Abstract
GadoTO, a MR contrast agent for the detection of cell death, consists of a nucleic acid-binding fluorophore attached to a gadolinium chelate.
Techniques for assessing cell death can be useful in analyzing diverse biological processes,1,2 or to provide guidance for the response to cancer chemotherapy,3 the success of organ transplantation,4 and the assessment of ischemia-damaged myocardium.5 Magnetic resonance-based techniques for assessing cell death can utilize the properties of hydrogen or carbon nuclei intrinsic to biological systems or take advantage of synthetic, water relaxation-enhancing magnetic probes. Among the former are techniques such as lipid proton MR spectroscopy,6 and diffusion weighted MRI,7 while the latter include annexinV-magnetic nanoparticles (MNP).8 We reasoned that a gadolinium probe binding dead (necrotic) cells might add substantially to MR methods for studying cell death, especially since it might be used along with our previously described annexinV-MNP which can image phosphatidylserine expression on apoptotic and dead cells in tissues by MRI.9
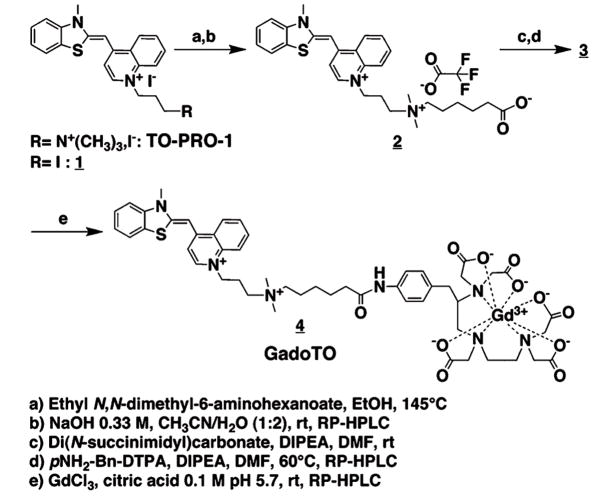
We here describe GadoTO (Scheme 1, 4), which for the first time uses a vital dye (TO-PRO-1) to target a paramagnetic metal (Gd) to dead cells.10 The binding of TO-PRO-1 to nucleic acids occurs through an intercalation of the thiazole orange (TO) moiety and electrostatic interaction between the negative phosphate backbone and cationic side chain in a non-sequence specific manner.11 TO-PRO-1 was selected as a nucleic acid-targeting dye in the design of our Gd-based probe for several reasons. First, we hypothesized that GadoTO, like TO-PRO-1, would be impermeable to healthy cells and selectively bind dead cells. Some DNA-binding, nucleus-staining dyes (e.g. Hoechst 33342) are permeable to healthy, apoptotic, and necrotic cells, and were deemed unsuitable for this application. Second, TO-PRO-1’s fluorescence (exc. 515 nm, em. 531 nm) would allow the facile tracking of GadoTO by fluorescence microscopy or flow cytometry, two widely used techniques to evaluate probe specificity. Third, unlike some vital dyes, a TO-PRO-1-like derivative could be readily synthesized affording a free carboxylate for the attachment of a Gd chelator while maintaining the two positive charges needed for DNA binding. The synthesis and characterization of GadoTO are detailed, and its accumulation in dead cells was measured by fluorescence and MR-based techniques. Conversely, a non-fluorescent and non-DNA binding Gd chelate, Gd-DTPA, had no effect on T1.
Scheme 1.
Synthesis and structure of GadoTO 4
The synthesis of GadoTO 4 was performed in three steps from 1 (Scheme 1). A 6-aminohexanoate linker was installed on the iodopropyl chain by nucleophilic substitution (step a). Saponification of the ethyl ester, followed by reverse phase HPLC purification, provided the intermediate 2, featuring the two quaternary amines of TO-PRO-1 and an additional carboxylate handle (b). The acid was converted into an activated N-hydroxysuccinimide ester (c) and coupled with p-aminobenzyldi-ethylenetriaminepentaacetic acid (p-NH2-Bn-DTPA) to provide the intermediate 3 (d). The reaction required heating at 60 °C and the use of a large excess of base due to the low nucleophilicity and reactivity of the p-toluidine moiety. The paramagnetic Gd(III) was complexed with 3 to obtain GadoTO 4 as confirmed by MS-ESI (e).
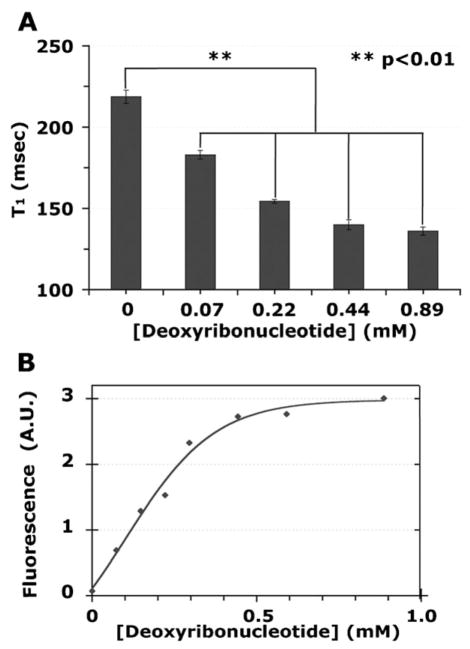
The interaction of GadoTO with DNA was examined by relaxometry and fluorescence. Increasing concentrations of DNA decreased the T1 (GadoTO 1 mM) from 220 to 135 msec (Fig. 1A) and produced a 50-fold increase in fluorescence (Fig. 1B). Samples from Fig. 1A were diluted for the more sensitive fluorescence measurements. To further characterize the interaction between GadoTO and DNA, we measured relaxation times as the concentrations of GadoTO were varied. With the addition of DNA, R1 increased from 4.23 ± 0.06 to 6.5 ± 0.2 (mM.sec)−1, while R2 increased from 4.80 ± 0.08 to 8.2 ± 0.1 (mM.sec)−1 (ESI) †. The R1 and R2 of GadoTO in the absence of DNA were similar to those of Gd-DTPA. Our data indicate that GadoTO binds to DNA and exhibits an increased fluorescence and relaxivity. The increased R1 likely reflects a slower tumbling time (τR) when GadoTO binds to DNA, since similar effects have been noted with many Gd chelates binding proteins.12 The half-maximal point for the T1 change was 0.089 mM(95%confidence interval 0.078–0.101) and was lower than the half-maximal concentration for fluorescence change (0.179 mM, 0.149–0.214). This may reflect differences in amplification of relaxivity (less than two fold) and fluorescence (fifty fold) upon DNA binding.
Fig. 1.
Binding of GadoTO to DNA by relaxometry and fluorescence.
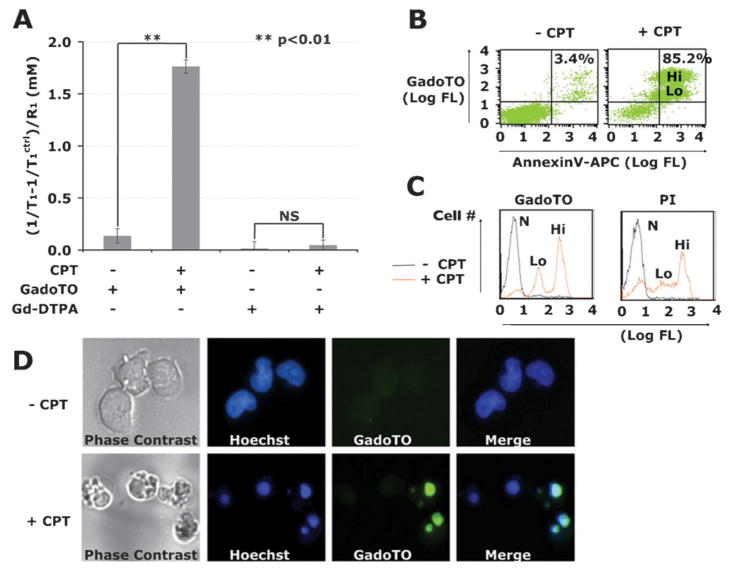
The interaction of GadoTO with camptothecin-treated (+CPT) and untreated (−CPT) Jurkat cells was studied by relaxometry, flow cytometry, and fluorescence microscopy. In all experiments, Jurkat T cells were treated with 10 μM CPT for 24 h, a common procedure for studying chemotherapy-induced cell death,13 and incubated with GadoTO (or the control chelate, Gd-DTPA). For relaxometry, cells were pelleted and lysed, and the T1 of cell lysates determined. Gd concentrations were obtained by the relation Δ(1/T1)/R1 (Fig. 2A). Accumulation of GadoTO was observed in CPT-treated cells while Gd-DTPA failed to accumulate. Dual-wavelength flow cytometry with suspended untreated cells indicated 3.4% cells bound both GadoTO and annexinV (upper right quadrant) (Fig. 2B). In our hands, healthy populations of various cell lines, like the Jurkat T cells employed here, typically have 1 to 10% of cells in the annexinV positive, PI positive quadrant. The binding of GadoTO to untreated cells was therefore limited and typical. Two populations of annexinV positive, GadoTO positive cells were noticeable with CPT treatment denoted as “Lo” or “Hi”. We compared the uptake of GadoTO with a “gold standard” vital dye, PI, using single channel flow cytometry (Fig. 2C). Three distinct populations of cells were apparent: non-binding/normal cells (N), low PI and low GadoTO binding cells (Lo), and high PI and high GadoTO binding cells (Hi). Binding of GadoTO and PI to CPT-treated cells were similar. The intracellular distribution of GadoTO was visualized by microscopy (Fig. 2D). Phase contrast (PC) images showed membrane blebbing with CPT-treated cells, absent in healthy cells. Hoechst 33342 uptake showed a diffuse, nuclear staining with healthy cells, while CPT-treated cells showed highly condensed nuclei. Healthy cells failed to take GadoTO, while most CPT-treated cells were positive for GadoTO confirming the presence of GadoTO in condensed nuclei of those cells.
Fig. 2.
Accumulation of GadoTO in CPT-treated cells by relaxometry and cytometry.
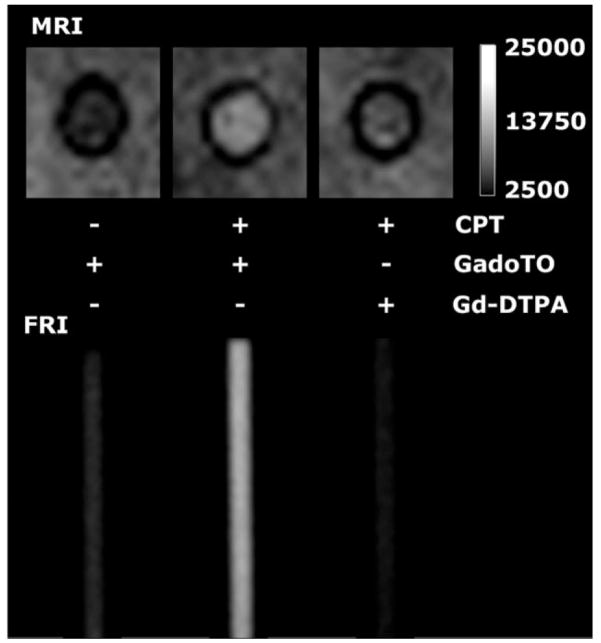
To demonstrate the ability of GadoTO to image cell death, cells were suspended in matrigel in 3 mm (ID) capillary tubes (Fig. 3). Tubes were imaged using a T1-weighted pulse sequence (MRI) or subjected to fluorescence reflectance imaging (FRI). CPT-treated cells took up GadoTO and were hyperintense (bright), consistent with the binding of GadoTO to DNA in dead/necrotic cells. FRI confirmed the uptake of GadoTO. The MRI of CPT-treated cells was notably brighter when cells were exposed to GadoTO instead of Gd-DTPA, indicating the important role played by the DNA-binding TO moiety of GadoTO in image brightness of necrotic cells.
Fig. 3.
Imaging necrotic cells by MRI and FRI in capillary tubes.
Hence, GadoTO is a multimodal, fluorescent/water relaxation rate enhancing compound that enables the detection of cell death by MR or fluorescence. Future studies with GadoTO in animal models appear justified.
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available: Experimental procedures. See DOI: 10.1039/b907375b
Notes and references
- 1.Majno G, Joris I. Am J Pathol. 1995;146:3. [PMC free article] [PubMed] [Google Scholar]
- 2.Blankenberg FG. Cancer Biol Ther. 2008;7:1525. doi: 10.4161/cbt.7.10.6934. [DOI] [PubMed] [Google Scholar]
- 3.Sen S. Biol Rev Cambridge Philos Soc. 1992;67:287. doi: 10.1111/j.1469-185x.1992.tb00727.x. [DOI] [PubMed] [Google Scholar]
- 4.Narula J, Acio ER, Narula N, Samuels LE, Fyfe B, Wood D, Fitzpatrick JM, Raghunath PN, Tomaszewski JE, Kelly C, Steinmetz N, Green A, Tait JF, Leppo J, Blankenberg FG, Jain D, Strauss HW. Nat Med. 2001;7:1347. doi: 10.1038/nm1201-1347. [DOI] [PubMed] [Google Scholar]
- 5.Korngold EC, Jaffer FA, Weissleder R, Sosnovik DE. Heart Failure Rev. 2008;13:163. doi: 10.1007/s10741-007-9068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blankenberg FG, Katsikis PD, Storrs RW, Beaulieu C, Spielman D, Chen JY, Naumovski L, Tait JF. Blood. 1997;89:3778. [PubMed] [Google Scholar]
- 7.Valonen PK, Lehtimaki KK, Vaisanen TH, Kettunen MI, Grohn OH, Yla-Herttuala S, Kauppinen RA. J Magn Reson Imaging. 2004;19:389. doi: 10.1002/jmri.20026. [DOI] [PubMed] [Google Scholar]
- 8.Schellenberger EA, Sosnovik D, Weissleder R, Josephson L. Bioconjugate Chem. 2004;15:1062. doi: 10.1021/bc049905i. [DOI] [PubMed] [Google Scholar]
- 9.Sosnovik DE, Schellenberger EA, Nahrendorf M, Novikov MS, Matsui T, Dai G, Reynolds F, Grazette L, Rosenzweig A, Weissleder R, Josephson L. Magn Reson Med. 2005;54:718. doi: 10.1002/mrm.20617. [DOI] [PubMed] [Google Scholar]
- 10.Lee LG, Chen CH, Chiu LA. Cytometry. 1986;7:508. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 11.Fechter EJ, Olenyuk B, Dervan PB. J Am Chem Soc. 2005;127:16685. doi: 10.1021/ja054650k. [DOI] [PubMed] [Google Scholar]
- 12.Lauffer RB. Magn Reson Q. 1990;6:65. [PubMed] [Google Scholar]
- 13.Poot M, Gibson LL, Singer VL. Cytometry. 1997;27:358. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.