Abstract
Autophagy is a major intracellular degradative pathway that is involved in many human diseases. The molecular mechanism of autophagy has been elucidated largely through studies on autophagy-related (Atg) proteins. One difficulty in understanding the mechanism of autophagy has been the lack of functional motifs in most of the Atg proteins. In the absence of this information, studies that have focused on the interactions between Atg proteins have shed light on their functions. However, in most studies, it is difficult to determine whether an interaction is direct or occurs through other Atg proteins, particularly in vivo. Here, we took advantage of a new reagent, a multiple knockout (MKO) strain lacking 24 ATG genes, and converted the strain into a yeast two-hybrid (Y2H) host strain. We introduced three reporter genes into the existing MKO strain, and analyzed known interactions in the new MKO Y2H strain background to verify its utility. We also probed a new interaction using the MKO Y2H strain, and our results suggest that Atg29 and Atg31 interact independently of other known Atg proteins, and this interaction may mediate the interaction between Atg17 and Atg29.
Keywords: autophagy, protein-protein interaction, Atg1 complex, Atg29, Atg31
Introduction
Macroautophagy (hereafter autophagy) is a complex intracellular degradative pathway that utilizes approximately 30 autophagy-related (Atg) proteins. Our understanding of autophagy has been greatly improved through research on these Atg proteins, and in particular many insights have come from studies concerning the interactions among them. For example, Atg16 was identified through a yeast two-hybrid (Y2H) screen using Atg12 as bait,1 and the interaction between Atg19 and precursor aminopeptidase I, together with that between Atg19 and Atg11 made it possible for us to understand the temporal order of the cargo packaging event in the cytoplasm to vacuole targeting pathway.2 Thus, different protein complexes held together by protein-protein interactions have helped us to dissect and connect different steps of autophagy. However, most of the previous studies did not address a question very important to understand complex formation—whether or not a particular interaction is direct. Two proteins may show a positive interaction by coimmunoprecipitation and Y2H analyses, but still may interact indirectly through other proteins in the complex.
Recently, we published a new reagent useful for determining in vivo whether an interaction is direct, or more precisely, whether the interaction is independent of other known Atg proteins. We created a yeast multiple knockout (MKO) strain lacking 24 ATG genes that are involved in the induction and vesicle formation steps in Saccharomyces cerevisiae.3 With regard to protein-protein interactions, the MKO strain (YCY123) has been useful for studying Atg9 self-interaction.4 To supplement the existing MKO strain and make protein-protein interaction analyses faster and more efficient, in this study we converted the MKO strain into a Y2H host strain. We integrated three reporter genes (GAL1-HIS3, GAL2-ADE2 and GAL7-lacZ5) into the MKO strain and tested those reporter genes with known interactions, such as some of those that occur within the Atg1 complex.6,7 After having validated the utility of this strain, we then extended our research to a previously unknown interaction. Using our MKO Y2H strain, we found that Atg29,8 and Atg31,9 interact independently of other known Atg proteins, an interaction that was further verified by coimmunoprecipitation. We also found that Atg29 and Atg17 did not interact with each other in the MKO Y2H strain, and by coimmunoprecipitation we found that in the absence of Atg31, the interaction between these two proteins was lost. These results suggest that the Atg29-Atg31 interaction may potentially mediate the interaction between Atg29 and Atg17.
Results
Construction of the MKO Y2H strain
In this study we created an MKO Y2H strain to provide a new tool for autophagy research. The MKO strain YCY123,3 was chosen as the starting strain. GAL4, GAL80 and ADE2 open reading frames (ORFs) were knocked out in YCY123. Three reporter genes were then introduced into the genome by sequential gene replacement as described previously.5 The HIS3 gene under the control of the GAL1 promoter (GAL1-HIS3) was inserted downstream of the LYS2 gene. The ade2Δ∷KanMX locus was replaced by the ADE2 gene under the control of the GAL2 promoter (GAL2-ADE2). Finally, the lacZ gene under the control of the GAL7 promoter (GAL7-lacZ) was inserted into the MET2 gene locus. The resulting strain, YCY148, is His+, Ade+ and lacZ+ in the presence of Gal4 activity.
Assessment of known interactions using the MKO Y2H strain
We tested the utility of the MKO Y2H strain and its three reporter genes by examining known interactions. First, we tested the GAL1-HIS3 reporter gene by checking the Atg9 self-interaction. Atg9 self-interacts in the absence of other Atg proteins in the MKO strain as assessed by coimmunoprecipitation.4 The same conclusion could be reached using Y2H analysis (Fig. 1A). The Y2H strain PJ69-4A5 (a “wild-type” strain that expresses all of the ATG genes) expressing activation domain (AD)-fused full-length Atg9 and binding domain (BD)-fused full-length Atg9 was used as a positive control. Cells expressing AD-Atg9 or BD-Atg9 alone (along with the appropriate empty vector) were used as negative controls. After 3 days on plates lacking histidine, cells that expressed AD-Atg9 and the empty BD vector showed only background levels of growth. Cells expressing BD-Atg9 and the empty AD vector also showed minimal growth, although the background level was higher. In contrast, both YCY148 and PJ69-4A cells expressing AD-Atg9 and BD-Atg9 were His+, indicating that Atg9 interacts with itself independently of other known Atg proteins by the Y2H approach.
Figure 1.
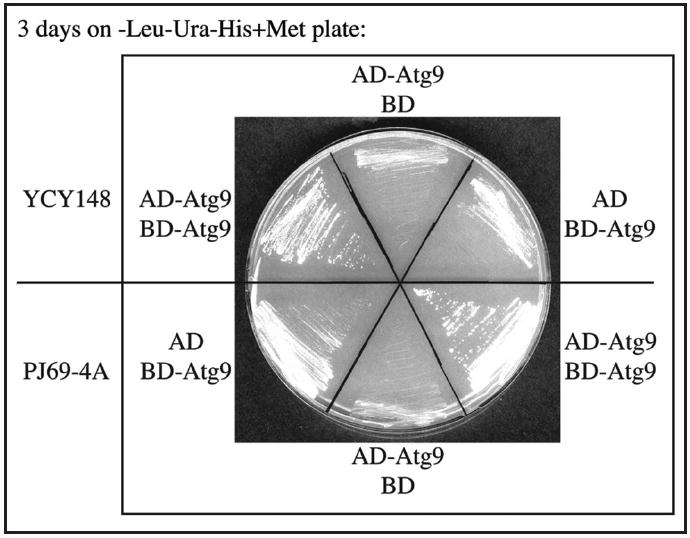
Validation of the MKO Y2H strain and assessment of the HIS3 reporter gene. The GAL1-HIS3 reporter gene was tested through Atg9 self-interaction. The MKO Y2H strain YCY148 and the “wild-type” Y2H strain PJ69-4A, which contains all of the ATG genes, were transformed with plasmids expressing AD-Atg9, BD-Atg9 or both. Interactions were assessed by cell growth on plates lacking leucine, uracil and histidine for 3 days.
Second, we tested the GAL2-ADE2 reporter gene by checking interactions between Atg1 and its known binding partners Atg11,10 Atg13,11 and Atg17.12 Because both AD-Atg1 and BD-Atg1 Y2H chimeric proteins autoactivate, in our experiments we used a Y2H plasmid expressing a truncated version of Atg1 lacking its N-terminal kinase domain (Atg1ΔK).13 Cell growth on plates lacking adenine was compared between YCY148 and PJ69-4A cells expressing AD-Atg11 and BD-Atg1ΔK, AD-Atg13 and BD-Atg1ΔK, or AD-Atg17 and BD-Atg1ΔK (Fig. 2A). After 3 days, both YCY148 and PJ69-4A cells expressing the above-mentioned plasmid combinations were positive on plates lacking adenine, whereas cells expressing AD-Atg11, AD-Atg13, AD-Atg17 or BD-Atg1ΔK alone were negative. In general, we found that growth of the cells was slower with the MKO Y2H strain; however, we note that, because cell growth of the MKO strain itself is slower than the corresponding wild-type strain,3 we cannot conclude that the strength of those interactions in the MKO Y2H strain is weaker than that in the PJ69-4A strain.
Figure 2.
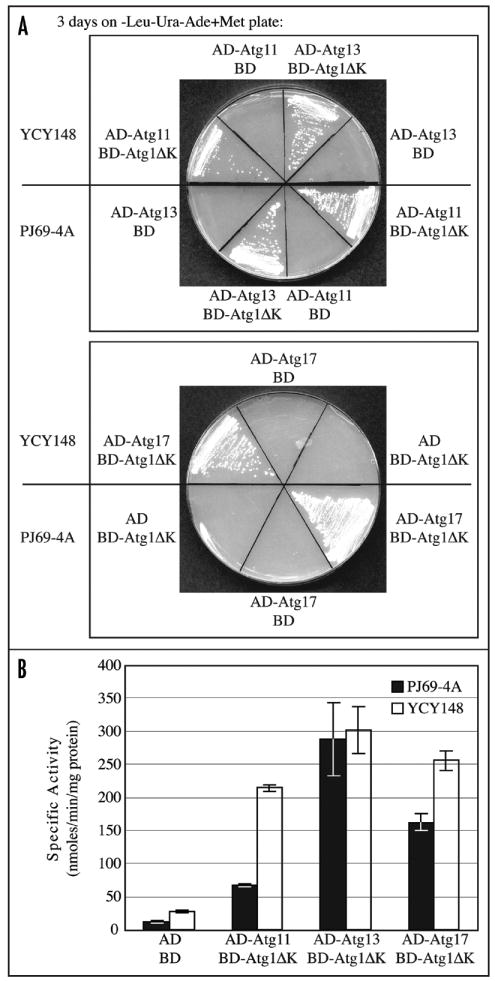
Assessment of two reporter genes in the MKO Y2H strain. (A) The GAL2-ADE2 reporter gene was tested via interactions between Atg1 lacking the N-terminal kinase domain (Atg1ΔK) and its known interaction partners Atg11, Atg13 and Atg17. The Y2H strains YCY148 and PJ69-4A were transformed with different plasmid combinations, and cell growth was monitored after 3 days on plates lacking leucine, uracil and adenine. (B) The GAL7-lacZ reporter gene was assessed via interactions between Atg1ΔK and its known interaction partners. The strains with plasmid combinations used in (A) were grown in minimal medium to maintain plasmid selection. The strength of the interaction for each set of proteins was quantified by measuring β-galactosidase activity from three independent experiments. The error bars represent the standard deviation.
Finally, to quantitatively compare the strength of the interactions among the Atg1 complex components, we measured β-galactosidase activity of YCY148 and PJ69-4A cells expressing AD-Atg11 and BD-Atg1ΔK, AD-Atg13 and BD-Atg1ΔK, or AD-Atg17 and BD-Atg1ΔK (Fig. 2B). Cells expressing empty AD and BD vectors were used as negative controls. All three combinations of Atg proteins showed interactions based on the measurement of β-galactosidase activity, with the Atg13-Atg1ΔK interaction being the strongest. By this assay the Atg11-Atg1, and Atg17-Atg1 interactions in YCY148 seemed to be stronger compared to the same interactions in PJ69-4A. It is possible that without other competing factors that are normally present within the Atg1 complex, Atg11 and Atg17 have a higher affinity for Atg1. Overall, the background level of β-galactosidase activity in YCY148 was slightly higher than that in PJ69-4A.
A new interaction identified using the MKO Y2H strain
Next, we used the MKO Y2H strain to explore a previously unknown interaction. Atg17, Atg29 and Atg31 are not required for the cytoplasm to vacuole targeting pathway, but they have partial or severe defects in nonselective autophagy, and in selective pexophagy and/or mitophagy.8,9,12,14 Atg17 interacts with both Atg29,15 and Atg31,9 which makes it likely that the three of them form a ternary complex. We wanted to know whether Atg29 and Atg31 interact with each other, and if so, whether this interaction is independent of other Atg proteins, such as Atg17.
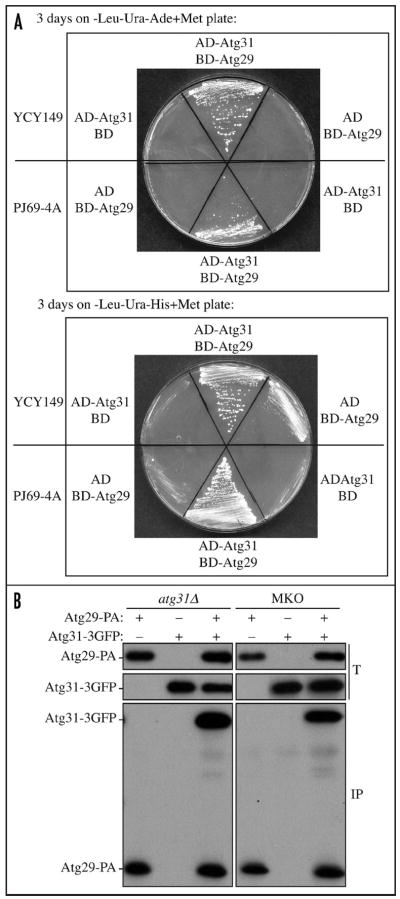
With our MKO Y2H strain, we found that an interaction between Atg29 and Atg31 exists in the absence of other known Atg proteins (Fig. 3A). Control cells expressing AD-Atg31 and the empty BD vector, or BD-Atg29 and the empty AD vector showed minimal growth on plates lacking histidine, and essentially no growth on plates lacking adenine. In contrast, cells expressing AD-Atg31 and BD-Atg29 together were able to grow in either PJ69-4A or the MKO Y2H strain YCY149 (YCY148 deleted for ATG31), suggesting that there is an interaction between Atg29 and Atg31, and the interaction exists in the absence of other known Atg proteins. To verify the results from the Y2H analysis, we performed a coimmunoprecipitation experiment in the MKO strain YCY132 (YCY123 atg31Δ). Endogenous promoter-driven Atg31-3GFP was coexpressed with endogenous promoter-driven Atg29 fused to protein A (PA) in both an ATG31 single-knockout strain (HCY111) and a MKO strain (YCY132). Protein extracts were generated under native conditions and subjected to affinity isolation using IgG-sepharose. We found that Atg29-PA coprecipitated Atg31-3GFP in both strains (Fig. 3B). As controls, we examined cells expressing Atg29-PA without Atg31-3GFP, or cells expressing Atg31-3GFP but not Atg29-PA, and we did not observe any coprecipitating Atg31-3GFP bands. Taken together, these results indicate that Atg29 interacts with Atg31 independently of other known Atg proteins. However, this result does not rule out the possibility that Atg29 and Atg31 may interact through other non-Atg proteins or unknown Atg proteins.
Figure 3.
Atg31 interacts with Atg29 in the absence of other known Atg proteins. (A) YCY149 and PJ69-4A cells were transformed with plasmids expressing AD-Atg31 and the empty BD vector, or AD-Atg31 and BD-Atg29, or the empty AD vector and BD-Atg29. The cells were grown on plates lacking leucine, uracil, and adenine or histidine for 3 days. (B) An atg31Δ strain (HCY111) or the MKO atg31Δ strain (YCY132) expressing endogenous promoter-driven Atg29-PA, Atg31-3GFP or both, were grown to mid-log phase and subjected to coimmunoprecipitation analysis. Note that ATG31-3GFP was integrated into the genome of HCY111 and YCY132, and we listed the derivatives of these two strains as YCY77 and YCY154 in Table 1. T, total lysates; IP, immunoprecipitates.
Atg29 and Atg17 interact through Atg31
Having established that Atg29 and Atg31 interact independently of other known Atg proteins, we asked whether the same was true for the Atg29 and Atg17 interaction. In contrast to the result with Atg29 and Atg31, we found that Atg29 and Atg17 did not interact in the MKO Y2H strain (Fig. 4A). Cells expressing AD-Atg17 and BD-Atg29 together showed no growth in the MKO Y2H strain YCY149, but were able to grow in the wild-type PJ69-4A strain. Control cells expressing AD-Atg17 and the empty BD vector, or BD-Atg29 and the empty AD vector showed no growth in either strain on plates lacking adenine. We also confirmed the Y2H result by coimmunoprecipitation. Atg17-3GFP was introduced into the ATG17 ORF of a wild-type strain and an atg31Δ strain through a plasmid-based integration. We found that Atg29-PA coprecipitated Atg17-3GFP in the wild-type strain, but not in the atg31Δ strain (Fig. 4B). As controls, we examined cells expressing Atg29-PA without Atg17-3GFP, or cells expressing Atg17-3GFP but not Atg29-PA, and we did not observe any coprecipitating Atg17-3GFP bands. These results suggest that Atg31 mediates the interaction between Atg29 and Atg17, and the Atg29-Atg31 interaction may be important for connecting Atg29 and Atg17.
Figure 4.
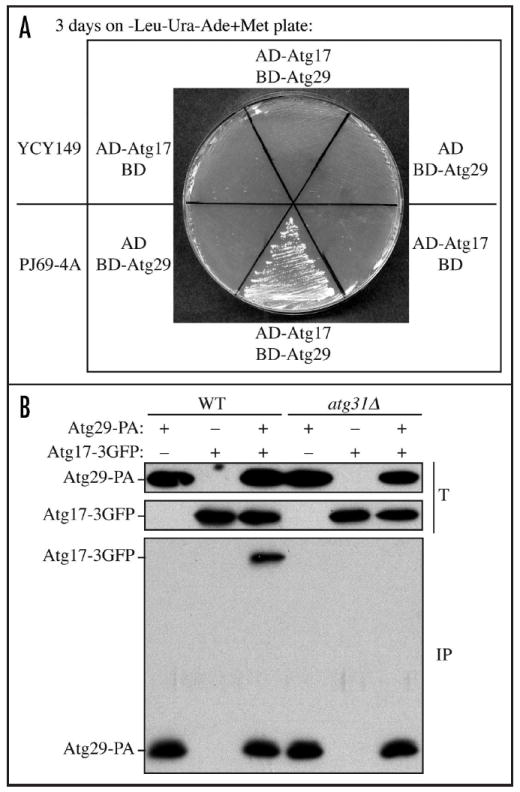
Atg29 interacts with Atg17 via Atg31. (A) YCY149 and PJ69-4A cells were transformed with plasmids expressing AD-Atg17 and the empty BD vector, AD-Atg17 and BD-Atg29, or the empty AD vector and BD-Atg29. The cells were grown on plates lacking leucine, uracil and adenine for 3 days. (B) The wild-type (SEY6210) or the atg31Δ (HCY111) strain expressing endogenous promoter-driven Atg29-PA, Atg17-3GFP or both, were grown to mid-log phase and subjected to coimmunoprecipitation analysis. Note that ATG17-3GFP was integrated into the genome of the wild-type and atg31Δ strains, and we listed the derivatives of these two strains as YCY80 and YCY81 in Table 1. T, total lysates; IP, immunoprecipitates.
Discussion
In this study, we created a Y2H version of the multiple ATG gene knockout strain, and used the strain to study known and unknown protein-protein interactions. This MKO Y2H strain is very useful and informative regarding questions about protein interactions in a multiprotein complex. Furthermore, it can be used along with the wild-type Y2H strain PJ69-4A, to provide information about the possible nature of an interaction. We found that consistent with published data, Atg9 self-interacts in the absence of other known Atg proteins by the Y2H approach (Fig. 1). Using this strain, we also found that Atg1 interacts with Atg11, Atg13 and Atg17 similar to previous reports (Fig. 2A and B). Additionally, we found that a new interaction between Atg29 and Atg31 exists in the absence of other known Atg proteins (Fig. 3); in contrast, the Atg29-Atg17 interaction was lost in the MKO strain (Fig. 4). These results suggest that the Atg29-Atg31 interaction may mediate the interaction between Atg29 and Atg17, and may be critical for autophagy induction.
Even though interactions indicated by cell growth sometimes seemed poorer in the MKO Y2H strain than in the wild-type Y2H strain, this was not necessarily reflective of a reduced interaction due to the overall slower growth rate of the MKO strain. Also, in a Y2H system, bait and prey proteins are vastly overexpressed and forced into the nucleus to allow interaction, which might result in false positives. For example, an Atg1-Atg17 interaction was not detected in the absence of Atg13 by coimmunoprecipitaion,16 but in our MKO Y2H strain these proteins showed a very strong interaction (Fig. 2A and B). Thus, it is worth noting that even though the MKO Y2H system shows a positive interaction, the same interaction may not be detected in the MKO strain by coimmunoprecipitation, similar to the case with wild-type strains. Therefore, data from the MKO Y2H strain need to be verified by other experiments. However, the MKO Y2H strain still has the merit of any yeast two-hybrid system for quick and effective in vivo analysis and screening.
Materials and Methods
Yeast strains and media
The yeast Saccharomyces cerevisiae strains used in this study are listed in Table 1. Knockout strains were constructed using the loxP/Cre system.17 For integration of the Atg31-3GFP fusion, the plasmid pATG31-3GFP(306) was linearized with SalI and integrated into the promoter region of the ATG31 gene locus. For integration of the Atg17-3GFP fusion, the plasmid pATG17-3GFP(306) was linearized with XbaI and integrated into the ORF of the ATG17 gene locus.7 Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal medium lacking appropriate amino acids.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| HCY111 | SEY6210 atg31Δ∷HIS3 | This study |
| KYY003 | YCY123 gal4Δ gal80Δ ade2Δ∷KanMX | This study |
| PJ69-4A | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2∷GAL1-HIS GAL2-ADE2 met2∷GAL7-lacZ | 5 |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | 20 |
| YCY77 | HCY111 ATG31-3GFP∷URA3 | This study |
| YCY80 | SEY6210 ATG17-3GFP∷URA3 | This study |
| YCY81 | HCY111 ATG17-3GFP∷URA3 | This study |
| YCY123 | SEY6210 atg1Δ, 2Δ, 3Δ, 4Δ, 5Δ, 6Δ, 7Δ, 8Δ, 9Δ, 10Δ, 11Δ, 12Δ, 13Δ, 14Δ, 16Δ, 17Δ, 18Δ, 19Δ, 20Δ, 21Δ, 23Δ, 24Δ, 27Δ, 29 Δ | 3 |
| YCY132 | YCY123 atg31Δ∷HIS3 | 21 |
| YCY147 | YCY123 gal4Δ gal80Δ GAL1-HIS3 GAL2-ADE2 | This study |
| YCY148 | YCY147 met2∷GAL7-lacZ | This study |
| YCY149 | YCY148 atg31Δ∷KanMX | This study |
| YCY154 | YCY132 ATG31-3GFP∷URA3 | This study |
Plasmids
Plasmids used for construction of the MKO Y2H strains—pGH1, pGAL2-ADE2 and pGAL7-lacZ—have been described previously.5 Y2H plasmids pAD-ATG9,4 pBD-ATG9,4 pGBD-ATG1ΔK,10 pAD-ATG11,4 pAD-ATG13,12 and pAD-ATG17,12 have been described previously. pATG17-3GFP(306) has also been described previously.7 To clone pGAL4(414), the GAL4 ORF with its 1 kb promoter and 0.5 kb terminator was amplified from yeast genomic DNA and cloned into XmaI and SacI sites of pRS414. pGAD-ATG31-C1 and pGBDU-ATG29-C1 were cloned as follows: The ATG29 ORF was amplified from genomic DNA using primers 5’ Y2H EcoRI-Atg29 (5’-GCG CGG GAA TTC ATG ATT ATG AAT AGT ACA AAC ACA GT-3’) and 3’ Y2H PstI-Atg29 (5’-GCG CGG CTG CAG TCA GAA TTG CAA TCT GTC CAT TAG C-3’). The PCR product was then digested with EcoRI and PstI, and cloned into the pGBDU-C15 vector linearized with EcoRI and PstI. Similarly, the ATG31 ORF was cloned into pGAD-C1,5 with EcoRI and PstI, using primers 5’ Y2H EcoRI-Atg31 (5’-GCG CGG GAA TTC ATG AAT GTT ACA GTT ACT GTT TAT GA-3’) and 3’ Y2H PstI-Atg31 (5’-GCG CGG CTG CAG TCA TAC GGA ATT GGA GAG CAT TTG TA-3’). For cloning pATG31-3GFP(306), the ATG31 ORF with its native promoter was cloned into the integration vector pPG5-3xGFP18 using primers 5’ XhoI-Atg31 (5’-CGC CGG GCT CGA GCG AAT CTT GTT TTG ACC CAA TCT TTG T-3’) and 3’ ClaI-Atg31 (5’-CGC CGG GAT CGA TGA TAC GGA ATT GGA GAG CAT TTG TAA TT-3’). For constructing pAtg29-PA(314), the full-length ATG29 ORF with its endogenous promoter was PCR-amplified and ligated into the XhoI and XmaI sites of pNopPA(314).19
Construction of the MKO Y2H strains
The MKO Y2H strains were created using multiple rounds of gene knockout, plasmid integration and 5-FOA-selected excision. The MKO strain YCY123,3 was used as the starting strain, in which GAL4 and GAL80 were knocked out and the markers for knockout were recovered using the loxP/Cre system.17 ADE2 was further deleted with a KanMX marker, yielding strain KYY003. The three reporter genes GAL1-HIS3, GAL2-ADE2 and GAL7-lacZ were integrated as described before,5 but with slight modifications. In brief, for integration of GAL1-HIS3, the strain KYY003 was transformed with the plasmid pGH1 linearized with PvuII, and spread on plates lacking uracil and histidine. Ura+ and His+ colonies were grown and plated on 5-FOA plates to select for excision of the vector sequences. Ura- and His- colonies were then transformed with pGAL4(414). A Trp+ and His+ transformant was selected and transformed with a 2.6 kb SmaI/HaeII fragment of pGAL2-ADE2. Trp+ and Ade+ transformants were selected. A colony that was Ade- without pGAL4(414) was selected and named YCY147. The YCY147 strain containing pGAL4(414) was transformed with the plasmid pGAL7-lacZ linearized with PacI, resulting in integration of the plasmid at the MET2 locus. Ura+, Trp+ and lacZ+ transformants were plated on 5-FOA plates to select for excision of the vector sequences. Ura-, Trp+ and lacZ+ colonies were selected, among which a colony that was lacZ- without pGAL4(414) was selected and named YCY148. ATG31 was knocked out with a KanMX marker in strain YCY148 using the loxP/Cre system, to generate strain YCY149.
Protein A affinity isolation
Cells were grown in 50 ml of SMD lacking the appropriate auxotrophic amino acids to mid-log phase. The cells (40 OD600 units) were collected, washed with water, and frozen at -80°C if desired. Cells were thawed on ice, resuspended in 4 ml lysis buffer (1x PBS, 200 mM Sorbitol, 1 mM MgCl2, 1% Tween 20, 2 mM PMSF and protease inhibitor cocktail), and broken by mixing with a vortex 6 times for 30 seconds each time, with 2 ml glass beads. Cell debris was removed by centrifugation and cell extracts were brought up to 4 ml. An aliquot (200 μl) of the cell extracts were saved as total lysate, with the rest being incubated with 100 μl IgG-sepharose beads for 1 h at 4°C. The beads were then washed with lysis buffer 6 times and the eluates were resolved by SDS-PAGE, transferred to PVDF membrane and probed with an antibody that binds protein A or anti-YFP (Clontech, 632381) antibody.
Enzyme assays
To select lacZ+ colonies after integration of the GAL7-lacZ reporter into the MET2 locus, a colony-lift filter assay was performed following a protocol from Clontech. β-galactosidase assays were performed as described previously.7
Acknowledgments
The authors thank Drs. Elizabeth A. Craig (University of Wisconsin), Stanley Fields (University of Washington) and Heesun Cheong for providing plasmids. This work was supported by National Institutes of Health Grant GM53396 (to D.J.K.).
Abbreviations
- AD
activation domain
- Atg
autophagy-related
- BD
binding domain
- MKO
multiple knockout
- ORF
open reading frame
- PA
protein A
- Y2H
yeast two-hybrid
References
- 1.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–96. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182:703–13. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–16. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamata T, Kamada Y, Suzuki K, Kuboshima N, Akimatsu H, Ota S, et al. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005;338:1884–9. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 9.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356:405–10. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–96. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–53. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–93. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the preautophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He C, Song H, Yorimitsu T, Monastyrska I, Yen W-L, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–35. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–48. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y, Klionsky DJ. New insights into autophagy using a multiple knockout strain. Autophagy. 2008;4:1073–5. doi: 10.4161/auto.6962. [DOI] [PMC free article] [PubMed] [Google Scholar]