Abstract
Prevailing notions of cerebral vascularization imply that blood vessels sprout passively into the brain parenchyma from pial vascular plexuses to meet metabolic needs of growing neuronal populations. Endothelial cells, building blocks of blood vessels, are thought to be homogeneous in the brain with respect to their origins, gene expression patterns and developmental mechanisms. These current notions that cerebral angiogenesis is regulated by local environmental signals contrast with current models of cell-autonomous regulation of neuronal development. Here we demonstrate that telencephalic angiogenesis in mice progresses in an orderly, ventral-to-dorsal gradient regulated by compartment-specific homeobox transcription factors. Our data offer new perspectives on intrinsic regulation of angiogenesis in the embryonic telencephalon, call for a revision of the current models of telencephalic angiogenesis and support novel roles for endothelial cells in brain development.
Brain development is supported by concomitant development of brain vasculature. However, blood vessels or endothelial cells are generally considered to lack molecular diversity in the embryonic or mature CNS1,2. Currently, it is believed that once the perineural plexuses on the pial surface (pial vessels) surrounding the neural tube are produced, cerebral vasculature develops further by passive vessel sprouting into the brain parenchyma in response to increased tissue mass and oxygen demand1,2. Although classical studies identified a ventral to dorsal temporal developmental angiogenesis gradient in the telencephalon3, the sequence of angiogenesis was considered to merely shadow neurogenesis and neuronal maturation. Similarly, although shared mechanisms regulating vascular and neuronal development have been recognized4, the canonical principles of neuronal development are not seen as applicable to CNS angiogenesis.
During early embryonic development, the neural tube, acting as a vessel patterning nexus, directs the formation of the perineural or pial vascular plexuses that encompass it5. Although regional identities emerging within the telencephalon are guided by region-specific transcription factors6–8, regional patterning signals have not been considered to instruct vascular development. It seemed unlikely to us that CNS angiogenesis would evolve to support the highly regulated CNS development without intrinsic regulation. If intrinsic CNS angiogenesis programs are revealed, valuable insights could emerge into mechanisms of CNS development and repair because both processes require concomitant remodeling of blood vessels and neuronal elements.
We show that angiogenesis in the embryonic telencephalon progresses along a ventral-to-dorsal gradient controlled by region-specific homeobox transcription factors, which also regulate development of telencephalic neuroepithelial domains and postmitotic neurons. Thus, telencephalic angiogenesis appears to follow the same fundamental regionalization principles that guide development of telencephalic compartments. By advancing this view, we anticipate a place for telencephalic angiogenesis within the province of segmental biology.
RESULTS
Telencephalic angiogenesis gradients
An unresolved issue concerning telencephalic angiogenesis is the origin of the telencephalic vasculature. Current models suggest that pial plexuses surrounding the neural tube sprout radial collaterals into the parenchyma to establish the vascular network2,9,10 in response to increasing metabolic demands of the growing neural tissue. We labeled blood vessels with biotinylated isolectin B4 in paraffin sections (Fig. 1a–c) or whole mounts (Fig. 1d–i). The pial vessels circumscribed the entire telencephalon as early as embryonic day (E) 9 (Fig. 1a). On the other hand, a spatially distinctive population of periventricular vessels was restricted to the ventral telencephalon, deep within the incipient marginal zones halfway between the pial and ventricular surfaces (Fig. 1a). The E9 dorsal telencephalon did not contain these periventricular vessels (Fig. 1a).
Figure 1.
Angiogenesis gradients in the embryonic telencephalon. (a–c) Isolectin-B4+ vessels in the E9 ventral telencephalon (arrows, a). The periventricular vessels extend into the dorsal telencephalon by E10 and E11 (b,c, arrows) producing a ventrolateral-to-dorsomedial gradient. Pial vessels (arrowheads) fully encircle the telencephalon by E9. (d) Isolectin-B4+ pial vessels covering a E10 dorsal telencephalon, cut open (at arrowheads) and mounted with the ventricular surface up. The basal vessel appears on the telencephalic floor (thick arrow). The dorsal telencephalon is unlabeled (thin arrow). (e–g) Isolectin B4–labeled periventricular vessels in an E11 whole mount appear in a single 20-µm focal plane (e) from which thin vessels emerge at right angles toward the pial surface (f) and contact the pial vessels (arrow in g), which appear in a different focal plane (g). (h,i) Isolectin B4–labeled prominent basal vessel (white star) in E11 ventral telencephalon whole mount unfurls into a periventricular vessel lattice. The broken line (i) indicates the advancing vessel front: the medial telencephalon has no periventricular vessels (white arrow). (j–l) Diagramatic representation of periventricular vessel development. The periventricular vessel network (red) originates from the basal vessel (red asterisk in j) in the telencephalon (peacock green) and grows in ventral-to-dorsal and lateral-to-medial directions. Dotted circle in j is expanded in k (purple, telencephalon; blue cross-hatching, basal forebrain) for a two-dimensional view of the periventricular network (yellow dotted circle) and the basal vessel (red asterisk in k). The boxed area in k represents i, with the medial aspects of the telencephalon devoid of periventricular vessels. (l) Ventral-to-dorsal and lateral-to-medial gradients of periventricular angiogenesis (broken red line with directional arrow). Blue dotted line, pial vessels. Scale bars: a, 100 µm (applies to a–d); e, 50 µm (applies to e–g); h, 100 µm; i, 50 µm.
By E10, a rich periventricular plexus had formed in the ventral telencephalon with branches encroaching upon the lateral pallium (Fig. 1b), and the periventricular network advanced into the dorsal telencephalon during E10 and E11 (Fig. 1b,c). By E11, the periventricular vessels formed a lattice pattern in the dorsal telencephalon, approaching its medial wall, and a ventrolateral to dorsomedial angiogenesis gradient was established (Fig. 1c,h–l). Other blood vessel markers such as laminin or platelet–endothelial cell adhesion molecule-1 (PECAM-1) confirmed that isolectin B4 labeled vessels reliably (Supplementary Fig. 1a–j online). The angiogenesis gradient was apparent at all rostrocaudal levels of the telencephalon (Supplementary Fig. 2a–d online). The periventricular plexus was restricted to an ∼20-µm-thick plane parallel to the pial surface and sandwiched between the ventricular and marginal zones in the E11 dorsal telencephalon (Fig. 1e). Narrow branches from the periventricular network joined the pial plexuses as fine, tapering vessels (Fig. 1e–g).
We found that the periventricular vessels in the ventral telencephalon originated from a prominent basal vessel located deep on the floor of the telencephalic vesicle in the basal ganglia primordium (Fig. 1d). The basal vessel unfurled into a plexus growing in a ventral-to-dorsal and lateral-to-medial direction, straddling the lateral ventricles (Fig. 1h–l). The basal vessel probably arises from pharyngeal arch arteries apparent in the cervical region by E9 (refs. 2,11). Based on earlier reports11, we suggest that the pial vessels develop into venous sinuses and the periventricular vessels into arterial networks. The tapering vessels joining the periventricular and pial vessels (Fig. 1e–g and Supplementary Fig. 3 online) may represent the earliest arterial-venous communication.
Ventral origin of dorsal periventricular vessels
In E10 telencephalon explants cultured for 2 h, periventricular vessels were restricted to the ventral telencephalon (Fig. 2a), as in vivo (Fig. 1b). When the E10 explants were cultured for 24 h, periventricular vessels extended into the dorsal telencephalon (Fig. 2b), in a manner resembling the in vivo E11 pattern. Pial vessels surrounded both the dorsal and ventral divisions in both sets of cultures (Fig. 2a,b), also recapitulating the in vivo pattern (Fig. 1b). Cell viability or explant health was not compromised (Supplementary Fig. 4 online).
Figure 2.
Dorsal periventricular vessels originate from the ventral telencephalon. (a) Isolectin-B4+ pial and periventricular vessels in the ventral telencephalon (arrow) of an E10 telencephalon explant cultured for 2 h. (b) After 24 h in culture, periventricular vessels enter the dorsal telencephalon (arrow). Arrowheads, pial vessels; LV, lateral ventricle. (c) Periventricular (arrow) and pial (arrowhead) vessels in E11 dorsal explant cultured for 2 h after removing the ventral telencephalon. Curved white arrow, ventral limit of the dorsal explant. (d) PAX6 immunohistochemistry confirms dorsal molecular identity of the explant. (e) Merged c and d. (f) There are pial vessels (arrowheads) but virtually no periventricular vessels in an E10 dorsal explant cultured for 24 h after removing the ventral telencephalon. Arrows indicate the few periventricular vessels. (g) PAX6 labeling confirms the explant’s dorsal molecular identity. (h) Merged f and g, showing the vessel-poor PAX6+ regions. White asterisks: cranial mesenchyme, other non-CNS tissues. (i) Periventricular vessel density (mean ± s.d.) in E10 dorsal explants cultured without the ventral explants (n = 30) is less than that in E11 dorsal-only (n = 25) or E10 whole explants (dorsal + ventral, n = 30). Data normalized to E10 whole explants. *P < 0.0001; E10 whole explants versus E10 dorsal explants. (j) An E10 whole explant cultured for 24 h after removing pial membranes (arrowhead) by microdissection. Isolectin B4 labeled periventricular vessels in the dorsal telencephalon (arrows) even in regions without intact pial membranes. (k) 6-hydroxydopamine-induced pial vessel degeneration in an E10 whole explant. Periventricular vessels appear in ventral and dorsal telencephalon. (l,m) Intact pial membrane in control (l) and disrupted pial membranes in 6-hydroxydopamine-treated (m) explants. Scale bars: a, 100 µm (applies to a–h,j,k); l, 25 µm (applies also to m).
Having established that the ventral-to-dorsal gradient of periventricular vessel development could be recapitulated in the explants, we examined whether dorsal pial vessels produced periventricular vessels in the immediate vicinity. We transected E10 or E11 telencephalon at its dorsal-ventral boundary (between the lateral ganglionic eminence and cortex) and cultured only the dorsal part with its pial membranes and pial vessels intact (Fig. 2c–h). If the dorsal pial vessels seeded dorsal periventricular vessels, as currently thought, then periventricular vessels should form in the dorsal explant in the absence of ventral telencephalon. However, the E10 dorsal explant cultured for 24 h, until E11, (Fig. 2f–i) had an 85–90% reduction in periventricular vessels (Fig. 2i). PAX6 immunohistochemistry, a marker of dorsal neuroepithelial cells12, confirmed that the dorsal telencephalon had maintained its molecular identity (Fig. 2d,g). These data suggest that dorsal periventricular vessels may arise from the ventral periventricular network at early embryonic stages.
To verify ventral origin of dorsal periventricular vessels, we removed dorsal pial membranes from E10 telencephalon explants and cultured the explants for 24 h. The periventricular vessels formed in such explants (Fig. 2j), suggesting that dorsal pial membranes were not necessary for dorsal periventricular vessel development. As meningeal cells rapidly degenerate upon exposure to 6-hydroxydopamine13, we exposed E10 telencephalon explants to 6-hydroxydopamine for 24 h. In these explants, although pial membranes were disrupted, periventricular vessels were seen in the dorsal and ventral telencephalon (Fig. 2k–m). Thus, dorsal pial vessels do not seed dorsal periventricular vessels.
Ventral to dorsal migration of periventricular vessels
How are dorsal periventricular vessels formed? As continuity with ventral telencephalon is essential for the development of the dorsal periventricular vessels (Fig. 2), vessels may migrate from the ventral to dorsal telencephalon. To test this possibility, we performed heterochronic explantations by culturing E11 ventral explants in contact with E10 dorsal explants (Fig. 3) to see whether the up-gradient or ‘more mature’ E11 ventral explant restored vascular pattern in the E10 dorsal explant, which contained pial but not periventricular vessels at explantation. The ventral explants were labeled with Qdot nanocrystals to verify ventral origin of the explants. Vascularization of the E10 dorsal explants that were in contact with the E11 ventral explants occurred (Fig. 3a,c and Supplementary Figure 5 online). E10 dorsal explants cultured alone did not have periventricular vessels (Fig. 2f), so we concluded that periventricular vessels in the dorsal explant originated (possibly migrated) from the ventral explants.
Figure 3.
Ventral-to-dorsal migration of endothelial cells. (a) When an E10 dorsal explant is cultured with an E11 ventral explant, isolectin-B4+ periventricular vessels (arrow) appear in the dorsal explant. (b) Pre-explantation Qdot labeling (asterisks) verifies the explant’s ventral origins. (c) Merged a and b. (d–g) When an E10 wild-type dorsal explant was cultured with an E11 Tie2-GFP ventral explant, isolectin-B4+ periventricular vessels (d) appeared in the E10 dorsal explant (arrow). (e) The E10 dorsal periventricular vessels contained GFP+ ventral endothelial cells (arrows). (f) PAX6 labeling confirms the explant’s dorsal identity. (g) Merged d–f. (h,i) E10 dorsal Tie2-GFP explant cultured B500 µm from E11 Tie2-GFP ventral explant for 24 h (h, bright field). Sections of the dorsal explant show GFP+ endothelial cells (arrows, i) in the endogenous pial vessels only. (j) Periventricular vessel density (mean ± s.d.) in E10 dorsal explants cultured with E11 ventral explants for 24 h approaches that of E10 whole explants (dorsal + ventral; n = 30), whereas E10 dorsal explants cultured without the ventral explant show significantly reduced vessel density (n = 30). Vessel densities were normalized to E10 whole explants. *P < 0.0001, E10 dorsal explants versus E10 dorsal explants cultured with E11 ventral explants. (k) Merged e and f showing GFP+ and PAX6+ endothelial cells in the dorsal explant. (l,m) GFP+ endothelial cell (l) from boxed area in k shows labeling with PAX6 (m). (n) Merged l and m. Scale bars: a, 100 µm (applies to a–g,i,k); n, 10 µm (applies to l–n).
Next, we cultured ventral explants from E11 Tie2-GFP mice (whose endothelial cells express GFP) in contact with E10 wild-type dorsal explants. After 24 h, the dorsal explants showed GFP+ endothelial cells (Fig. 3d,e,g and Supplementary Fig. 6 and Supplementary Fig. 7 online) confirming ventral origin of dorsal endothelial cells. It should be noted that the GFP in Figure 3e labels endothelial cells only, whereas the isolectin B4 in Figure 3d labels endothelial cells and other vessel components. Therefore, Figure 3d has more label than Figure 3e. Notably, vessel densities (that is, isolectin-B4+ profiles) in E10 wild-type dorsal explants cultured with E11 Tie2-GFP ventral explants equaled that in an intact E10 wild-type explant cultured for 24 h (Fig. 3j), suggesting that the heterochronic transplantation recapitulated the dorsal vessel formation seen in the intact explants. We also established that endothelial cells from an E10 ventral explant migrated into E10 dorsal explants, as in the intact telencephalon (data not shown).
When dorsal and ventral explants from E10 Tie2-GFP mice were cultured at a distance of 500 µm for 24 h (Fig. 3h), GFP+ endothelial cells did not enter the dorsal explant: GFP+ endothelial cells in the dorsal explant were restricted to pial plexuses (Fig. 3i). Thus, contact between the ventral and dorsal explants was required for endothelial cell migration. Although some VEGF isoforms can regulate vessel development through long-distance diffusible mechanisms4, such mechanisms may not be sufficient for endothelial cell migration across the ventral-dorsal telencephalic boundary. The exact site of contact was not critical; a ventral explant in contact with the medial or lateral edge or at any point along the pial or ventricular surfaces of the dorsal explant initiated periventricular vascularization of the dorsal explant.
Endothelial cells express homeobox transcription factors
The ventral-to-dorsal angiogenesis gradient corresponds to the transverse neurogenesis gradient with respect to direction, but it is in advance of the latter temporally14–17. The direction of endothelial cell migration resembles tangential migration of GABA neurons, but the endothelial cell migration begins earlier18,19. Neurogenesis and GABA neuron migration are regulated by region-specific homeobox transcription factors, which bestow cell-autonomous regional identities7,20,21. These observations raised the possibility that telencephalic endothelial cells also may express region-specific homeobox transcription factors.
We considered the three ventral genes Nkx2-1 (hereinafter referred to as Nkx2.1), Dlx1, Dlx2 and one dorsal gene Pax6 (Fig. 4). Nkx2.1 is expressed selectively in cells of the ventral telencephalon (preoptic area, medial ganglionic eminence, septum and amygdala)12. Dlx1 and Dlx2 are selectively expressed in ventral progenitors (basal ganglia and septum)7,22,23. Pax6 is expressed in dorsal progenitors24–27. We found that NKX2.1 and DLX2 were expressed in ventral but not dorsal periventricular endothelial cells (Fig. 4a–c,f–h). PAX6 was expressed in the dorsal but not ventral periventricular endothelial cells (Fig. 4k–m). The pial endothelial cells were negative for NKX2.1, DLX2 and PAX6.
Figure 4.
Endothelial cells express homeobox transcription factors. (a) NKX2.1 immunohistochemistry (red) in an E11 Tie2-GFP telencephalon labels medial ganglionic eminence (MGE) cells and highlights MGE borders (arrows). Lateral ganglionic eminence (LGE) shows some labeling. (b) A GFP+ endothelial cell in an E11 Tie2-GFP ventral telencephalon expresses NKX2.1 (c) No NKX2.1 labeling in the E11 Nkx2.1−/− telencephalon, confirming antibody specificity. (d) Immunoblot of NKX2.1 in nuclear and cytosolic fractions (NF and CF, respectively) of cultured E13 mouse brain endothelial cells. (e) Cultured E13 mouse brain endothelial cell labeled with isolectin B4, NKX2.1 and DAPI, showing nuclear (white) and cytoplasmic (yellow) NKX2.1. (f) DLX2 immunohistochemistry (red) in E11 Tie2-GFP telencephalon labels lateral ganglionic eminence (LGE) cells. (g) GFP+ endothelial cell in E11 Tie2-GFP ventral telencephalon expresses DLX2. (h) No DLX2 in E11 Dlx1−/−Dlx2−/− telencephalon, confirming antibody specificity. (i) Immunoblot of DLX2 in nuclear and cytosolic fractions (NF and CF, respectively) of cultured E13 mouse brain endothelial cells. (j) Cultured E13 mouse brain endothelial cell labeled with isolectin B4, DLX2 and DAPI, showing nuclear (white) and cytoplasmic (yellow) DLX2 (k) PAX6 immunohistochemistry (red) in E11 Tie2-GFP mouse telencephalon labels dorsal (arrow) telencephalon. (l) GFP+ endothelial cells in E11 Tie2-GFP dorsal telencephalon express PAX6. (m) Decreased PAX6 staining in E11 SeyDey mutant telencephalon. (n) Immunoblot of PAX6 in nuclear and cytosolic fractions (NF and CF, respectively) of cultured E13 mouse brain endothelial cells. (o) Cultured E13 mouse brain endothelial cells labeled with isolectin B4, PAX6 and DAPI, showing PAX6 in the nucleus (white) and cytoplasm (yellow). (p) Ventral (Dlx1/2, medium blue region; Nkx2.1, overlap of medium blue and dark blue regions) and dorsal (Pax6, red) homeobox gene expression defines telencephalic periventricular vessel domains. (q) FACS analysis of GFP+ endothelial cells from Tie2-GFP brains. PE, phycoerythrin. (r) Relative expression (mean ± s.d.) of Nkx2.1, Dlx1, Dlx2 and Pax6 mRNA in GFP+ endothelial cells from E17 (EC1) or E13 (EC2) brains. Data normalized to E13. Scale bars: c, 100 µm (applies to a,c,f,h,k,m); e, 10 µm (applies to b,e,g,j,l,o).
In E13 mouse brain primary endothelial cell cultures, NKX2.1, DLX2 and PAX6 were expressed in cytosolic and nuclear fractions (Fig. 4d,i,n). Immunohistochemistry confirmed the nuclear and cytoplasmic expression (Fig. 4e,j,o). Western blot confirmed NKX2.1, DLX2 and PAX6 expression in human umbilical vein endothelial cells (Supplementary Fig. 8 online). Labeling with an antibody to the endothelial cell nuclear marker FLI1 confirmed nuclear localization of NKX2.1, DLX2 and PAX6 in endothelial cells of the E11 telencephalon (Supplementary Fig. 9 online). Finally, GFP+ endothelial cells from E13 or E17 Tie2-GFP mouse brains (Fig. 4q) showed Nkx2.1, Dlx1, Dlx2 and Pax6 mRNA expression (Fig. 4r).
Neuroepithelial cells in the telencephalic ventricular pseudostratified epithelium are tethered to one another and restricted to a given compartment throughout development28. However, GABA neurons of the basal forebrain are not restricted to the compartment of origin but migrate across compartmental boundaries18,19, despite expressing compartment-specific homeobox genes29. Which of these two models applies to telencephalic endothelial cells? We found that ventral endothelial cells that normally express Dlx1 and Dlx2 (Dlx1/2) and Nkx2.1 and normally do not express Pax6 turned on Pax6 upon entering the dorsal telencephalon explant (Fig. 3k–n). As endothelial cells divide extensively during migration, cell divisions may facilitate dynamic suppression and expression of genes in the daughter cells under environmental influence.
Ventral homeobox genes regulate telencephalic angiogenesis
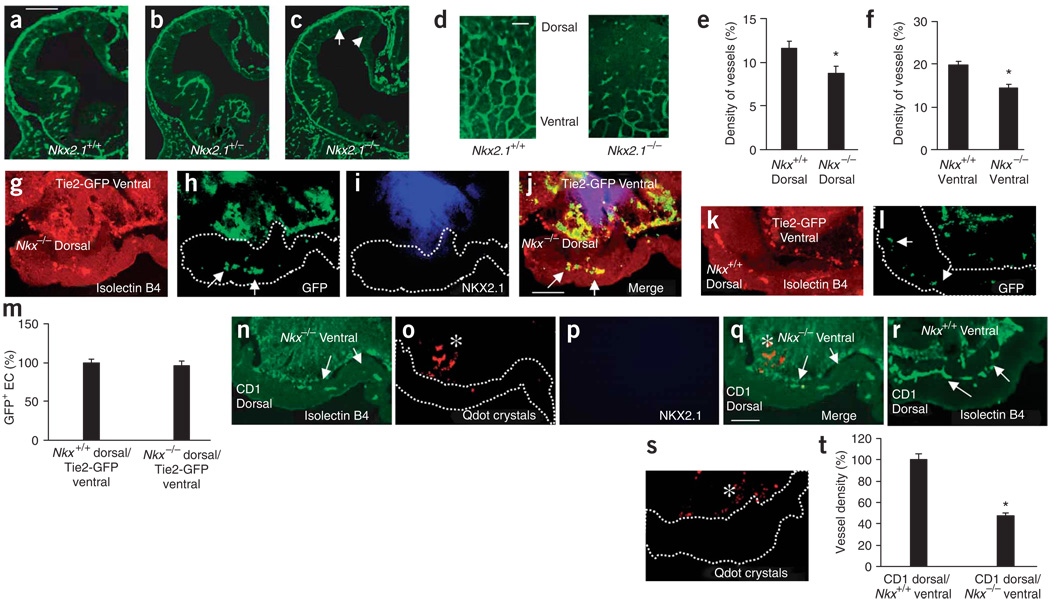
Because ventral endothelial cells expressed Nkx2.1 and Dlx1/2, and because these genes regulate neuroepithelial cell proliferation and GABA neuron migration12,30,31, we analyzed angiogenesis in the telencephalons of mice with mutations in these genes (Fig. 5 and Fig. 6). In E11 Nkx2.1−/− mice, periventricular vessel density was significantly decreased in the dorsal and ventral telencephalon (Fig. 5a–c,e,f and Supplementary Fig. 2e–h). The pial vessels appeared unaffected. In E11 Nkx2.1−/− brain whole mounts, fewer vessels crossed into the dorsal telencephalon (Fig. 5d). The reduction in Nkx2.1−/− dorsal telencephalon vessel density persisted even at E13 and E17 (Supplementary Fig. 10a–f online).
Figure 5.
Nkx2.1 and Dlx1/2 regulate telencephalic angiogenesis. (a–c) Isolectin-B4+ blood vessels in E11 Nkx2.1+/+ (a), Nkx2.1+/− (b) and Nkx2.1−/− (c) telencephalon, showing fewer periventricular vessels in Nkx2.1−/−, especially in the dorsal and medial regions (arrows). (d) Impaired angiogenesis in E11 Nkx2.1 −/− dorsal telencephalon whole mount. (e,f) Periventricular vessel density (mean ± s.d.) was significantly reduced in the dorsal (e, n = 15, *P = 0.0066) and ventral (f, n = 15, *P = 0.0025) E11 Nkx2.1−/− telencephalon. (g–j) Cocultured E10 Nkx2.1−/− dorsal and E11 Tie2-GFP ventral explants show isolectin-B4+ vessels in the Nkx2.1−/− dorsal explant (g) that contain GFP+ endothelial cells (h, arrows), confirming the vessels’ ventral origin. (i) NKX2.1 labeling confirms E11 explant’s ventral identity. (j) Merged g–i. (k,l) Cocultured E10 Nkx2.1+/+ dorsal and E11 Tie2-GFP ventral explants show isolectin-B4+ vessels (k) in the Nkx2.1+/+ dorsal explant, which contain GFP+ endothelial cells (l, arrows), confirming the vessels’ ventral origins. (m) Comparable vascularization of Nkx2.1−/− and Nkx2.1+/+ dorsal explants (n = 8 each) cultured with Tie2-GFP ventral explants (n = 8). (n) Cocultured CD1 wild-type E10 dorsal and E11 Nkx2.1−/− ventral explants show isolectin-B4+ vessels in the dorsal explant (white arrows). (o) Pre-explantation Qdot labeling (asterisk) identifying the Nkx2.1−/− ventral explant. (p) No NKX2.1 labeling in the Nkx2.1−/− ventral explant. (q) Merged n–p. (r) Cocultured CD1 wild-type E10 dorsal and E11 Nkx2.1+/+ ventral explants show isolectin-B4+ vessels in the dorsal explant (arrows). (s) Pre-explantation Qdot labeling (white asterisk) identifying Nkx2.1+/+ ventral explant. (t) Periventricular vessel density (mean ± s.d.) in the E10 CD1 wild-type dorsal explant cultured with E11 Nkx2.1−/− ventral explant (n = 7) decreased by 53% (*P < 0.0001) compared with culturing with Nkx2.1+/+ ventral explants (n = 7). Scale bars: a, 100 µm (applies to a–c,g–l); d, 50 µm; q, 100 µm (applies to n–s).
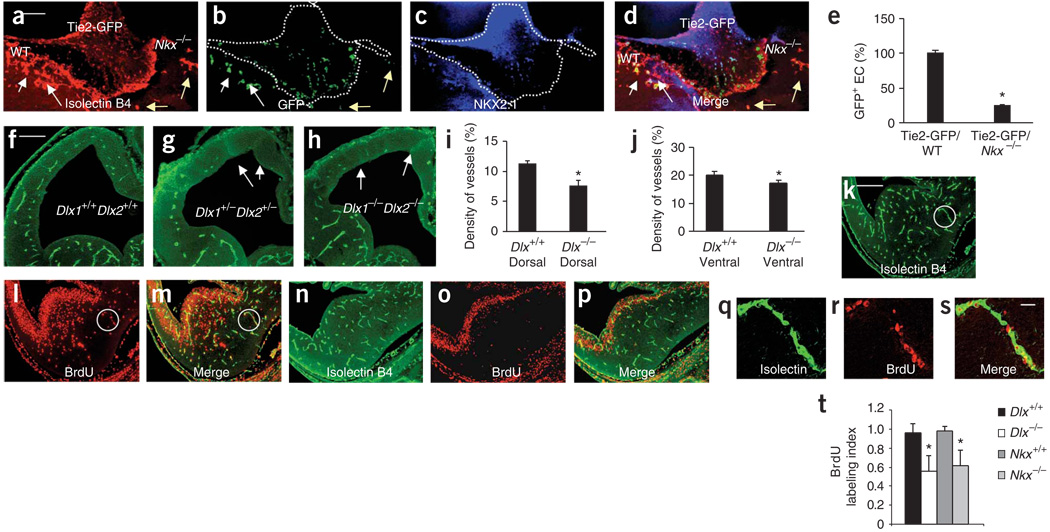
Figure 6.
Ventral homeobox genes regulate endothelial cell migration and proliferation. (a) E11 Tie2-GFP ventral explant was cultured with wild-type CD1 and Nkx2.1−/− ventral explants. Isolectin-B4+ vessels entered the Tie2-GFP and the CD1 explants (white arrows) in greater numbers than the Nkx2.1−/− explant (yellow arrows). (b) GFP+ endothelial cells (arrows) in the isolectin-B4+ vessels. (c) NKX2.1 labeled the Tie2-GFP and CD1 but not Nkx2.1−/− explant. (d) Merged a–c. (e) Decrease of ∼75% (n = 7, *P < 0.0001) in GFP+ cell density in the Nkx2.1−/− explant versus CD1 explant. (f–h) Isolectin-B4+ vessels in E11 Dlx1+/+Dlx2+/+ (f), Dlx1+/−Dlx2+/− (g) and Dlx1−/−Dlx2−/− (h) telencephalon. Periventricular vessel numerical density (mean ± s.d.) is significantly decreased in the dorsal (i, n = 15, *P = 0.0034) and ventral (j, n = 15, *P = 0.049) telencephalon of Dlx1−/−Dlx2−/− mice (Dlx−/−) compared to Dlx1+/+Dlx2+/+ littermates (Dlx+/+). (k–m) Isolectin B4 (k) and BrdU (l) labeling in E12 Nkx2.1+/+ telencephalon, and isolectin B4 and BrdU double labeling in the dorsal and ventral telencephalon (m). (n–p) Isolectin B4 (n) and BrdU (o) labeling in E12 Nkx2.1−/− telencephalon. Isolectin B4 and BrdU double labeling (p) is decreased compared to that in the Nkx2.1+/+ telencephalon. (q–s) Isolectin B4–labeled vessels (q), BrdU labeled nuclei (r) and isolectin B4–BrdU double labeling (s) from circled regions in k–m. (t) Significant reduction in ventral BrdU labeling index (mean ± s.d.; *P < 0.0001) in the Dlx1−/−Dlx2−/− (Dlx−/−; n = 14) and Nkx2.1−/− (n = 14) versus wild-type littermates. Scale bars: a, 100 µm (applies to a–d); f, 100 µm (applies to f–h); k, 100 µm (applies to k-p); s, 12 µm (applies to q–s).
Prevailing concepts suggest that when increases in tissue mass outpace vascular supply the resulting anoxia induces new vessel growth10, offering a non-cell-autonomous explanation for angiogenesis. However, a pimonidazole hydrochloride assay for cellular oxygenation did not show differences between E10 or E11 Nkx2.1−/− and Nkx2.1+/+ littermates (Supplementary Fig. 11 online). Moreover, the expression of hypoxia-inducible factor-1 Hif1a or adult hemoglobin Hba mRNA in endothelial cells from Nkx2.1−/− and Nkx2.1+/+ littermates was not significantly different (Supplementary Fig. 12 online). Thus, differences in oxygenation could not have caused changes in angiogenesis in the Nkx2.1−/− telencephalon. Cortical thickness, one measure of tissue mass, also was not affected in E11 Nkx2.1−/− mutants (data not shown).
To examine whether the loss of Nkx2.1 was sufficient to impair telencephalic angiogenesis, we performed heterochronic explantations using ventral or dorsal telencephalon from E10 or E11 Nkx2.1−/−, Tie2-GFP and wild-type CD1 mice. If loss of Nkx2.1 from the endothelial cells is sufficient to impair migration, Nkx2.1−/− endothelial cell migration into Nkx2.1+ explants should be significantly reduced. On the other hand, if the loss of Nkx2.1 from the environment is sufficient to impair endothelial cell migration, Nkx2.1+/+ endothelial cell migration into Nkx2.1−/− explants should be significantly decreased. If the loss of Nkx2.1 from both the endothelial cells and the environment is essential for the phenotype, then endothelial cell migration should be decreased under both conditions.
When E11 Tie2-GFP ventral explants were cultured with E10 Nkx2.1+/+ dorsal explants (we used E10 explants because endogenous endothelial cells would not have entered the dorsal explant at this stage), GFP+ endothelial cells migrated into the dorsal explant. When E11 Tie2-GFP ventral explants were cultured with E10 Nkx2.1−/− dorsal explants, GFP+ endothelial cells migrated to the dorsal explant (Fig. 5g–j) implying that ventral blood vessels could enter the dorsal telencephalon in the absence of Nkx2.1 in the dorsal explant. This finding was as expected, as Nkx2.1 is not normally expressed in the dorsal telencephalon. However, when Nkx2.1−/− E11 ventral explants were cultured with E10 CD1 (wild type) dorsal explants, there was a 50% decrease in dorsal vessel density (Fig. 5n–q,t) suggesting that Nkx2.1 expression in the ventral telencephalon was required for the normal migration of vessels. Thus, apparently normal neuronal development in the CD1 or Nkx2.1+/+ dorsal telencephalon was not sufficient to promote Nkx2.1−/− ventral endothelial cell migration, implying that Nkx2.1 may act in a cell-autonomous fashion.
We cultured E11 Tie2-GFP ventral explants with E10 Nkx2.1−/−and E10 wild-type CD1 ventral explants so that GFP+ ventral endothelial cells could ‘choose’ between Nkx2.1 or wild-type CD1 ventral explants. GFP+ cells migrated into the wild-type CD1 but not Nkx2.1−/− ventral explants (Fig. 6a–e). Thus, Nkx2.1 seems to be required in the ventral telencephalon to promote endothelial cell migration.
The vessel density was also decreased significantly in the Dlx1−/− Dlx2−/− dorsal and ventral telencephalon (Fig. 6f–j, Supplementary Fig. 2i–l). We examined whether the decrease in vessel density in the Nkx2.1−/− and Dlx1−/− Dlx2−/− mouse telencephalon was due to decreased proliferation of endothelial cells. Endothelial cell bromodeoxyuridine (BrdU) labeling index was significantly decreased in the ventral telencephalon of E12 Nkx2.1−/− and Dlx1 Dlx2−/− mice (Fig. 6k–t). Thus, Dlx1/2 and Nkx2.1 are also required for ventral periventricular endothelial cell proliferation.
Impaired angiogenesis in the dorsal homeobox mutant Pax6Sey-Dey
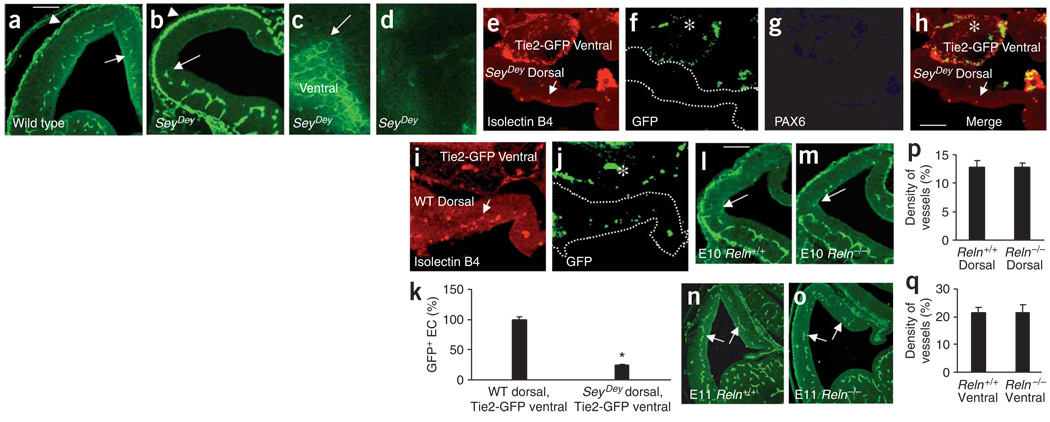
We analyzed heterozygous Pax6Sey-Dey mice (hereinafter referred to as SeyDey), which have a deletion of the Pax6 chromosomal region (Fig. 7). The phenotype of the heterozygote suggests that a minimum amount of Pax6 activity is necessary for normal development26,27. We observed a marked decrease in dorsal periventricular vessels in E11 SeyDey mutant heterozygous mice compared to wild type littermates (Fig. 7a,b). The pial vasculature appeared normal (Fig. 7a,b). The ventral periventricular vascular lattice was intact and extended up to the cortico-striatal boundary (Fig. 7c) but degraded and disappeared beyond this boundary in the dorsal telencephalon (Fig. 7c,d). The impairment was apparent at all rostrocaudal levels (Supplementary Fig. 2m–p) and persisted at E13 and E17 (Supplementary Fig. 10g–l). Thus, Pax6 is required for normal development of periventricular vessels in the dorsal telencephalon.
Figure 7.
Telencephalic angiogenesis in SeyDey mutant mice. (a,b) Isolectin-B4+ vessels in E11 wild-type (a) and SeyDey mutant (b) littermates. Periventricular vessel gradient ‘stops’ at the ventral-dorsal boundary in SeyDey mutant sections (white arrow, b) and whole mounts (c, white arrow). (d) Dorsal telencephalon whole mount from a SeyDey mutant, without periventricular vessels. (e) E10 SeyDey mutant dorsal explant cultured with E11 Tie2-GFP ventral explant shows virtually no isolectin B4 labeling (arrow, the few labeled vessels). (f) GFP+ endothelial cells in the Tie2-GFP ventral explant (asterisk) only. (g) PAX6 is absent in the SeyDey mutant dorsal explant. (h) Merged e–g. (i) E11 Tie2-GFP ventral explant cultured with E10 SeyDey wild-type dorsal explant shows isolectin-B4+ vessels (arrows). (j) GFP+ endothelial cells in the ventral (asterisk) and dorsal explant. (k) GFP+ cell numbers (mean ± s.d.; percentage of total GFP+ cells) were significantly reduced (*P = 0.0001) in the cultured SeyDey mutant versus SeyDey wild-type dorsal explant (n = 8 each). (l–o) Isolectin-B4+ periventricular blood vessel distribution is unaffected in the reeler mutant at E10 (l,m) and E11 (n,o). The periventricular vessels approach the ventral-dorsal boundary at E10 (l,m; arrows) and extend to medial edge of dorsal telencephalon (n,o; arrows) at E11. (p,q) Isolectin-B4+ vessel density (mean ± s.d.) in the dorsal (p, n = 18) and ventral (q, n = 18) telencephalon is not significantly different between E11 reeler mutants and wild-type littermates. Data normalized to wild-type values. Scale bars, 100 µm: a applies to a–d; h applies to e–j; l applies to l–o.
When E11 Tie2-GFP ventral explants were cultured with E10 SeyDey mutant dorsal explants, there was a 70–80% reduction in the numbers of endothelial cells entering the dorsal explant (Fig. 7e–h), suggesting that normal Pax6 expression in the dorsal telencephalon is required for formation of dorsal periventricular vessels. The expression of Hif1a or Hba mRNA in SeyDey or wild-type endothelial cells was not significantly different (Supplementary Fig. 12), suggesting that changes in tissue oxygenation did not contribute to impairment of angiogenesis.
Telencephalic angiogenesis in the reeler mouse
As mentioned earlier, the classical view of angiogenesis suggests that neuronal maturation drives CNS angiogenesis. This view is especially strongly held for cerebral cortical angiogenesis3,9. We sought direct evidence for independent regulation of vascular and neuronal development by examining periventricular vessel developmental gradients in the reeler mouse.
The Reln gene is expressed in both dorsal and ventral telencephalon and affects neuronal positioning in many brain regions32,33. Periventricular vessel developmental gradient was unaffected in the E10 or E11 Reln−/− telencephalon (Fig. 7l–o) and the density of isolectin B4–labeled vessels appeared normal in the dorsal or ventral telencephalon (Fig. 7p,q). Thus, angiogenesis may proceed according to an agenda independent of the neuronal developmental agenda, at least early in embryonic development.
Cell-autonomous roles of Nkx2.1 and Pax6
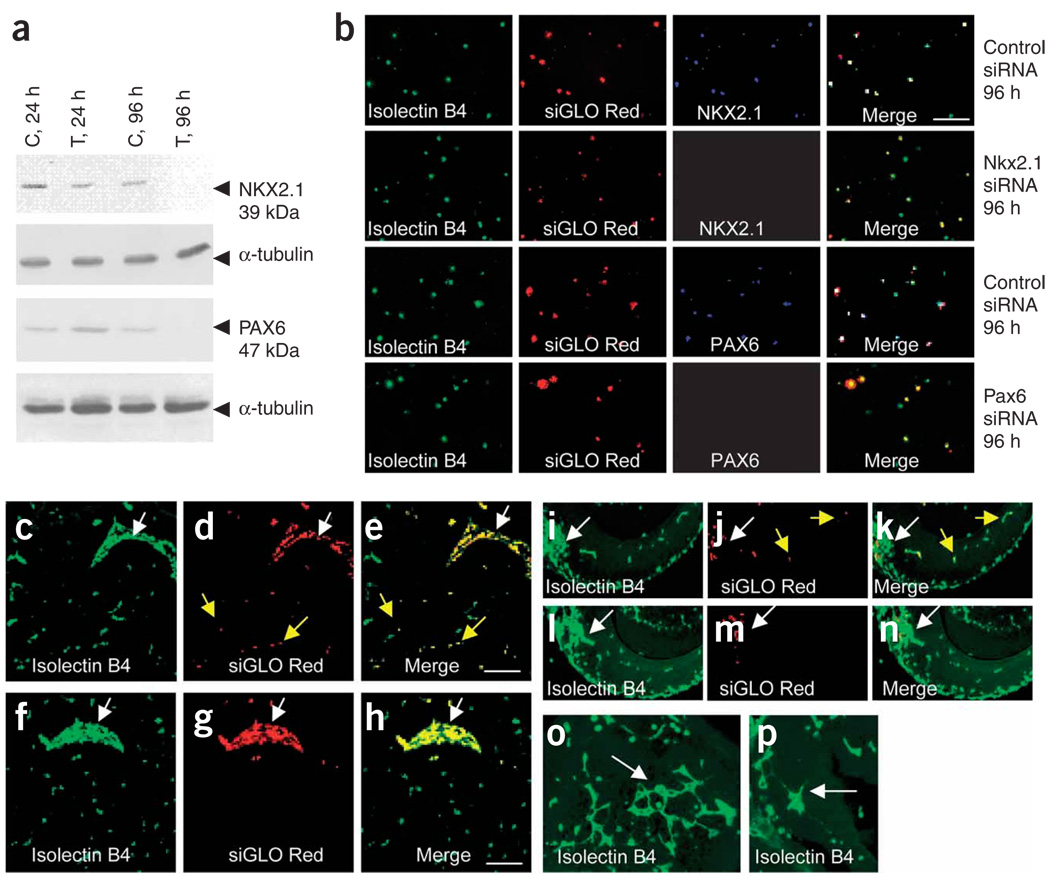
The data described above suggest but do not explicitly prove a cellautonomous role for ventral or dorsal transcription factors in telencephalic angiogenesis. To address this issue, we used small interfering RNA (siRNA) technology. We cotransfected E13 mouse brain–derived endothelial cells with siRNA constructs for one ventral (Nkx2.1) and one dorsal (Pax6) transcription factor. Nontargeting siRNA constructs served as controls. Western blot confirmed virtually complete knockdown of NKX2.1 and PAX6 within 96 h (Fig. 8a). These data were verified by immunocytochemistry (Fig. 8b). Next, we transplanted Nkx2.1 siRNA–transfected or control-transfected ventral endothelial cells into E11 wild-type CD1 ventral telencephalon explants. Control-transfected endothelial cells migrated from the site of transplantation into the host ventral explant in 24 h (Fig. 8c–e). On the other hand, virtually all the Nkx2.1 siRNA transfected endothelial cells were restricted to the site of transplantation (Fig. 8f–h, Supplementary Fig. 13a online). When we transplanted Pax6 siRNA–transfected or control–transfected dorsal endothelial cells into E11 CD1 dorsal telencephalon explants, control endothelial cells migrated from the site of transplantation into the dorsal explant (Fig. 8i–k). However, virtually all Pax6 siRNA–transfected dorsal endothelial cells were restricted to the site of transplantation (Fig. 8l–n, Supplementary Fig. 13b). The BrdU labeling index was also significantly decreased in endothelial cells treated with either Nkx2.1 siRNA or Pax6 siRNA (Supplementary Fig. 13c) suggesting that knockdown of these transcription factors affected cell proliferation, as in the intact brains of the mutants (Fig. 6k–t). In further experiments, we transplanted endothelial cells derived from E13 Nkx2.1−/−, SeyDey or wild-type littermates into E11 CD1 ventral telencephalon explants. Nkx2.1−/− or SeyDey endothelial cells showed less migration than wild-type endothelial cells (Supplementary Fig. 14 online), supporting the siRNA data. Collectively, these data offer proof that Nkx2.1 and Pax6 act in a cell-autonomous manner to regulate telencephalic angiogenesis.
Figure 8.
Cell autonomous regulation of endothelial cell migration by Nkx2.1 and Pax6. (a) Virtually complete knockdown of NKX2.1 and PAX6 in endothelial cells after 96 h of NKX2.1 siRNA or PAX6 siRNA transfection (T) versus control (C) transfections. (b) Immunohistochemistry for NKX2.1 and PAX6 confirms the knockdown. A siGLO Red transfection indicator ascertains transfection and identifies transfected cells. Transfected endothelial cells are isolectin-B4+ and siGLO-Red+. However, Nkx2.1 siRNA–transfected cells are NKX2.1−, whereas control siRNA–transfected cells are NKX2.1+. PAX6 expression is downregulated in Pax6 siRNA–transfected cells but not in control siRNA–transfected cells. (c,d) Control siRNA transfected endothelial cells transplanted into E11 CD1 wild-type ventral telencephalon and double-labeled with isolectin B4 (c) and siGLO Red (d) migrated from the transplantation site (arrow in c) into the explant (yellow arrows). (e) Merged c and d. (f,g) Virtually all of the Nkx2.1 siRNA–transfected endothelial cells, identified by isolectin B4 (f) and siGLO Red (g) labeling, were restricted to the site of transplantation (arrow in f). (h) Merged f and g. (i–k) Control siRNA–transfected endothelial cells transplanted into E11 CD1 wild-type dorsal telencephalon explants and double labeled with isolectin B4 (i) and siGLO Red (j) migrated from the transplantation site (white arrows) into the explant (i–k, yellow arrows). (l–m) Virtually all the Pax6 siRNA–transfected endothelial cells, identified by isolectin B4 (l) and siGLO Red (m) labeling, were restricted to the site of transplantation. (o,p) Control siRNA–transfected endothelial cells in ventral (o) and dorsal (p) explants form intricate vessel patterns (white arrows). Such patterns were never seen after Nkx2.1 or Pax6 siRNA–transfected endothelial cell transplantation. Scale bars, 100 µm: e applies to c–e,i–n; h applies to f–h,o–p.
What may be the signaling mechanisms perturbed by the loss of Nkx2.1 and Pax6 in the endothelial cells that may contribute to the impaired proliferation and migration? To address this issue, we examined expression of vascular endothelial growth factor (isoform A; Vegfa) and its receptors Kdr (also known as Flk1) and Flt1, as well as brain derived neurotrophic factor (Bdnf), in endothelial cells isolated from Nkx2.1−/− and SeyDey telencephalons. There was a significant (P < 0.05) decease in Vegfa, Kdr and Bdnf mRNA (but not Flt1 mRNA) in the Nkx2.1−/− and SeyDey endothelial cells (Supplementary Fig. 15 online), suggesting that the loss of the transcription factors altered the expression of molecules critical for endothelial cell development.
DISCUSSION
Our data present new insights into the regulation of angiogenesis in the embryonic mouse telencephalon. The idea that telencephalic angiogenesis is a passive process merely responding to increases in tissue mass and metabolism and merely shadowing gradients of neurogenesis or differentiation may require reevaluation. We show that telencephalic angiogenesis is under significant intrinsic regulation by homeobox transcription factors Dlx1/2, Nkx2.1 and Pax6, which also regulate neurogenesis and neuronal migration. Our data unify the principles of neuronal and endothelial cell development at fundamental, cellautonomous levels beyond the currently recognized commonalities at the level of extracellular signaling molecules and receptors.
A new model of telencephalic angiogenesis
The current model of CNS angiogenesis (Supplementary Fig. 16a online) proposes that pial and periventricular vessels arise from the same population of endothelial cells and implies that developmental gradients apply equally to both populations10. According to this model, pial vessels extend radial branches toward the ventricle, form new branches upon arrival in the periventricular region, reverse direction to grow back to the pia, and finally branch into plexuses. This model does not account for our findings of distinct developmental schedules for pial and periventricular vessels nor for the selective effects of Nkx2.1, Dlx1/2 or SeyDey mutations on periventricular vessels. Similarly, it is difficult to explain the pia-directed ‘reverse’ growth of the vessels proposed in the current model on the basis of the trophic effects of VEGF (refs. 9,34) produced by neuroepithelial cells at the ventricular border.
We propose an alternative model that can support the findings reported here and also fit with the notion of VEGF-guided vessel growth (Supplementary Fig. 16b). In this model, pial and periventricular vessels develop along independent schedules. The pial vessels do not display developmental gradients. The periventricular vessels are branches of the basal vessel located in the basal ganglia primordium. The basal vessel is likely to arise from the pharyngeal arch arteries that are apparent in the cervical region by E9 (refs. 2,11). The basal vessel produces periventricular branches, which form an orderly lattice in the ventral telencephalon. The periventricular vessel network propagates into the dorsal telencephalon, producing a periventricular lattice in this region, under the control of homeobox transcription factors.
Compartmental influences on telencephalic angiogenesis
Although our data show that Nkx2.1, Dlx1/2 and Pax6 regulate development of telencephalic vasculature in a cell-autonomous fashion, how these transcription factors regulate endothelial cell proliferation or migration remains unclear. Some insights into this issue are provided by our data showing that loss of Nkx2.1 and Pax6 alters expression of molecules associated with endothelial cell development (Supplementary Fig. 15).
Regarding other potential mediators of the transcription factors’ influences on angiogenesis, it is well known that sonic hedgehog (Shh) induces Nkx2.1 expression35,36 and that Shh has pivotal roles in both neurogenesis and angiogenesis37. In Nkx2.1−/− mutants, at E10.5 and E11.5, Shh expression is virtually undetectable in the basal telencephalon12. In the Dlx1−/−Dlx2−/− mutant, there is increased expression of delta1 (Notch ligand) in an expanded anatomical domain of Notch signaling in the subcortical telencephalon38. This might also affect vascular patterns, as Notch signaling can cause endothelial-cell contact inhibition. Pax6+/− mice have an eye phenotype in which the cornea is spontaneously vascularized, with deficiency in a secreted isoform of VEGF receptor Flt1 (ref. 39). It is possible that impaired development of telencephalic vasculature in the SeyDey mice is a result of impaired VEGF signaling. The role of Nkx2.1 and Pax6 in angiogenesis is not restricted to the CNS but seems more generalized and affects the lung, eye and cardiopulmonary system39–41.
A role for angiogenesis in neuronal development
What may be the neurobiological significance of our findings that endothelial cell development is intrinsically regulated and that it occurs in spatially and temporally regulated gradients? The development of telencephalic periventricular vascular networks predates development of neuronal networks42, radial units of the dorsal telencephalon43 or striosome-matrix compartments of the ventral telencephalon44. Thus, the periventricular networks are temporally and spatially well positioned to shape development of the incipient cortical radial units and guide tangential migration of GABA neurons18,31. Progression of the periventricular endothelial cell gradient coincides spatially and temporally with the appearance of mitotic figures in the telencephalic subventricular zone45,46, raising the possibility that the two events may be linked mechanistically. Finally, an unresolved issue in developmental neurobiology is how inductive morphogenetic gradients propagate over relatively long distances in the embryonic brain. We speculate that the timing and route of migration of periventricular endothelial cells and the dynamic up- or downregulation of gene expression accompanying cross-border migration render these cells ideal for morphogen synthesis, delivery and establishment of morphogenetic gradients.
In summary, our data provide new conceptual and mechanistic insights into telencephalic angiogenesis in the developing brain. We suggest reconsideration of current concepts that blood vessel development merely shadows neuronal development and propose that telencephalic angiogenesis has intrinsically programmed growth patterns that may have critical functions in promoting neural development.
METHODS
Mice
We used timed pregnant CD1 mice (Charles River), Tie2-GFP mice (B6.Cg-Tg (TIE2GFP) 287Sato/1J; Jackson Laboratories), Nkx2.1+/− mice (heterozygous breeders from S. Anderson; see Acknowledgments) and Pax6 heterozygotes (B6EiC3Sn a/A-Pax6Sey-Dey/J; Jackson). Nkx2.1 homozygotes and heterozygotes from an Nkx2.1+/− × Nkx2.1+/− cross were genotyped by PCR47. Pax6 mutants were identified by eye phenotype. Tie2-GFP+ embryos were identified by the presence of GFP fluorescence and verified by genotyping. Studies were approved by the animal care and use committee of Massachusetts General Hospital.
Explant culture
Embryos (E10 or E11) were collected by hysterotomy of deeply anesthetized dams (ketamine, 50 mg per kilogram body weight and xylazine, 10 mg/kg intraperitoneally (i.p.)) and decapitated. Intact telencephalon or explants of dorsal or ventral telencephalon were transferred to polycarbonate membrane filters (8-µm pore size; Gibco-Life Technologies) in sterile six-well plates and incubated with Neurobasal medium containing 2% B27 supplement and penicillin-streptomycin-glutamine mix (Gibco). Explants were maintained for 24 h at 37 °C, with 5% CO2 and 95% humidity. Explant viability was tested with a LIVE/DEAD Viability/Cytotoxicity Kit (Invitrogen). For 6-hydroxydopamine hydrochloride (6-OHDA) treatment, 10 µg/ml of 6-OHDA (Sigma) was added to the culture medium. Explants were embedded flat for paraffin wax histology. Blood vessels were labeled in 15-µm-thick sections with biotinylated isolectin B4 (1:50, Sigma) and with polyclonal PAX6 antibody (1:100, Covance) to identify dorsal telencephalon.
Heterochronic explantation
We used E11 donor (ventral) and E10 host (dorsal) CD1 embryos. E11 ventral tissue was labeled for 1 h with the Qtracker 655 Cell Labeling Kit (Invitrogen) to deliver red fluorescent nanocrystals into the cytoplasm of live cells. Using fine tungsten needles, we inserted the Qdotlabeled E11 ventral tissue into the E10 dorsal explant. During this process, the grafted tissue was visualized with a fluorescence microscope (Nikon Eclipse E400) to confirm appropriate positioning (Supplementary Fig. 5). When Tie2 GFP+ mice were used, E11 ventral explants were directly inserted into E10 CD1 dorsal explants (Supplementary Fig. 6). All explants were maintained for 24 h and processed for paraffin histology. Transplantation experiments using Nkx2.1 and SeyDey embryos were performed using the same procedures as above. SeyDey E10 and E11 embryos were identified by their eye phenotype. Nkx2.1−/− and Nkx2.1+/+ transplants were identified post hoc by genotyping and subsequently used for immunohistochemistry.
Immunohistochemistry
E9, E10 and E11 brains were fixed in 4% paraformaldehyde or a zinc fixative (BD Pharmingen) for 24 h and processed for paraffin histology. Blood vessels were labeled in 20-µm-thick sections with biotinylated isolectin B4 (1:50, Sigma), followed by secondary antibody streptavidin–Alexa Fluor 488 conjugate (Molecular Probes). We used rabbit polyclonal (1:100) and mouse monoclonal (1:30) antibodies to NKX2.1 (TTF1, both from Abcam) and DLX2 (Abcam). For PAX6, we used a rabbit polyclonal (1:100, Covance) or a mouse monoclonal antibody (1:30, Synaptic Systems). Rabbit polyclonal antibody to laminin (1:50, Sigma), mouse monoclonal antibody to PECAM-1 (1:30, BD Pharmingen), rabbit polyclonal antibody to FLI1 (1:100, Abcam) and rabbit polyclonal antibody to VEGF (1:100, Santa Cruz Biotechnology) were also used.
For whole mount preparations, E9, E10 and E11 brains were fixed in 4% paraformaldehyde at 4 °C overnight. Brains were washed in PBS, permeabilized with 0.5% Triton X-100 in 1% BSA at 4 °C overnight and incubated with biotinylated isolectin B4 (1:40, Sigma) in antibody diluent (BD Pharmingen) with 1% Triton X-100 at 4 °C overnight. After six PBS washes, brains were incubated with Alexa 488–streptavidin conjugates diluted 1:200 in antibody diluent (BD Pharmingen) with 0.5% Triton X-100 for 6 h at 4 °C. The neural tube or brain was cut open along the roof plate, cut again in a caudal to rostral direction and opened like a book. These were mounted with Vectashield HardSet mounting medium (Vector Laboratories) with the ventricular surface facing the viewer and examined using a Zeiss Pascal LSM 5 laser confocal microscope.
Bromodeoxyuridine labeling
We injected pregnant dams with a single dose of bromodeoxyuridine (BrdU) (50 µg per g body weight, i.p.; Sigma) 2 h before death45. Embryos were removed and decapitated, and embryonic heads were immersed in zinc fixative (BD Pharmingen) for 24 h and processed for paraffin wax histology. BrdU immunohistochemistry was performed on coronal, 20-µm sections. Double labeling was performed with a mouse monoclonal antibody to BrdU (BD Pharmingen) and biotinylated isolectin B4 (1:50, Sigma) or rabbit polyclonal Von Willebrand factor (DAKO).
Pimonidazole assay
We administered pimonidazole hydrochloride (Hydroxyprobe-1, Chemicon; 60 mg/kg, i.p.) to pregnant dams carrying E10 or E11 mice. One hour later, we collected embryos and processed them for paraffin histology. Coronal, 5-µm-thick sections were stained with Hydroxyprobe-1 monoclonal antibody from the Hydroxyprobe-1 kit (Chemicon).
Fluorescence-activated cell sorting
GFP+ endothelial cells were isolated from Tie2-GFP brains according to published protocol48. Approximately 3 × 107 cells were sorted using the FACSAria cell sorter (Becton Dickinson). GFP+ fractions were collected and GFP fluorescence verified before RNA isolation.
Real-time PCR
RNA was extracted from samples collected by fluorescence-activated cell sorting (FACS) using the PicoPure RNA Isolation kit (Arcturus). Reverse transcription was performed using a Transcriptor First Strand cDNA synthesis kit (Roche). PCR was performed on an ABI Prism 7500 (Applied Biosystems) machine. Taqman primers with 6-carboxyfluorescein probes for Nkx2.1, Dlx1, Dlx2, Pax6, Vegfa, Kdr, Flt1, Bdnf, Hif1a and Hba were purchased from Applied Biosystems. The housekeeping gene β2-microglobulin was used as a control. The relative gene expression among different samples was determined according to published methodology49.
Primary mouse brain endothelial cell culture
E13 embryos were dissected and the telencephalon from each was removed and cut open along the roof plate in a caudal to rostral direction. Ventral and dorsal counterparts were separated by an incision at the dorsoventral boundary. Tissue pieces were minced into 1- to 2-mm fragments with a scalpel, rinsed in PBS and incubated at 37 °C for 0.5 h in prewarmed PBS with trypsin (0.25%), DNase I (1 mg/ml) and EDTA (0.5 mM). Cells were dissociated, spun down, resuspended in warm 10% FCS-DMEM and filtered through a sterile 40-µm nylon mesh. Cells were collected, resuspended in 1 ml red blood cell lysing buffer (Sigma), overlaid onto DMEM and centrifuged. The cell pellet was resuspended in 1 ml of 10% FCS-DMEM.
A rat monoclonal antibody to mouse CD31 (PECAM-1) (BD Pharmingen) was covalently coupled to prewashed Dynabeads coated with sheep antibody to rat IgG (Invitrogen) at 4 °C overnight and washed three times with 2% FCS in PBS. We then added 1 ml of cell suspension to the tube containing the washed beads. After 30 min at 4 °C with occasional agitation, bead-bound cells were recovered with a magnetic field, washed five times in 10% FCS-DMEM and once with FCS-free DMEM, and then digested for 10 min at 37 °C in 1 ml of trypsin/EDTA (GIBCO) to release the beads. Bead-free cells were centrifuged in 10% FCS-DMEM and resuspended in endothelial cell culture medium (BIO-COAT endothelial cell growth environment, BD Biosciences). Cells were cultured in 75-cm2 BIOCOAT Collagen Type 1 flasks (BD Biosciences), fed every 2 d by complete medium exchange and trypsinized for subculture at 80% confluency.
Some cells (1 × 103) were plated onto fibronectin-coated coverslips (BD Biosciences) and used for NKX2.1, DLX2 and PAX6 immunohistochemistry. Other cells were harvested for cytosolic and nuclear extraction using the nuclear/cytosol fractionation kit (BioVision). Extracts were run on 12% polyacrylamide gels and transferred to nitrocellulose membranes (Osmonics). Blots were probed with rabbit polyclonal PAX6 antibody (Covance, 1:400), rabbit polyclonal DLX2 antibody (Abcam; 1:500) and a rabbit polyclonal NKX2.1 antibody (TTF1, Abcam; 1:200).
siRNA preparation and transfection
We used predesigned siRNA constructs for Nkx2.1 (ON-TARGETplus SMARTpool, L-041979-01, Dharmacon), Pax6 (ON-TARGETplus SMARTpool, L-062890-00, Dharmacon) and a control nontargeting siRNA (D-001206-14-05, Dharmacon). A siGLO Red transfection indicator (Dharmacon) was cotransfected with the siRNA. We found that optimal conditions involved transfecting endothelial cells, maintained in DMEM, at 80% confluency in six-well plates, using 2 µM siRNA, 1 µM siGLO Red and DharmaFECT transfection reagent 1 according to manufacturer’s protocols. Cells were collected every 24, 48 and 96 h after transfection and processed for immunohistochemistry for NKX2.1 and PAX6. Some cells were harvested and lysed in RIPA buffer (SIGMA) with protease inhibitor cocktail (Roche). Forty micrograms of cellular protein were resolved on 12% polyacrylamide gels, transferred to nitrocellulose membranes and probed with NKX2.1 and PAX6 antibodies. Blots were stripped and probed with antibody to α-tubulin (1:5000, Abcam) to estimate total protein amounts.
Vessel density
A stereological point grid was superimposed on digital images of biotinylated isolectin-B4+ vessels. The ratio between points falling on blood vessels and on brain tissue was calculated for each section and average values were obtained for a given brain.
Statistics
We analyzed statistical significance of differences between groups by two-tailed Student’s t-test (Prism; GraphPad Software). Results were expressed as mean ± s.d. and statistical significance was reported at P < 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Anderson (Weill-Cornell Medical College) for sharing with us the Nkx2.1+/− line. We gratefully acknowledge technical assistance by I. Bagayev (confocal microscope), L. Zagachin (real-time PCR) and M. Waring (FACS). This work was supported by RO1NS43246, RO1DA020796 and P30NS045776 to P.G.B.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 2.Kurz H. Physiology of angiogenesis. J. Neurooncol. 2000;50:17–35. doi: 10.1023/a:1006485716743. [DOI] [PubMed] [Google Scholar]
- 3.Strong LH. The early embryonic pattern of internal vascularization of the mammalian cerebral cortex. J. Comp. Neurol. 1964;123:121–138. doi: 10.1002/cne.901230111. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 5.Hogan KA, Ambler CA, Chapman DL, Bautch VL. The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development. 2004;131:1503–1513. doi: 10.1242/dev.01039. [DOI] [PubMed] [Google Scholar]
- 6.Keynes R, Lumsden A. Segmentation and the origin of regional diversity in the vertebrate nervous system. Neuron. 1990;4:1–9. doi: 10.1016/0896-6273(90)90438-l. [DOI] [PubMed] [Google Scholar]
- 7.Puelles L, Rubenstein JLR. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggests a neuromeric organization. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 8.Keynes RJ, Stern CD. Mechanisms of vertebrate segmentation. Development. 1988;103:413–429. doi: 10.1242/dev.103.3.413. [DOI] [PubMed] [Google Scholar]
- 9.Plate KH. Mechanisms of angiogenesis in the brain. J. Neuropathol. Exp. Neurol. 1999;58:313–320. doi: 10.1097/00005072-199904000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg DA, Jin K. From angiogenesis to neuropathology. Nature. 2005;438:954–959. doi: 10.1038/nature04481. [DOI] [PubMed] [Google Scholar]
- 11.Hiruma T, Nakajima Y, Nakamura H. Development of pharyngeal arch arteries in early mouse embryo. J. Anat. 2002;201:15–29. doi: 10.1046/j.1469-7580.2002.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 13.Supèr H, Martínez A, Soriano E. Degeneration of Cajal-Retzius cells in the developing cerebral cortex of the mouse after ablation of meningeal cells by 6-hydroxydopamine. Devel. Brain Res. 1997;98:15–20. doi: 10.1016/s0165-3806(96)00155-1. [DOI] [PubMed] [Google Scholar]
- 14.Bayer SA, Altman J. Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog. Neurobiol. 1987;29:57–106. doi: 10.1016/0301-0082(87)90015-3. [DOI] [PubMed] [Google Scholar]
- 15.Bayer SA, Altman J. Neocortical Development. New York: Raven Press; 1991. [Google Scholar]
- 16.Takahashi T, Nowakowski RS, Caviness VS., Jr The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J. Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS., Jr Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J. Neurosci. 1999;19:10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 19.De Carlos JA, López-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J. Neurosci. 1996;16:6146–6156. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubenstein JL, et al. Genetic control of cortical regionalization and connectivity. Cereb. Cortex. 1999;9:524–532. doi: 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- 21.Anderson S, Mione M, Yun K, Rubenstein JLR. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb. Cortex. 1999;9:646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- 22.Eisenstat DD, et al. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J. Comp. Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 24.Stoykova A, Fritsch R, Walther C, Gruss P. Forebrain patterning defects in Small eye mutant mice. Development. 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- 25.Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill RE, et al. Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 27.Schedl A, et al. Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 28.Fishell G, Mason CA, Hatten ME. Dispersion of neural progenitors within the germinal zones of the forebrain. Nature. 1993;362:636–638. doi: 10.1038/362636a0. [DOI] [PubMed] [Google Scholar]
- 29.Ghanem N, et al. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J. Neurosci. 2007;27:5012–5022. doi: 10.1523/JNEUROSCI.4725-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nery S, Corbin JG, Fishell G. Dlx2 progenitor migration in wild type and Nkx2.1 mutant telencephalon. Cereb. Cortex. 2003;13:895–903. doi: 10.1093/cercor/13.9.895. [DOI] [PubMed] [Google Scholar]
- 31.Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J. Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: a developmental study based upon (3H) thymidine autoradiography. Devel. Brain Res. 1982;4:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 33.Rakic P, Caviness VS., Jr Cortical development: view from neurological mutants two decades later. Neuron. 1995;14:1101–1104. doi: 10.1016/0896-6273(95)90258-9. [DOI] [PubMed] [Google Scholar]
- 34.Breier G, Albrecht U, Sterrer S, Risau W. Expression of vascular endothelial growth factor during embryonic angiogenesis and endothelial cell differentiation. Development. 1992;114:521–532. doi: 10.1242/dev.114.2.521. [DOI] [PubMed] [Google Scholar]
- 35.Ericson J, et al. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81:747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 36.Shimamura K, Martinez S, Puelles L, Rubenstein JLR. Patterns of gene expression in the neural plate and neural tube subdivide the embryonic forebrain into transverse and longitudinal domains. Dev. Neurosci. 1997;19:88–96. doi: 10.1159/000111190. [DOI] [PubMed] [Google Scholar]
- 37.Cohen MM., Jr Craniofacial anomalies: clinical and molecular perspectives. Ann. Acad. Med. Singapore. 2003;32:244–251. [PubMed] [Google Scholar]
- 38.Yun K, et al. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- 39.Ambati BK, et al. Corneal avascularity is due to soluble VEGF receptor-1. Nature. 2006;443:993–997. doi: 10.1038/nature05249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kusakabe T, et al. Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol. Endocrinol. 2006;20:1796–1809. doi: 10.1210/me.2005-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan B, et al. Inhibition of distal lung morphogenesis in Nkx2.1(−/−) embryos. Dev. Dyn. 2000;217:180–190. doi: 10.1002/(SICI)1097-0177(200002)217:2<180::AID-DVDY5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Ari Y, Khalilov I, Represa A, Gozlan H. Interneurons set the tune of developing networks. Trends Neurosci. 2004;27:422–427. doi: 10.1016/j.tins.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 44.Gerfen CR. The neostriatal mosaic: striatal patch-matrix organization is related to cortical lamination. Science. 1989;246:385–388. doi: 10.1126/science.2799392. [DOI] [PubMed] [Google Scholar]
- 45.Bhide PG. Cell cycle kinetics in the embryonic mouse corpus striatum. J. Comp. Neurol. 1996;374:506–522. doi: 10.1002/(SICI)1096-9861(19961028)374:4<506::AID-CNE3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 46.Sheth AN, Bhide PG. Concurrent cellular output from two proliferative populations in the early embryonic mouse corpus striatum. J. Comp. Neurol. 1997;383:220–230. [PubMed] [Google Scholar]
- 47.Kimura S, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 48.Motoike T, et al. Universal GFP reporter for the study of vascular development. Genesis. 2000;28:75–81. doi: 10.1002/1526-968x(200010)28:2<75::aid-gene50>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 49.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.