Abstract
Ecdysteroids initiate molting and metamorphosis in insects via a heterodimeric receptor consisting of the ecdysone receptor (EcR) and ultraspiracle (USP). The EcR–USP heterodimer preferentially mediates transcription through highly degenerate pseudo-palindromic response elements, resembling inverted repeats of 5′-AGGTCA-3′ separated by 1 bp (IR-1). The requirement for a heterodimeric arrangement of EcR–USP subunits to bind to a symmetric DNA is unusual within the nuclear receptor superfamily. We describe the 2.24 Å structure of the EcR–USP DNA-binding domain (DBD) heterodimer bound to an idealized IR-1 element. EcR and USP use similar surfaces, and rely on the deformed minor groove of the DNA to establish protein–protein contacts. As retinoid X receptor (RXR) is the mammalian homolog of USP, we also solved the 2.60 Å crystal structure of the EcR–RXR DBD heterodimer on IR-1 and found the dimerization and DNA-binding interfaces to be the same as in the EcR–USP complex. Sequence alignments indicate that the EcR–RXR heterodimer is an important model for understanding how the FXR–RXR heterodimer binds to IR-1 sites.
Keywords: ecdysone/EcR/nuclear receptor/RXR/USP
Introduction
Ecdysteroids are arthropod-specific hormones that function as the major inducing and coordinating signals responsible for the widespread changes in gene expression associated with molting and metamorphosis in insects. In Drosophila, the ring gland secretes two major ecdysteroids, α-ecdysone and 20-deoxymakisterone A, which are largely inactive (Riddiford, 1996; Gilbert et al., 1997). 20-Hydroxyecdysone (20E), which is the product of the α-ecdysone conversion in the peripheral tissues (Gilbert et al., 2002), is thought to be a major signal for the coordinated programming of gene expression patterns responsible for the complete metamorphosis of the organism from larva to fly (Ashburner, 1973, 1974). The study of 20E-controlled gene activities has been pivotal in formulating modern concepts of steroid hormone-controlled gene activities and has functioned as a paradigm for steroid hormone activity (Riddiford, 1993, 1996). Signaling by 20E in Drosophila is mediated by the ecdysone receptor complex, a heterodimer of the ecdysone receptor (EcR; NR1H1) and ultraspiracle (USP; NR2B4) proteins, both of which are members of the nuclear receptor family (Koelle et al., 1991; Yao et al., 1992, 1993; Thomas et al., 1993; Grad et al., 2001). Although USP has no known ligands, it is the Drosophila homolog of the mammalian retinoid X receptors (RXRs) (Oro et al., 1990), important members of the nuclear receptor family that bind to 9-cis retinoic acid and form heterodimeric complexes with other nuclear receptors, including the thyroid hormone receptor (TR), vitamin D3 receptor (VDR) and the all-trans retinoic acid receptor (RAR) (Mangelsdorf et al., 1995).
Because there are only two types of consensus half-sites (5′-AGGTCA-3′ and 5′-AGAACA-3′) used by essentially all nuclear receptors (Glass, 1994), target selectivity appears to rely on the geometry and spacing of the half-sites, and not just the half-site sequences. Importantly, the DNA-binding domains (DBDs) typically generate the same pattern of DNA selectivity and subunit dimerization as the full-length receptors (Ożyhar et al., 1991; Ożyhar and Pongs, 1993; Mader et al., 1993; Perlmann et al., 1993; Zechel et al., 1994b, Grad et al., 2001). There are exceptions to this rule, such as the VDR, whose DBD does not generate the same pattern of subunit dimerization as the intact receptor (Shaffer and Gewirth, 2002).
The RXR heterodimers generally target direct repeat DNA sites, which differ in their inter-half-site spacing, described by a 1–5 rule (Umesono and Evans, 1989; Umesono et al., 1991; Mangelsdorf and Evans, 1995). A set of recent crystal structures of receptor DBDs on direct repeats has revealed how RXR and other non-steroid receptors bind to their cognate sites selectively. Specifically, the structures of RXR–RXR/DR-1, RXR–RAR/DR-1, RevErb–RevErb/DR-2, VDR–VDR/DR-3 and RXR–TR/DR-4 have been determined thus far, in each case showing DBDs forming non-symmetric ‘head to tail’ interactions (Rastinejad et al., 1995, 2000; Zhao et al., 1998, 2000; Sierk et al., 2001; Shaffer and Gewirth, 2002).
In contrast, vertebrate steroid receptors, such as the glucocorticoid receptor (GR), the estrogen receptor (ER), the progesterone receptor (PR) and the mineralocorticoid receptor (MR), each form homodimers on their response elements. These response elements are inverted repeats with the same 3 bp (IR-3), causing their DBDs to form symmetric ‘head to head’ interactions (Luisi et al., 1991; Schwabe et al., 1993). Natural ecdysone response elements (EcREs) are imperfect inverted repeats of AGGTCA sequences separated by 1 bp (IR-1) (Riddihough and Pelham, 1987; Cherbas et al., 1991; Ożyhar et al., 1991; D’Avino et al., 1995; Fisk and Thummel, 1995; Horner et al., 1995; Lehmann et al., 1995, 1997; Hall and Thummel, 1998; Vögtli et al., 1998). Therefore, the requirements for DNA binding for EcR–USP are distinct from those associated with both the RXR heterodimers and the steroid receptor homodimers. Besides the EcR–USP, only the RXR–FXR heterodimer is known to target IR-1 response elements preferentially (Laffitte et al., 2000), where FXR is the nuclear receptor that binds directly to bile acids (Chawla et al., 2001; Mi et al., 2003).
Here, we describe the co-crystal structure of EcR–USP/IR-1 at 2.24 Å resolution. Importantly, RXR is the mammalian homolog of USP (Oro et al., 1990), and these two receptors can also substitute for each other in stimulating high affinity DNA binding of EcR to IR-1 in vitro (Yao et al., 1992, 1993; Thomas et al., 1993). In addition, RXR and USP have both been shown to support ecdysone-responsive trans-activation equally in transfection assays in mammalian cell lines (Vögtli et al., 1998). Moreover, USP has been shown to be capable of substituting for RXR in forming heterodimers with TR, RAR and VDR (Yao et al., 1992; Thomas et al., 1993; No et al., 1996). Therefore, we also studied the 2.60 Å crystal structure of the EcR–RXR/IR-1 complex, to see to what extent the protein–protein and protein–DNA interactions were related to those in the EcR–USP/IR-1 complex. The structures together show how the DBDs form their protein–protein interactions in a manner that is highly discriminatory for the 1 bp spacing and inverted repeat geometry of the IR-1 site. Importantly, the dimerization surfaces used to engage EcR on DNA are essentially the same in USP and RXR but distinct from those observed in other RXR heterodimers or steroid receptor homodimers. Because FXR is closely related in sequence to EcR along its DBD and also binds as an RXR heterodimer to IR-1 (Laffitte et al., 2000), our current structure of the EcR–RXR/IR-1 complex serves as a useful model for understanding how FXR and RXR are likely to cooperate in DNA binding.
Results
Overall architecture of the EcR–USP and EcR–RXR complexes
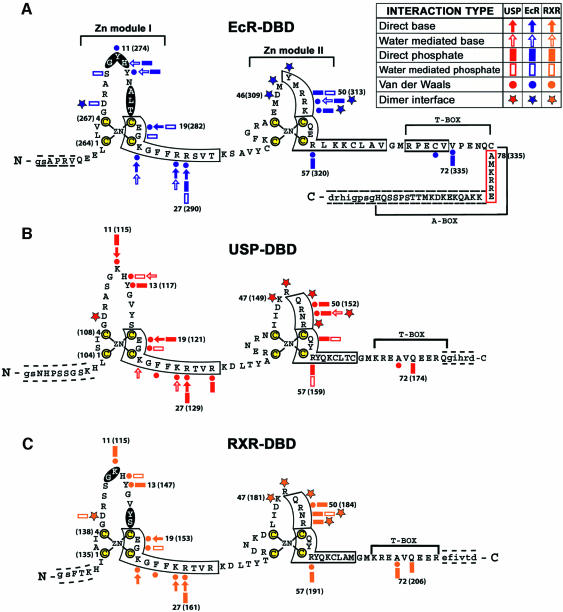
An 86 residue fragment of the Drosophila USP protein (residues 94–179) and a 109 residue fragment of Drosophila EcR (residues 256–364) were individually purified and used for co-crystallization on an IR-1 element (see Figure 1A, B and D, and Materials and methods). In the absence of EcR-DBD, USP-DBD can bind as a monomer to IR-1 (Niedziela-Majka et al., 2000). Furthermore, in the absence of USP-DBD, EcR-DBD can bind IR-1, but primarily as homodimer (Niedziela-Majka et al., 2001). However, when both DBDs are present in solution, the heterodimeric EcR–USP complex on IR-1 is formed with greater affinity and in a synergistic manner (Niedziela-Majka et al., 2001). A number of natural EcREs have been identified to date, with most of these being imperfect palindromes of PuG(G/T)TCA with a single base pair acting as the spacer between the half-sites (Riddihough and Pelham, 1987; Cherbas et al., 1991; Ożyhar et al., 1991; Antoniewski et al., 1993,1994, 1996; D’Avino et al., 1995; Lehmann et al., 1995, 1997; Vögtli et al., 1998). Because idealized IR-1 elements form higher affinity EcREs, we carried out crystallization studies using the IR-1 sequence in which both half-sites were identical 5′-AGGTCA-3′ consensus sequences, as shown in Figure 1D (Vögtli et al., 1998; Niedziela-Majka et al., 2000). The crystals of the EcR–USP-DBD heterodimer on DNA diffract to 2.24 Å resolution and contain a single complex in their asymmetric units.
Fig. 1. The protein and DNA constructs used in crystallization and their contacts. (A) Drosophila EcR-DBD (blue), (B) Drosophila USP-DBD (red) and (C) human RXRα-DBD (gold) protein sequences. The yellow colored cysteines are those used for zinc coordination. The α-helices are boxed and the β-sheets are shaded. The numbering is relative to the first conserved cysteine, with the authentic numbers appearing in parentheses (Mangelsdorf et al., 1995; Oro et al., 1990; Koelle et al., 1991). A key to the different interactions is provided within the figure. Dashed lines indicate N- and C-terminal residues not visible in the electron density maps. Cloning artifacts from the expression vector are indicated by lower case letters. The red boxed sequences in (A) indicate the A-box helix seen in the EcR–RXR complex. (D) The idealized IR-1 response element used in the EcR–USP complex, and (E) in the EcR–RXR complex. The symbols used are the same as in (A–C). Red symbols indicate contacts derived from USP, blue symbols indicate those from EcR, and gold indicates those from RXR.
The crystallization of the EcR–RXR/DNA complex relied on the same EcR protein construct (Figure 1A) and the same IR-1 DNA sequence (Figure 1E) used for the EcR–USP/IR-1 complex. The RXR-DBD used was an 80 residue DBD fragment derived from the human RXRα (residues 130–209), which contained five N-terminal residues preceding the N-terminal zinc-binding cysteine, the entire 66 residue core DBD and nine residues at the C-terminus beyond the GM amino acids (residues 65 and 66 in Figure 1C) that constitute the C-terminal boundary of the core DBD regions of all nuclear receptors. These crystals contain a single complex of EcR–RXR and IR-1 within their asymmetric units, and diffracted to 2.60 Å resolution. This structure was solved using multiple anomalous diffraction (MAD) phasing derived using the anomalous signal from the zinc ions of the DBDs. The refined coordinates were used subsequently as the search model to solve the structure of the EcR–USP/DNA complex (described above) by molecular replacement. For clarity, we rely on a common numbering scheme for EcR, USP and RXR from hereon, with residue numberings beginning with the N-terminal most cysteine coordinating the Zn-I module. Table I summarizes the crystallographic data and refinement statistics for both complexes.
Table I. Data collection and refinement statistics.
| EcR–USP | EcR–RXR |
|||
|---|---|---|---|---|
| Zn-peak | Zn-inflection | Zn-remote | ||
| Data collection | ||||
| Wavelength(Å) | 1.2834 | 1.2834 | 1.2837 | 1.2155 |
| Space group | P212121 | P212121 | ||
| Resolution(Å) | 30.0–2.24 | 30.0–2.60 | 30.0–2.60 | 30.0–2.60 |
| Mean I/σ(I) | 22.0 (5.8) | 23.2 (5.5) | 27.0 (3.7) | 27.5 (6.7) |
| Completeness (%) | 99.6 (97.8) | 94.2 (75.4) | 98.1 (97.6) | 96.6 (86.6) |
| <Redundancy> | 7.5 | 9.8 | 10.5 | 10.4 |
| Unique reflections | 15 928 | 11 884 | 12 008 | 12 403 |
| Rsyma (%) | 8.8 | 10.2 | 8.3 | 8.1 |
| Refinement statistics | ||||
| Resolution (Å) | 20.00–2.24 | 15.00–2.60 | ||
| R-factor | 23.0 | 23.2 | ||
| Rfreeb | 26.4 | 28.2 | ||
| Average B-factors/no. of atoms | ||||
| Protein (Å2) | 41.9 | 62.9 | ||
| DNA (Å2) | 46.4 | 61.4 | ||
| Zinc (Å2)/zinc atoms | 45.4/4 | 58.6/4 | ||
| Solvent (Å2)/solvent atoms | 54.6/233 | 58.8/102 | ||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.008 | 0.011 | ||
| Bond angles (°) | 1.30 | 1.60 | ||
| Dihedral angles (°) | 20.5 | 21.6 | ||
| Improper angles (°) | 1.33 | 1.36 | ||
Numbers in parentheses are for the highest resolution shell.
aRsym = Σ|Ih – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry-equivalent reflections.
bRfree was calculated using 7% of the data excluded from refinement.
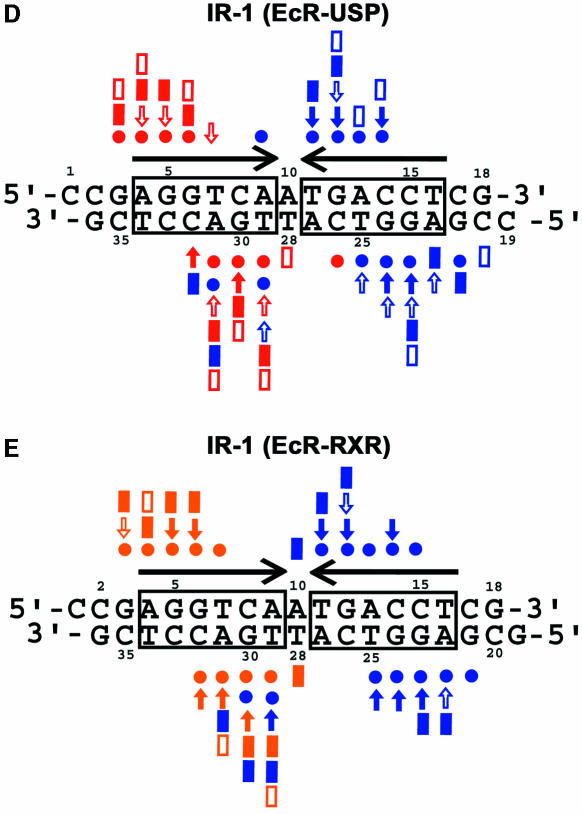
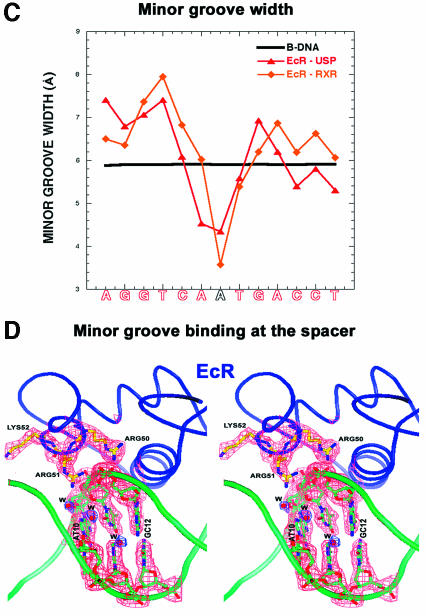
Stereo views of both structures and the protein–DNA contacts are shown in Figure 2A and B, and a superposition of the EcR–USP/IR-1 and EcR–RXR/IR-1 complexes is shown in Figure 2C. Overall, the EcR–USP and the EcR–RXR complexes show an r.m.s.d. of 0.67 Å, when calculated over the entire DNA and all their Cα atoms. A side-by-side comparison of these two complexes indicates that the subunit arrangements, the protein–protein interactions that stabilize the heterodimer, and the protein–DNA interactions that stabilize each subunit on its respective half-site are generally well preserved in these two structures (Figures 2A and B, and 3A–C). Furthermore, the EcRE DNA structure in both complexes conforms to B-DNA geometry, lacking significant distortions, except at the spacer where there is similar minor groove narrowing in both structures (see Figure 4C).
Fig. 2. Overview of the heterodimeric protein complexes on DNA. Stereo views of (A) the EcR–USP/IR-1 and (B) the EcR–RXR/IR-1 complex. Shown are side chains involved in direct base or phosphate contacts or in zinc coordination. The residue numbers and the base pair numbers are the same as those used in Figure 1. The two 5′-AGGTCA-3′ half-sites are in lilac, the spacer is in gold, and the flanking base pairs are in gray. (C) Superposition of the EcR–USP/IR-1 and EcR–RXR/IR-1 complexes. Shown are USP (red) and EcR (dark blue) DBDs bound to their IR-1 (green), as well as RXR (gold) and EcR (light blue) bound to their IR-1 (gray). The central base pair that acts as a spacer is in magenta, and the zinc ions are in green. Arrows indicate the relative orientation of the two half-sites.
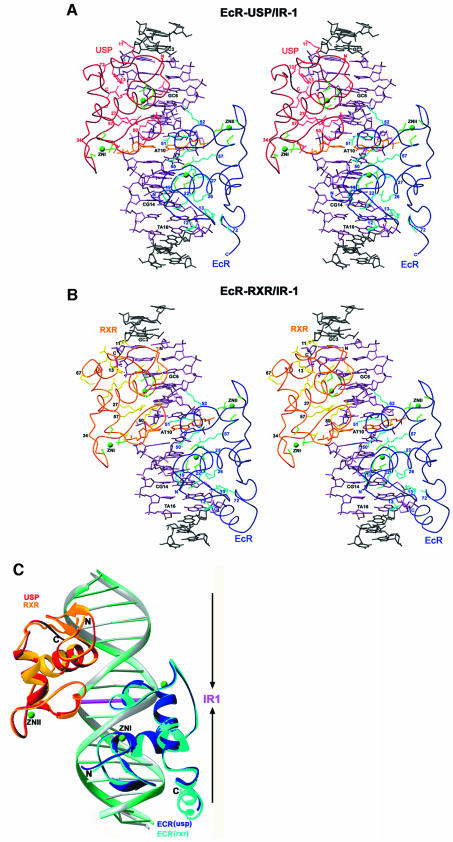
Fig. 3. Stereo views of the recognition α-helices and their contacts to DNA. Shown are interactions with the major grooves for (A) EcR (blue), (B) USP (red), (C) RXR (gold) with a composite omit electron density map (2Fo – Fc) shown for the side chains and the DNA base pairs, and (D) RXR from the RAR–RXR/DR-1 complex (yellow). In all the cases, the side chains that make direct or water-mediated contacts to the bases are shown in green, the water molecules that mediate the contacts between the protein and DNA are yellow/magenta spheres, and hydrogen bonds are shown as dotted lines. The orientation of the half-site is indicated by arrows.
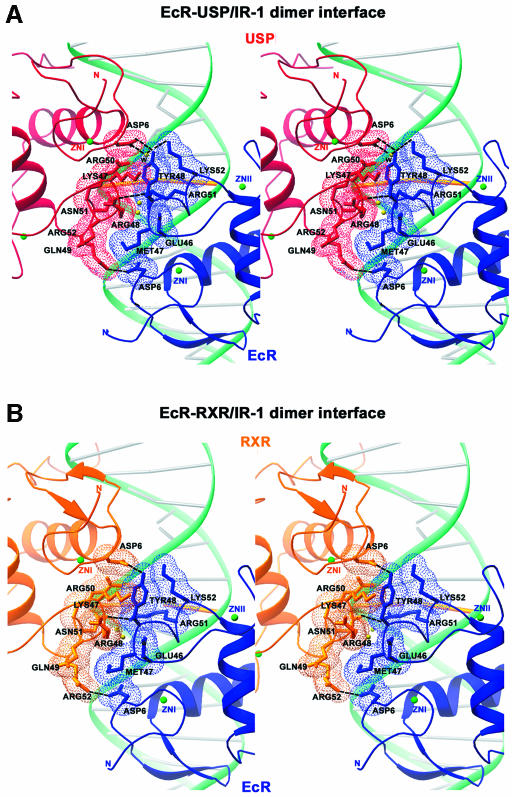
Fig. 4. The subunit interactions and role of DNA minor groove in dimerization. (A) Stereo view of the dimerization contacts between USP (red) and EcR (blue). (B) Stereo view of the dimerization contacts between RXR (gold) and EcR (blue). The side chains of the residues that make the dimer contacts are shown together with dotted surfaces indicating van der Waals complementary surfaces and dotted lines indicating hydrogen bonds. Also shown are the backbone DNA in green, the base pairs in gray and the spacer base pair in gold. The N- and C-termini of the two monomers are labeled, and the zincs are indicated by green spheres. (C) The minor groove width of the response element as a function of IR-1 sequence. The red and orange lines correspond to the EcR–USP and EcR–RXR DNAs, respectively. Both structures show a sharp minimum in the width at the spacer, the location in which interaction between the two proteins is supported. The minor groove width values were derived from the program CURVES (Lavery and Sklenar, 1988), which records the inter-phosphate distances after generating a smoothed curve through the phosphate atoms. (D) Stereo view of the electron density at the minor groove. Also shown are the contacts made by EcR to the phosphate backbone of the DNA together with the associated water molecules (W) located in the spine of hydration along the minor groove.
Because the DNA is nearly symmetric, the same Zn-I and Zn-II modules of EcR and USP/RXR are brought to the subunit interface. These regions of EcR are distinct in sequence from their counterparts in USP/RXR, and thus the dimer interface uses different residues from each of the two DBDs despite the pseudo-symmetric interactions between the EcR and USP/RXR subunits. This observation is in contrast to those made in the case of GR–GR/IR-3 and ER–ER/IR-3 crystal structures, in which DBD interactions use identical residues from each subunit’s Zn-II sites to form perfectly symmetric interactions (Luisi et al., 1991; Schwabe et al., 1993).
The A-box region in EcR, which is the C-terminal extension (CTE) of the core DBD, forms an ordered helical structure in the EcR–RXR complex, but this conformation is not seen in the EcR–USP complex where this region has disordered density instead. Similarly, the Zn-II regions in the EcR–USP/RXR complexes, which are observed here to contain short α-helices that foster dimerization and phosphate DNA binding, may assume different conformations in the uncomplexed DBDs. This was the case observed in the GR, where similar helical conformations in the Zn-II regions were seen in the GR homodimer DBD complex on IR-3 DNA, but not seen in the monomeric GR-DBD off DNA (Luisi et al., 1991; van Tilborg et al., 1995).
The protein–DNA contacts
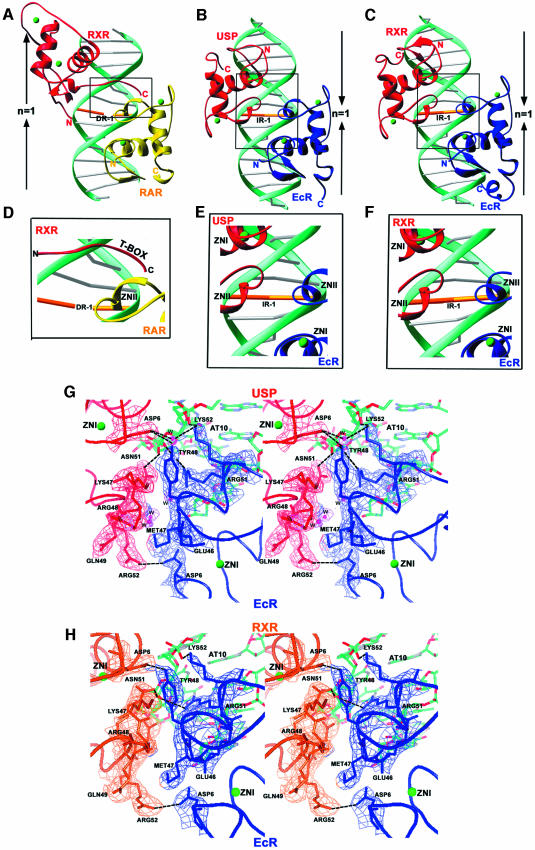
In the EcR–USP complex, a total of 3110 Å2 of water-accessible surface is buried in DNA binding, 1580 Å2 of which is derived from the binding of the EcR subunit, and 1530 Å2 from the binding of USP. In the EcR–RXR complex, a slightly larger area of 3250 Å2 is buried, in part due to the contribution of the EcR C-terminal A-box helix, which is not ordered in the EcR–USP complex. These values are in line with those associated with DR-1 response element binding by RXR–RAR, which buries 3200 Å2 (Rastinejad et al., 2000), and involves similarly sized protein subunits, DNA sequences and 1 bp spacing (Figure 5A–C).
Fig. 5. Comparison of the RXR–RAR complex on DR-1 with those of EcR–USP and EcR–RXR on IR-1. Side-by-side comparison of (A) RAR–RXR/DR-1 with (B) EcR–USP/IR-1 and (C) EcR–RXR/IR-1 complexes. The arrows indicate the relative orientation of the half-sites associated with inverted repeat and direct repeat response elements. Boxed regions are shown in close-up views to indicate the dimer interfaces in (D) RAR–RXR/DR-1, (E) EcR–USP/IR-1 and (F) EcR–RXR/IR-1. Also shown are stereo views of the composite omit (2Fo – Fc) electron density map at the dimer interface of (G) EcR–USP/IR-1 and (H) EcR–RXR/IR-1.
Interestingly, the pattern of hydrogen bonds between the recognition helices and the AGGTCA half-sites varies to some extent between EcR, USP, RXR and the previously characterized RXR subunit of the RXR–RAR/DR-1 heterodimer (Figure 3) (Rastinejad et al., 2000). However, the same set of ‘hook’ residues are used consistently in all of these cases, with their exact hydrogen-bonded partners on the DNA base pairs differing slightly (Figure 3). These hook residues, consisting of Glu19, Gly20, Lys22, Arg/Lys26 and Arg27 in EcR, USP and RXR, form their base-specific contacts. Importantly, the central AT spacer base pair is involved in phosphate contacts only, and not major groove recognition at the base pairs (Figure 1D and E). This observation is consistent with the notion that the spacer’s influence is limited to setting the correct binding site geometry in the response element (Ożyhar and Pongs, 1993).
The dimer interfaces
Dimerization of DBDs is of major functional significance for RXR, its partners and the steroid receptors, in part because dimerization allows the formation of sufficiently extended and complementary protein surfaces for high affinity and selective DNA binding. Figures 1, 4, and 5G and H show how binding of one subunit at its half-site leads to facilitated binding of the adjacent subunit via protein–protein interactions. Due to the single base pair spacer, the relative orientation between the subunits is distinct from complexes involving steroid receptors GR and ER, and as such brings into contact the Zn-I modules of both proteins in addition to their Zn-II modules. Moreover, there are clear distinctions with previously studied heterodimers of RXR-DBD on direct repeats, in which the upstream subunit relies on its Zn-II region and the downstream subunit relies on its T- and/or A-boxes for protein–protein contacts (Figure 5A–H) (Khorasanizadeh and Rastinejad, 2001; Rastinejad, 2001).
In the current structures, neither subunit relies on its T- or A-boxes for dimerization, with these regions instead being pointed towards the 3′- and 5′-flanking sequences of the IR-1 element. The involvement of USP T-box sequence in interactions with 5′-flanking sequence has been suggested previously by Grad et al. (2001) who analyzed interactions of derivatives of the natural 20E response element with a USP-DBD protein in which the C-terminal region was deleted. However, a deletion removing the A-box of EcR disrupted the DNA binding of the EcR–USP (Niedziela-Majka et al., 2000), which cannot be explained by our crystallographic results. Any additional role for the EcR A-box in dimerization cannot be ruled out, as our structure allows us to visualize only seven out of 25 A-box residues.
In the EcR–USP complex, a total of 560 Å2 of water-accessible surface area is buried between the subunits, whereas all previously studied DBD homo- and heterodimers bury a smaller surface, in the range 300–480 Å2. Figures 4A and B, and 5G and H show the detailed dimer interface in the EcR–USP and EcR–RXR complexes, respectively. These dimerization interfaces rely on hydrogen bonds between subunit residue side chains as well as significant complementary surfaces relying on van der Waals contacts. Essentially all the contacts between the subunits are conserved between the EcR–USP and EcR–RXR complexes.
Cooperation between EcR and USP/RXR
Nuclear receptor DBDs do not form homo- or heterodimers in the absence of DNA (Hard et al., 1990; Mader et al., 1993; Perlmann et al., 1993; Schwabe et al., 1993; Zechel et al., 1994a). Receptor homo- or heterodimer formation through DBDs is strictly dependent and enhanced by the cognate DNA-binding sites. In the heterodimeric complexes studied here, the structures suggest that there are three mechanisms by which the IR-1 appears cooperatively to enhance the dimerization between the EcR and the USP/RXR homologs. First, the same Zn-II regions involved in the formation of the dimer interface are also used extensively for making DNA contacts (Figures 1A–C, and 5G and H). In particular, residues Arg51 and Lys52 from EcR and residue Asn51 of USP are simultaneously involved in both dimerization and DNA binding functions. This implies that DNA binding and subunit dimerization are mutually supportive.
A second mechanism by which the DNA enhances the dimer interactions can be seen in Figures 4A and B, and 5G and H, showing how the subunit interfaces are in part embedded in the minor groove. Figure 4C and D shows that a significant minor groove distortion is associated with the spacer AT base pair, this being the convergence point of the protein–protein interactions. Importantly, these minor groove widths represent sharp departures from standard B-DNA values, and are associated with both the EcR–USP and EcR–RXR structures. In particular, there is a 4.3 Å minor groove width in the EcR–USP DNA and a <4.0 Å width in the EcR–RXR DNA. The reliance on minor groove distortions to stabilize dimer binding is reminiscent of the RXR–RAR/DR-1 and the RevErb–RevErb/DR-2 complexes on their cognate DNA targets (Rastinejad et al., 2000; Sierk et al., 2001).
A third mechanism for subunit cooperation is evident in Figure 1D and E, which shows that the EcR-DBD footprint on DNA extends well beyond its own AGGTCA site to reach over both its 3′-flanking sequences and a large portion of the USP half-site. In total, the EcR footprint in the USP complex extends over a region totaling 13 bp, and to 12 bp in the RXR complex (Figure 1D and E). This is consistent with mutational studies which identified base pairs within the USP half-site that were found to be critical not for the USP-DBD binding (as a monomer) but for effective heterodimer formation (Grad et al., 2001). This extended binding mode exhibited by EcR may contribute to the cooperativity of subunit association, by reducing the conformational flexibility at the USP site and as such pre-paying the entropic costs associated with adjacent site binding by USP. A similar mechanism based on tandem site stabilization has been suggested as the basis for the cooperation between the POU domains of Oct-1 on DNA, as well as the binding of the RXR–RAR heterodimer on DNA (Klemm and Pabo, 1996; Rastinejad et al., 2000).
Comparison with other nuclear receptor DBDs
Figure 5A–F shows how RXR-DBD and USP-DBD, derived from the RXR–RAR, EcR–USP and EcR–RXR crystal structures, form their respective dimerization contacts on DR-1 and IR-1 elements. Importantly, the C-terminal extension (T-box) of the RXR-DBD, used extensively in heterodimeric interactions with RAR on the DR-1 complex, is not used similarly in the EcR complex on IR-1 (Figure 5D–F). Moreover, the Zn-I and Zn-II regions used in both the EcR–USP and EcR–RXR complexes on IR-1 are not used for heterodimerization with RAR on DR-1. Therefore, RXR/USP are able to use distinct regions of their DBDs for forming dimerization contacts on DR-1 and IR-1. These crystallographic observations are supported by previously reported biochemical studies that rely on gel shift experiments with RXR, RAR, USP and EcR DBDs (Lee et al., 1993; Zechel et al., 1994a, b; Niedziela-Majka et al, 2000). Specifically, when the T-box residues of USP (KREAVQEERQR) are deleted from the DBD, USP is still able to form its cooperative interactions with EcR on IR-1 (Niedziela-Majka et al., 2000), as predicted from the structures shown in Figure 5B, C, E and F. However, a deletion within the T-box of mRXR (removing the residues EERQR and beyond) prevents RXR from forming a protein–protein interaction with RAR on DR-1 (Zechel et al., 1994a, b), consistent with its importance in the RXR–RAR/DR-1 complex (Figure 5A and D). A similar T-box mutation in hRXR-DBD (removing the sequences VQEERQR and beyond) also diminishes the ability of RXR to form cooperative homodimers on DR-1 (Lee et al., 1993).
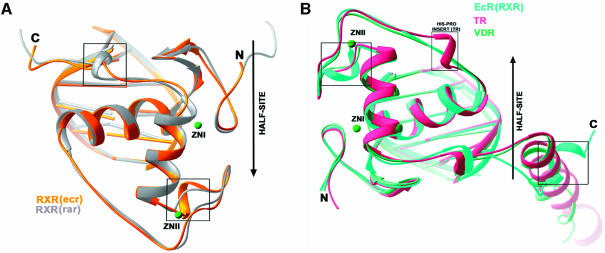
To see how the RXR-DBD subunits, derived from the EcR–RXR and RAR–RXR DBD complexes (Figure 5A–C) adjust their conformations to adapt their partners and DNA elements, we superimposed their backbone structures on half-sites in Figure 6A. The boxed areas in Figure 6A show the major differences in conformation, which correspond to two regions both of which are involved in the formation of their respective heterodimeric complexes. These are the C-terminal extension of the DBD, used for heterodimeric interactions with RAR on the DR-1 complex, and the Zn-II region used in heterodimeric interactions in the EcR complex. Interestingly, the T-box of RXR, which has also been shown to be an α-helix in the absence of DNA (Holmbeck et al., 1998), adopts a conformation not seen in the current structures or in any of the RXR–RXR and RXR–RAR complexes previously observed bound to DR-1. These differences provide further evidence that adaptable surfaces in nuclear receptor DBDs readily undergo structural rearrangements that help accommodate their association with their dimerization partners and response elements.
Fig. 6. Comparison of EcR complexes with other DBD–DNA complexes. (A) RXR on its 5′-AGGTCA-3′ site from EcR–RXR/IR-1 is superimposed onto RXR on its downstream 5′-AGGTCA-3′ half-site from RXR–RAR/DR-1. (B) EcR from the EcR–RXR/IR-1 complex is superimposed onto TR (TR–RXR/DR-4) and VDR (VDR–VDR/DR-3), the latter of which are from downstream half-sites in direct-repeats. While the overall fold of the receptors and their placement of the half-site DNA elements are very similar, the notable differences (indicated by boxes) can be attributed to the use of some protein regions for dimerization contacts.
As noted above, the EcR subunit associated with the RXR complex described here showed an α-helical structure in its A-box region. To see how the EcR-DBD compares with other steroid and non-steroid DBD structures previously reported to contain α-helical A-boxes within their C-terminal extensions, we superimposed their half-complexes in Figure 6B. This comparison used only the VDR (from the VDR homodimer) (Shaffer and Gewirth, 2002) and TR (from TR–RXR) (Rastinejad et al., 1995), as similar helical regions had not been observed in other DBD co-crystal structures. The VDR and TR subunits, which are both positioned in the downstream half-site of direct repeat complexes, use their respective C-terminal helices to form protein–protein and protein–DNA interfaces that stabilize their respective complexes. In the case of EcR, this helical region is not near the dimer interface and is instead directed 5′ to the half-site. Moreover, the helical region was visualized here only in the EcR–RXR complex and not in the EcR–USP complex.
Discussion
While the binding of a heterodimer to a symmetric response element has been observed with other transcription factors, this form of subunit association has never before been observed in the nuclear receptor family. As there is a high level of amino acid sequence conservation between RXR and USP DBDs, together with the observation that RXR and USP can substitute for each other in DNA binding with EcR (Yao et al., 1992; Vögtli et al., 1998), we also studied the crystal structure of the EcR–RXR/IR-1 complex. USP and RXR differ in only six residues, and our findings indicate that structural determinants for EcR and DNA binding are well conserved in USP and RXR (Figure 7).
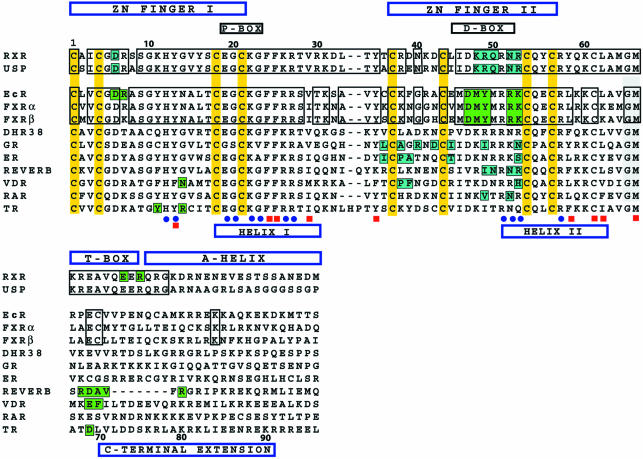
Fig. 7. Sequence comparison of nuclear receptor DBDs and the conservation of their protein–protein and protein–DNA contacts. The zinc-coordinating cysteines are in yellow, and the regions corresponding to the Zn-I, Zn-II, helix I (recognition helix), helix-II and CTEs are labeled. Blue dots indicate residues that are involved in specific DNA binding, and red squares are the residues that form the hydrophobic core that stabilizes the protein. The residues involved in dimer contacts are shaded in blue (RXR and USP) and green (EcR). Importantly, all of the DNA-binding residues are fully conserved between RXR and USP, and between EcR, and FXRα and FXRβ.
Like EcR, FXR also forms heterodimers with RXR that bind to IR-1 sites (Forman et al., 1995; Laffitte et al., 2000). Our phylogenetic analysis, using members of the superfamily and focused strictly on the portion of receptors including the DBDs and 30 residues at the CTE, indicate that EcR is most closely related to FXR. Within the core DBD of 66 residues, which contains all of the determinants of EcR binding to DNA and USP, there are only eight non-conserved residues between EcR and FXR (Figure 7). None of the non-conserved residues in FXR fall at sites used for response element binding or subunit dimerization by EcR in its IR-1 complexes. Moreover, Figure 7 shows that the critical interacting residues in EcR are equally shared with both hFXRα and mFXRβ, but not with other receptors. Therefore, it is likely that the current structure of EcR–RXR closely mimics the mode of subunit interactions used by the FXR–RXR/IR-1 complex.
In addition, the Drosophila receptor DHR38 is also known to form heterodimers with USP (Henrich et al., 1994; Sutherland et al., 1995; Baker et al., 2003). However, DHR38 appears much more closely related to the human NGFI-B than to either EcR or FXR. The sequence alignment in Figure 7 shows that the determinants of dimerization with USP used by EcR are not at all conserved in DHR38. These findings would suggest that USP–DHR38 either use other types of DNA response elements, or form an altogether different set of protein–protein interactions than described here in the EcR complexes. Figure 7 also shows that while the known determinants of DBD dimerization in GR, ER, RevErb, VDR, RAR and TR involve to some extent the Zn-II regions, the residues used differ in sequence and position.
The nuclear receptor USP is required in multiple tissues of the organism at various stages of metamorphosis. All these factors together implicate USP as a regulator of multiple pathways in Drosophila. It is also known that mutations in USP result in complex embryonic and adult phenotypes, making it a vital factor involved in Drosophila development. This pleiotropy could be based on the interaction of USP with other factors involved in development, reminiscent of the role of mammalian RXR in regulating multiple pathways. It is interesting to note that while these two receptor homologs do not share the same ligand-binding properties, they nevertheless each act as a common dimerization partner for multiple other nuclear receptors in flies or mammals.
Because EcR relies on both an inverted repeat target and heterodimerization, this receptor has characteristics typical of both steroid and non-steroid receptors, and may be a clear evolutionary link between vertebrate steroid and non-steroid receptors. Apart from the degenerate palindromic elements, the EcR dimer also activates transcription in vivo through direct repeats (D’Avino et al., 1995). Since it is evident that EcR is functional only in the presence of USP, this implies that the two subunits are capable of dimerizing on a variety of response elements. At a physiological level, this versatility could be the mechanism through which they regulate multiple pathways. At a molecular level, this further supports the notion that flexible and adjustable dimerization surfaces in DBDs are responsible for their cooperation on various response elements.
Materials and methods
Crystallization and data collection
The DBDs of EcR, USP and RXR were expressed in Escherichia coli as fusions with GST, using pGEX-2T (USP, EcR) and pGEX-4T (RXR) vectors (Pharmacia) (Rastinejad et al., 1995; Niedziela-Majka et al., 1998). The proteins and the two oligonucleotide strands corresponding to IR-1 were purified as described previously (Zhao et al., 2000; Niedziela-Majka et al., 2001). Samples of EcR–RXR co-crystallization contained DNA and the two proteins at concentrations of 0.5 mM each. Crystals were grown using hanging drops at 9°C by addition of 2 µl of the complex to an equal volume of a solution containing 12–14% PEG 3350, 50 mM MgSO4, 50 mM Li2SO4, 5 mM dithiothreitol (DTT), 10 mM MgCl2, 0.1 M MES pH 5.6. Crystals were streaked through a cryo-solvent solution containing the reservoir solution supplemented with 20% glycerol, and flash frozen in liquid nitrogen. Diffraction data were collected on these frozen crystals under a stream of liquid nitrogen at 100 K, at the Argonne National Laboratory at beamline SBC-CAT 19ID. The DBDs contain two zinc ions each, which were used to obtain MAD data at three different wavelengths (1.2834, 1.2837 and 1.2155 Å) corresponding to the zinc peak, inflection and a remote wavelength. The crystals diffracted to a resolution of 2.60 Å and crystallized in the orthorhombic space group (P212121, a = 55.62, b = 60.25, c = 115.00).
The EcR–USP complex was crystallized with the same IR-1 response element. Crystals were grown using hanging drops at 9°C by addition of 2 µl of the complex (0.5 mM) to an equal volume of a solution containing 4–8% PEG 3350, 100 mM NaCl, 5 mM DTT, 10 mM MgCl2 and 0.1 M MES pH 5.6. Crystals were streaked through a cryo-solvent solution containing the reservoir solution containing 20% glycerol, and flash frozen in liquid nitrogen. X-ray diffraction data were collected on these frozen crystals under a stream of liquid nitrogen, at the BNL-X4A beamline. The crystals diffracted to 2.24 Å and crystallized in orthorhombic space group (P212121, a = 50.37, b = 59.98, c = 113.57). All the data collected were integrated and scaled using HKL2000 (Otwinowski and Minor, 1997).
Structure determination
Phases were calculated from the data obtained from the EcR–RXR crystals using SOLVE (Terwilliger and Berendzen, 1999) and resulted in a figure of merit of 0.61 (for data to 3.0 Å resolution). SOLVE identified the positions of all four zinc ions in the complex. RESOLVE and DM (CCP4) were used for solvent flattening and density modification, improving the quality of the map. These phases were used to calculate an initial 3.0 Å electron density map, which was readily interpretable in terms of the DNA duplex and the backbone chains of both the proteins. A partial model consisting of the core DBDs of both the proteins (with side chains for the conserved residues) and most of the DNA duplex was built using the program O (Jones et al., 1991). The model was partially refined in CNS (Brünger et al., 1998) by rigid body refinement, simulated annealing and energy minimization. The refined model provided improved maps which made it possible to distinguish between the two proteins. All the side chains in the core DBDs were gradually added and the model was refined to R-values of 38 and 32% (Rfree and R-factor). This model was used as the search model for molecular replacement to obtain the EcR–USP structure. The entire complex including the zinc atoms, the two subunits and the DNA was used as the search model for molecular replacement. Molecular replacement was carried out using MOLREP (CCP4) and gave a clear solution with a correlation coefficient of 0.40 and an R-factor of 45%.
Both models were refined in CNS with successive rounds of rigid-body refinement, simulated annealing and energy minimization (Brünger et al., 1998). Initially tight restraints were imposed for Watson–Crick DNA base pairing and the planarity of the atoms in the bases (Parkinson et al., 1996). In addition, all the residues in the 5′-subunit in the EcR–RXR/IR-1 model were changed to correspond to those in USP. Improved maps helped visualize residues beyond the core DBD in all the proteins. The tight restraints subsequently were released, followed by multiple rounds of manual rebuilding and refinement in CNS including individual B-factor refinement. Solvent molecules were added to the model using Arp waters (Cowtan, 1994), and the model refined to R-values as shown in Table I.
Acknowledgments
Acknowledgements
We thank Christine Wright and Li-Zhi Mi for helpful discussions, Anita Niedziela-Majka and Iwona Grad for sharing reagents, and Craig Ogata and Randy Abramowitz for assistance with data collection at BNL-X4A. This study was supported by NIH grant GM55217 (F.R.), a State Committee for Scientific Research grant (Poland) 3 P04B 009 23 (A.O.), and a fellowship from the Rett Syndrome Research Foundation (J.M.H.).
References
- Antoniewski C., Laval,M. and Lepesant,J.A. (1993) Structural features critical to the activity of an ecdysone receptor binding site. Insect Biochem. Mol. Biol., 23, 105–114. [DOI] [PubMed] [Google Scholar]
- Antoniewski C., Laval,M., Dahan,A. and Lepesant,J.A. (1994) The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol. Cell. Biol., 14, 4465–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C., Mugat,B., Delbac,F. and Lepesant,J.A. (1996) Direct repeats bind the EcR/USP receptor and mediate ecdysteroid responses in Drosophila melanogaster. Mol. Cell. Biol., 16, 2977–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. (1973) Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. I. Dependence upon ecdysone concentration. Dev. Biol., 35, 47–61. [DOI] [PubMed] [Google Scholar]
- Ashburner M. (1974) Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev. Biol., 39, 141–157. [DOI] [PubMed] [Google Scholar]
- Baker K.D. et al. (2003) The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell, 113, 731–742. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chawla A., Repa,J.J., Evans,R.M. and Mangelsdorf,D.J. (2001) Nuclear receptors and lipid physiology: opening the X-files. Science, 294, 1866–1870. [DOI] [PubMed] [Google Scholar]
- Cherbas L., Lee,K. and Cherbas,P. (1991) Identification of ecdysone response elements by analysis of the Drosophila Eip28/29 gene. Genes Dev., 5, 120–131. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- D’Avino P.P., Crispi,S., Cherbas,L., Cherbas,P. and Furia,M. (1995) The moulting hormone ecdysone is able to recognize target elements composed of direct repeats. Mol. Cell. Endocrinol., 113, 1–9. [DOI] [PubMed] [Google Scholar]
- Fisk G.J. and Thummel,C.S. (1995) Isolation, regulation and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc. Natl Acad. Sci. USA, 92, 10604–10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman B.M. et al. (1995) Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell, 81, 687–693. [DOI] [PubMed] [Google Scholar]
- Gilbert L.I., Song,Q. and Rybczynski,R. (1997) Control of ecdysteroidogenesis: activation and inhibition of prothoracic gland activity. Invert. Neurosci., 3, 205–216. [DOI] [PubMed] [Google Scholar]
- Gilbert L.I., Rybczynski,R. and Warren,J.T. (2002) Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol., 47, 883–916. [DOI] [PubMed] [Google Scholar]
- Glass C.K. (1994) Differential recognition of target genes by nuclear receptor monomers, dimers and heterodimers. Endocr. Rev., 15, 391–407. [DOI] [PubMed] [Google Scholar]
- Grad I., Niedziela-Majka,A., Kochman,M. and Ożyhar,A. (2001) Analysis of Usp DNA binding domain targeting reveals critical determinants of the ecdysone receptor complex interaction with the response element. Eur. J. Biochem., 268, 3751–3758. [DOI] [PubMed] [Google Scholar]
- Hall B.L. and Thummel,K.E. (1998) The RXR homolog ultraspiracle is an essential component of the Drosophila ecdysone receptor. Development, 125, 4709–4717. [DOI] [PubMed] [Google Scholar]
- Hard T. et al. (1990) Solution structure of the glucocorticoid receptor DNA-binding domain. Science, 249, 157–160. [DOI] [PubMed] [Google Scholar]
- Henrich V.C., Szekely,A.A., Kim,S.J., Brown,N.E., Antoniewski,C., Hayden,M.A., Lepesant,J.A. and Gilbert,L.I. (1994) Expression and function of the ultraspiracle (usp) gene during development of Drosophila melanogaster. Dev. Biol., 165, 38–52. [DOI] [PubMed] [Google Scholar]
- Holmbeck S.M., Dyson,H.J. and Wright,P.E. (1998) High-resolution solution structure of the retinoid X receptor DNA-binding domain. J. Mol. Biol., 284, 533–539. [DOI] [PubMed] [Google Scholar]
- Horner M.A., Chen,T. and Thummel,C.S. (1995) Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev. Biol., 168, 490–502. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S. and Rastinejad,F. (2001) Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci., 26, 384–390. [DOI] [PubMed] [Google Scholar]
- Klemm J.D. and Pabo,C.O. (1996) Oct-1 POU domain–DNA interactions: cooperative binding of isolated subdomains and effects of covalent linkage. Genes Dev., 10, 27–36. [DOI] [PubMed] [Google Scholar]
- Koelle M.R., Talbot,W.S., Segraves,W.A., Bender,M.T., Cherbas,P. and Hogness,D.S. (1991) The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell, 67, 59–77. [DOI] [PubMed] [Google Scholar]
- Laffitte B.A., Kast,H.R., Nguyen,C.M., Zavacki,A.M., Moore,D.D. and Edwards,P.A. (2000) Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J. Biol. Chem., 275, 10638–10647. [DOI] [PubMed] [Google Scholar]
- Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Kliewer,S.A., Provencal,J., Wright,P.E. and Evans,R.M. (1993) Structure of the retinoid X receptor α DNA binding domain: a helix required for homodimeric DNA binding. Science, 260, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Lehmann M. and Korge,G. (1995) Ecdysone regulation of the Drosophila Sgs-4 gene is mediated by the synergistic action of ecdysone receptor and SEBP 3. EMBO J., 14, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Wattler,F. and Korge,G. (1997) Two new regulatory elements controlling the Drosophila Sgs-3 gene are potential ecdysone receptor and fork head binding sites. Mech. Dev., 62, 15–27. [DOI] [PubMed] [Google Scholar]
- Luisi B.F., Xu,W.X., Otwinowski,Z., Freedman,L.P., Yamamoto,K.R. and Sigler,P.B. (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature, 352, 497–505. [DOI] [PubMed] [Google Scholar]
- Mader S., Chen,J.Y., Chen,Z., White,J., Chambon,P. and Gronemeyer,H. (1993) The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J., 12, 5029–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J. and Evans,R.M. (1995) The RXR heterodimers and orphan receptors. Cell, 83, 841–850. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L.Z., Devarakonda,S., Harp,J.M., Han,Q., Pellicciari,R., Willson,T.M., Khorasanizadeh,S. and Rastinejad,F. (2003) Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell, 11, 1093–1100. [DOI] [PubMed] [Google Scholar]
- Niedziela-Majka A., Rymarczyk,G., Kochman,M. and Ożyhar,A. (1998) Pure, bacterially expressed DNA-binding domains of the funtional ecdysteroid receptor capable of interacting synergestically with the hsp 27 20-hydroxyecdysone response element. Protein Expr. Purif., 14, 208–220. [DOI] [PubMed] [Google Scholar]
- Niedziela-Majka A., Kochman,M. and Ożyhar,A. (2000) Polarity of the ecdysone receptor complex interaction with the palindromic response element from the hsp27 gene promoter. Eur. J. Biochem., 267, 507–519. [DOI] [PubMed] [Google Scholar]
- No D., Yao,T.P. and Evans,R.M. (1996) Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA, 93, 3346–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oro A.E., McKeown,M. and Evans,R.M. (1990) Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature, 347, 298–301. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Ożyhar A. and Pongs,O. (1993) Mutational analysis of the interaction between ecdysteroid receptor and its response element. J. Steroid Biochem. Mol. Biol., 46, 135–145. [DOI] [PubMed] [Google Scholar]
- Ożyhar A., Strangmann-Diekmann,M., Kiltz,H.H. and Pongs,O. (1991) Characterization of a specific ecdysteroid receptor–DNA complex reveals common properties for invertebrate and vertebrate hormone-receptor/DNA interactions. Eur. J. Biochem., 200, 329–335. [DOI] [PubMed] [Google Scholar]
- Parkinson G., Wilson,C., Gunasekera,A., Ebright,Y.W., Ebright,R.E. and Berman,H.M. (1996) Structure of the CAP–DNA complex at 2.5 angstroms resolution: a complete picture of the protein–DNA interface: atomic hydrogen bond in sequence-specific protein DNA recognition. J. Mol. Biol., 260, 395–408. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Rangarajan,P.N., Umesono,K. and Evans,R.M. (1993) Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev., 7, 1411–1422. [DOI] [PubMed] [Google Scholar]
- Rastinejad F. (2001) Retinoid X receptor and its partners in the nuclear receptor family. Curr. Opin. Struct. Biol., 11, 33–38. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Perlmann,T., Evans,R.M. and Sigler,P.B. (1995) Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature, 375, 203–211. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Wagner,T., Zhao,Q. and Khorasanizadeh,S. (2000) Structure of the RXR–RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J., 19, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford L.M. (1993) Hormone receptors and the regulation of insect metamorphosis. Receptor, 3, 203–209. [PubMed] [Google Scholar]
- Riddiford L.M. (1996) Juvenile hormone: the status of its ‘status quo’ action. Arch. Insect Biochem. Physiol., 32, 271–286. [DOI] [PubMed] [Google Scholar]
- Riddihough G. and Pelham,H.R.B. (1987) An ecdysone response element in the Drosophila hsp27 promoter. EMBO J., 6, 3729–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J.W., Chapman,L., Finch,J.T. and Rhodes,D. (1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell, 75, 567–578. [DOI] [PubMed] [Google Scholar]
- Shaffer P.L. and Gewirth,D.T. (2002) Structural basis of VDR–DNA interactions on direct repeat response elements. EMBO J., 21, 2242–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierk M.L., Zhao,Q. and Rastinejad,F. (2001) DNA deformability as a recognition feature in the reverb response element. Biochemistry, 40, 12833–12843. [DOI] [PubMed] [Google Scholar]
- Sutherland J.D., Kozlova,T., Tzertzinis,G. and Kafatos,F.C. (1995) Drosophila hormone receptor 38: a second partner for Drosophila USP suggests an unexpected role for nuclear receptors of the nerve growth factor-induced protein B type. Proc. Natl Acad. Sci. USA, 92, 7966–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H.E., Stunnenberg,H.G. and Stewart,A.F. (1993) Heterodimerization of the Drosophila ecdysone receptor with retinoid X receptor and ultraspiracle. Nature, 362, 471–475. [DOI] [PubMed] [Google Scholar]
- Umesono K. and Evans,R.M. (1989) Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell, 57, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami,K.K., Thompson,C.C. and Evans,R.M. (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid and vitamin D3 receptors. Cell, 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilborg M.A., Bonvin,A.M., Hard,K., Davis,A.L., Maler,B., Boelens,R., Yamamoto,K.R. and Kaptein,R. (1995) Structure refinement of the glucocorticoid receptor–DNA binding domain from NMR data by relaxation matrix calculations. J. Mol. Biol., 247, 689–700. [DOI] [PubMed] [Google Scholar]
- Vögtli M., Elke,E., Imhof,M.O. and Lezzi,M. (1998) High level transactivation by the ecdysone receptor complex at the core recognition motif. Nucleic Acids Res., 26, 2407–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.P., Segraves,W.A., Oro,A.E., McKeown,M. and Evans,R.M. (1992) Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell, 71, 63–72. [DOI] [PubMed] [Google Scholar]
- Yao T.P., Forman,B.M., Jiang,Z., Cherbas,L., Chen,J.D., McKeown,M., Cherbas,P. and Evans,R.M. (1993) Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature, 366, 476–497. [DOI] [PubMed] [Google Scholar]
- Zechel C., Shen,X.Q., Chambon,P. and Gronemeyer,H. (1994a) Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J., 13, 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel C., Shen,X.Q., Chen,J.Y., Chen,Z.P., Chambon,P. and Gronemeyer,H. (1994b) The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J., 13, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Khorasanizadeh,S., Miyoshi,Y., Lazar,M.A. and Rastinejad,F. (1998) Structural elements of an orphan nuclear receptor–DNA complex. Mol. Cell, 1, 849–861. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Chasse,S.A., Devarakonda,S., Sierk,M.L., Ahvazi,B. and Rastinejad,F. (2000) Structural basis of RXR–DNA interactions. J. Mol. Biol., 296, 509–520. [DOI] [PubMed] [Google Scholar]