Introduction
An organism is made up of a myriad of intricate structures, each with its own shape and designated place within the whole. The development of an organism thus requires the proper formation and correct positioning of each of these individual parts. Accordingly, a major focus of developmental research has been an understanding of the mechanisms underlying the establishment of positional information, which serves as the basis for both the creation of individual structures and for the correct placement of tissues within the entire organism. After many fruitful years of research into this area, many aspects of patterning are becoming clear. But positional information is an abstract code while the organism is a physical reality, the code being interpreted in the process of morphogenesis. If we are really to understand the flow of the structural information that proportions, shapes, and positions structures, we need to understand the process of interpretation of positional information.
One role of positional information is the specification of the tissue-specific programs of gene expression. While tissue-specific gene expression is the major determinant of the functional specialization of different tissues, other aspects of the tissue such as size and shape are regulated differently. Consider your arm and leg: they consist of similar tissues that are organized into similar elements and laid out in an analogous fashion, yet the differences between them are profound and of obvious importance. The development of these highly related structures likely relies upon the same combination of cell behaviors, and the differences between the structures presumably reflect subtle differences in the time, magnitude, and position of these common processes. Similarly, the formation of all organs and structures of the body will rely to a major extent on the precise control of basic cellular processes such as cell movements, shape changes, and division. While these phenomena can be controlled by regulation of gene expression, the format of this regulation is different from that usually emphasized in developmental studies of tissue-specific gene expression.
Morphogenesis is far too complex to study holistically, but the regulation of some of the individual cell behaviors that contribute to morphogenesis can be approached. One of the cell behaviors whose control we are beginning to understand at a molecular level is cell proliferation. The lessons learned from studies of the cell cycle are generalizable and provide a paradigm for an understanding of many aspects of cell behavior required for morphogenesis; we will discuss here the regulation of several such behaviors.
There are several steps involved in the link between patterning information and cell behavior: the interpretation of complex patterning information by a regulatory factor, the transduction of a signal produced by this factor, and, finally, the regulation of specific cell biological phenomena in response to this transduction. In some cases, such as that involving the mitotic regulator Cdc25String, a single factor completes the link between patterning and cell behavior. In others, the individual steps are carried out by distinct factors, introducing increasing complexity to the link between patterning and cell behavior.
Lessons from string: a Direct Connection between Patterning and the Cell Cycle
The early fly embryo develops as a syncytium and is subdivided by the expression of numerous localized transcription factors. At the time of cellularization, when the ∼5000 nuclei of the embryo are partitioned into a simple monolayer of cells, each cell or local group of cells inherits a unique assortment of transcription factors. This assortment, whose singularity reflects combinatorial interpretation of the quantitative as well as qualitative differences in the factors, provides cells of the now cellular blastoderm with positional addresses that will ultimately drive cell biological phenomena in complex but stereotypical patterns. The code of patterning information also provides temporal information, because the particular assortment of transcription factors in a cell changes with time. Consequently, an event triggered by a particular assortment of factors will occur at a specific time in a specific place.
Following cellularization, the embryo undergoes gastrulation and other major rearrangements of the embryonic tissues. These movements are guided by the precise spatial and temporal programming of cellular events. During these tissue rearrangements, cells divide. These cell divisions are tightly regulated and follow a stereotypical pattern (Foe, 1989) that is determined by the patterning information subdividing the embryo (Edgar et al., 1994). Studies of the mechanism that schedules these mitoses have provided a paradigm for understanding how patterning information can be translated into cell behavior.
The first 13 cycles of Drosophila development do not contain gap phases and consist solely of alternating S phases and mitoses. All the factors required for mitosis and S phase are presumably present at high levels during these cycles, allowing immediate entry into each phase following completion of the other. During the 14th cell cycle, however, Cdc25String, a phosphatase that removes inhibitory phosphorylation from Cdc2, becomes limiting and prevents immediate entry into mitosis, thereby creating a G2 phase (O'Farrell et al., 1989; Edgar and Datar, 1996). Cdc25String is thereafter expressed in an intricate spatio-temporal pattern that precisely anticipates mitosis (Edgar and O'Farrell, 1989). Its expression is the only unsatisfied requirement for mitosis at this stage: providing Cdc25String by heat shock induction during this G2 phase provokes precocious and rapid entry into mitosis (Edgar and O'Farrell, 1990). Studies of the string promoter have also shown that Cdc25String expression is directly controlled by patterning information (Edgar et al., 1994). Cdc25String thus provides a direct link between patterning information and the cell cycle (Figure 1A).
Figure 1.
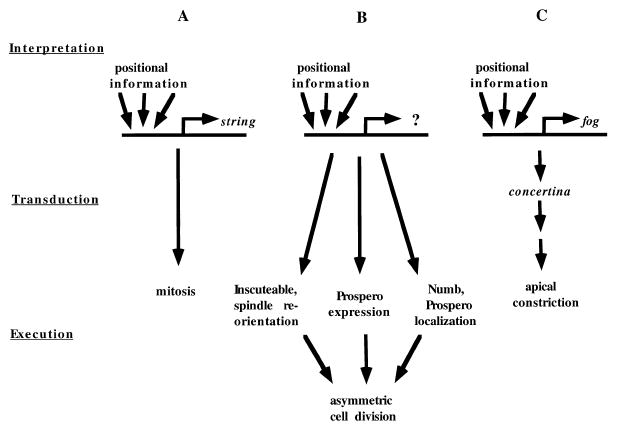
Linking Patterning Information to Cell Behavior
The three levels involved in the connection between patterning information and cell behavior—interpretation of positional information, transduction of a signal arising from that interpretation, and execution of the signal through the control of cell behavior—can take various forms.
(A) Cdc25String expression is directly controlled by positional information; this information takes the form of combinations of transcription factors that bind to the string promoter and drive its expression in specific ways. Because Cdc25String directly controls the cell cycle, no transduction of the interpretation step is required to affect cell behavior. Cdc25String thus provides a direct link between patterning information and cell behavior.
(B) Although the details of the circuitry linking patterning information and the generation of asymmetry during cell division are unclear, it appears that multiple activities required for the asymmetry are coordinated by an unknown upstream regulator. One of these downstream activities is the expression of Inscuteable, which may directly control mitotic spindle orientation. In this situation, it is not known if any transduction is required to link the interpretation of patterning information and the regulation of cell behavior.
(C) The apical constriction of cells that drives the tissue movements facilitating gastrulation is regulated by expression of Folded gastrulation (Fog). Although no analysis of the folded gastrulation promoter has been published, its pattern of expression is consistent with the possibility that it, like Cdc25String, is acting to interpret patterning information. Because Folded gastrulation is a secreted molecule, it is most likely not causing apical constrictions itself, but is instead probably generating a signal that is transduced ultimately to bring about the observed cellular effects.
Analysis of the large and complex string promoter, and of the effects of mutations in various patterning genes on Cdc25String expression, have begun to define how the interpretation of patterning information occurs (Edgar et al., 1994). Individual elements of the string promoter can drive the expression of reporter genes in particular regions of the embryo that comprise subsets of the normal pattern of string expression. In addition, mutations in individual patterning genes can affect specific domains of the normal complement of Cdc25String expression. For example, in embryos that are mutant in twist, a gene encoding a transcription factor that is usually expressed in prospective mesoderm, there is a specific absence of string expression and cell division in the mesodermal cells. Together, these results suggest that each transcription factor, perhaps at specific levels and in combination with other factors, can bind to specific elements within the string promoter and drive a subset of normal Cdc25String expression; because each cell (or small domain of cells) has a unique assortment of these factors, the subsets will combine to produce Cdc25String in unique patterns.
Uncovering the mode of regulation of string expression and its direct role in controlling the cell cycle has led to the recognition that the string gene acts as an interface between patterning information and cell behavior. The “interpretation” of one spatial pattern (i.e., the combinations of transcription factors) to create another spatial pattern (i.e. Cdc25String expression) underlies the programming of mitosis. While the spatial pattern of localized transcription factors represents an abstract code of patterning information, the pattern of Cdc25String expression corresponds to the observed pattern of mitosis.
We would like to point out several features of this format of regulation. First and foremost, it is generalizable; that is, any cellular process can be controlled by the expression or activation of a limiting gene product specific to that process. The code of patterning information can thus be independently interpreted by many different factors, each specialized to trigger a particular morphogenetic event. Second, because the execution of even a complex process like mitosis can be controlled by a single limiting gene product, the developmental programming can be distilled down to the regulation of a single limiting factor, simplifying the circuitry required to control the process. Finally, the genes that carry out this interpretation can be rather widely expressed (e.g. Cdc25String is expressed in all but a few nondividing tissues), yet can regulate local morphogenetic events by their precise schedule of expression or activation.
Some features seen in the control of mitosis by Cdc25String are likely to be general, such as the importance of an interpretation step in which numerous patterning inputs are integrated. Other features, however, are likely to be more variable. For example, in the case of string, a single gene, rather than an elaborate cascade of regulators, links the patterning information to cell behavior. Cdc25String, which is itself the interpreter of patterning information, acts directly to control the cell cycle by relieving Cdc2 of its inhibitory phosphorylation, thereby allowing entry into mitosis. There is no transduction of the signal though intermediary gene products, as Cdc25String provides all aspects of the patterning–cell cycle link. An additional source of variation in this type of regulation may be found in the form of the patterning information that is being interpreted. The pattern of Cdc25String expression (as well as numerous other regulators acting during Drosophila embryogenesis) is controlled by particular combinations of transcription factors; this mode of regulation presumably reflects the prominent role of transcription factors in the patterning of the early fly embryo. We expect that in many developmental contexts, however, patterning information will take other forms, such as combinations of signaling events that lead to the post-translational activation of key regulators of cell biological phenomena.
Below, we consider additional examples of the link between patterning and morphogenesis, each of which can be considered in terms of steps outlined above: integration of pattern information, transduction of a signal resulting from this integration, and execution of a cell biological process. These examples begin to reveal the diversity of these processes while demonstrating the power of the general format of this regulation.
inscuteable Links Positional Information and Division Orientation
The orientation of the spindle during cell division often plays an essential role in the organization of tissues (Horvitz and Herskowitz, 1992). For example, it can determine the position of daughter cells following division or facilitate the asymmetric inheritance of molecules that direct daughter cells to adopt distinct fates. The connection between morphology and the placement of daughter cells by oriented cleavage is most evident in plants, where the cells do not move at all and birth position is key to the final morphology (see, for example, Di Laurenzio et al., 1996). But there are also beautiful correlations between cell division programs and the emergence of complex structures in numerous organisms, and their experimental perturbation can disturb development (e.g. early divisions in C. elegans [Priess and Thomson, 1987]). Consequently, the programming of division orientation and asymmetry makes fundamental contributions to morphogenesis. Work in Drosophila has begun to reveal how these processes are controlled.
Most of the cell divisions in the early embryo occur with spindles oriented parallel to the surface of the embryo. In neuroblast precursors of the central nervous system, however, the spindles are oriented perpendicular to the surface. The behavior of neuroblasts in a region of the embryo called the procephalic neurogenic region (PNR) first deviates from the surrounding cells at mitosis, when their spindles rotate to assume a perpendicular position. Elsewhere, neuroblasts segregate from the ectodermal epithelium (in a process called delamination) prior to the elaboration of a perpendicularly oriented spindle. Spindle orientation in both of these groups of neuroblasts is determined by a recently identified protein, Inscuteable (Kraut and Campos-Ortega, 1995; Kraut et al., 1996), which provides another example of a link between patterning and cell behavior. Inscuteable is expressed in a complex pattern in fly embryos that correlates precisely with spindle reorientation, and inscuteable mutants fail to establish the perpendicular orientation of their spindles.
Aspects of the inscuteable phenotype differ between the two types of neuroblasts. In neuroblasts of the PNR, the mutant spindle assumes an orientation similar to that of the surrounding ectodermal cells and is parallel to the surface of the embryo. In delaminated neuroblasts that are no longer in contact with the ectoderm, however, loss of Inscuteable results in randomly oriented spindles. This difference in the mutant phenotype between these two types of neuroblasts presumably reflects either cell type differences between them or differences in their environment, i.e. the presence or absence of contact with surrounding ectoderm. In either case, the two mutant patterns are interestingly reminiscent of the two types of phenotypes seen in mutants of the BUD genes involved in bud site selection in the yeast S. cerevisiae, in which one class of mutants displays an abnormal but still ordered pattern of site selection, while others position their buds randomly (Chant and Herskowitz, 1991; Chant et al., 1991).
In addition to its expression pattern and mutant phenotype, ectopic expression of Inscuteable has provided evidence for its role as a key regulator of spindle orientation. In those experiments, Inscuteable has been shown to be sufficient to cause spindle reorientation in ectodermal cells outside of the PNR. Inscuteable expression is thus acting to control spindle orientation, perhaps by providing a function that links existing cellular asymmetries to an apparatus that guides centrosome localization and consequent spindle orientation.
These cell divisions that are perpendicular to the surface are asymmetric in that the two daughters of the division follow different fates. This differentiation in fate specification is guided by the unequal segregation of at least two regulatory molecules, Numb and Prospero. Numb is expressed throughout the embryo, and is localized near the surface of the cell. In most cells, it is equally partitioned to daughter cells during division. Specifically in those cells with a reoriented spindle, however, Numb is localized to the basal surface, near one of the centrosomes, and is asymmetrically distributed upon division (Knoblich et al., 1995). This asymmetry is important for the identity of the cells: if Numb is absent, then both daughter cells adopt the fate of the daughter normally not receiving Numb. Overexpression of Numb, however, causes both daughters to contain Numb, and both cells consequently adopt the fate of the daughter normally inheriting Numb (Rhyu et al., 1994). Prospero is expressed in most or all neuronal precursor cells (Doe et al., 1991; Vaessin et al., 1991), and is selectively partitioned into the basally-located daughter cell (Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995) where, like Numb, it plays a role in directing the developmental fate of the daughter cells.
Both Numb and Prospero fail to localize properly in the neuroblasts of an inscuteable mutant, demonstrating that Inscuteable function is required for Numb and Prospero localization. Nevertheless, at least some of the activities involved in this asymmetry do not depend on Inscuteable. For example, in an inscuteable mutant, Prospero continues to be expressed in cells that ordinarily would have had perpendicularly oriented spindles. In addition, ectopic expression of Inscuteable in ectodermal cells is not sufficient for the asymmetric localization of Numb; apparently, despite ubiquitous Inscuteable, an activity required for Numb localization is restricted to cells that usually localize Numb. This Inscuteable-independent activity is not provided by Prospero, since Numb localization is independent of Prospero function. It thus appears that three distinct factors—Inscuteable, Prospero, and another function required for asymmetric localization of Numb—are localized to the cells that undergo asymmetric and perpendicular cleavage.
Because of its complex pattern of expression, Inscuteable may be acting to interpret patterning information and thereby provide, like Cdc25String, a direct link between patterning and cell behavior. No analysis of the inscuteable promoter has been published, however, and an unequivocal identification of Inscuteable as an interpreter of patterning information depends upon such an analysis. The Inscuteable-independent presence of additional activities in the same cells as Inscuteable suggests the possibility that an upstream regulator is jointly regulating events involved in asymmetric division and oriented division. In this case, the upstream regulator would be the interpreter of patterning information, and Inscuteable would be acting in response to the signal generated by that upstream regulator to control spindle orientation (Figure 1B). Alternatively, the simultaneous presence of multiple activities involved in asymmetric division and oriented division could result from the independent but identical, or nearly identical, interpretation of patterning information by regulators specific to each activity, including Inscuteable. This latter means of regulation would be particularly useful if, in other contexts, spindle orientation is controlled independently of asymmetric inheritance of specific molecules.
Folded gastrulation: Generalizing the Model
The control over the timing and orientation of mitosis provides examples of how cell behavior can be directed by patterning information. We think that the paradigm revealed from these studies is equally valid for understanding other aspects of cell behavior. The regulation of the folding of the epithelium that underlies much of gastrulation provides one such example of the regulation of a non-cell cycle aspect of morphogenesis.
Following cellularization, the early Drosophila embryo initiates a series of foldings and invaginations that are driven by localized changes in the shape of cells. For example, many cells along the ventral surface of the embryo constrict apically, causing that region of the embryo to invaginate and form a structure called the ventral furrow. This constriction is driven by the expression of a molecule called Folded gastrulation (Costa et al., 1994). Although some cells do change shape and furrow formation still occurs in folded gastrulation mutants, these events are reduced and abnormal. Increased expression of Folded gastrulation is sufficient to drive the apical constrictions outside of the normal territories of furrow formation.
In this example, although the connection between patterning information and cell behavior is very specific, it may not be direct (Figure 1C). While Folded gastrulation is possibly acting as the interpreter of positional information, as it is expressed in precise patterns in the embryo that mark the positions of folds, it is probably not directly controlling cell behavior. Folded gastrulation is a secreted molecule that presumably initiates a signal, transmitted via a receptor and transduction pathway that includes the Gα protein encoded by concertina (Parks and Wieschaus, 1991), to alter the cytoskeleton and direct the apical constrictions.
Setting the Stage for the Regulation of Specific Cell Behaviors
The final step in the connection between patterning information and the control of cellular behavior is the expression or activation of a factor directly controlling the behavior. This is made possible because these factors have become limiting, thereby setting the stage for dramatic effects upon their appearance. Thus, the process of regulating cellular events by the expression of key regulators must begin prior to this expression. For example, string message and protein must be eliminated during cycle 14 to allow it single-handedly to control entry into mitosis upon its expression. In other situations, the setting of the stage may not be nearly so simple. For instance, a G1 phase is introduced into the embryonic cell cycle during cycle 17, and cell cycle progression is subsequently controlled by the joint regulation of expression of a number of S phase functions including Cyclin E (Knoblich et al., 1994; Duronio and O'Farrell, 1995). The creation of a G1 phase, allowing this regulatory mode, involves the alteration of expression or activity of a number of factors (Follette and O'Farrell, 1997), including E2F (Duronio and O'Farrell, 1994) and its targets, Cyclin E (Knoblich et al., 1994), and a newly isolated inhibitor of Cyclin E, Dacapo (de Nooij et al., 1996; Lane et al., 1996).
Conclusion
We have described here a general format for the regulation of cellular behaviors that involves little more than the efficient use of known mechanisms. The step that we have called interpretation of patterning information is entirely analogous to steps in the established cascade of regulatory factors that refine pattern information. For example, the regulation of string expression in a spatial and temporal program by combinatorial action of localized transcription factors is analogous to, if perhaps somewhat more complicated than, the regulation of even skipped by pair-rule genes (Lawrence, 1992). Indeed, there are numerous examples of pattern interpretation within the cascade of pattern regulators. The subsequent steps of this general format for regulation of morphogenesis are similarly less than novel. We have simply suggested that linear cascades can transduce the interpreted information finally to drive the expression or stimulate the activity of a rate-limiting factor that triggers a fairly complex event, such as mitosis in the case of Cdc25String. Nonetheless, we suggest that the recognition of this format might have fairly profound consequences, because it appears to be widely used in development, and because it can direct research to biological controls that will provide important insight into the flow of information that governs morphogenesis.
The generality of the format we have described here is hinted at by a number of examples in which localized gene expression directs events important for morphogenesis. For example, programmed cell death during Drosophila embryogenesis, which appears to be required for aspects of morphogenesis, is determined by specific patterns of expression of certain key genes (White et al., 1994, 1996; Grether et al., 1995; Chen et al., 1996). In addition, localized transcription drives cytoplasmic branching events during the development of the Drosophila tracheal system (Guillemin et al., 1996; Samakovlis et al., 1996). We suggest that both of these examples, and almost certainly many more, will follow the general rules we have outlined here.
The localized expression of interpreter genes differs from tissue-specific gene expression in that it does not typify any particular tissue, but rather guides the formation of structures by its specific schedule. The conceptual impact of this might best be illustrated by a proposal. Homeotic genes do not direct the formation of different tissues, but rather guide distinctions between structures such as the different Drosophila appendages. Because the different appendages include homologous tissues, yet are uniquely shaped, we suggest that homeotic genes will largely influence the details of expression of interpreter genes rather than direct tissue-specific gene expression. The resulting modulations in the schedules of expression of various interpreter genes will then guide the formation of the distinctive morphologies of different body parts.
Like the different appendages of Drosophila, arms and legs are comprised of similar tissues that are arranged in similar but distinct patterns. The close similarities between these structures suggests that their development will rely in large part on the expression of the same genes. As with Drosophila appendages, we suggest that the distinctions between arms and legs will be guided by the schedules of expression of a large number of interpreter genes, each responsible for directing one aspect of the cellular behaviors that underlie morphogenesis.
This perspective, though by no means simple, provides a format to begin a dissection of morphogenesis. If we identify the integration steps, outline the transduction steps, and define the limiting activity whose regulation finally determines whether a particular process is on or off, then we can perhaps begin to understand how development can build an arm or a leg out of the same tissues, using the same biology.
Acknowledgments
We are grateful to Mary Maxon, for assistance with Figure 1, and to Megan Grether, and all the members of the O'Farrell laboratory, for comments and discussion.
References
- Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chant J, Corrado K, Pringle JR, Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991;65:1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76:1075–1089. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor, dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/s0092-8674(00)81819-x. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH. Developmental control of a G1-S transcriptional program in Drosophila. Development. 1994;120:1503–1515. doi: 10.1242/dev.120.6.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH. Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Lehman DA, O'Farrell PH. Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development. 1994;120:3131–3143. doi: 10.1242/dev.120.11.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes Dev. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Follette PJ, O'Farrell PH. Cdks and the Drosophila cell cycle. Curr Opin Genet Dev. 1997 doi: 10.1016/s0959-437x(97)80104-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Guillemin K, Groppe J, Dücker K, Treisman R, Hafen E, Affolter M, Krasnow MA. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development. 1996;122:1353–1362. doi: 10.1242/dev.122.5.1353. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Kraut R, Campos-Ortega JA. inscuteable, a neural precursor gene of Drosophila, encodes a candidate for a cytoskeleton adaptor protein. Dev Biol. 1996;174:65–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- Lawrence PA. The Making of a Fly: the Genetics of Animal Design. Oxford: Blackwell Scientific Publications; 1992. [Google Scholar]
- O'Farrell PH, Edgar BA, Lakich D, Lehner CF. Directing cell division during development. Science. 1989;246:635–640. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- Parks S, Wieschaus E. The Drosophila gastrulation gene concertina encodes a G alpha-like protein. Cell. 1991;64:447–458. doi: 10.1016/0092-8674(91)90652-f. [DOI] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell. 1987;48:241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Hacohen N, Manning G, Sutherland DC, Guillemin K, Krasnow MA. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development. 1996;122:1395–1407. doi: 10.1242/dev.122.5.1395. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]


