Abstract
High copy number and random segregation confound genetic analysis of the mitochondrial genome. We developed an efficient selection for heritable mitochondrial genome (mtDNA) mutations in Drosophila, thereby enhancing a metazoan model for study of mitochondrial genetics and mutations causing human mitochondrial disease. Targeting a restriction enzyme to mitochondria in the germline compromised fertility, but escaper progeny carried homoplasmic mtDNA mutations lacking the cleavage site. Among mutations eliminating a site in the cytochrome c oxidase gene, mt:CoIA302T was healthy, mt:CoIR301L was male sterile but otherwise healthy, and mt:CoIR301S exhibited a wide range of defects, including growth retardation, neurodegeneration, muscular atrophy, male sterility, and reduced life span. Thus, germline expression of mitochondrial restriction enzymes creates a powerful selection and has allowed direct isolation of mitochondrial mutants in a metazoan.
Atypical animal cell contains hundreds to thousands of copies of the mitochondrial genome (mtDNA), which encodes 13 essential subunits of the electron transport chain complexes and RNAs (2 rRNAs and 22 tRNAs) required for mitochondrial translation (1, 2). It is not clear how the genetic integrity of this amitotically distributed genome is maintained. Due to a high mutation rate, lack of recombination, and the presence of coresident genomes to protect mutant genomes from selection, mtDNA mutations should readily accumulate in cells (3). Indeed, a dramatic accumulation of mitochondrial mutations in somatic tissues appears to contribute to age-related disorders in humans (4), and numerous mutations on human mtDNA have been linked to maternally inherited diseases (1, 2). The mitochondrial genome has not been very amenable to functional genetic studies in metazoans (5, 6). Successful generation of trans-mitochondrial mice by hybridizing mtDNA-depleted embryonic stem cells and enucleated donor cells containing mtDNA mutations (6) has been constrained by the limited collections of donor cells carrying mtDNA mutations.
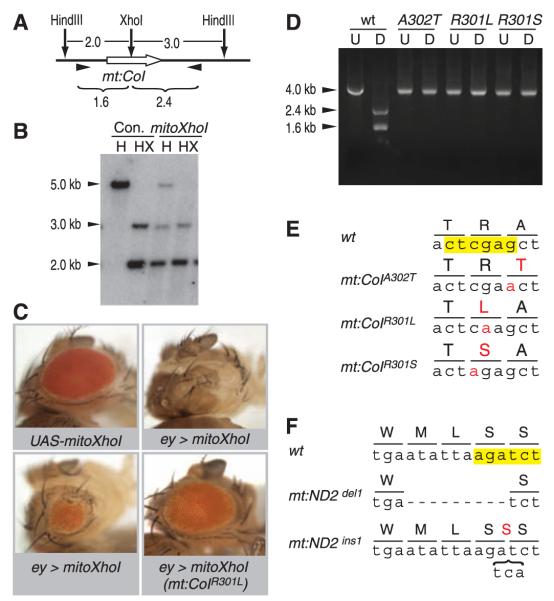
Targeting restriction enzymes to mitochondria can selectively compromise propagation of mtDNA that has a site targeted by the enzyme (7). We reasoned that expression of a restriction enzyme targeting wild-type mtDNA would create a selection allowing rare mutations lacking the restriction site to take over (fig. S1). XhoI has a single recognition site in the cytochrome c oxidase subunit I locus (mt:CoI) in the Drosophila melanogaster mitochondrial genome (fig. S2). It overlaps codons specifying three amino acid residues in a conserved region of CoI (Fig. 1E and fig. S3). pMT-mitoXhoI, which encodes XhoI fused to a mitochondrial targeting leader at its N terminus and a Myc tag at its C terminus (fig. S4), was used to establish S2 cell lines. MitoXhoI protein was specifically localized to mitochondria (fig. S4). Upon induction, the majority of the mtDNA was cleaved at the XhoI site (Fig. 1, A and B), demonstrating that MitoXhoI can efficiently target the wild-type mitochondrial genome.
Fig. 1.
Manipulation of mtDNA with a mitochondrially targeted XhoI (MitoXhoI). (A) Restriction map of a mitochondrial genomic region spanning cytochrome c oxidase subunit 1 (mt:CoI, thick white arrow). The size of each fragment is given in kb. (B) Total DNA was extracted from control (Con.) and mitoXhoI-expressing (mitoXhoI) S2 cells, digested with HindIII (H) or HindIII and XhoI (HX), and hybridized with probe against the mt:CoI gene. (C) UAS-mitoXhoI, when expressed under the control of eyeless-Gal4 (ey>mitoXhoI), caused eye ablation or small eye, which was suppressed in flies containing XhoI-resistant mtDNA [ey>mitoXhoI(mt: CoIR301L)]. (D) mtDNAswere amplified using the pair of primers denoted as arrow-heads in (A). U, undigested polymerase chain reaction (PCR) product; D, PCR product digested with XhoI. There are no detectable wild-type bands in any of the mutants. (E) Recovered mutant mtDNAs with mutations in the XhoI site (yellow). Mutated nucleotides and changed amino acid residues are highlighted in red. (F) Recovered mutant mitochondrial genomes with mutations affecting the BglII site (yellow). Dashed line represents a 9-nucleotide deletion. Bracket represents a 3-nucleotide insertion encoding an extra serine residue (red).
To test the consequence of MitoXhoI expression in animals, we generated transgenic flies expressing mitoXhoI under the UASp promoter. Ubiquitous expression of UAS-mitoXhoI activated by a tub-GAL4 driver blocked hatching of 95% of the embryos, and the remaining 5% died as first instar larvae. Selective expression of UAS-mitoXhoI in the eye primordium using an eyeless-GAL4 driver ablated or greatly reduced the eye (Fig. 1C).
To select for heritable mitochondrial mutants, we used nanos-GAL4 to activate UAS-mitoXhoI expression in germline cells, reasoning that those cells carrying XhoI-resistant mtDNA would survive to produce progeny carrying the mutant mtDNA (fig. S5). Most of the UAS-mitoXhoI/+; nanos-Gal4/+ flies were sterile, but about 1% of the females gave a few escaper progeny. Escapers all carried XhoI-resistant mtDNA (Fig. 1D), and no wild-type genomes were detected, which suggests that each mutant was homoplasmic. Sequencing identified single base pair mutations that eliminated the XhoI site (Fig. 1E). Three different sequence variants were recovered, mt:CoIA302T, mt:CoIR301L,and mt:CoIR301S, all resulting in a single amino acid change (Fig. 1E). No secondary mutations were seen in these mutants, and each mutant was maternally inherited and conferred complete resistance to MitoXhoI in the eye (Fig. 1C).
We investigated whether any of these mt:CoI mutations were pathogenic. Only mt:CoIA302T behaved as wild-type flies in the phenotypic analyses we conducted (table S1). mt:CoIR301L flies were healthy, except that the males were sterile (table S1). Spermatogenesis appeared normal, and sperm were motile but reduced in number (movies S1 and S2). Sperm were transferred to the female during mating but were not stored in the sperm storage organs. Despite being sterile, male mt:CoIR301L flies had similar cytochrome c oxidase activity and slightly higher adenosine triphosphate (ATP) levels than wild-type flies (fig. S6). It is not clear how a mitochondrial mutation selectively compromises sperm function, but, because transmission of mtDNA occurs through the female, it has been argued that such mutations can be sustained in a population (8) and could make a substantial contribution to male sterility (9). The mt:CoIR301S allele exhibited markedly more severe phenotypes. The mutant had an extended larval phase (about 10 days at 25°C), and the mechanosensory bristles on the thorax were missing or thinned and shortened (Fig. 2, A and B). The males were sterile without any mature sperm, which suggests a defect in spermatogenesis. Female flies were fertile but produced only 20% as many progeny as did wild-type flies. The mt-CoIR301S flies had about half the normal cytochrome c oxidase activity and significantly reduced ATP levels (fig. S6).
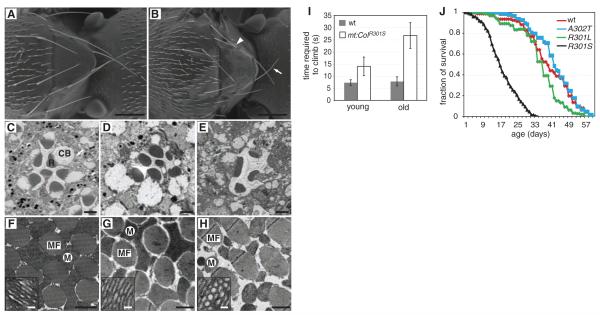
Fig. 2.
Characterization of mt:CoIR301S flies. (A and B) Wild-type (A) and mt:CoIR301S (B) fly thoraxes showing the short and thin (arrow) or missing bristles (arrowhead) in mt:CoIR301S flies. Scale bars, 250 μm. (C to E)Retinal morphology of 2-week old w1118 (C), 1-day old mt:CoIR301S (D), and 2-week old mt:CoIR301S (E). R, rhabdomere; CB, cell body. Scale bars, 2 μm. (F to H) Cross-section of indirect flight muscle of 2-week-old w1118 (F), 1-day-old mt:CoIR301S (G) and 2-week-old mt:CoIR301S (H). Inserts show high-magnification images of inner mitochondrial membrane structure. MF, myofibrils; M, mitochondria. Scale bars, 1 μm in large view and 50 nm in inserts. (I) Climbing abilities of wild-type and mt:CoIR301S flies at different ages were assayed by measuring the time required (seconds) to climb 10cm. The 2- to 3-day-old mt:CoIR301S flies have weaker climbing ability (14.0 ± 3.8 s) than wild-type flies of the same age (7.3 ± 1.2 s). The defects were exacerbated in 2-week-old flies (26.7 ± 5.3 s). Results are means ± SD (n = 3 for each data point). (J) Life span of wild-type (w1118) and mitochondrial DNA mutant flies. mt:CoIR301S flies showed greatly reduced longevity, with a median life span of 17 days. The median life span of wild-type flies is 41 days. Neither mt:CoIA302T nor mt:CoIR301L flies displayed any significant alterations in life span compared with wild-type flies.
Because most mtDNA diseases in humans show neuropathy and myopathy, we examined the morphology of retina and indirect flight muscles to determine whether mt:CoIR301S flies had similar defects. Young mt:CoIR301S flies had the full complement of ommatidia components, although some rhabdomeres had slight morphogenetic defects (Fig. 2, C and D). However, the ommatidia of aged mt:CoIR301S, but not wild-type flies, were disorganized and the rhabdomeres were shrunken or completely lost (Fig. 2E), indicating age-dependent degeneration of photoreceptor neurons.
Transmission electron microscopy (TEM) of flight muscle in wild-type and young mt:CoIR301S flies revealed orderly muscle fibers and fused mitochondria with long and tubular cristae (Fig. 2, F and G). However, two weeks after adult eclosion, the mt:CoIR301S mitochondria were small and fragmented, contained many vesicular structures, and did not completely fill the intermyofibril space (Fig. 2H). Similar mitochondrial morphological defects were also reported in flies with a mutation in the mitochondrial ATP6 locus (10). Moreover, when tested in a climbing assay, mt:CoIR30lS flies showed mobility defects enhanced by age, consistent with age-dependent neurodegeneration and myopathy (Fig. 2I). Beyond these age-dependent neurological and muscular dysfunctions, mt:CoIR301S flies had substantially reduced longevity (Fig. 2J).
The pathologies associated with human mitochondrial DNA diseases often develop with age (1), much like the degenerative changes that we find for the mt:CoIR301S allele. Because the mutations responsible for human mitochondrial disease usually affect only a fraction of the mitochondrial genomes (heteroplasmy), the progressive increase in the severity of the phenotype might be attributed to an increase in the load of mutant genomes in the affected tissues or to an inherent feature of the mitochondrial defect. Because the mutations we have selected are homoplasmic, the age-dependent degenerative phenotype must be inherent to the mutation.
There are 31 single-cutting restriction enzymes sites in Drosophila mtDNA, distributed within eight protein-encoding genes, two rRNAs and three tRNAs (table S2). To test the generality of our approach, we produced a UASp-regulated mitoBglII transgene that targets a single BglII site in the mt:ND2 locus. After germline expression, we recovered two lines carrying mutations in mt:ND2. In contrast to the single base changes recovered in mt:CoI, one mt:ND2 mutation is an in-frame deletion and the other is a three-nucleotide insertion (Fig. 1F). Use of the full panel of available enzymes will target the majority of the protein coding genes in Drosophila mtDNA and allow generation of multiple alleles, although we expect that alleles that completely eliminate respiratory activity will not be recovered as a result of lethality.
In the described selection for resistance to germline expression of mitochondrial restriction enzymes, roughly one out of several thousand germline precursor cells (PGCs) survived to produce progeny. Mice have about 50 PGCs, and this population is greatly expanded before definitive germ cell differentiation (11). Furthermore, one could employ the previously developed “mtDNA mutator mouse” carrying an error-prone mitochondrial DNA polymerase (12) to increase the frequency of enzyme-resistant mitochondrial genomes. If the frequency of mtDNA mutations in mice is similar to that in flies, generation of novel homoplasmic mitochondrial mutant mice by the described approach should be practical.
Supplementary Material
Fig. S1. Hypothesis of mitochondrially targeted restriction enzyme-based selection for mtDNA mutations. (a) Linearization of mtDNA by a restriction enzyme (red arrowhead) will block mtDNA replication, and thereby cause mtDNA loss. (b) In the presence of a mitochondrially-targeted restriction enzyme that can digest wild type mtDNA (green circle), the cells that do not contain any enzyme-resistant mitochondrial genome will lose all of their mtDNA and be eliminated. Those cells harboring enzyme resistant mtDNAs (red circle) will survive the selection by repopulating their mitochondria with these mutant mtDNAs.
Fig. S2. The mitochondrial genome of Drosophila melanogaster. The restriction enzymes used in this study and their relative positions are labeled.
Fig. S3. (A) Alignment of partial amino acid sequences of CoI proteins of Drosophila (Dm), human (Hs), mouse (Mm) and S. cerevisiae (Sc). Conserved amino acid residues are shaded in black. Asterisks label 3 amino acids that may be affected by mutations on XhoI site in mt:CoI gene.
Fig. S4. Targeting restrction enzyme into mitochondria. (A) Schematic view of mitoXhoI fusion gene. (B) S2 cells expressing mitoXhoI under the control of a metallothionein promoter were induced with 100 μM CuSO4, labeled with MitoTracker (green) and stained with anti-myc antibody (red). Scale bar, 10 μm.
Fig. S5. Cross scheme for selecting Drosophila lines carrying XhoI-resistant mitochondrial genomes.
Fig. S6. Biochemical deficiencies of wild type and mtDNA mutants. mt:CoIR301S flies showed dramatically reduced COX activity (0.48±0.04 of wild type) and ATP levels (0.67±0.02 of wild type). There were no significant changes in mt:CoIA302T and mt:CoIR301L flies compared to wild type.
Table S1. Phenotypic comparison of mt:CoI mutants in this study with other mutants in electron transport chain complexes in Drosophila (A)
Table S2. The restriction enzymes with one site in the Drosophila mitochondrial genome
Acknowledgments
We thank members of O’Farrell laboratory for comments on the manuscripts; M.-L. Wong and M. Sepanski for technical help with TEM; M. Fuller and A. W. Shermoen for help with examination of spermatogenesis and sperm in female reproductive organs; and Bloomington Stock Center for providing some Drosophila stocks. This work is supported by an NIH RO1 grant and a Sandler Family Supporting Foundation award to P.H.O. H.X. is supported by a postdoctoral fellowship from the Larry L. Hillblom Foundation.
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/321/5888/575/DC1 Materials and Methods Figs. S1 to S6 Tables S1 and S2 References Movies S1 and S2
References and Notes
- 1.Wallace DC. Annu. Rev. Genet. 2005;39:359. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RW, Turnbull DM. Nat. Rev. Genet. 2005;6:389. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstrom CT, Pritchard J. Genetics. 1998;149:2135. doi: 10.1093/genetics/149.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor RW, et al. J. Clin. Invest. 2003;112:1351. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayona-Bafaluy MP, Fernández-Silva P, Enríquez JA. Mitochondrion. 2002;2:3. doi: 10.1016/s1567-7249(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 6.Khan SM, Smigrodzki RM, Swerdlow RH. Am. J. Physiol. Cell Physiol. 2007;292:C658. doi: 10.1152/ajpcell.00224.2006. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava S, Moraes CT. Hum. Mol. Genet. 2001;10:3093. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- 8.Frank SA, Hurst LD. Nature. 1996;383:224. doi: 10.1038/383224a0. [DOI] [PubMed] [Google Scholar]
- 9.Spiropoulos J, Turnbull DM, Chinnery PF. Mol. Hum. Reprod. 2002;8:719. doi: 10.1093/molehr/8.8.719. [DOI] [PubMed] [Google Scholar]
- 10.Celotto AM, et al. J. Neurosci. 2006;26:810. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam PP, Snow MH. J. Embryol. Exp. Morphol. 1981;64:133. [PubMed] [Google Scholar]
- 12.Trifunovic A, et al. Nature. 2004;429:417. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Hypothesis of mitochondrially targeted restriction enzyme-based selection for mtDNA mutations. (a) Linearization of mtDNA by a restriction enzyme (red arrowhead) will block mtDNA replication, and thereby cause mtDNA loss. (b) In the presence of a mitochondrially-targeted restriction enzyme that can digest wild type mtDNA (green circle), the cells that do not contain any enzyme-resistant mitochondrial genome will lose all of their mtDNA and be eliminated. Those cells harboring enzyme resistant mtDNAs (red circle) will survive the selection by repopulating their mitochondria with these mutant mtDNAs.
Fig. S2. The mitochondrial genome of Drosophila melanogaster. The restriction enzymes used in this study and their relative positions are labeled.
Fig. S3. (A) Alignment of partial amino acid sequences of CoI proteins of Drosophila (Dm), human (Hs), mouse (Mm) and S. cerevisiae (Sc). Conserved amino acid residues are shaded in black. Asterisks label 3 amino acids that may be affected by mutations on XhoI site in mt:CoI gene.
Fig. S4. Targeting restrction enzyme into mitochondria. (A) Schematic view of mitoXhoI fusion gene. (B) S2 cells expressing mitoXhoI under the control of a metallothionein promoter were induced with 100 μM CuSO4, labeled with MitoTracker (green) and stained with anti-myc antibody (red). Scale bar, 10 μm.
Fig. S5. Cross scheme for selecting Drosophila lines carrying XhoI-resistant mitochondrial genomes.
Fig. S6. Biochemical deficiencies of wild type and mtDNA mutants. mt:CoIR301S flies showed dramatically reduced COX activity (0.48±0.04 of wild type) and ATP levels (0.67±0.02 of wild type). There were no significant changes in mt:CoIA302T and mt:CoIR301L flies compared to wild type.
Table S1. Phenotypic comparison of mt:CoI mutants in this study with other mutants in electron transport chain complexes in Drosophila (A)
Table S2. The restriction enzymes with one site in the Drosophila mitochondrial genome