Abstract
Notch signals are important for lymphocyte development but downstream events that follow Notch signaling are not well understood. Here, we report that signaling through Notch modulates the turnover of E2A proteins including E12 and E47, which are basic helix–loop–helix proteins crucial for B and T lymphocyte development. Notch-induced degradation requires phosphorylation of E47 by p42/p44 MAP kinases. Expression of the intracellular domain of Notch1 (N1-IC) enhances the association of E47 with the SCFSkp2 E3 ubiquitin ligase and ubiquitination of E47, followed by proteasome-mediated degradation. Furthermore, N1-IC induces E2A degradation in B and T cells in the presence of activated MAP kinases. Activation of endogenous Notch receptors by treatment of splenocytes with anti-IgM or anti-CD3 plus anti-CD28 also leads to E2A degradation, which is blocked by the inhibitors of Notch activation or proteasome function. Notch-induced E2A degradation depends on the function of its downstream effector, RBP-Jκ, probably to activate target genes involved in the ubiquitination of E2A proteins. Thus we propose that Notch regulates lymphocyte differentiation by controlling E2A protein turnover.
Keywords: basic helix–loop–helix/cell fate/development/Notch/ubiquitination
Introduction
In mammals, the Notch receptor family consists of four members, Notch1–4, which are expressed in a variety of tissues (Callahan and Raafat, 2001; Allman et al., 2002). Interaction of Notch receptors with the Jagged and delta-like ligands triggers proteolytic cleavage at their transmembrane domain and the translocation of the intracellular (IC) domain to the nucleus. Notch-IC then interacts with the CSL family of transcription factors such as RBP-Jκ/CBF1 and activates gene expression (Jarriault et al., 1995; Tamura et al., 1995; Allman et al., 2002). Notch signal transduction is also modulated by a variety of regulators that act at different steps along the pathway (Allman et al., 2002), in particular Deltex functions in a CSL-independent manner. Deltex is thought to positively regulate Notch signaling in Drosophila (Matsuno et al., 1995), but Deltex overexpression appears to antagonize Notch function in mouse lymphopoiesis (Izon et al., 2002).
With regard to lymphopoiesis, Notch1 is crucial for the B versus T cell fate decision, as well as subsequent decisions concerning αβ versus γδ T cells and CD4 versus CD8 T cells (MacDonald et al., 2001; Allman et al., 2002). Inducible disruption of the Notch-1 gene completely blocked the differentiation of T cells without any effect on the development of B cells and myeloid cells (Radtke et al., 1999). Thymuses of these Notch1 knockout mice were primarily populated with B cells. Wilson et al. (2001) further concluded that multipotent lymphoid progenitor cells derived from the bone marrow can adopt a B-cell fate in the thymus without Notch1. Complementarily, overexpression of a constitutively active form of Notch1-IC (N1-IC) blocked B-cell development and promoted ectopic differentiation of T cells in the bone marrow (Pui et al., 1999). Thus these findings suggest that Notch1 inhibits B-cell differentiation but promotes T-cell ontogeny. However, the downstream events involved in Notch-mediated T-cell lineage determination are not understood.
Ordentlich and colleagues have provided evidence to suggest that the basic helix–loop–helix (bHLH) transcription factor E47 is a downstream target of Notch signaling and that activation of Notch1 or 2 inhibits its transcriptional activity (Ordentlich et al., 1998). E47 and E12 are alternative splicing products of the E2A gene, and they differ only in the bHLH DNA binding domain (Murre et al., 1989; Sun and Baltimore, 1991). E2A, HEB and E2-2 genes encode a family of bHLH proteins with similar functions, collectively called E proteins (Massari and Murre, 2000). They are important for both B- and T-cell development. Lack of the E2A gene alone is sufficient to block B-cell development at the earliest progenitor stage, while ablation of both E2A and HEB genes is necessary for the complete arrest of T-cell development (Bain et al., 1994; Zhuang et al., 1994; Barndt et al., 1999). The E proteins can be inhibited by the Id proteins, and Id overexpression in B or T cells severely impairs B- or T-cell differentiation (Sun, 1994; Heemskerk et al., 1997; Kim et al., 1999). Therefore it is plausible that the suppression of B-cell development caused by N1-IC overexpression might be mediated through inhibition of E proteins. However, it is puzzling why overexpression of N1-IC did not impair T-cell development since E proteins are also essential for T cells. Perhaps different cellular environments in B and T cells influence the effect of Notch signals.
To understand how N1-IC inhibits the transcriptional activity of E47, we found that it largely accelerates degradation of E47 proteins through a ubiquitin-mediated and proteasome-dependent mechanism. We then showed that the ubiquitination and degradation of E2A proteins induced by Notch signals are tightly regulated by the level of p42/p44 MAP kinases, which phosphorylate E2A proteins. Therefore signals from the Notch and MAP kinase pathways act in concert to regulate the level of E2A proteins, which may directly impact on the differentiation of B and T lymphocytes.
Results
Activated Notch1 inhibits the transcriptional activity of E47 by reducing the level of E47 protein
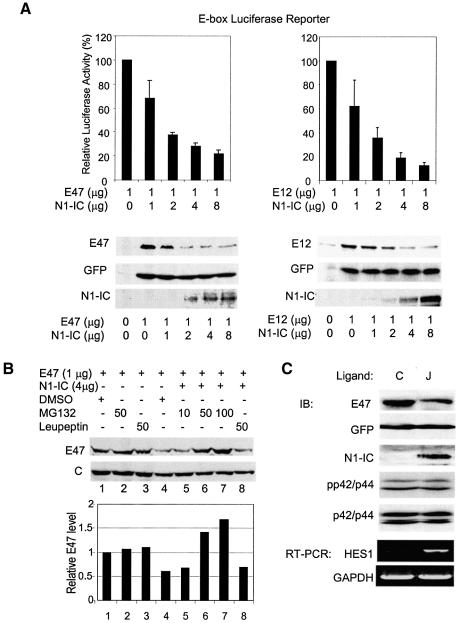
Consistent with previous work (Ordentlich et al., 1998), we observed a dose-dependent inhibitory effect of N1-IC on both E47- and E12-activated gene expression (Figure 1A). In transient transfection assays, E47 or E12 was coexpressed in NIH3T3 cells together with or without N1-IC, along with plasmids carrying E-box luciferase and CMV-LacZ reporter genes. Without N1-IC, expression of E47 or E12 typically leads to about 100- to 150-fold activation of reporter activity in NIH3T3 cells. With increasing amounts of N1-IC, the transcriptional activities of E47 and E12 were reduced by up to 80% and 90%, respectively. As a control, we measured the amounts of E47 and E12 present in the transfected cells. Surprisingly, we found that amounts of these proteins diminished in parallel with their transcriptional activities. This suggests that the apparent inhibition of transcriptional activities of E47 and E12 might be due to lower amounts of proteins present in the transfected cells, probably resulting from an increased rate of protein degradation. Indeed, we found that E47 degradation was completely blocked in a dose-dependent manner by treatment with a proteasome inhibitor, MG132, but not by a protease inhibitor, leupeptin (Figure 1B). Thus these results suggest that activation of Notch-signaling pathways may trigger E47 degradation through a proteasome-dependent pathway.
Fig. 1. N1-IC inhibits E47 and E12 transcriptional activity by accelerating their degradation. (A) Dose responses. NIH3T3 cells were transiently transfected with 5 µg, 1 µg each of E-box luciferase and β-galactosidase reporter plasmids along with 1 µg of E47 or E12 plus increasing amounts of N1-IC expressing constructs. Luciferase activities were assayed 36 h later and normalized to those of β-galactosidase. Transcriptional activity of E47 in each sample is presented relative to that in the absence of N1-IC with standard deviations from at least three independent experiments. For protein analysis, indicated amounts of E47 or E12 and N1-IC were cotransfected with a GFP expression vector. Whole lysates were probed with anti-E47, GFP and N1-IC antibodies. (B) A proteasome inhibitor blocks the degradation of E47 protein induced by N1-IC. Immunoblot assays were carried out with NIH3T3 cells transfected with 1 µg of E47 expressing plasmid with or without 4 µg of the N1-IC construct and treated with the indicated amounts of MG-132 proteasome inhibitor, leupeptin or dimethylsulfoxide vehicle for 3 h prior to harvest. C indicates loading control. (C) Activation of endogenous Notch receptors enhances degradation of E47. The NIH3T3 fibroblasts expressing Notch ligand, Jagged-1 (J) or not (C), were infected with a retroviral construct producing both E47 and GFP. Cells were harvested 3 days after infection. Whole-cell lysates were analyzed using immunoblotting (IB) with antibodies against E47 and GFP. Nuclear extracts were used for detecting N1-IC. Another aliquot of the infected cells was used to isolate RNA for RT–PCR assays of HES1 and GAPDH mRNA.
To determine whether activation of endogenous Notch receptors could also cause E47 degradation, we activated endogenous Notch receptors with their ligand, Jagged-1, generated by infecting NIH3T3 cells with a retrovirus expressing Jagged-1. Since these NIH3T3 cells expressed endogenous Notch1, cells carrying the Jagged-1-expressing construct but not the empty vector would contain activated Notch1. This was shown not only by the presence of intracellular domain of Notch1 in the nucleus but also by the upregulation of a Notch target gene, HES-1 (Figure 1C). We then infected the Jagged-1-expressing cells or control cells with the retrovirus producing both E47 and GFP and cultured the cells to confluence. In Jagged-1-expressing cells, the level of E47 was reduced by 55% compared with the control cells, whereas the GFP levels were comparable in both cell lines (Figure 1C).
Mapping the sequence in E47 responsible for Notch1-mediated degradation
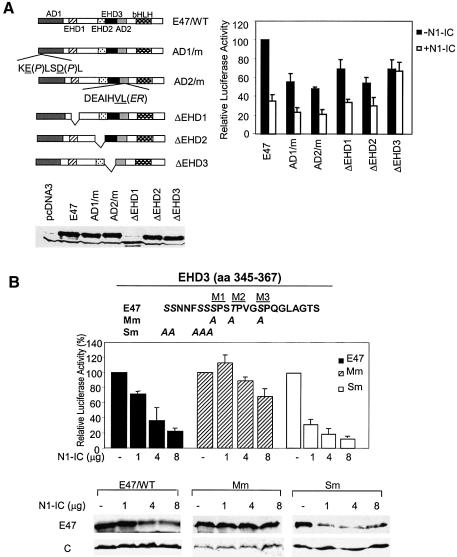
E12 and E47 contain two transcription activation domains, AD1 and AD2, whose sequences are conserved among all E proteins. AD1 located at the N-terminus was shown to have a helical structure (Massari et al., 1996). AD2 was initially thought to contain a loop–helix motif but was further narrowed down to the helix (Aronheim et al., 1993; Quong et al., 1993). Mutations in the helix of AD1 or AD2 completely abolished the transcriptional activity when assayed separately as GAL4 fusion proteins (Quong et al., 1993; Massari et al., 1996). The E proteins also contain two additional highly conserved regions (designated as E protein homology domains, EHD1 and EHD2) (Figure 2A). To delineate the sequences responsible for Notch1-induced degradation, we created a series of constructs expressing E47 deletion or point mutants in these conserved regions. These proteins were expressed at similar levels in NIH3T3 cells (Figure 2A). In the absence of N1-IC, point mutations in either AD1 or AD2 resulted in about 50% reduction of transcriptional activity, suggesting that both activation domains contribute to the activity of E47 (Figure 2A). Although EHD1 and EHD2 were not identified as activation domains from the GAL4 fusion studies, their deletion led to 30–40% reduction of activity, which would imply that these sequences are involved in maintaining the full function of E47. Alternatively, deletion of these regions might change the conformation of E47 and decrease its transcriptional activity. Deletion of EHD3, originally called the loop of AD2, also decreased E47 activity slightly. In the presence of N1-IC, the transcriptional activity of wild-type E47 was inhibited by 60%. Similar inhibition was seen in all the deletion or point mutants except ΔEHD3. ΔEHD3 exhibited similar transcriptional activity with or without N1-IC, thus suggesting that EHD3 is responsible for Notch-mediated inhibition of E47 transcription activity (Figure 2A).
Fig. 2. A functional domain mediating Notch1-induced inhibition. (A) Analysis of deletion mutants. In a schematic representation of E47 and mutants, activation and EHD domains are marked with patterned boxes and labeled. Sequences of the mutations in AD1 and AD2 are shown below the boxes with mutated residues underlined and followed by the substituting residues in parentheses. Expression of wild-type and mutant E47 in NIH3T3 cells was confirmed using immunoblots with antibodies against E47. One microgram of E47 or mutant-expressing plasmid was cotransfected with or without 2 µg of N1-IC-expressing constructs along with reporters into NIH3T3 cells and assayed as described for (Figure 1A). (B) Identification of MAP kinase sites in EHD3. Sequence of EHD3 is shown with MAP kinase sites labeled as M1–3. In the Mm and Sm constructs, serine or threonine shown in italic are substituted with alanine. NIH3T3 cells were cotransfected with 1 µg of E47 or mutant construct with reporter constructs and the indicated amounts of N1-IC construct. The reporter activities and protein levels were analyzed as described in Figure 1A.
Examination of the amino acid sequence of EHD3 revealed three potential MAP kinase phosphorylation sites, M1–3. We then created the Mm mutant, in which the serine or threonine residues in M1–3 were replaced with alanine (Figure 2B). As a control, we generated the Sm mutant by changing two clusters of serine into alanine. Wild-type or mutant E47 constructs were cotransfected into NIH3T3 cells together with E-box luciferase and CMV-LacZ reporters plus increasing amounts of N1-IC. Although the Mm and Sm mutations did not cause significant loss in their intrinsic transcription activity, both the activity and protein level of E47 and Sm were dramatically reduced by N1-IC whereas those of Mm remained unchanged (Figure 2B). These results suggest that the MAP kinase sites in EHD3 are crucial for Notch-induced E47 degradation. Since the M1 site is also mutated in the Sm mutant, the data indicate that M1 is not essential for Notch1-induced degradation.
MAP kinase-mediated phosphorylation is necessary but not sufficient for Notch-induced degradation
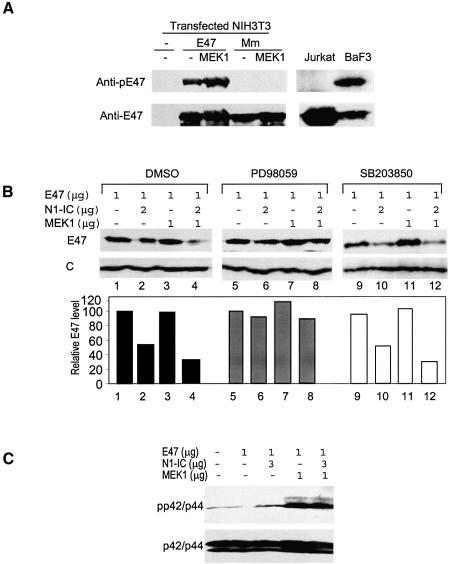
To determine if E47 is phosphorylated at EHD3, antibodies were generated using a phosphopeptide with the sequence covering the M2 site. Wild-type E47 but not the Mm mutant reacted to the antibodies, suggesting that E47 is at least phosphorylated at the M2 site (Figure 3A). Cotransfection with a constitutively active form of MEK1, which activates p42/p44 MAP kinases, increased the amount of phospho-E47. Furthermore, endogenous E2A proteins are phosphorylated in BaF3 B cells but not in Jurkat T cells (Figure 3A), which correlates with their levels of active p42/p44 (see Figure 5B).
Fig. 3. MAP kinase mediated phosphorylation of E47 is necessary but not sufficient for Notch1-induced degradation. (A) Phosphorylation of E47. Whole-cell lysates of NIH3T3 cells transfected with E47 or Mm with or without MEK1 were probed with anti-phospho-M2 peptide (SSPSpTPVGSPQG) and then anti-E47 antibodies. Whole-cell lysates from Jurkat T and BaF3 B cells were analyzed similarly. (B) MAP kinases are necessary but not sufficient for Notch1-mediated degradation of E47 proteins. PD98059 or SB203850 were added at a final concentration of 50 µM for 2 h after transfection for 34 h. Protein levels were determined as described above. E47 levels normalized against loading controls are shown in the bar graph as relative levels to that in cells expressing E47 alone under each treatment. (C) MAP kinase activation by MEK1 and N1-IC. Transfected NIH3T3 cells were harvested 36 h post transfection and whole-cell lysates were used in immunoblotting with antibodies against phospho-p42/44 and p42/p44.
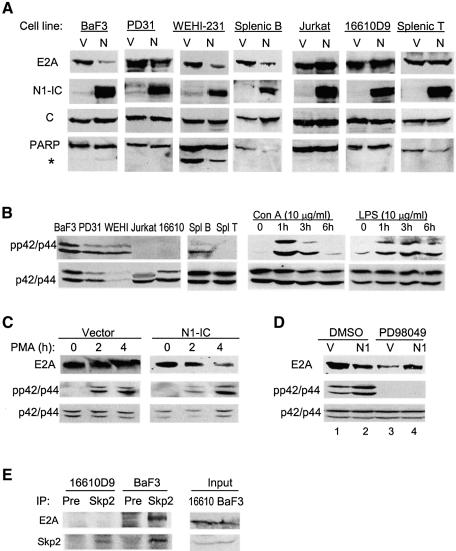
Fig. 5. MAP kinase activities are required for Notch1-induced E2A degradation. (A) The indicated cell lines or cultures were infected with N1-IC-expressing retrovirus (N) or vector control (V). Infected cells were sorted for GFP fluorescence. Endogenous E2A protein levels were detected using anti-E47 antibodies. The same membranes were reprobed with antibodies as indicated. The asterisk indicates the cleavage product of PARP. (B) Left: Levels of activated MAP kinases in cells used in (A). Right: kinetics of MAP kinase activation in total splenocytes treated with Con A and LPS. Whole-cell lysates were analyzed. Whole-cell lysates from vector or N1-IC-infected 16610D (C) or BaF3 (D) cells were analyzed with the indicated antibodies with or without treatment with 10 ng/ml PMA for 2 and 4 h (C) or 50 µM PD98049 for 3 h (D). (E) Interaction between endogenous E2A and Skp2. Co-IP was performed with antibodies against Skp2 or a preimmune serum and whole-cell lysates from the indicated cells. The precipitates were probed for E2A and Skp2.
If phosphorylation of E47 by MAP kinases is a prerequisite for its degradation, further activation of MAP kinases should facilitate E47 degradation while inhibition should prevent it. As shown in Figure 3B, MEK1 facilitated Notch-induced E47 degradation (compare lanes 2 and 4) but it alone failed to cause E47 degradation (lane 3). Conversely, treatment of the transfected cells with a MEK1/2 inhibitor, PD98059, stabilized E47 protein in the presence of N1-IC or N1-IC plus MEK1 (lanes 5–8). In contrast, the inhibitor of the p38 MAP kinase, SB203850, did not block E47 degradation (lanes 9–12). It thus appears that Notch1-induced E47 degradation depends on its phosphorylation mediated by p42/p44 MAP kinases. However, activation of MAP kinases alone is not sufficient to trigger E47 degradation since coexpression of E47 and MEK1 did not accelerate E47 turnover. Although overexpression of N1-IC resulted in a slight activation of p42/p44 MAP kinases in NIH3T3 cells, that effect was much less than that by the active form of MEK1 (Figure 3C). Furthermore, activation of endogenous Notch receptors does not cause activation of p42/p44 MAP kinases even though it leads to E47 degradation (Figure 1C). These results would argue that N1-IC induces E47 degradation by altering other cellular functions besides activation of MAP kinases.
N1-IC stimulates E47 ubiquitination
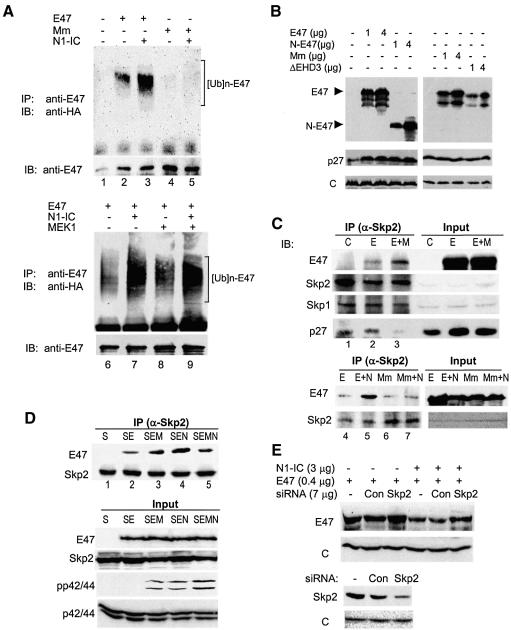
Dependence on substrate phosphorylation is characteristic of ubiquitin-mediated protein degradation (Karin and Ben Neriah, 2000; Harper, 2001). To test if E47 is ubiquitinated and if Notch1 enhances the ubiquitination of E47, we performed in vivo ubiquitination assays (Figure 4A). Wild-type E47 or the Mm mutant was coexpressed with or without N1-IC in 293T cells along with a HA-tagged ubiquitin construct (Treier et al., 1994). The immunoprecipitates of anti-E47 antibodies were blotted with an anti-HA antibody. Although wild-type E47 was basally ubiquitinated, N1-IC dramatically increased the level of ubiquitination (lanes 2 and 3). In contrast, Mm was not ubiquitinated with or without N1-IC (lanes 4 and 5). Again, MEK1 did not have a significant effect on E47 ubiquitination (compare lanes 6 and 8), suggesting that phosphorylation of E47 alone is not sufficient to enhance the ubiquitination.
Fig. 4. N1-IC enhances ubiquitination of E47 protein in vivo by facilitating its association with the SCFSkp2 complex. (A) In vivo ubiquitination assay. Seven micrograms of HA-ubiquitin-expressing construct were transfected into 293T cells with 1 µg of E47 or Mm ± 3 µg of N1-IC or 1 µg of MEK1 together or individually as indicated. Transfected cells were treated 24 h later with MG132 at a final concentration of 4 µM overnight. Immunoprecipitation (IP) was performed with 800 µg of whole-cell lysates and anti-E47 antibodies. The precipitates were blotted with an anti-HA monoclonal antibody and then with anti-E47 antibodies. (B) Overexpression of E47 rescues p27 protein. Immunoblot assays were performed with 293T cells transfected with indicated constructs using antibodies against E47 and loading control. (C) Interaction between E47 and endogenous Skp2. Three micrograms of E47 (E) or Mm mutant (Mm) construct were cotransfected with or without 3 µg of MEK1 (M) or 9 µg of N1-IC (N) construct into 293T cells for 28 h as labeled at the top of each lane. Anti-Skp2 immunoprecipitates from whole-cell lysates were probed with antibodies against E47 and subsequently with antibodies against indicated proteins. (D) Interaction between E47 and exogenous Skp2. Co-IP assay was performed with 293T cells transfected with constructs indicated by their single-letter codes as described above except that 3 µg of the Skp2 (S) construct was included in all samples. An aliquot of total lysates was probed for pp42/44 and p42/44 in addition to E47 and Skp2. (E) Inhibition of E47 degradation by siRNA against Skp2. NIH3T3 cells were cotransfected with indicated constructs. Two days after transfection, whole-cell extracts were used for immunoblot assays of indicated proteins.
With regard to the machinery for E47 ubiquitination, we obtained a hint from our previous fortuitous finding that E47 overexpression caused accumulation of the cyclin-dependent kinase inhibitor p27. As shown in Figure 4B, the p27 level was significantly increased by overexpression of wild-type E47 or the N-terminal half of E47 (N-E47) including EHD3, but lacks the bHLH domain. Since N-E47 cannot activate transcription, E47 likely rescues the p27 protein by competing for the machinery involved in p27 degradation. The p27 protein is known to be ubiquitinated by the SCFSkp2 E3 ubiquitin ligase complex (Harper, 2001; Jackson and Eldridge, 2002). The SCF complex consists of Skp1, Cul1, Rbx1 and a member of the F-box family of proteins, Skp2. F-box proteins usually interact with substrates in a phosphorylation-dependent manner.
To determine whether E47 binds to endogenous Skp2, 293T cells were transfected with constructs expressing E47 alone or together with MEK1 (Figure 4C). Anti-Skp2 immunoprecipitates were probed with antibodies against E47, Skp2, Skp1 and p27. Indeed, antibodies against Skp2 brought down not only Skp2 and Skp1 but also E47 (lane 2). Activation of MAP kinases by MEK1 increased the amounts of E47 coprecipitated with Skp2 (lane 3), perhaps by facilitating the phosphorylation of overexpressed E47. Interestingly, the amount of p27 coprecipitated with Skp2 decreased as that of E47 increased (Figure 4C). Also, N1-IC dramatically enhanced the interaction between Skp2 and wild-type E47 but not Mm (Figure 4C, lanes 4–7). To examine the interaction between E47 and Skp2 further, a construct expressing Skp2 was cotransfected with E47 ± MEK1 and/or N1-IC (Figure 4D). Interaction between Skp2 and E47 was readily detectable and enhanced by coexpression with MEK1 or N1-IC. Intriguingly, expression of MEK1 plus N1-IC, rather than further stimulating the interaction, consistently decreased it compared with either alone. One explanation may be that MEK1 and N1-IC act through a similar mechanism to facilitate the interaction, e.g. activation of MAP kinases. Indeed, we found that in 293T cells N1-IC increased the level of phospho-p42/p44 to a similar extent as done by MEK1 (Figure 4D). Expression of both MEK1 and N1-IC led to a superactivation of MAP kinases, which may have an adverse effect on E47-Skp2 interaction through an unknown mechanism. Together, these results suggest that N1-IC or MEK1 promotes the interaction between E47 and Skp2 by activation of MAP kinases in 293T cells. However, because N1-IC dramatically stimulates E47 ubiquitination whereas MEK1 does not, N1-IC may act at additional points to facilitate E47 ubiquitination beyond the interaction between E47 and Skp2.
To demonstrate further the role of Skp2 in E47 degradation, we have expressed siRNA against Skp2 along with E47 and N1-IC in NIH3T3 cells (Figure 4E). Expression of the siRNA against Skp2, but not a control siRNA, decreased the level of Skp2 and prevented N1-IC-induced E47 degradation. Interestingly, siRNA against Skp2 also increased the E47 level slightly in the absence of N1-IC, which is consistent with the idea that Skp2 mediates the constitutive ubiquitination of E47 even without Notch signaling.
Notch1-induced E2A degradation in B and T cells depends on MAP kinase activities
To determine whether activated Notch causes the degradation of endogenous E2A proteins, we examined the effect of N1-IC on E2A proteins in B and T cells. B- and T-cell lines were infected with retroviral constructs expressing N1-IC and GFP or GFP alone. Levels of endogenous E2A proteins in infected cells were then determined using immunoblot assays with antibodies against E47 (Figure 5A). The same membranes were probed with antibodies against TFIIH for loading controls or N1-IC to confirm its expression. E2A protein levels were reduced by 65–80% when N1-IC was expressed in BaF3, PD31 and WEHI-231 cells, which represent B cells at different stages. Furthermore, primary splenocytes stimulated with lipopolysaccharide (LPS) were infected with the same retroviruses. Infected B cells were isolated and the level of E2A proteins was also reduced by 80% in N1-IC-expressing cells compared with vector-infected cells (Figure 5A). The decrease in E2A levels in these B cells is not due to apoptosis induced by N1-IC, because no significant cleavage of PARP, an indication of caspase activation during apoptosis (Tewari et al., 1995), was observed in N1-IC expressing cells (Figure 5A).
Surprisingly, expression of N1-IC not only did not decrease but also slightly increased E2A levels in Jurkat and 16610D9 T cells (Figure 5A). The E2A level in primary splenic T cells infected with the retrovirus after Con A stimulation was also unchanged (Figure 5A). These results might have suggested that Notch signals could not cause E2A degradation in T cells. However, since E2A degradation depends on phosphorylation by MAP kinases, we examined the levels of activated p42/p44 in the B and T cells cultured under the same conditions as those for the measurement of E2A protein level. We found that all of the T cell samples had very low levels of activated MAP kinases while B cell samples had plenty of such enzymes (Figure 5B). Consistently, phosphorylated E2A proteins were detected in BaF3 cells but not in Jurkat cells (Figure 3A). Although Con A treatment of splenocytes rapidly activated MAP kinases in T cells, they began to be inactivated by 3 h after stimulation (Figure 5B). In contrast, LPS stimulation of B cells led to a sustained activation of MAP kinases. Therefore, it is possible that the inability of N1-IC to induce E2A degradation in T cells was due to a lack of MAP kinase activities.
To test if E2A can be degraded in T cells once MAP kinases are activated, we treated 16610D9 T cells infected with N1-IC or control retroviruses with PMA for 2 and 4 h. We found that MAP kinases were activated and E2A proteins were degraded in N1-IC expressing cells but not in control cells (Figure 5C). Conversely, when BaF3 B cells were treated with the MEK inhibitor, PD98059, N1-IC induced degradation of E2A protein was diminished (compared lane 3 to 4 in Figure 5D), even though the inhibitor appeared to reduce the overall level of E2A for unknown reasons (lane 1 versus 3 in Figure 5D). Furthermore, we examined the interaction between endogenous E2A and Skp2 proteins in these T and B cells (Figure 5E). E2A is specifically bound to Skp2 in BaF3 but not 16610D9 cells, which correlates with their levels of MAP kinase activities and susceptibilities to degradation.
E2A degradation in T cells by endogenous Notch signals
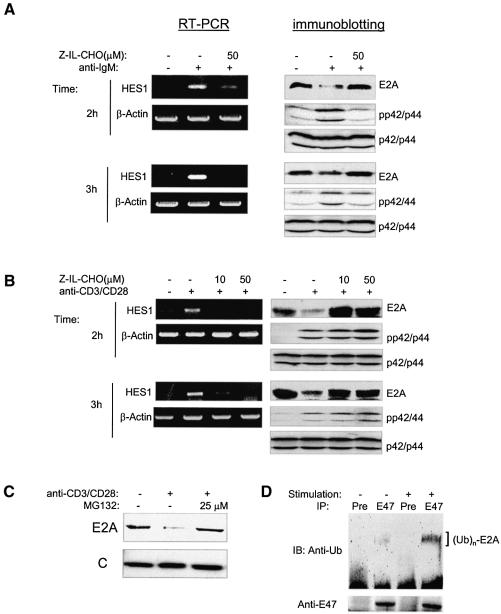
To examine the effect of endogenous Notch signals on E2A degradation, we used splenic B- and T-cell culture systems recently developed by H.M.Jack and B.Osborne, respectively (personal communications). Treatment of splenocytes co-cultivated with splenic stromal cells in the presence of anti-IgM or anti-CD3 plus anti-CD28 antibodies for 2 and 3 h activated the Notch-signaling pathway as indicated by HES1 gene upregulation (Figure 6A and B). Furthermore, this activation is diminished by treatment with an inhibitor of γ-secretases, Z-IL-CHO, which blocks the release of intracellular domains of Notch receptors to the nucleus upon ligand binding. Because MAP kinases are also activated by anti-IgM or anti-CD3/CD28 stimulation, we tested whether E2A proteins become degraded under this condition. Indeed, we found that E2A protein levels decreased significantly (Figure 6A and B). Like the effect on HES1 expression, treatment with Z-IL-CHO completely recovered the level of E2A proteins (Figure 6A and B), which would suggest that E2A degradation induced by anti-IgM or anti-CD3/CD28 is indeed mediated through Notch-signaling pathways. We also found that E2A proteins were rescued by the proteasome inhibitor MG132 in splenocytes cocultured with anti-CD3/CD28, suggesting that E2A proteins are degraded through a proteasome-dependent pathway (Figure 6C). Furthermore, we have detected an increased amount of ubiquitinated E2A proteins in splenocytes stimulated with antibodies against IgM, or CD3 plus CD28, which activate MAP kinases and Notch-signaling pathways in B or T cells (Figure 6D).
Fig. 6. E2A degradation by endogenous Notch signals. Activation of Notch-signaling pathways and E2A degradation in splenic cultures treated with antibodies against (A) IgM or (B) CD3 and CD28. Total RNA was isolated from splenocytes cultured under the indicated conditions for 2 or 3 h. Expression of HES1 and β-actin was measured using RT–PCR. Whole-cell lysates from splenocytes cultured under the same conditions were used for immunoblot assays with antibodies against the indicated proteins. Z-IL-CHO or an equal volume of the vehicle was added to the indicated cultures. (C) Proteasome inhibitor blocks E2A degradation. MG132 was included in the 2 h culture as described in (B). (D) Ubiquitination of E2A proteins in a 3 h culture of splenocytes treated with 25 µM MG132 plus a mixture of antibodies against CD3, CD28 and IgM as described in Materials and methods; 108 cells were used for each IP with a preimmune serum (pre) or anti-E47 (E47) antibodies. Immunoblots of the precipitates were performed with the indicated antibodies.
Notch signals leading to E2A degradation
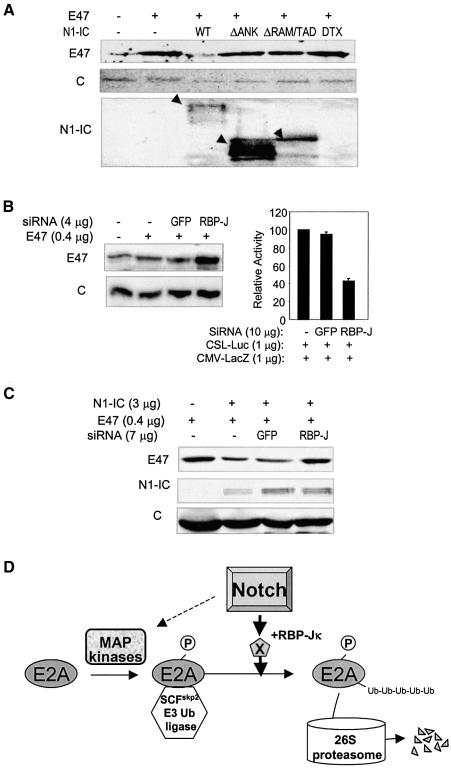
Notch signal transduction involves CSL-dependent and CSL-independent pathways. For the CSL-dependent pathway, Notch-IC binds to RBP-Jκ/CBF1 via its RAM domain while its ankyrin repeats and transcription activation domain recruit coactivators (Jarriault et al., 1995; Tamura et al., 1995; Callahan et al., 2001; Allman et al., 2002). The end result of this signaling pathway is the transcriptional activation of target genes. On the other hand, a CSL-independent pathway involves the interaction between Notch-IC and Deltex through the ankyrin repeats (Diederich et al., 1994). To determine the pathway through which Notch induces E47 degradation, we tested the effects of N1-IC deletion mutants and Deltex on E47 turnover. While wild-type N1-IC induced a dramatic reduction in the level of E47 protein, the mutants lacking the RAM and transcription activation domains or the ankyrin repeats failed to cause E47 degradation although they were expressed at higher levels (Figure 7A). Likewise, overexpression of wild-type Deltex had no effect on E47 stability even though it was shown to inhibit the transcriptional activity of E47, perhaps through a different mechanism (Ordentlich et al., 1998).
Fig. 7. Notch signals leading to E2A degradation. (A) Analyses of N1-IC deletion mutants and Deltex. NIH 3T3 cells were transfected with 1 µg of E47 and 4 µg of N1-IC or Deltex constructs. Protein levels were analyzed 2 days later as described above. Wild-type and mutant N1-IC proteins are indicated with arrowheads. (B) Jagged-1-expressing NIH3T3 cells were transfected with indicated constructs. Protein (left) and luciferase and β-galactosidase activities (right) were measured as described. (C) NIH3T3 cells were transfected with the indicated constructs and protein levels were analyzed using immunoblots. (D) A model for Notch-induced E2A degradation. E2A proteins are phosphorylated by MAP kinases, which allows association with the SCFskp2 E3 ubiquitin ligase. The ubiquitinated E2A proteins are degraded by the 26S proteasome. Notch signals enhance the ubiquitination of E2A proteins, probably by activating expression of their downstream target genes involved in the ubiquitination reaction. Notch signals also activate MAP kinases in some cell types.
To confirm the involvement of the CSL-dependent pathway, we used siRNA to knock down the level of endogenous RBP-Jκ/CBF1 in NIH3T3 cells. Expression of siRNA against RBP-Jκ, but not that for GFP, rescued the E47 protein coexpressed in Jagged-1-expressing NIH3T3 cells (Figure 7B). As a control, the ability of the siRNA to knock down RBP-Jκ was determined by cotransfecting siRNA constructs with a CSL-luciferase report gene, which monitors RBP-Jκ–dependent transcription. Indeed, the siRNA against RBP-Jκ inhibited reporter expression by 60%. Similarly, NIH3T3 cells were cotransfected with E47, N1-IC and siRNA constructs, and siRNA against RBP-Jκ also diminished N1-IC-induced E47 degradation (Figure 7C). Taken together, these results suggest that Notch-induced degradation of E2A protein is mediated through a CSL-dependent pathway, probably by activating transcription of unknown downstream genes.
Discussion
Mechanisms of Notch-induced E2A degradation
Notch-induced degradation of E2A (E12/E47) proteins follows a typical pattern for ubiquitin-mediated proteolysis (Pickart, 2001). We have shown that E47 degradation depends on its phosphorylation by MAP kinases (Figure 7). Since MAP kinase activation is controlled by a variety of cellular signals, this dependence on phosphorylation could be utilized to regulate the degradation of E2A proteins. Without p42/p44 MAP kinase activities, E2A would not be degraded even in the presence of Notch signals. In our experimental systems, such as NIH3T3 cells, MAP kinase activities already exist due to stimulation by serum, which in turn allow constitutive ubiquitination and degradation of E47. Overexpression of E47 probably overwhelms basal levels of MAP kinase activities such that expression of an active MEK1 further increases MAP kinase activities and phospho-E47 levels. These phosphorylated E47 molecules can then associate with the SCFSkp2 E3 ubiquitin ligase complex.
The active form of Notch1 facilitates the association between E47 and SCFSkp2, in part by activating MAP kinases in 293T cells (Figure 4D). However, it is important to note that N1-IC dramatically enhances the ubiquitination of E47, whereas MEK1 fails to do so (Figure 4A). This would suggest that Notch signals facilitate E47 ubiquitination through mechanisms in addition to the interaction between E47 and Skp2. How Notch signals stimulate E47 ubiquitination is not understood. Because we have found that neither Deltex nor an N1-IC deletion mutant lacking the RAM and transcriptional activation domains could induce E47 turnover and knocking down RBP-Jκ rescues E47 proteins, it is possible that N1-IC activates expression of genes involved in E2A ubiquitination (Figure 7D). However, current knowledge about downstream target genes activated by Notch signals could not explain how Notch1 increases E47 ubiquitination. A complete understanding of the molecular mechanisms underlying Notch-induced E47 degradation will follow the development of a more comprehensive database for Notch target genes and a better understanding of the E3 ligase complexes responsible for E47 ubiquitination.
Notch-induced E2A degradation and the B versus T lineage choice
One of the major decisions during lymphopoiesis is whether multipotent progenitor cells become B or T lymphocytes. Several lines of evidence have implicated Notch as an important decision-maker (MacDonald et al., 2001; von Boehmer, 2001; Allman et al., 2002) It is believed that strong Notch signals are essential for T-cell development and are delivered in the thymus. Inducible disruption of the Notch1 or RBP-Jκ gene completely blocks T-cell development, suggesting that Notch signals may promote T-cell differentiation by turning on their downstream target genes (Radtke et al., 1999; Tomita et al., 1999; Han et al., 2002). More interestingly, lack of Notch1 or RBP-Jκ results in an accumulation of B cells in the thymus (Radtke et al., 1999; Wilson et al., 2001). This would imply that a parallel function of Notch1 is to prevent multipotent progenitors from differentiating along the B lineage. Consistent with this finding, aberrant activation of the Notch-signaling pathway inhibits B-cell development in the bone marrow (Pui et al., 1999). How Notch signals exert these differential effects on B and T cells remains largely unknown.
Since E2A proteins are crucial for early stages of B- and T-cell development, modulation of E2A function could be an effective means for controlling lymphocyte differentiation. In this report, we have shown that activation of the Notch-signaling pathway induces the degradation of E2A proteins in a MAP-kinase-dependent manner. Intriguingly, high levels of p42/p44 MAP kinase activities were found in all the B-cell lines and cultures we tested, and thus E2A proteins become readily degraded upon signaling through Notch pathways. In contrast, MAP kinase activities were very low in the T cells we tested and only increased transiently by various treatments. Consequently, MAP kinase activities become limiting factors for E2A degradation in T cells in the presence of Notch signals. These observations raise the question of whether B and T cells use different mechanisms to activate or inactivate their MAP kinases such that the activities are more sustained in B cells and transient in T cells. Under this assumption, one might envision that in the thymus, where abundant Notch signals exist, the progenitor cells choosing the B-cell fate would lose E2A proteins due to the presence of MAP kinase activities and thus abort the differentiation process. In contrast, differentiation into T cells proceeds because Notch signals are unable to induce E2A degradation without MAP kinase activities. This hypothesis would suggest that the fate decision made by progenitor cells is independent of Notch signals, but Notch selects for those adopting a T-cell fate and encourages their further development along the T lineage. This is consistent with findings in Drosophila, where Notch signaling adjusts cellular responses to differentiation stimuli but does not dictate a particular cell fate (Artavanis-Tsakonas et al., 1999).
Whether and how B and T cells differentially control their MAP kinases remains to be understood. However, it is possible that these two types of cells have different modes of regulation for MAP kinases. MAP kinases in B cells are shown to be activated by stimulation with IL-7 (Fleming and Paige, 2001), which is available in the thymus. T-cell progenitors do not have activated MAP kinases until they have formed functional pre-TCR (L.Nie and X.H.Sun, unpublished data). The kinetics of activation in T cells may also be different from that in B cells as we have seen when splenocytes were treated with Con A or LPS (Figure 5B). Furthermore, a new population of progenitor cells in the thymus, termed early T lineage progenitor (ETP), has recently been described (Allman et al., 2003). These cells give rise to T cells independently of the conventional ‘common lymphoid progenitors’ (CLPs) and have lower potential to generate B cells. ETPs are distinct from CLPs, particularly in their lack of response to IL-7 stimulation. The different responses to IL-7 may result in different levels of MAP kinase activities in ETPs and CLPs. Thus we postulate that T cells in the thymus are principally derived from ETPs, which can produce high levels of E2A proteins upon differentiation since they have low levels of MAP kinases. Differentiation from CLP or its equivalent is normally suppressed due to the degradation of E2A proteins in the presence of MAP kinases and Notch signals. In Notch1-deficient mice, CLPs retain their E2A proteins and can differentiate into B cells in the thymus even though T-cell development is arrested due to the lack of promoting effect from Notch signals. Owing to the scarcity of these progenitor cells, it is not feasible to use them to test our hypothesis directly using biochemical approaches. However, key support could come from examination of E2A knockin mice carrying the Mm mutation such that the E2A proteins would be resistant to Notch-induced degradation in all cell types, thus allowing B cells to grow in the thymus.
In summary, this study led to the discovery of a novel function of the Notch-signaling pathway, namely induction of ubiquitin-mediated and proteasome-dependent protein degradation. By controlling E2A function through its turnover, as well as the expression of Id proteins such as Id3 (Reynaud-Deonauth et al., 2002), Notch signals can exert their influence on lymphocyte differentiation at many levels. Since E2A proteins also play crucial roles in the differentiation of other cell types by forming heterodimers of cell-type-specific bHLH proteins, placing E2A downstream of the Notch-signaling pathway may help to explain how Notch receptors facilitate cell fate determination during mammalian development.
Materials and methods
Cell lines and transfection assays
Lymphoid cell lines were cultured in RPMI-1640 supplemented with 10% FBS and 50 µM 2-mercaptoethanol. BaF3 cells were cultured in the same medium plus 20% WEHI-3 conditioned medium for interleukin 3. Jagged-1-expressing and control NIH3T3 cell lines were generous gifts from Dr T.Kadesch, University of Pennsylvania. Cells were cotransfected with desired constructs along with E-box luciferase and CMV-LacZ reporter constructs using a calcium phosphate precipitation method. The cells were harvested 36 h post-transfection and assayed for luciferase and β-galactosidase activities by using a luciferase assay kit (Promega, Madison, WI) or β-galactosidase assay kit (Tropix, Bedford, MA). Production of photons by either reaction was measured with a luminometer (EG & G Berthold, Nashua, NH). Luciferase activity in transfected cells was normalized with that of β-galactosidase.
Plasmids and retroviral constructs
Point and deletion mutants of E47 were generated by PCR-assisted mutagenesis from the parental plasmid E47-pcDNA3 (Park and Sun, 1998). The E47 truncation mutant, N-E47, was generated by inserting an EcoRI–XhoI fragment into the pcDNA3 vector. The N1-IC–pcDNA3 construct was a gift from Dr T. Kadesch. MIGR1 and MIGR1/N1-IC retrovirus constructs and N1-IC deletion mutants as well as Deltex constructs (Pui et al., 1999; Izon et al., 2002) were kindly provided by Dr W.Pear, University of Pennsylvania. The constructs expressing a constitutively active form of MEK1 and T7-tagged Skp2 were a gift from Dr K.-L.Guan, University of Michigan (Sugimoto et al., 1998) and Dr H.Zhang, Yale University School of Medicine, respectively. Oligonucleotides containing hairpin structures of sequences from RBP-Jκ, GFP and Skp2 were cloned into the pBS/U6 vector (Sui et al., 2002). The top-strand sequences of the oligonucleotides are: RBP-Jκ, TCGAGGACGACCC TGTATCACAACGAGTACTGGTTGTGATA CAGGGTCGTCTTTTT; GFP, TCGAG GCGATGCCACCTACGGC AAGTTCAGAGACTTGCCGTAGGTGGCATCGCCTTTT. The sequence for the siRNA against Skp2 is as described (Zheng et al., 2002).
Retroviral transduction
Retroviral stocks were prepared by transfecting Phoenix-E cells as previously described (Pear et al., 1993; Pui et al., 1999) and culture supernatants were collected 48 h post transfection. To transduce B- and T-cell lines, 2 × 106 exponentially growing cells were spin-infected as described (Engel and Murre, 1999). The cells were then cultured in fresh media for 36 h and sorted for GFP expression. The sorted cells were analyzed directly for 16610D9 cells or after culturing for 24–48 h for all other cell lines. To transduce splenocytes, spleens were disrupted and red blood cells were lysed with NH4Cl. Cells were incubated in RPMI-1640 medium with 10% fetal bovine serum (FBS) and 0.1 mM non-essential amino acids, 10 µg/ml Con A or LPS for 24 h. The cells were spin-infected as described above. The infected cells were stained with anti-B220-PE or CD4-PE plus CD8-PE monoclonal antibodies and sorted for GFP and PE using flow cytometry 36 h post infection. Sorted cells were lysed and analyzed using immunoblots.
Protein analysis
Typically, cells were collected for protein analysis 36 h after transfection except that cells used for in vivo ubiquitination assays were treated with 4 µM MG132 12 h prior to harvest. Antibodies used for immunoblots were from the following sources: E47, Notch1-IC, Skp2, GFP and TFIIH (Santa Cruz Biotechnology, Santa Cruz, CA); unphosphorylated and phosphorylated p42/p44 MAP kinases (Cell Signaling Technology, Beverly, MA); Poly (ADP-ribose) polymerase and p27 (BD Bioscience, San Diego, CA); ubiquitin (Zymed, San Francisco, CA). The immunoreactive proteins were detected using enhanced chemiluminescence reagents (Amersham, Piscataway, NJ) and quantified using a LumiImager (FUJIFILM Medical Systems USA Inc., Stamford, CT). Loading controls were carried out by probing for TFIIH or p42/p44.
For immunoprecipitation assays, cells were lysed in a lysis buffer (150 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.5% NP40, 1 mM dithiothreitol, 20 mM NaF, 1 mM Na3VO4 and a cocktail of protease inhibitors) and sonicated. The supernatants obtained after centrifugation were precleared with protein-A+G agarose beads. Five micrograms of antibodies were used to incubate with the supernatants for 2 h or overnight at 4°C. The precipitates were collected by incubating with protein-A+G agarose beads followed by several washes. The precipitates bound to the beads were analyzed by immunoblotting with appropriate antibodies.
Splenic culture
Spleen tissues were dispersed by gently grinding against a cell strainer. Splenocyte suspension was collected and incubated on Petri dishes at 37°C for 1 h after hypotonic lysis of red blood cells. Stromal cells were prepared by trypsinization of tissue debris for 30 min. They were then seeded on 12-well cell culture plate and splenocytes were added onto stromal cell layer at a density of 2 × 107 cells/ml. The cultures were treated with anti-IgM (4 µg/ml) or anti-CD3 plus anti-CD28 at 2 µg/ml each for different lengths of time ± 10 or 50 µM γ-secretase inhibitor Z-IL-CHO (Calbiochem, San Diego, CA).
Acknowledgments
Acknowledgements
We would like to thank Dr Thomas Kadesch, Dr Warren Pear, Dr Kun-Liang Guan, Dr Hui Zhang and Dr Dirk Bohmann for reagents and advice. We are grateful to Dr Paul W.Kincade and Dr Sankar Ghosh for critical reading of the manuscript. We thank Mei-Ying Xiong for technical assistance. The work was supported by grants from the National Institute of Health to X.-H.S. [AI33597, CA77553 and RR15577 (COBRE Award)].
References
- Allman D., Punt,J.A., Izon,D.J., Aster,J.C. and Pear,W.S. (2002) An invitation to T and more: notch signaling in lymphopoiesis. Cell, 109, S1–S11. [DOI] [PubMed] [Google Scholar]
- Allman D., Sambandam,A., Kim,S., Miller,J.P., Pagan,A., Well,D., Meraz,A. and Bhandoola,A. (2003) Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol., 4, 168–174. [DOI] [PubMed] [Google Scholar]
- Aronheim A., Shiran,R., Rosen,A. and Walker,M.D. (1993) The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc. Natl Acad. Sci. USA, 90, 8063–8067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand,M.D. and Lake,R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776. [DOI] [PubMed] [Google Scholar]
- Bain G. et al. (1994) E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell, 79, 885–892. [DOI] [PubMed] [Google Scholar]
- Barndt R., Dai,M.F. and Zhuang,Y. (1999) A novel role for HEB downstream or parallel to the pre-TCR signaling pathway during αβ thymopoiesis. J. Immunol., 163, 3331–3343. [PubMed] [Google Scholar]
- Callahan R. and Raafat,A. (2001) Notch signaling in mammary gland tumorigenesis. J. Mammary Gland Biol. Neoplasia, 6, 23–36. [DOI] [PubMed] [Google Scholar]
- Diederich R.J., Matsuno,K., Hing,H. and Artavanis-Tsakonas,S. (1994) Cytosolic interaction between Deltex and Notch ankyrin repeats implicates Deltex in the Notch signaling pathway. Development, 120, 473–481. [DOI] [PubMed] [Google Scholar]
- Engel I. and Murre,C. (1999) Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc. Natl Acad. Sci. USA, 96, 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming H.E. and Paige,C.J. (2001) Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity, 15, 521–531. [DOI] [PubMed] [Google Scholar]
- Han H., Tanigaki,K., Yamamoto,N., Kuroda,K., Yoshimoto,M., Nakahata,T., Ikuta,K. and Honjo,T. (2002) Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol., 14, 637–645. [DOI] [PubMed] [Google Scholar]
- Harper J.W. (2001) Protein destruction: adapting roles for Cks proteins. Curr. Biol., 11, R431–R435. [DOI] [PubMed] [Google Scholar]
- Heemskerk M.H., Blom,B., Oda,K., Stegmann,A.P., Bakker,A.Q., Weijer,K., Res,P.C. and Spits,H. (1997) Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J. Exp. Med., 186, 1597–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izon D.J. et al. (2002) Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity, 16, 231–243. [DOI] [PubMed] [Google Scholar]
- Jackson P.K. and Eldridge,A.G. (2002) The SCF ubiquitin ligase: an extended look. Mol. Cell, 9, 923–925. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Brou,C., Logeat,F., Schroeter,E.H., Kopan,R. and Israel,A. (1995) Signalling downstream of activated mammalian Notch. Nature, 377, 355–358. [DOI] [PubMed] [Google Scholar]
- Karin M. and Ben Neriah,Y. (2000) Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol., 18, 621–663. [DOI] [PubMed] [Google Scholar]
- Kim D., Peng,X.C. and Sun,X.H. (1999) Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol. Cell. Biol., 19, 8240–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald H.R., Wilson,A. and Radtke,F. (2001) Notch1 and T-cell development: insights from conditional knockout mice. Trends Immunol., 22, 155–160. [DOI] [PubMed] [Google Scholar]
- Massari M.E. and Murre,C. (2000) Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol., 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M.E., Jennings,P.A. and Murre,C. (1996) The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol. Cell. Biol., 16, 121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K., Diederich,R.J., Go,M.J., Blaumueller,C.M. and Artavanis-Tsakonas,S. (1995) Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development, 121, 2633–2644. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw,P.S. and Baltimore,D. (1989) A new DNA binding and dimerization motif in immunoglobin enhancer binding, daughterless, MyoD and myc proteins. Cell, 56, 777–783. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Lin,A., Shen,C.P., Blaumueller,C., Matsuno,K., Artavanis-Tsakonas,S. and Kadesch,T. (1998) Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol., 18, 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.T. and Sun,X.H. (1998) The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J. Biol. Chem., 273, 7030–7037. [DOI] [PubMed] [Google Scholar]
- Pear W.S., Nolan,G.P., Scott,M.L. and Baltimore,D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA, 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanism underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Pui J.C. et al. (1999) Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity, 11, 299–308. [DOI] [PubMed] [Google Scholar]
- Quong M.W., Massari,M.E., Zwart,R. and Murre,C. (1993) A new transcriptional-activation motif restricted to a class of helix–loop–helix proteins is functionally conserved in both yeast and mammalian cells. Mol. Cell. Biol., 13, 792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F., Wilson,A., Stark,G., Bauer,M., van Meerwijk,J., MacDonald,H.R. and Aguet,M. (1999) Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity, 10, 547–558. [DOI] [PubMed] [Google Scholar]
- Reynaud-Deonauth S., Zhang,H., Afouda,A., Taillefert,S., Beatus,P., Kloc,M., Etkin,L.D., Fischer-Lougheed,J. and Spohr,G. (2002) Notch signaling is involved in the regulation of Id3 gene transcription during Xenopus embryogenesis. Differentiation, 69, 198–208. [DOI] [PubMed] [Google Scholar]
- Sugimoto T., Stewart,S., Han,M. and Guan,K.L. (1998) The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J., 17, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui G., Soohoo,C., Affar,e.B., Gay,F., Shi,Y., Forrester,W.C. and Shi,Y. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.-H. (1994) Constitutive expression of the Id1 gene impairs mouse B cell development. Cell, 79, 893–900. [DOI] [PubMed] [Google Scholar]
- Sun X.-H. and Baltimore,D. (1991) An inhibitory domain of E12 prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell, 64, 459–470. [DOI] [PubMed] [Google Scholar]
- Tamura K., Taniguchi,Y., Minoguchi,S., Sakai,T., Tun,T., Furukawa,T. and Honjo,T. (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H). Curr. Biol., 5, 1416–1423. [DOI] [PubMed] [Google Scholar]
- Tewari M., Quan,L.T., O’Rourke,K., Desnoyers,S., Zeng,Z., Beidler,D.R., Poirier,G.G., Salvesen,G.S. and Dixit,V.M. (1995) Yama/CPP32β, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell, 81, 801–809. [DOI] [PubMed] [Google Scholar]
- Tomita K., Hattori,M., Nakamura,E., Nakanishi,S., Minato,N. and Kageyama,R. (1999) The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev., 13, 1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., Staszewski,L.M. and Bohmann,D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell, 78, 787–798. [DOI] [PubMed] [Google Scholar]
- von Boehmer H. (2001) Coming to grips with Notch. J. Exp. Med., 194, F43–F46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., MacDonald,H.R. and Radtke,F. (2001) Notch1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med., 194, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. et al. (2002) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell, 10, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Soriano,P. and Weintraub,H. (1994) The helix–loop–helix gene E2A is required for B cell differentiation. Cell, 79, 875–884. [DOI] [PubMed] [Google Scholar]