/metastasis is largely contributed by loss of E-cadherin function, because E-cadherin is essential for the maintenance of adherent junctions between neighboring cells, thus confers physical integrity on epithelial cells (Beavon, 2000; Guarino, 2007). E-cadherin loss has been shown to increase cell invasion in multiple in vitro models, and has been correlated with increased metastasis in several epithelial tumor types (Strathdee, 2002). Therefore, E-cadherin is considered a suppressor of tumor invasion.
Given that ErbB2 overexpression alone in DCIS is not sufficient for progression to IBC, we explored whether 14-3-3ζ overexpression in DCIS may serve as a “second hit” that cooperates with ErbB2 to drive a subset of ErbB2-overexpressing DCIS progression into IBC.
Results
ErbB2 and 14-3-3ζ co-overexpression in DCIS is associated with increased invasion potential
To investigate whether 14-3-3ζ overexpression cooperates with ErbB2 to drive a subset of ErbB2-overexpressing DCIS progression to IBC, we initially examined DCIS samples from 25 patients for whom up to 7-years of follow-up data was available. We analyzed the expression of ErbB2 and 14-3-3ζ by immunohistochemistry (IHC) staining. Fourteen of the 25 cases (56%) showed a high level of ErbB2 expression (Table 1), consistent with previous reports of ErbB2 overexpression in 50-60% of DCIS cases (Nofech-Mozes et al., 2005). Eight of the 25 (32%) exhibited high levels of both ErbB2 and 14-3-3ζ (Table 1 and Figure S1). Strikingly, four of these eight patients had disease recurrence with distant site metastasis, whereas none of the 17 DCIS patients whose tumors did not overexpress both proteins developed distant metastasis (Table 1). Thus, ErbB2 and 14-3-3ζ co-overexpression in this small cohort significantly (P<0.05) correlated with distant site metastasis, suggesting that 14-3-3ζ cooperates with ErbB2 to promote the progression from DCIS to IBC and metastasis.
Table 1. Expression of ErbB2 and 14-3-3ζ in 25 DCIS cases and the incidence of metastatic recurrence after 7-years of follow-up.
ErbB2 and 14-3-3ζ expression levels were examined by IHC staining in 25 DCIS patients’ samples. ErbB2 and 14-3-3ζ co-overexpression (3+/3+) in this cohort significantly (P<0.05) correlated with metastatic disease recurrence (Fisher's exact test ).
| 14-3-3ζ (<3+) |
14-3-3ζ (3+) |
Total | |
|---|---|---|---|
| No. of cases (No. of metastatic recurrence) | |||
| ErbB2 (<3+) |
5 (0) |
6 (0) |
11 (0) |
| ErbB2 (3+) |
6 (0) |
8 (4) |
14 (4) |
| Total | 11(0) | 14 (4) | 25 (4) |
MCF10A, a non-transformed human MEC line, is an excellent in vitro model in 3D culture for studying breast cancer progression as it forms well-organized acinar structures which mimic the normal mammary end bud in vivo (Debnath et al., 2003). Here, we used the MCF10A 3D culture model system to study whether and how 14-3-3ζ cooperates with ErbB2 to gain invasiveness. We established multiple stable MCF10A sublines overexpressing ErbB2 (10A.ErbB2), HA-tagged 14-3-3ζ (10A.14-3-3ζ), or both ErbB2 and HA-tagged 14-3-3ζ (10A.ErbB2.ζ), with 10A.Vec as the control (Figure 1A). We found that only the 10A.ErbB2.ζ cells formed soft agar colonies, whereas 10A.ErbB2, 10A.14-3-3ζ, and 10A.Vec MECs did not (Figure 1B). The data indicated that ErbB2 or 14-3-3ζ overexpression alone was not sufficient to induce a full transformation in MCF10A MECs, but ErbB2 and 14-3-3ζ co-overexpression could cooperatively induce full transformation – an important step for cancer invasion/metastasis.
Figure 1. ErbB2 and 14-3-3ζ co-overexpression is associated with increased invasion potential in an in vitro DCIS model.
(A) Establishment of MCF10A stable cell lines overexpressing ErbB2 alone, 14-3-3ζ alone or both ErbB2 and 14-3-3ζ. Multiple stable clones were established for each sublines, and most experiments were repeated with different clones to rule out clonal effects. Immunoblot analysis of one representative of each subline for the indicated proteins is shown.
(B) Co-overexpression of ErbB2 and 14-3-3ζ led to anchorage-independent growth of MCF10A cells in soft agar assay.
(C) Distinct acinar structures of MCF10A sublines in 3D culture. Top: Phase-contrast images of acinar structures (19 days); bar: 200 μm. Inset: Amplified view of individual 10A.ErbB2.ζ cell invading into surrounding matrigel; bar: 100 μm.
(D) Loss of basement membrane integrity in 10A.ErbB2.ζ acini. MCF10A sublines were cultured in 3D matrigel for 28 days and then stained for laminin V (red) and DAPI (blue). Representative images were shown. Bar: 100 μm.
Strikingly, the four sublines showed distinct acinar structures when grown in 3D matrigel (Figure 1C and 1D, Figure S2). 10A.ErbB2 cells formed highly proliferative, but non-invasive, DCIS-like structures characterized by impaired proliferation suppression and luminal cell apoptosis resistance, similar to a previous report (Muthuswamy et al., 2001). 10A.14-3-3ζ cells developed into abnormal acinar structures with no lumen formation, but no growth advantage, as we recently reported (Danes et al., 2008). 10A.ErbB2.ζ cells, however, demonstrated severe disruption of the acinar architecture, characterized by increased acinar size and no lumen formation (Figure 1C and 1D). The most distinct feature of the 10A.ErbB2.ζ acini was the gain of invasive capacity, as many cells escaped from 10A.ErbB2.ζ acini and invaded the surrounding matrix (Figure 1C and 1D). An important feature of the non-invasive DCIS is the intact basement membrane that surrounds it, while invasive carcinomas are defined by loss of basement membrane integrity (Rizki and Bissell, 2004). Indeed, we observed that individual cells in 10A.ErbB2.ζ acini were patched by diffuse basement membrane protein laminin V, whereas laminin V formed a continuous basement membrane layer surrounding acini from 10A.ErbB2, 10A.14-3-3ζ, and 10A.Vec MECs (Figure 1D). Together, co-overexpression of ErbB2 and 14-3-3ζ in MCF10A MECs conferred invasiveness, while overexpression of ErbB2 or 14-3-3ζ alone did not.
Invasion is the collective effect of ErbB2-mediated increase of cell migration and 14-3-3ζ–mediated decrease of cell-cell adhesion via EMT
Tumor cell invasion is a multistep process, of which the key events include increased migration, increased protease secretion, and altered adhesion to allow dissemination from primary tumor sites (Liotta and Stetler-Stevenson, 1991). We detected no significant difference in matrix metalloproteinase levels among the four MCF10A sublines (data not shown). However, migration and wound healing assays showed that both 10A.ErbB2 and 10A.ErbB2.ζ cells had increased cell motility, whereas 10A.14-3-3ζ cells had a low motility similar to that of 10A.Vec (Figure 2A and 2B). Thus, the increased cell motility was largely contributed by ErbB2 overexpression, not by 14-3-3ζ overexpression. Multiple ErbB2 downstream signaling pathways can be involved in ErbB2-mediated cell motility, including PI3K, PAK1, Rac1, and Src activation (Feldner and Brandt, 2002). We found that Src phosphorylation is specifically increased in the two ErbB2 overexpressing MCF10A sublines compared to the two ErbB2 low-expressing MCF10A sublines (Figure S3A, B). Moreover, treatment with a Src kinase inhibitor (Saracatinib, or AZD0530) significantly inhibited the motility of 10A.ErbB2 and 10A.ErbB2.ζ cells, while Rac1 and PI3K inhibitors had no significant effect (Figure S3C).
Figure 2. ErbB2 overexpression led to increased cell migration, and 14-3-3ζ overexpression led to decreased cell-cell adhesion as a result of EMT.
(A) Transwell migration assay of the indicated MCF10A sublines. Cells that migrated to the bottom of the chamber were counted in five fields under 20X magnification. Experiments were done three times with triplicates, error bar represents SEM.
(B) Wound healing assay of the indicated MCF10A sublines. Wound closures were photographed at 0 and 6 hours after wounding. Bar: 200 μm.
(C) The four MCF10A sublines exhibited different morphologies in 2D culture. Bar: 100 μm.
(D) IFS of EMT markers in MCF10A sublines. E-cadherin (top, red), vimentin (bottom, red), DAPI (blue). Bar: 100 μm.
(E) Immunoblot analysis of the indicated EMT markers in MCF10A sublines. Expression of epithelial cell markers (E-cadherin, β-catenin, α-catenin, p120 catenin) and mesenchymal cell marker (N-cadherin) were examined by immunoblot analysis in both 2D and 3D culture cell lysates.
Reduced cell-cell adhesion is another prerequisite for individual cell invasion, and EMT has been implicated in tumor invasion partly by reducing cell-cell adhesion (Guarino, 2007). In contrast to 10A.Vec and 10A.ErbB2 cells that had a cobblestone-like epithelial morphology in 2D culture, 10A.ErbB2.ζ and 10A.14-3-3ζ cells displayed a spindle-like shape and exhibited a scattered distribution, indicating loss of cell-cell contact and EMT (Figure 2C). The morphological changes during EMT are driven by a number of molecular and cellular alterations, including loss or decrease of epithelial cell markers (e.g., E-cadherin, β-catenin, α-catenin, p120-catenin) and de novo expression of mesenchymal markers (e.g., N-cadherin, vimentin, fibronectin). Certainly, we found that 10A.Vec and 10A.ErbB2 cells expressed high levels of E-cadherin, β-catenin, α-catenin, and p120-catenin, but minimal levels of N-cadherin and vimentin. 10A.ErbB2.ζ and 10A.14-3-3ζ cells, however, showed E-cadherin loss, dramatically reduced β-catenin, α-catenin, and p120-catenin, and de novo expression of N-cadherin and vimentin (Figures 2D and 2E). Similarly, 14-3-3ζ overexpression in HMEC-hTERT cells, immortalized by the telomerase reverse transcriptase catalytic subunit, also led to EMT (Figure S4). Thus, 14-3-3ζ overexpression contributed to the loss of cell-cell adhesion and the EMT phenotype. Together, a collective effect of ErbB2-mediated increase of cell migration and 14-3-3ζ-mediated decrease of cell-cell adhesion conferred 10A.ErbB2.ζ acini invasiveness.
E-cadherin loss, a key event of EMT, is mediated by ZFHX1B in 10A.ErbB2.ζ cells
We next investigated how 14-3-3ζ overexpression led to E-cadherin loss, a key event of EMT resulting in decreased cell-cell adhesion. RT-PCR analysis showed that E-cadherin mRNA level was dramatically lower in 10A.ErbB2.ζ and 10A.14-3-3ζ cells than in 10A.Vec and 10A.ErbB2 cells (Figure 3A). E-cadherin mRNA loss could result from hypermethylation of its promoter (Strathdee, 2002), but we detected no significant differences in E-cadherin promoter methylation status among the four MCF10A sublines (data not shown). Another major mechanism of E-cadherin mRNA loss is direct transcriptional repression by repressors, including snail, slug, twist, E12, E47, ZFHX1B (also named SIP1) and deltaEF1 (Peinado et al., 2004). These transcriptional repressors have been found to induce EMT in vitro, and their overexpression in a variety of human tumors is associated with increased tumor invasion/metastasis and poor prognosis. We examined the expression levels of snail, slug, twist, E12, E47, and deltaEF1 and found they were not significantly different among the four MCF10A sublines (Figure 3B). Interestingly, expression of ZFHX1B was dramatically higher in 10A.ErbB2.ζ and 10A.14-3-3ζ cells than in 10A.Vec and 10A.ErbB2 cells at both mRNA and protein levels (Figure 3C).
Figure 3. EMT phenotype in 10A.ErbB2.ζ and 10A.14-3-3ζ cells was mediated by ZFHX1B upregulation.
(A) E-cadherin mRNA was dramatically reduced in 10A.ErbB2.ζ and 10A.14-3-3ζ cells.
(B) Expression of EMT regulators in the four MCF10A sublines. Top: Immunoblot analysis of EMT regulators, including snail, twist, and slug. Bottom: RT-PCR analysis of EMT regulators, including E12, E47, and delta EF1.
(C) RT-PCR and immunoblot analysis of ZFHX1B levels in the four MCF10A sublines. MDA-MB-435 cells served as the positive control for ZFHX1B protein expression.
(D) 14-3-3ζ overexpression led to transcriptional repression of E-cadherin promoter activity. Top: Schematic representation of the luciferase reporter driven by E-cadherin proximal promoter containing two ZFHX1B binding E-box motif: pGL3.Ecad. Bottom: Relative luciferase activity of pGL3.Ecad in the four MCF10A sublines, P<0.05. Error bars indicate SEM.
(E) Downregulation of ZFHX1B in 14-3-3ζ–overexpressing MCF10A sublines by siRNA partially relieved suppression of E-cadherin promoter-driven luciferase activity. Top: Control siRNA and ZFHX1B siRNA were transfected into 10A.ErbB2.ζ cells and 10A.14-3-3ζ cells. After 48 hours, downregulation of ZFHX1B was examined by RT-PCR with GAPDH as internal control. Bottom: Cells were pre-transfected with ZFHX1B siRNA or control siRNA. After 48 hours, pGL3.Ecad was co-transfected with pRL.TK plasmid as transfection efficiency control. Relative luciferase activity was determined 36 hours later. Error bars indicate SEM.
(F) A reverse correlation between ZFHX1B and E-cadherin expression in breast cancer cell lines. RNA was extracted from four E-cadherin–negative breast cancer cell lines (BT549, Hs578T, MDA-MB-231, MDA-MB-435), five E-cadherin–positive breast cancer cell lines (MCF-7, BT474, T47D, MDA-MB-361, MDA-MB-453), and a non-transformed breast epithelial cell line (MCF10A), then reverse transcribed into cDNA, followed by PCR with ZFHX1B specific primers. GAPDH served as loading control.
ZFHX1B is a two-handed zinc-finger protein that binds to the E-boxes in the E-cadherin proximal promoter to represses E-cadherin transcription (Comijn et al., 2001). To examine whether the E-cadherin loss in 10A.ErbB2.ζ and 10A.14-3-3ζ cells was due to transcriptional repression by the upregulated ZFHX1B, a fragment of E-cadherin promoter (containing two consensus ZFHX1B binding motifs: CANNTG) was cloned upstream of a luciferase reporter plasmid (pGL3.Ecad) and its activity was compared among the MCF10A sublines (Figure 3D). Indeed, pGL3.Ecad luciferase activities were significantly repressed (P<0.05) in 10A.ErbB2.ζ and 10A.14-3-3ζ cells versus 10A.Vec and 10A.ErbB2 cells (Figure 3D). Moreover, the repression of E-cadherin promoter-driven luciferase activity was partially relieved in 10A.ErbB2.ζ and 10A.14-3-3ζ cells when ZFHX1B expression was inhibited by small interfering RNA (siRNA; Figure 3E). Therefore, ZFHX1B upregulation contributed to the transcriptional repression of E-cadherin in 10A.ErbB2.ζ and 10A.14-3-3ζ cells. In addition, examination of ZFHX1B expression in six E-cadherin–positive and four E-cadherin–negative breast cancer cell lines showed a general correlation between ZFHX1B expression and E-cadherin loss (Figure 3F).
ZFHX1B is upregulated by 14-3-3ζ through upregulation of TGFβ receptor I and activation of TGFβ/Smads pathway
Next, we investigated the mechanism of ZFHX1B upregulation in 10A.ErbB2.ζ and 10A.14-3-3ζ cells. Since TGFβ/Smads pathway activation was shown to induce EMT and was also known to be involved in ZFHX1B upregulation (Zavadil and Bottinger, 2005),we examined whether ZFHX1B upregulation by 14-3-3ζ might be due to increased TGFβ/Smads signaling. Expression of the TβRI protein, but not RNA, was dramatically increased in 10A.ErbB2.ζ and 10A.14-3-3ζ cells, whereas TβRII protein levels were similar among the four MCF10A sublines (Figure 4A). Consistently, we also observed increased TβRI level in 14-3-3ζ–overexpressing HMEC-hTERT-HA-14-3-3ζ cells accompanied by upregulation of ZFHX1B (Figure S4C). The increased TβRI protein levels led to increased TGFβ/Smads activation, as indicated by the increased nuclear phospho-smad2/smad3 and total smad2/smad3 levels in 10A.ErbB2.ζ and 10A.14-3-3ζ cells (Figure 4B). Moreover, chromatin immunoprecipitation assay (CHIP) detected binding of nuclear smad3 to the ZFHX1B promoter in 10A.ErbB2.ζ and 10A.14-3-3ζ cells, but not in 10A.Vec or 10A.ErbB2 cells (Figure 4C). These data indicate that 14-3-3ζ-mediated TGFβ/Smads activation contributed to ZFHX1B transcriptional upregulation. Indeed, blocking 14-3-3ζ by siRNA reduced TβRI protein expression, which also led to reduced ZFHX1B expression (Figure 4D and Figure S5A).
Figure 4. 14-3-3ζ overexpression led to TGFβ/Smads pathway activation in MCF10A cells via increasing TβRI expression.
(A) Immunoblot and RT-PCR analysis of TβRI and TβRII expression in MCF10A sublines.
(B) Immunoblot analysis of nuclear p-smads (p-smad2/p-smad3), T-smad2/3, with lamin C as nuclear fraction loading control.
(C) Chromatin immunoprecipitation assay with the indicated antibodies showed smad3 binding with ZFHX1B promoter in 10A.ErbB2.ζ and 10A.14-3-3ζ cells (arrows), not in 10A.Vec and 10A.ErbB2 cells (empty arrows).
(D) Knockdown of 14-3-3ζ by siRNA reduced TβRI and ZFHX1B expression in 10A.ErbB2.ζ and 10A.14-3-3 ζ cells. TβRI level and ZFHX1B level were analyzed by immunoblot and RT-PCR, respectively, 48 hours after siRNA transfection.
(E) 14-3-3ζ inhibited TβRI ubiquitination. 10A.ErbB2.ζ and 10A.ErbB2 cells were co-transfected with vectors expressing myc-TβRI and HA-ubiquitin (left), 10A.ErbB2.ζ and Hela cells were co-transfected with myc-TβRI, HA-ubiquitin, and control/14-3-3ζ siRNA (middle), Hela cells were co-transfected with myc-TβRI, HA-ubiquitin, and pcDNA3.Vec/pcDNA3.14-3-3ζ (right).After 48 hours, cell lysates were collected and subjected to immunoprecipitation and immunoblot with myc and HA antibodies.
(F) 10A.ErbB2 and 10A.ErbB2.ζ cells were treated with DMSO or 20 μg/ml MG132 for 6 hours; TβRI levels were analyzed by immunoblot.
(G) 14-3-3ζ associated with TβRI. 10A.ErbB2.ζ and 10A.14-3-3ζ cell lysates were immunoprecipitated by anti-HA antibody, followed by immunoblot analysis of TβRI.
(H) Schematic representation of different TβRI mutants.
(I) 14-3-3ζ binds to TβRI at its kinase domain between amino acid 210 and 370. 10A.ErbB2.ζ cells were infected with lentivirus expressing different TβRI mutants as in (H). Then the cell lysates were subjected to pull-down assay with GST or GST-14-3-3ζ, followed by immunoblot with TβRI antibody.
TβRI protein level is mainly regulated by its internalization, followed either by trafficking back to the cell membrane after engulfed in early endosome, or by ubiquitination-mediated degradation when engulfed in lipid-raft-caveolae-1 vesicles (Di Guglielmo et al., 2003; Lin et al., 2004). To investigate the mechanisms of 14-3-3ζ-mediated TβRI protein upregulation, we first investigated whether it is contributed by reduced TβRI ubiquitination. Indeed, ubiquitination of Myc-tagged TβRI in 10A.ErbB2.ζ cells was reduced compared to 10A.ErbB2 cells when HA-tagged ubiquitin was coexpressed (Figure 4E, left). 14-3-3ζ knockdown by siRNA in 10A.ErbB2.ζ cells and in Hela cells (endogenous 14-3-3ζ) led to a consistent increase in TβRI ubiquitination, while TβRI ubiquitination was inhibited when 14-3-3ζ was overexpressed (Figure 4E, middle and right). Furthermore, treatment with MG132, a proteasome inhibitor, led to greater accumulation of TβRI in 10A.ErbB2 cells than in 10A.ErbB2.ζ cells (Figure 4F), indicating a more rapid TβRI ubiquitination and proteasome-mediated degradation in 14-3-3ζ low-expressing 10A.ErbB2 cells. Next, we examined whether 14-3-3ζ inhibited TβRI ubiquitination and degradation by binding to TβRI. Indeed, 14-3-3ζ and TβRI coexisted in the same complex (Figure 4G) and the binding region is between amino acid 210 and 370 in the kinase domain of TβRI (Figure 4H, I). Immunofluorescence staining (IFS) also detected diffuse staining of both 14-3-3ζ and TβRI proteins both in the cytosol and on the cell membrane (Figure S5B). The data are consistent with previous reports that TβRI is constantly recycled between membrane and cellular vesicles, resulting in ~20% localization to the cell membrane and ~80% remaining in the cytosol (Di Guglielmo et al., 2003). Most importantly, the binding of 14-3-3ζ protects TβRI from degradation because the TβRI-210 that cannot bind to 14-3-3ζ has a much shorter half-life compared to the TβRI-370 that binds to 14-3-3ζ (Figure S5C). Furthermore, when 14-3-3ζ expression is knocked down by siRNA, the half-life of TβRI-370 is dramatically reduced, while the half-life of TβRI-210 is not affected (Figure S5D, and data not shown). These results indicated that overexpressed 14-3-3ζ in 10A.ErbB2.ζ and 10A.14-3-3ζ cells bound to TβRI, and inhibited the proteasome-mediated TβRI degradation, leading to increased TβRI protein level and TGFβ/Smads pathway activation.
To further confirm the role of TGFβ/Smads pathway activation in the induction of the EMT phenotypes, we treated the 10A.Vec and 10A.ErbB2 cells with TGFβ1 to activate the TGFβ/Smads pathway. The treatment induced smad3 phosphorylation, ZFHX1B upregulation, and morphological features of EMT, with corresponding downregulation of E-cadherin, and upregulation of vimentin and fibronectin (Figure S6). Therefore, activation of the TGFβ/Smads pathway was sufficient to induce EMT in MCF10A cells.
Inhibition of TGFβ receptor activation partially reversed EMT and inhibited the invasiveness of 10A.ErbB2.ζ acini
To determine whether activation of the TGFβ/Smads pathway is required for the EMT and invasive phenotype of the 10A.ErbB2.ζ cells, we inhibited TGFβ/Smads pathway activation by treating 10A.ErbB2.ζ cells with a TGFβ receptor I/II kinase inhibitor, LY2109761 (Melisi et al., 2008). LY2109761 treatment reduced smad2/3 phosphorylation and total smad3, but had no significant effect on the phosphorylation of Akt (p-Akt) or p42-MAPK (p-P42) (Figure 5A). Interestingly, LY2109762-treated 10A.ErbB2.ζ cells adhered to neighboring cells to form cell islands, indicating improved cell-cell adhesion (Figure 5B, left). More importantly, the invasive phenotype of 10A.ErbB2.ζ acini in 3D matrigel culture was dramatically inhibited by LY2109761 treatment compared to control treatment (Figure 5B, middle). In contrast, LY2109761 treatment had no significant impact on acini development and maintenance in the other MCF10A sublines (Figure S7). Consistent with the partial reversal of EMT morphology of the cells in 2D culture and reduced invasiveness in 3D culture, there was increased epithelial protein expression, such as E-cadherin and α-catenin, after LY2109761 treatment. E-cadherin was specifically located in the membrane regions forming cell-cell contacts, a prerequisite for adherent junction formation (Figure 5B, right). Prolonged treatment also led to decreased mesenchymal protein expression (Figure 5C). Collectively, these data indicate that 14-3-3ζ-mediated TGFβ/Smads pathway activation plays a critical role in the EMT phenotype and gain of invasiveness in 10A.ErbB2.ζ cells.
Figure 5. Inhibition of TGFβ/Smads pathway partially reversed EMT of 14-3-3ζ–overexpressing MCF10A cells and inhibited invasion of 10A.ErbB2.ζ cells.
(A) LY2109761 efficiently inhibited smad2/3 phosphorylation in 10A.ErbB2.ζ cells. Cells were treated with TGFβ1 (5 ng/ml) or TGFβ1 (5 ng/ml) plus TβR inhibitor LY2109761 (10 μM) for 60 minutes, then analyzed by immunoblot with indicated antibodies (left). Cells were treated with increasing dose of LY2109761, then analyzed by immunblot for p-Akt, T-Akt, p-P42, T-P42 and β-actin (right).
(B) Treatment of 10A.ErbB2.ζ cells with TGFβ receptor inhibitor partially reversed the EMT morphology in 2D culture and inhibited the invasiveness of acini in 3D culture. Left: Cells were treated with LY2109761 for 48 hours, and images were photographed under a phase-contrast microscope. Middle: 10A.ErbB2.ζ cells were cultured on 3D matrigel for 13 days to form invasive acinar structures, and then treated with DMSO or LY2109761 for 6 days. Right: 10A.ErbB2.ζ cells were treated with DMSO or LY2109761 and subjected to immunofluorescence staining of E-cadherin. Bar: 100 μm. Cells lysates were also collected after treatment, and subjected to immunoblot with E-cadherin, α-catenin and β-actin antibodies.
(C) Prolonged LY2109761 treatment (15days) reduced mesenchymal protein expression (fibronectin, vimentin).
(D) E-cadherin expression was stably re-introduced into 10A.ErbB2.ζ cells.
(E) E-cadherin restoration led to increased epithelial protein expression (α-catenin, β-catenin, p120-catenin) and decreased mesenchymal protein expression (N-cadherin, vimentin).
(F) E-cadherin restoration inhibited 10A.ErbB2.ζ cell invasion. Phase-contrast microscopy of acinar structures of 10A.ErbB2.ζ.Vec and 10A.ErbB2.ζ.Ecad cells in 3D culture. Bar: 200 μm.
Re-introduction of E-cadherin in 10A.ErbB2.ζ cells inhibits invasion
Inhibition of TGFβ/Smads pathway by LY2109761 partially recovered E-cadherin expression that inhibited the invasion of 10A.ErbB2.ζ acini, indicating that E-cadherin loss was a key event in the gain of invasiveness during EMT. To further determine the critical role of E-cadherin loss in invasion, we restored E-cadherin expression in the 10A.ErbB2.ζ cells (named 10A.ErbB2.ζ.Ecad) to levels similar to those in the 10A.Vec cells (Figure 5D). The restored E-cadherin expression led to the recovery of other epithelial proteins, such as α-catenin, β-catenin, and p120-catenin, and reduced mesenchymal proteins, such as N-cadherin and vimentin (Figure 5E). Moreover, the cells with recovered E-cadherin expression showed a dramatic increase in cell adhesion (Figure S8A, S8B). Importantly, 10A.ErbB2.ζ.Ecad cells formed acinar structures with fewer individual cells invading into surrounding matrigel, in contrast to the highly invasive acinar structures of 10A.ErbB2.ζ.Vec cells (Figure 5F and S8C). Thus, re-expression of E-cadherin in 10A.ErbB2.ζ cells efficiently increased cell-cell adhesion and inhibited, at least partially, the invasive phenotype in 3D culture. Therefore, E-cadherin loss played a critical role in inducing invasiveness of 10A.ErbB2.ζ cells.
14-3-3ζ overexpression is associated with high levels of TβRI expression in both human DCIS and IBC
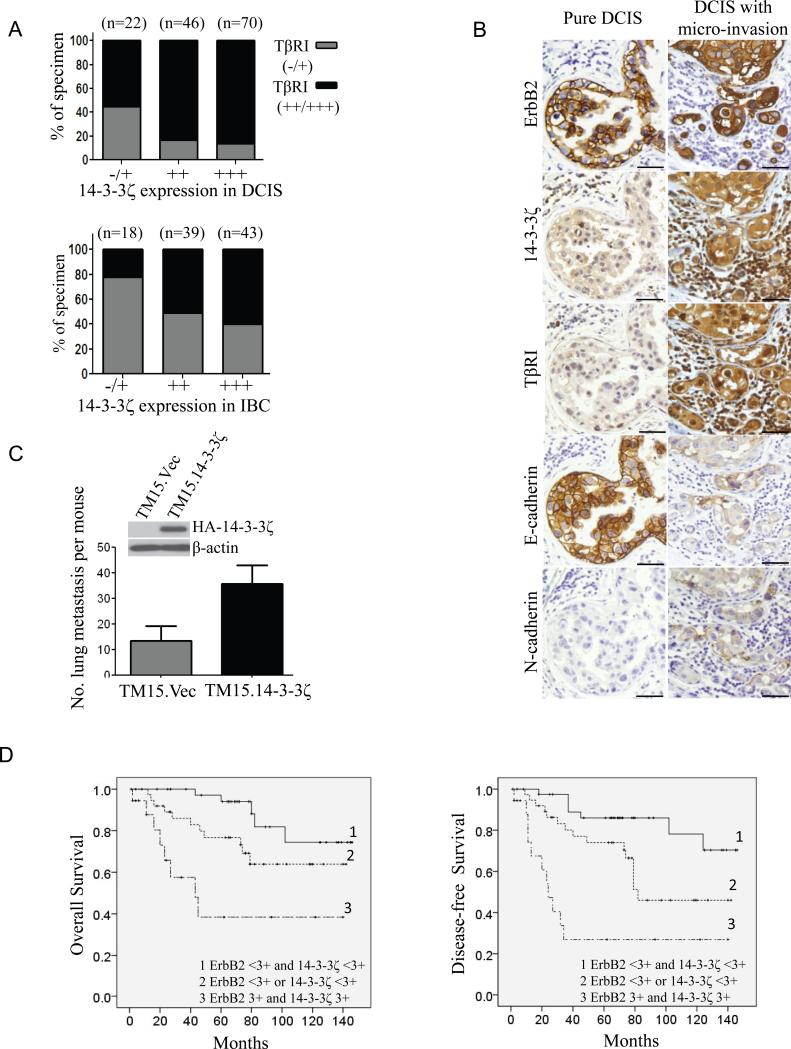
We have identified TβRI upregulation as a major mechanism of 14-3-3ζ overexpression-induced invasiveness in MCF10A.ErbB2.ζ cells. To evaluate the biological relevance of these findings, we investigated whether there is a correlation between TβRI and 14-3-3ζ expression in patients’ samples. Since we did not have enough of the DCIS samples shown in Table 1 remaining for these staining, we stained 138 DCIS samples from patients with recently diagnosed disease and 100 invasive breast cancers with clinical follow-up. We found that 14-3-3ζ overexpression significantly (P<0.05) correlated with increased TβRI levels in both populations (Figure 6A). Moreover, IHC staining for 14-3-3ζ, TβRI, ErbB2, E-cadherin, vimentin, and N-cadherin on the DCIS samples showed that co-overexpression of 14-3-3ζ and TβRI significantly (P<0.05) correlated with (at least two) EMT marker alterations (reduced expression of epithelial marker E-cadherin, and expression of mesenchymal markers vimentin and/or N-cadherin; Table S1 and Figure S9). Importantly, high 14-3-3ζ and TβRI expression levels plus two EMT marker alterations in DCIS were significantly (P< 0.05) associated with high-grade DCIS phenotype, which correlates with a higher risk of invasive recurrence (Table S2). Representative images of multiple markers’ expressions in a pure low-grade DCIS and in a DCIS sample with micro-invasion are shown in figure 6B. Together, 14-3-3ζ overexpression in DCIS lesions correlated with TβRI upregulation and induced EMT that could contribute to a higher risk of invasive recurrence.
Figure 6. Co-overexpression of ErbB2 and 14-3-3ζ promotes invasion and metastasis.
(A) 140 DCIS (top) and 107 IBC (bottom) samples were subjected to IHC staining for TβRI and 14-3-3ζ, of which 138 DCIS and 100 IBC cases were satisfactory for analysis. Chi-square test indicated a correlation between TβRI and 14-3-3ζ expression in both cohorts, P<0.05.
(B) Representative IHC staining for ErbB2, 14-3-3ζ, TβRI, E-cadherin, and N-cadherin in a DCIS case with microinvasion and a low-grade DCIS. Bar: 50 μm.
(C) 14-3-3ζ overexpression in TM15 cells increased lung metastasis. TM15 cells were stably transfected with HA-14-3-3ζ or empty vector. TM15.Vec and TM15.14-3-3ζ cells were injected into mammary fat pad of nude mice (four mice each group). When the primary tumors reached the size of 150 mm3, mice were sacrificed and their lungs underwent histological analysis to measure metastasis, P<0.05. Error bar indicates SEM. Experiment was repeated twice and similar results were observed.
(D) Co-overexpression of ErbB2 and 14-3-3ζ in IBC correlated with higher rates of death and disease recurrence. Tumor samples from 107 breast cancer patients were IHC-stained for ErbB2 and 14-3-3ζ, Kaplan-Meier plots of overall survival (left) and disease-free survival (right) according to ErbB2 and 14-3-3ζ expression are shown. P<0.05 (log-rank test).
Co-overexpression of ErbB2 and 14-3-3ζ is associated with higher metastatic potential in mice and increased metastatic disease recurrence and death in breast cancer patients
The above findings demonstrated that co-overexpression of ErbB2 and 14-3-3ζ increased the invasiveness of MECs in 3D culture. To determine whether co-overexpression of ErbB2 and 14-3-3ζ may increase invasion/metastasis in vivo, we stably overexpressed 14-3-3ζ in TM15 cells, a mouse mammary tumor cell line from a MMTV-Cre/flox-neoNeuNT mouse that expresses the transforming neu under an endogenous promoter. We established the TM15.14-3-3ζ cell line with TM15.Vec as our controls. The two sublines were injected into mammary fat pads of nude mice to establish xenografts and mice were monitored for metastatic lesions. Mice injected with the TM15.14-3-3ζ cells definitely had more lung metastasis than mice with TM15.Vec cells (Figure 6C). To further investigate the impact of co-overexpression of ErbB2 and 14-3-3ζ on breast cancer progression, especially metastatic disease recurrence and death of breast cancer patients, we performed IHC analysis to examine ErbB2 and 14-3-3ζ expression in 107 cases of IBC in consecutive slides. Remarkably, 23 of the 107 patients had breast tumors co-overexpressing both ErbB2 and 14-3-3ζ, and these patients had significantly shorter overall survival (P<0.05) and disease-free survival (P<0.05) than patients whose tumors overexpressed either one or neither (Figure 6D). In addition, in this patient cohort, multivariate analysis demonstrated that co-overexpression of ErbB2 and 14-3-3ζ in breast tumors can predict poor prognosis (Table S3). Since a majority of these patients died of recurrent metastatic disease, these data indicated that breast cancers overexpressing both ErbB2 and 14-3-3ζ are more aggressive and have greater metastatic potential.
Discussion
14-3-3ζ is a biomarker for patients with ErbB2-overexpressing DCIS who have a higher risk of progression to IBC
Both clinical and experimental data support that ErbB2 overexpression plays a critical role in DCIS, but is not sufficient to drive progression of the non-invasive DCIS to IBC. It has been puzzling as to what other alterations may cooperate with ErbB2 to allow a subgroup of ErbB2-overexpressing DCIS to progress to life-threatening invasive/metastatic breast tumors. Here, we identified 14-3-3ζ as a molecule that, when co-overexpressed with ErbB2, increases the potential of DCIS to progress to IBC.
Individual tumor cell invasion is a highly complicated process that requires malignant cells to obtain at least both the “capability” (migration) and the “freedom” (dissemination) to escape from the constraint of tissue structure. We found that ErbB2 overexpression alone promoted cell migration via Src activation, but not invasion, whereas 14-3-3ζ overexpression alone had no effect on cell motility but was sufficient to reduce cell-cell adhesion via inducing EMT. Therefore, the increased invasive potential in cells overexpressing both the ErbB2 and the 14-3-3ζ proteins is the collective effect of ErbB2-mediated increase in migration plus 14-3-3ζ-mediated decrease in cell-cell adhesion (Figure 7A). This finding is likely to have broader implications. Other genetic or epigenetic alterations that facilitate the loss/reduction of cell-cell adhesion, either by inducing EMT, like 14-3-3ζ, or by other mechanisms, may also promote the ErbB2-overexpressing DCIS to progress to IBC. More comprehensive investigations through non-biased analysis of both appropriate animal models and human patient samples will significantly advance our understanding of the critical step in the transition from DCIS to IBC. More importantly, for the clinical management of DCIS, evaluation of multiple proteins, including ErbB2 and 14-3-3ζ, could facilitate the identification of patients at higher risk of progressing to IBC, therefore influence the therapeutic decision.
Figure 7. Models represent how co-overexpression of ErbB2 and 14-3-3ζ promotes invasion.
(A) A model of how co-overexpression of ErbB2 and 14-3-3ζ promotes invasion. ErbB2 overexpression increased cell proliferation and motility; 14-3-3ζ overexpression decreased cell-cell adhesion via induction of EMT. Collectively, co-overexpression of ErbB2 and 14-3-3ζ promoted cell invasion. Co-overexpression of ErbB2 and 14-3-3ζ may promote progression from DCIS to IBC via this mode of cooperation. ↑: increase; ↓: decrease; −: no significant effect.
(B) A model of 14-3-3ζ–mediated E-cadherin repression. 14-3-3ζ interacts with and stabilizes TβRI, leading to smad2/3 phosphorylation and translocation to the nucleus, where smads bind to ZFHX1B promoter to increase its transcription. ZFHX1B then represses E-cadherin transcription by binding to its promoter.
14-3-3ζ contributes to the increased invasive ability of ErbB2-overexpressing MECs by inducing EMT
Accumulating evidence supports the role of EMT in promoting tumor invasion (Guarino, 2007). Pathological examination shows that malignant cells have often detached from the tumor mass at the periphery or at the invading front of the tumor. Moreover, EMT has recently been associated with “cancer stem cell” traits, suggesting a role for EMT in the initiation of recurrent tumors from disseminating cancer cells (Mani et al., 2008). However, the involvement of EMT in invasion and metastasis under a clinical setting remains controversial because of the transient and elusive nature of EMT in vivo. In this study, we detected deregulation of EMT markers more frequently in DCIS overexpressing 14-3-3ζ and TβRI, which significantly associated with higher grade DCIS that had a greater risk of developing invasive recurrence. These findings strongly support the involvement of EMT in DCIS progression toward invasive/metastatic disease. Clearly, further studies in larger cohorts are needed and may guide the design of strategies for intervention in the progression from non-invasive DCIS to life-threatening IBC.
EMT-mediated invasion has been largely attributed to the loss of E-cadherin, a tumor invasion suppressor (Beavon, 2000). Indeed, restoration of E-cadherin expression increased cell-cell adhesion and reduced invasion in 3D culture of the invasive 10A.ErbB2.ζ cells (Figure 5). A key mechanism of E-cadherin loss downstream of 14-3-3ζ overexpression is ZFHX1B upregulation (Figure 3). ZFHX1B, like other E-cadherin transcriptional repressors, has been implicated in regulation of EMT during embryogenesis (Van de Putte et al., 2003), and elevated level of ZFHX1B mRNA has been reported to associate with metastasis of ovarian (Elloul et al., 2005), gastric (Rosivatz et al., 2002), and pancreatic tumors (Imamichi et al., 2007). Our findings that ZFHX1B suppressed E-cadherin in 10A.ErbB2.ζ and 10A.14-3-3ζ cells and that high level of ZFHX1B expression correlated with E-cadherin loss in multiple breast cancer cell lines indicate a role for ZFHX1B in breast cancer cell invasion.
14-3-3ζ overexpression promotes TGFβ/Smads pathway activation
14-3-3ζ upregulated ZFHX1B by binding to TβRI and inhibiting the ubiquitin-proteasome pathway-mediated TβRI degradation, resulting in increased TβRI level, which subsequently led to TGFβ/Smads pathway activation and ZFHX1B upregulation (Figure 7B). Interestingly, overexpression of 14-3-3ζ in 293T cells has no discernable effect on ubiquitination of receptor interacting protein (RIP) (data not shown), which indicates that the effect of 14-3-3ζ on TβRI ubiquitination is selective rather than an overall deregulation of the ubiquitination machinery. Furthermore, 14-3-3 protein binding can both positively and negatively regulate the stability of distinct target proteins. For example, 14-3-3ζ has been previously found to promote MDMX's ubiquitination and degradation (LeBron et al., 2006). One possible explanation for the different effects of 14-3-3 binding is that the binding on different target proteins may either expose or mask additional signaling motif that is essential for triggering the degradation process. Further investigation is needed to elucidate the detailed mechanism.
There are seven 14-3-3 isoforms and 14-3-3ζ can form heterodimers with other 14-3-3 isoforms. Therefore, it is possible that overexpression of other isoforms may have an effect on TβRI ubiquitination. Consistently, Schistosoma mansoni 14-3-3ε was found to interact with SmRK1, a divergent type I TGFβ receptor, and positively regulated its signaling (McGonigle et al., 2001). On the other hand, despite of the highly conserved sequence and tertiary structure of 14-3-3 proteins, they appear to have distinct binding specificity and affinity to various target proteins. For example, 14-3-3σ has a unique tumor suppressor function partially by directly binding and stabilizing p53 in response to DNA damage, whereas none of other 14-3-3 isoforms share this mode of regulation (Yang et al., 2003). Therefore, further systematic studies are clearly needed to investigate the effect of other 14-3-3 isoforms on the TGFβ/Smads pathway.
The TGFβ/Smads pathway can both positively and negatively regulate tumor development (Bachman and Park, 2005). On the one hand, TGFβ/Smads pathway is a tumor suppressor prior to and during early tumor progression, mainly through inhibiting proliferation (Bachman and Park, 2005). Consistently, 10A.14-3-3ζ cells with increased TβRI expression proliferated at a slower rate than 10A.Vec cells (data not shown), and formed smaller acini than 10A.Vec cells. The inhibition of proliferation may result from upregulation of cell cycle inhibitors downstream of TGFβ/Smads activation in the non-transformed MCF10A cells. On the other hand, the overexpressed ErbB2 in 10A.ErbB2.ζ cells can activate various downstream signals to counter the growth-inhibitory effect of TGFβ/Smads activated by 14-3-3ζ. However, during the later stages of tumor progression, the TGFβ/Smads pathway can function as a tumor invasion promoter via induction of EMT (Bachman and Park, 2005). Intriguingly, 14-3-3ζ overexpression alone in MCF10A cells led to TGFβ/Smads pathway activation and EMT (as in 10A.ErbB2.ζ cells), although without increased invasion. These data indicate that 14-3-3ζ-mediated EMT is necessary, but not sufficient, to promote cell invasion, because of its lack of intrinsic migration ability, whereas migration is promoted by ErbB2 overexpression in 10A.ErbB2.ζ cells that become invasive. Our findings are consistent with a previous report that ErbB2 activation can cooperate with TGFβ treatment to promote invasion (Seton-Rogers et al., 2004). Conversely, bitransgenic mice that expressed MMTV-neu and a soluble antagonist of TGFβ had a significant reduction of metastasis (Yang et al., 2002). Our findings on the synergistic effect of ErbB2 overexpression and 14-3-3ζ-mediated activation of TGFβ/Smads pathway shed light on molecular mechanisms of gain of invasiveness during ErbB2-overexpressing DCIS progression, which is contributed by ErbB2-induced motility and proliferation plus 14-3-3ζ-mediated loss of cell-cell adhesion via inducing EMT. Recently, the TGF/Smads pathway was implicated to play a critical role in the “communication” of MECs with their “natural invasion suppressors” myoepithelial cells (Hu et al., 2008). The impact of ErbB2 and 14-3-3ζ co-overexpression on myoepithelial cells will be investigated in future studies.
Our findings that ErbB2 and 14-3-3ζ co-overexpression in DCIS predicts a higher risk of progression to IBC also provide molecular targets for designing combination therapies to intervene in DCIS progression. Targeting 14-3-3ζ may be challenging at the current stage because 14-3-3ζ regulates many important proteins that are essential for homeostasis. Identification of the TGFβ/Smads pathway as a downstream event of 14-3-3ζ overexpression in promoting invasion represents an opportunity for therapeutic intervention. Currently, the TGFβ/Smads pathway is under intensive investigation as a therapeutic target (Dumont and Arteaga, 2003; Yingling et al., 2004). Given the dichotomous role of the TGFβ/Smads pathway in tumor development, it is critical to dissect the TGFβ/Smads downstream signals and their crosstalk with other signaling networks, such as ErbB2 signaling, in order to specifically activate its tumor-suppressing role or specifically inhibit its tumor-promoting role. Our findings suggest the potential therapeutic benefit of inhibiting the TGFβ/Smads pathway in the context of ErbB2 and 14-3-3ζ co-overexpressing breast cancers.
Experimental Procedures
Tissue specimens
Twenty five DCIS and 107 invasive breast cancer specimens were obtained from the Cancer Hospital, FuDan University (Shanghai, China). Additional 138 DCIS were collected at The University of Texas M. D. Anderson Cancer Center (MDACC, Houston, TX). Patient samples were collected and processed in compliance with protocols approved by the MDACC Institutional Review Board and by the Cancer Hospital/Cancer Institute, FuDan University Institutional Review Board. Detailed clinic-pathological information of these patients cohort is provided in Table S4-6.
Cells, constructs, antibodies, and reagents
The MCF10A cell line was a kind gift from Dr. Robert Pauley (Karmonos Cancer Institute, Detroit, MI) and was cultured in 3D culture as previously described (Debnath et al., 2003). The HMEC-hTERT cell line was kindly provided by Dr. Victoria Seewaldt (Duke University, Durham, NC). ErbB2, HA-14-3-3ζ, and E-cadherin genes were cloned into pLPCX, pLNCX2, and pLHCX vectors (Clontech), respectively. Retroviral infection was done as previously described (Danes et al., 2008). Stable clones were selected with 400 μg/ml neomycin, 800 ng/ml puromycin, and 100 μg/ml hygromycin, respectively. Multiple stable clones were used to rule out potential clonal effects. HA-14-3-3ζ was also cloned into pRV3 (with GFP in the backbone) and pLOVE lentiviral vectors. Lentivirus production and infection, and reagents and antibodies used, are described in Supplemental Procedures.
Soft agar colony formation assay, cytoplasm and nuclear protein fractionation, immunoblot, immunohistochemistry, immunofluorescence staining, immunoprecipitation, RNA extraction, RT-PCR, and quantitative RT-PCR, luciferase reporter assay, migration assay, wound healing assay, cell adhesion assay, and cell aggregation assay
The detailed procedures are described in Supplemental Procedures. IFS analysis of monolayer cell cultures was done as previously described (Debnath et al., 2003). For IFS of 3D cultures, acini were embedded in sucrose and frozen in Tissue-Tek OCT (Sakura Finetek, Torrance, CA), and 5-μm frozen sections were cut and subjected to analysis (Weaver et al., 1997)
siRNA transfection and ChIP
ON-TARGET plus SMART Pool siRNA for 14-3-3ζ, ON-TARGET plus control siRNA, and ZFHX1B siRNA were purchased from Dharmacon. Transfection was done as previously described (Danes et al., 2008). ChIP assay was performed with ChIP™-IT kit from Active Motif according to the manufacturer's instructions. The DNA pulled downed by antibodies was amplified with ZFHX1B promoter-specific primers.
Cell adhesion assay and Cell aggregation assay
For the cell adhesion assay, 96-well plate was coated with fibronectin (10ng/ml) before use. 10,000 cells were re-suspended as single cell suspension in 200 μl DMEM/F12 media containing 0.5% BSA, and added to the coated plate for incubation at 37°C for one hour. Non-adherent cells were washed away with DMEM/F-12 media containing 0.1% BSA. Adherent cells were detected by MTS assay according to manufacturer's instruction. For aggregation assay, 6-well plate was coated with poly 2- hydroxylthyl methacrylate (polyHEMA) before use. 50,000 cells were resuspended to single cell suspension in 500 μl DMEM/F12 containing 0.5% BSA, added to the polyHEMA coated plates and incubated at 37°C on a rotating platform (150 rpm) for 30 mins. Cells were then fixed with methanol and photographed under phase-contrast microscope.
Statistical analysis
Statistical tests used to analyze data included Fisher's exact test, log-rank test, Chi-square test, and Student's t-test. Multivariate statistical analysis was performed using a Cox regression model. Statistical analysis was performed using SPSS for Windows (16.0 SPSS Inc.), and GraphPad Prism (Prism 5.0, GraphPad Software Inc.) packages. A P-value <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank J.M. Shu, Dr. J.P. Issa, and Dr. X. Lin (M. D. Anderson Cancer Center), and Dr. Y. Higashi (Osaka University) for their technical support and ZFHX1B antiserum. Dr. D. Yu is the Nylene Eckles Distinguished Professor in Breast Cancer Research at M. D. Anderson Cancer Center. W. Treekitkarnmongkol is partially supported by the Royal Golden Jubilee Program, Thailand Research Fund. This work is supported by NIH grants P30-CA 16672 (MDACC), RO1-CA109570, RO1-CA119127, PO1-CA099031 project 4, and P50 CA116199 project 4, DOD Center of Excellence grant subproject W81XWH-06-2-0033, DOD Synergistic Award W81XWH-08-1-0712, and Susan G. Komen Breast Cancer Foundation Promise Grant KG091020 (D. Yu). The authors have no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Significance
More than 90% of breast cancer-related deaths are caused by metastasis not primary tumor. To effectively reduce cancer mortality, it is extremely important to predict the risk of, and to intervene in, the critical transition from non-invasive DCIS to life-threatening IBC. Here, we discovered that 14-3-3ζ overexpression is a “second hit” or “risk factor” facilitating a subset of ErbB2-overexpressing DCIS transition into IBC and identified molecular mechanisms/pathways through which ErbB2 and 14-3-3ζ co-overexpression promotes invasion. This study identified biomarkers that allow selection of high-risk DCIS patients for more aggressive treatments at early stages of cancer development, while saving low-risk patients from ablative clinical procedures. Moreover, it provided promising targets for future intervention strategies to prevent DCIS progression to IBC.
References
- Andrechek ER, Laing MA, Girgis-Gabardo AA, Siegel PM, Cardiff RD, Muller WJ. Gene expression profiling of neu-induced mammary tumors from transgenic mice reveals genetic and morphological similarities to ErbB2-expressing human breast cancers. Cancer research. 2003;63:4920–4926. [PubMed] [Google Scholar]
- Bachman KE, Park BH. Duel nature of TGF-beta signaling: tumor suppressor vs. tumor promoter. Current opinion in oncology. 2005;17:49–54. doi: 10.1097/01.cco.0000143682.45316.ae. [DOI] [PubMed] [Google Scholar]
- Beavon IR. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur J Cancer. 2000;36:1607–1620. doi: 10.1016/s0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. The New England journal of medicine. 2004;350:1430–1441. doi: 10.1056/NEJMra031301. [DOI] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Molecular cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- Danes CG, Wyszomierski SL, Lu J, Neal CL, Yang W, Yu D. 14-3-3 zeta down-regulates p53 in mammary epithelial cells and confers luminal filling. Cancer research. 2008;68:1760–1767. doi: 10.1158/0008-5472.CAN-07-3177. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods (San Diego, Calif. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nature cell biology. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Dumont N, Arteaga CL. Targeting the TGF beta signaling network in human neoplasia. Cancer cell. 2003;3:531–536. doi: 10.1016/s1535-6108(03)00135-1. [DOI] [PubMed] [Google Scholar]
- Elloul S, Elstrand MB, Nesland JM, Trope CG, Kvalheim G, Goldberg I, Reich R, Davidson B. Snail, Slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103:1631–1643. doi: 10.1002/cncr.20946. [DOI] [PubMed] [Google Scholar]
- Feldner JC, Brandt BH. Cancer cell motility--on the road from c-erbB-2 receptor steered signaling to actin reorganization. Experimental cell research. 2002;272:93–108. doi: 10.1006/excr.2001.5385. [DOI] [PubMed] [Google Scholar]
- Guarino M. Epithelial-mesenchymal transition and tumour invasion. The international journal of biochemistry & cell biology. 2007;39:2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, Richardson A, Violette S, Nikolskaya T, Nikolsky Y, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi Y, Konig A, Gress T, Menke A. Collagen type I-induced Smad-interacting protein 1 expression downregulates E-cadherin in pancreatic cancer. Oncogene. 2007;26:2381–2385. doi: 10.1038/sj.onc.1210012. [DOI] [PubMed] [Google Scholar]
- LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Stetler-Stevenson WG. Tumor invasion and metastasis: an imbalance of positive and negative regulation. Cancer research. 1991;51:5054s–5059s. [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle S, Beall MJ, Feeney EL, Pearce EJ. Conserved role for 14-3-3epsilon downstream of type I TGFbeta receptors. FEBS letters. 2001;490:65–69. doi: 10.1016/s0014-5793(01)02133-0. [DOI] [PubMed] [Google Scholar]
- Melisi D, Ishiyama S, Sclabas GM, Fleming JB, Xia Q, Tortora G, Abbruzzese JL, Chiao PJ. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Molecular cancer therapeutics. 2008;7:829–840. doi: 10.1158/1535-7163.MCT-07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature cell biology. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofech-Mozes S, Spayne J, Rakovitch E, Hanna W. Prognostic and predictive molecular markers in DCIS: a review. Advances in anatomic pathology. 2005;12:256–264. doi: 10.1097/01.pap.0000184177.65919.5e. [DOI] [PubMed] [Google Scholar]
- Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. The International journal of developmental biology. 2004;48:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- Perin T, Canzonieri V, Massarut S, Bidoli E, Rossi C, Roncadin M, Carbone A. Immunohistochemical evaluation of multiple biological markers in ductal carcinoma in situ of the breast. Eur J Cancer. 1996;32A:1148–1155. doi: 10.1016/0959-8049(96)00037-8. [DOI] [PubMed] [Google Scholar]
- Provenzano E, Hopper JL, Giles GG, Marr G, Venter DJ, Armes JE. Biological markers that predict clinical recurrence in ductal carcinoma in situ of the breast. Eur J Cancer. 2003;39:622–630. doi: 10.1016/s0959-8049(02)00666-4. [DOI] [PubMed] [Google Scholar]
- Ringberg A, Anagnostaki L, Anderson H, Idvall I, Ferno M. Cell biological factors in ductal carcinoma in situ (DCIS) of the breast-relationship to ipsilateral local recurrence and histopathological characteristics. Eur J Cancer. 2001;37:1514–1522. doi: 10.1016/s0959-8049(01)00165-4. [DOI] [PubMed] [Google Scholar]
- Rizki A, Bissell MJ. Homeostasis in the breast: it takes a village. Cancer cell. 2004;6:1–2. doi: 10.1016/j.ccr.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Rosivatz E, Becker I, Specht K, Fricke E, Luber B, Busch R, Hofler H, Becker KF. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. The American journal of pathology. 2002;161:1881–1891. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, Brugge JS. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science (New York, NY. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Strathdee G. Epigenetic versus genetic alterations in the inactivation of E-cadherin. Seminars in cancer biology. 2002;12:373–379. doi: 10.1016/s1044-579x(02)00057-3. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Seminars in cancer biology. 2006;16:203–213. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. American journal of human genetics. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. The Journal of cell biology. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker E, Yaffe MB. 14-3-3 Proteins--a focus on cancer and human disease. Journal of molecular and cellular cardiology. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Yang HY, Wen YY, Chen CH, Lozano G, Lee MH. 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Molecular and cellular biology. 2003;23:7096–7107. doi: 10.1128/MCB.23.20.7096-7107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Dukhanina O, Tang B, Mamura M, Letterio JJ, MacGregor J, Patel SC, Khozin S, Liu ZY, Green J, et al. Lifetime exposure to a soluble TGF-beta antagonist protects mice against metastasis without adverse side effects. The Journal of clinical investigation. 2002;109:1607–1615. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov. 2004;3:1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- Yu D, Hung MC. Overexpression of ErbB2 in cancer and ErbB2-targeting strategies. Oncogene. 2000;19:6115–6121. doi: 10.1038/sj.onc.1203972. [DOI] [PubMed] [Google Scholar]
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.