Abstract
Nurr1, an orphan nuclear receptor, plays an essential role in the generation and maintenance of dopaminergic neurons in the brain. Rare mutations in Nurr1 are associated with familial Parkinson’s disease, but the underlying basis for this relationship has not been established. Here, we demonstrate that Nurr1 unexpectedly functions to inhibit expression of pro-inflammatory neurotoxic mediators in both microglia and astrocytes. Reduced Nurr1 expression results in exaggerated inflammatory responses in microglia that are further amplified by astrocytes, leading to the production of factors that cause death of tyrosine hydroxylase-expressing neurons. Nurr1 exerts anti-inflammatory effects by docking to NF-κB-p65 on target inflammatory gene promoters in a signal-dependent manner. Subsequently, Nurr1 recruits the CoREST corepressor complex, resulting in clearance of NF-κB-p65 and transcriptional repression. These studies suggest that Nurr1 protects against loss of dopaminergic neurons in Parkinson’s disease in part by limiting the production of neurotoxic mediators by microglia and astrocytes.
INTRODUCTION
Parkinson’s disease (PD) is the most prevalent movement disorder among people over 65 years old. Denervation of dopaminergic neurons in the substantia nigra (SN) results in severely debilitating motor symptoms such as bradykinesia, resting tremor and rigidity (Farrer, 2006; Fearnley and Lees, 1991). Although the etiologies of most common forms of PD remain poorly understood, the disease is generally associated with an inflammatory component that is manifested in part by the presence of activated microglia (central nervous system-resident macrophages) and elevated serum or cerebrospinal fluid levels of pro-inflammatory factors (Block et al., 2007; McGeer and McGeer, 2008; Nagatsu and Sawada, 2005). Several lines of evidence suggest that inflammatory mediators such as tumor necrosis factor (TNF)α, nitric oxide (NO) and Interleukin (IL)-1β derived from non-neuronal cells, including microglia, modulate the progression of PD (Brown, 2007; Hartmann et al., 2003; Whitton, 2007). Whether inflammation is an initiating factor of PD in humans is unclear, but intracranial infusion of bacterial lipopolysaccharide (LPS), a ligand for Toll-like receptor (TLR)4 and a potent activator of microglia, is sufficient to induce the loss of TH+ neurons in rodents (Meredith et al., 2008). LPS-induced inflammation can also synergize with α-Synuclein and Parkin mutations associated with familial PD to potentiate the loss of tyrosine hydroxylase (TH)+ neurons in animal models (Frank-Cannon et al., 2008; Gao et al., 2008). These observations are consistent with the possibility that environmental factors, such as infection, may interact with common but less penetrant susceptibility genes to influence the onset of most commonly observed sporadic PD cases (Tansey et al., 2007).
TLRs induce inflammatory gene expression by regulating the activities and expression of signal-dependent transcription factors that include members of the NF-κB, AP-1 and IRF families (Kawai and Akira, 2007). The TLRs play essential roles in innate immune responses to microbial pathogens based on their ability to recognize pathogen-associated molecular patterns (Akira et al., 2006; Medzhitov, 2007). More recently, genetic loss-of-function experiments in mice have shown that TLRs contribute to the pathogenesis of a number of diseases in which inflammation is known to play a pathogenic role, including atherosclerosis, inflammatory bowel disease, and liver fibrosis (Atkinson, 2008). These results are consistent with TLRs being able to signal in response to the generation of endogenously derived ligands, such as components of necrotic cells.
Nurr1 (NR4A2) belongs to the nuclear receptor (NR)4 family of orphan nuclear receptors and is known to function as a constitutively active transcription factor by binding to target genes as a monomer, homodimer or heterodimer with retinoid X receptors (RXRs) (Aarnisalo et al., 2002; Maira et al., 1999; Wang et al., 2003). Deletion of the Nurr1 gene in mice results in a severe reduction in dopaminergic neurons and perinatal lethality (Zetterstrom et al., 1997), consistent with an essential role for Nurr1 in the development and/or maintenance of dopaminergic neurons. Human mutations resulting in reduced expression of Nurr1 are associated with late-onset familial PD (Le et al., 2003), indicating that Nurr1 may play a protective role.
Nurr1 is also expressed in non-neuronal cell types and Nurr1 mRNA is induced by inflammatory stimuli, including LPS, in macrophages (Barish et al., 2005; Pei et al., 2005). Intriguingly, recent observations suggest that members of the NR4 family can function as both activators and repressors of cell type-specific inflammatory responses. For instance, Nurr1 suppresses expression of inflammatory response genes in human macrophages that are implicated in the development of arthrosclerosis (Bonta et al., 2006). In contrast, Nurr1 promotes the development of mouse and human Th17 T cells that contribute to the pathogenesis of multiple sclerosis (Doi et al., 2008). However, the molecular mechanisms by which Nurr1 controls transcriptional activation or repression of inflammatory responses have not been established, and the potential impact of Nurr1 function on the inflammatory component of PD has not been evaluated.
Here, we present evidence that Nurr1 plays an essential role in both microglia and astrocytes as a signal-dependent transcriptional repressor of genes that encode pro-inflammatory neurotoxic factors. Nurr1 functions as a key component of a negative feedback loop in both microglia and astrocytes by recruiting CoREST co-repressor complexes to NF-κB target genes. CoREST complexes, in turn, mediate the turnover of NF-κB and restore activated gene expression to a basal state. Loss of Nurr1 function in extraneuronal cells of the SN results in exaggerated and prolonged inflammatory responses that accelerate the loss of dopaminergic neurons in response to LPS or overexpression of a mutant form of α-Synuclein (A30P) associated with familial PD (Kruger et. al., 1998).
RESULTS
Nurr1 protects TH+ neurons from LPS-induced inflammation in vivo
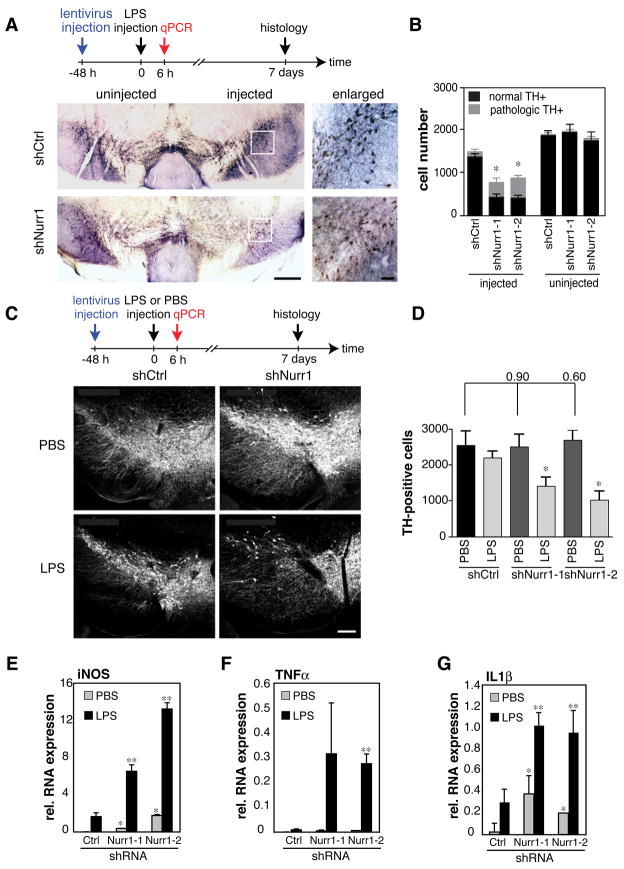
Studies demonstrating that Nurr1 mRNA is upregulated by LPS in macrophages (Barish et al., 2005; Pei et al., 2005) raised the question of whether it might also be expressed in non-neuronal cells and influence the development of PD. Analysis of Nurr1 protein and mRNA levels in primary human and mouse microglia, primary human astrocytes, and the BV2 microglia cell line demonstrated significant protein expression under basal conditions and induction of Nurr1 mRNA in microglia in response to LPS (Fig. S1A–E and data not shown). Similarly, Nurr1 protein colocalized with the microglia marker F4/80 (Fig. S2E) and Nurr1 mRNA was induced approximately 2-fold in the SN 6h following stereotaxic injection of LPS (Fig. S1F). To investigate the potential role of Nurr1 in PD pathology, we evaluated the impact of reducing Nurr1 expression. Since Nurr1-deficient mice die shortly after birth, we performed stereotaxic injections of lentiviruses encoding two independent shRNAs against Nurr1 (shNurr1-1 and shNurr1-2) or control shRNA (shCtrl) into the SN of adult wild-type mice (Fig S2A). shNurr1-1 and -2 efficiently and specifically reduced Nurr1 mRNA and protein expression in the SN, as determined by qPCR and immunostaining (Fig. S2B–E). A comparison of lentivirus-directed GFP expression with cell-specific markers indicated preferential transduction of non-neuronal cells, including microglia and astrocytes (Fig. S2G). The lentiviral injection was followed two days later by injection of LPS into the same coordinates. We then analyzed the magnitude of the inflammatory response by qPCR 6h after LPS injection and quantified TH+ neurons by immunohistochemistry (IHC) 7 days after LPS injection.
Loss of TH+ neurons following LPS injection normally takes 2–3 weeks (Meredith et al., 2008). However, stereological analysis revealed a significant decrease in TH+ neurons in the SN of shNurr1 lentivirus-injected mice compared to shCtrl-injected animals after only 7 days of LPS treatment (Fig. 1A and B). Interestingly, a pathological morphology of TH+ neurons with reduced or absent processes and alterations in the size and shape of the cells was observed more often in the shNurr1 groups (Supp Fig. 2E and Fig. 1B). In addition, pathological TH+ cells were observed close to activated microglia (Fig. S2F). The accelerated loss of TH+ neurons following Nurr1 knockdown required LPS injection, as it was not observed in buffer (PBS)-injected animals (Fig. 1C and D). In addition, LPS injection was associated with detection of caspase-3 cleavage, suggesting that loss of TH+ cells was due to cell death rather than to loss of TH expression (Fig. S2G) (Sakurada et al., 1999). Reduction of Nurr1 expression in the SN also resulted in exaggerated expression of inflammatory mediators in response to LPS injection, including iNOS, TNFα and IL1β (Fig. 1E–G).
Fig. 1. Nurr1 suppresses LPS-induced inflammation and loss of TH+ neurons.
A. TH-DAB staining of a representative brain section of mice injected with shCtrl- or shNurr1-lentivirus and LPS is shown at AP −3.3 mm. Regions indicated by a rectangle in the injected side of the brain are enlarged in the right panels. Scale bars: 200 μm, right panels and 50 μm, left panels. B. Quantification of TH+ cell numbers in the shNurr1 groups compared to the shCtrl-injected and the uninjected side. Bars indicate normal (black) and pathological (gray) TH+ neurons. See Supplemental Experimental Procedures for the definition of normal/pathological TH+ neurons. Asterisk, p<0.01 compared to the numbers from shCtrl-lentivirus-injected. (n=5) C. Fluorescence-TH staining of a representative brain section of mice injected with shCtrl- or shNurr1-lentivirus followed by PBS or LPS. Experimental diagram is indicated at the top. Scale bar 200 μm. D. Quantification of TH+ cell numbers in the setting of Nurr1 knockdown followed by LPS or PBS injection. Asterisk, p<0.002 compared to PBS injection (n=4). E–G. Expression of iNOS (E), TNFα (F) and IL1β (G) mRNA in Nurr1-knockdown SN 6 hours after LPS injection as determined by qPCR and normalized to HPRT expression (n=4). Error bars represent SD. Asterisk, p<0.01 compared to shCtrl/PBS-injected; **, p<0.01 compared to shCtrl/LPS-injected samples.
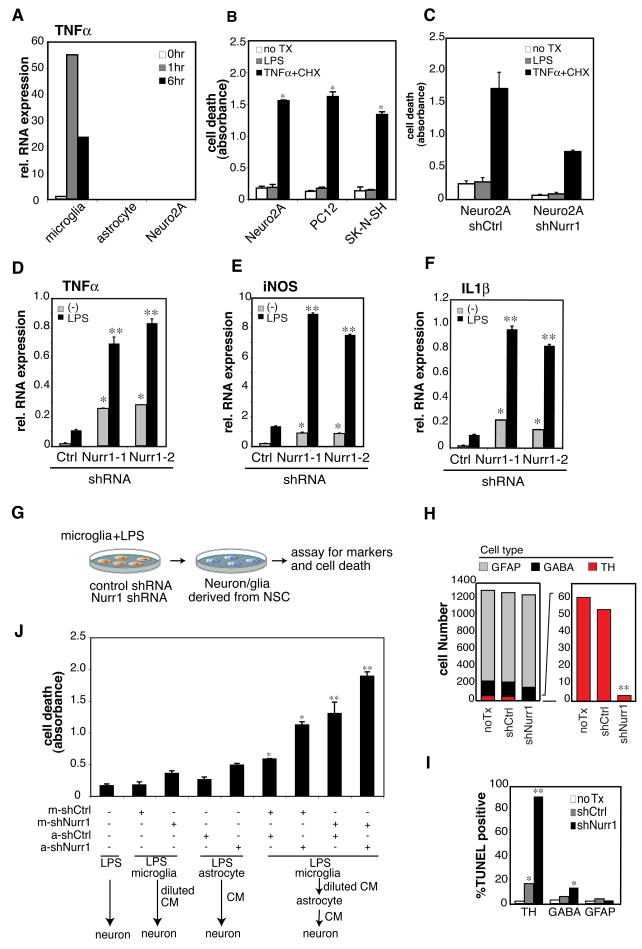
Fig. 2. Microglia initiate LPS-mediated inflammation and astrocytes propagate the production of neurotoxic factors.
A. Expression of TNFα mRNA upon LPS stimulation in primary microglia, astrocytes and Neuro2A cells. B. Effect of LPS or TNFα + cycloheximide (CHX) on viability of the indicated neuronal cell lines determined using a TUNEL ELISA assay. *, p<0.01 compared to untreated sample (no Tx) (white). C. Effect of Nurr1 knockdown in Neuro2A cells on sensitivity to TNFα + CHX assessed by TUNEL ELISA assay. D–E. Effect of Nurr1 knockdown inBV2 cells on LPS-induced expression of TNFα (D), iNOS (E) and IL1β (F) mRNA.*, p<0.01 compared to no stimulation (Ctrl); **, p<0.01 compared to LPS stimulation of shCtrl-BV2 cells. G. Scheme of conditioned media (CM) and cell death assay. CMs were harvested from shCtrl- and shNurr1-BV2 cells that were stimulated with LPS for 24h. Neurons or glial cells were assayed for specific markers by immunostaining and for cell death by TUNEL assay. H. Effect of CM from LPS-treated shCtrl-BV2 cells and a mixture of the CMs from shNurr1-1- and shNurr1-2-infected BV2 cells (shNurr1) on neurons and glial cells derived from in vitro differentiated neural stem cells (NSC). TUNEL assay was performed on TH-, GABA- or GFAP-positive cells derived from mouse NSC. Numbers of TUNEL-negative live cells are shown. TH-positive cells are indicated in red. I. The percentages of TUNEL-positive population are shown. *, p<0.01 and **, p<0.001 compared to no treatment (no TX). J. Effect of Nurr1 knockdown in microglia and astrocytes on the production of neurotoxic factors. Primary mouse microglia and astrocytes were infected with shCtrl- or shNurr1-lentivirus. Cells were treated with LPS for 2h and washed extensively with PBS. Cells were cultured for another 24h to generate CM. For sequential CM assay, CMs harvested from microglia were cultured with lentivirus-infected astrocytes for 24 hours. Then, CMs were harvested and tested for effect on viability of Neuro2A cells.
We also examined whether Nurr1 exerted neuro-protective effects in the context of overexpression of an α– Synuclein mutant (A30P) associated with familial PD by combining stereotaxic injection of shNurr1- or shCtrl-lentivirus with a lentivirus encoding mutant α –Synuclein (A30P). A30P expression alone caused weak inflammation in the SN, whereas reduction of Nurr1 expression in the context of A30P expression resulted in a dramatic increase in expression of numerous inflammatory response genes, including TNFα and IL1β, and significant loss of TH+ neurons (Fig. S3A–C and data not shown). In concert, these experiments indicate that Nurr1 limits inflammatory responses in the CNS and protects TH+ neurons from LPS- and α-Synuclein (A30P)-induced toxicity.
Glia-mediated inflammation contributes to the death of TH+ neurons
To define the cell types responsible for LPS-mediated inflammation in the SN, we evaluated the responses of human and mouse microglia, astrocytes and neurons to LPS. These experiments demonstrated that microglia are orders of magnitude more responsive than astrocytes or neurons, exemplified by the pattern of TNFα induction in primary mouse and human microglia and astrocytes and the neuronal Neuro 2A (mouse neuroblastoma) cell line (Fig. 2A, Fig. S4F). These results are consistent with the expression patterns of TLR4, co-receptors and down-stream signaling molecules in neurons and glial cells (Fig. S4A–E). Although, TLR4 expression was virtually absent from the neuronal cell lines examined, we tested whether LPS could directly induce the death of these cells. Three different neuronal cell lines, Neuro2A, SK-N-SH and PC12, were incubated with LPS for 24h. No significant cell death was observed by TUNEL assay or caspase-3 cleavage, in contrast to the effects of TNFα plus cyclohexamide (CHX) treatment (Fig. 2B and Fig. S4G). In addition, knockdown of Nurr1 in Neuro2A cells did not increase the sensitivity to LPS or death signaling (TNFα plus CHX) as determined by TUNEL assay (Fig. 2C).
Based on these results, we evaluated the consequences of reducing Nurr1 expression in microglia on LPS responses. Knockdown of Nurr1 expression in BV2 microglia using specific lentivirus-encoded shRNAs led to significant increases in LPS-dependent expression of inflammatory mediators, including TNFα, iNOS and IL-1β (Fig. 2D-F). Similar results were observed in the primary mouse (Fig. S5 and S6A-C) and human (data not shown) microglia. To explore whether loss of Nurr1 in microglia resulted in secretion of mediators exhibiting preferential toxicity for TH+ neurons, we knocked down Nurr1 expression using lentivirus-encoded shRNAs in BV2 cells and tested the activity of conditioned media (CM) after LPS stimulation on in vitro differentiated neurons and glial cells derived from mouse neuronal stem cells (NSC). CM from shNurr1-BV2 cells resulted in the death of nearly all TH+ neurons, with a significantly smaller effect on gamma-aminobutyric acid (GABA)-positive neurons and no significant effect on glial fibrillary acidic protein (GFAP)-positive astroglial cells (Fig. 2G–I).
Experiments using neuron and glia co-culture in vitro suggest that activation of innate immunity in the CNS can trigger neuronal death (Lehnardt et al., 2003). Since NSC-derived neurons always co-exist with astrocytes, it is possible that astrocytes contributed to the neurotoxic effect of the microglia CM. To explore this possibility, we performed sequential CM experiments employing isolated primary microglia and astrocytes and using Neuro2A cells as a read-out for neurotoxicity. Primary murine astrocytes and microglia were infected with shCtrl- or shNurr1-lentivirus used for the injection into the SN. Cells were then stimulated with LPS and CM was harvested as described in Figure 2G. CM of microglia infected with shNurr1 induced significant cell death in Neuro2A cultures, whereas CM of astrocytes infected with shNurr1 had much less effect on the death of Neuro2A cells. Intriguingly, sequential conditioning of media from microglia to astrocytes indicated that astrocytes significantly amplified the production of neurotoxic factors when exposed to microglia-conditioned media (Fig. 2J lane 2 to lane 6 and 7). This effect was further increased when expression of Nurr1 was reduced in astrocytes (Fig. 2J lane 3 to lane 8 and 9). We conclude that microglia are the initial responders to LPS-mediated inflammation and that astrocytes amplify the production of neurotoxic factors after the microglial activation. The knockdown of Nurr1 in either microglia or astrocytes increases the toxicity of CM, suggesting that Nurr1 inhibits the production of neurotoxic factors in both cell types.
Nurr1-mediated transrepression requires GSK3β-dependent recruitment of Nurr1 monomers to p65
Chromatin immunoprecipitation (ChIP) experiments indicated that Nurr1 was recruited to LPS-responsive promoters following LPS treatment, exemplified by the TNFα promoter (Fig. 3A), suggesting that it was acting locally to repress transcription. Two different general mechanisms of NR-mediated repression have been described: active repression, involving sequence-specific DNA binding, and transrepression, involving tethering of NRs to negatively regulated target genes via protein-protein interactions (Glass and Ogawa, 2006). A mutant of Nurr1 (Nurr1C280A/E281A, CEAA), defective for sequence-specific DNA binding and unable to activate NGFI-B responsive element (NBRE)-luciferase, a reporter for Nurr1 monomer-binding, was fully able to repress iNOS induction by LPS (Fig. 3B and Fig. S6D). In addition, mutations directed at the heterodimerization (I-box) domain (Aarnisalo et al., 2002) of Nurr1 (Nurr1K555A/L556A/L557A, KLL) that prevented its ability to activate a Nurr1/RXR-dependent (DR5)-promoter did not interfere with Nurr1-mediated repression of iNOS (Fig. 3B). On the other hand, this I-box mutation increased transcriptional activation of Nurr1 monomers through the NBRE element, as previously reported (Aarnisalo et al., 2002) (Fig. S6D).
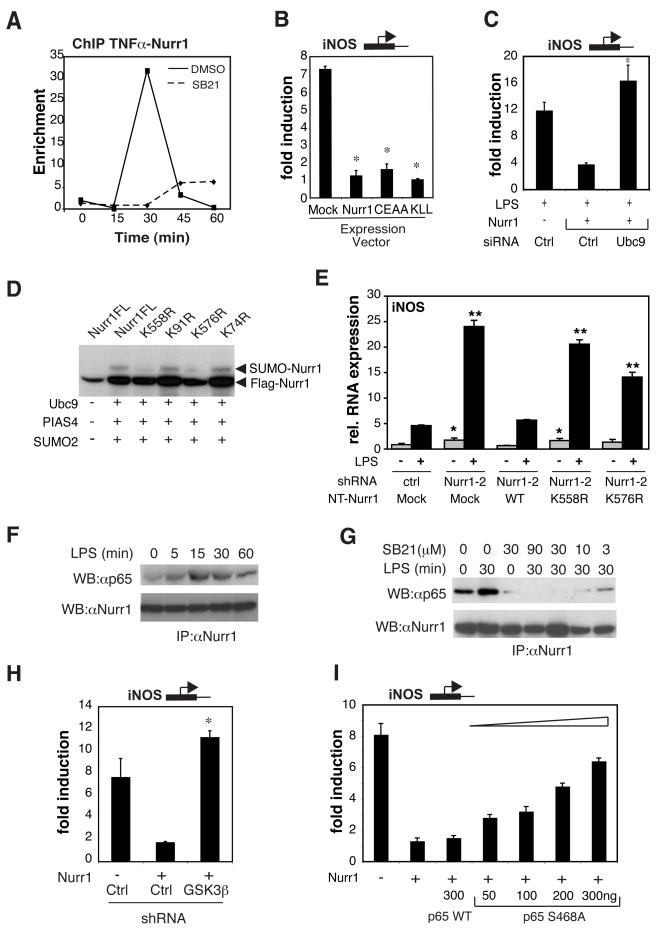
Fig. 3. Nurr1 acts as an RXR-independent, GSK3.
β-dependent transrepressor for NF-κB. A. ChIP assay of Nurr1 on the TNFα-promoter in response to LPS and effect of the GSK3β-specific inhibitor SB216763 (SB21). BV2 cells were pre-incubated with DMSO or 30 μM SB21 for 1h followed by LPS stimulation for the indicated times before ChIP assay. Data are displayed as fold enrichment over control IgG. B. Repression activities of Nurr1 mutants. RAW264.7 cells were transfected with wild type Nurr1, P-box mutant (CEAA) and I-box mutant (KLL). iNOS-promoter activity in RAW264.7 cells in response to LPS was measured by luciferase-reporter assay. *, p<0.01 compared to control (Mock). C. Effect of knockdown of Ubc9 on Nurr1 repression of iNOS-promoter activity. *, p<0.01 compared to Nurr1 with control siRNA. D. Identification of the predominant SUMOylation sites of Nurr1. Flag-tag mutants of Nurr1 were transfected into Hela cells. SUMOylation assay was performed as described in Supplemental Experimental Procedure. E. Effect of reconstitution of Nurr1 shRNA-2 BV2 cells with non-targeted (NT) WT and mutant forms of Nurr1 that are not recognized by shNurr1-2. Endogenous iNOS mRNA levels are shown relative to levels in untreated BV2 cells transduced with control shRNA and mock Nurr1 lentivirus. *, p<0.01 compared to mock control cells. F. Effect of LPS stimulation on interaction of Nurr1 and p65 in BV2 cells. Lysates of BV2 cells stimulated with LPS for the indicated times were immunoprecipitated with anti-Nurr1 antibody and Western blots were developed with anti-p65 antibody. G. Effect of SB21 on Nurr1/p65 interaction. BV2 cells were incubated with SB21 at the indicated concentrations for 1h prior to stimulation with LPS. IP and Western blotting were performed as described in F. H. Effect of siRNA against GSK3β on Nurr1-mediated repression of iNOS-promoter activity. Nurr1 expression vector was transfected into RAW264.7 cells together with control siRNA or siRNA against GSK3β *, p<0.01 compared to Nurr1 with control siRNA. I. Effect of S468A mutation of p65 on Nurr1-mediated repression of iNOS-promoter activity.
SUMOylation of NRs has recently been established to play important roles in transrepression (Pascual et al., 2005). Since it is known that Nurr1 interacts with the protein inhibitor of activated STAT (PIAS) 4 (Galleguillos et al., 2004), which is a SUMO E3 ligase, we examined whether SUMOylation is also involved in Nurr1-mediated repression. As shown in Figure 3C, knockdown of Ubc9, an essential E2 enzyme for SUMOylation (Hay, 2005), reversed Nurr1-mediated repression of iNOS, suggesting that SUMOylation is required. Next, we confirmed that Nurr1 could be SUMOylated with SUMO2 and SUMO3 using PIAS4 as an E3 ligase (Fig. S6E and F) and found that IL1β stimulation could induce SUMOylation of Nurr1 in the absence of overexpression of PIAS4 (Fig. S6F). Mutational studies demonstrated that lysine 558 and, to a lesser extent lysine 576, are essential SUMO sites of Nurr1 (Fig. 3D). Since both K558R and K576R mutants are located in the ligand binding domain and close to the I-box and RXR is not required for repression activity (Fig. 3B and Fig. S6D), we hypothesize that SUMOylation is required for monomerization of Nurr1. Consistent with this, K558R and K576R mutants were less able to activate the NBRE reporter and preferentially activated the DR5 reporter (Fig. S6G). In addition, reconstitution of BV2 microglia cells expressing the Nurr1 shRNA-2 with a non-targeted (NT) form of WT Nurr1 reversed hyperactivation of the endogenous iNOS and TNFα genes, while the non-targeted forms of the SUMOylation mutants (Nurr1 K558R and K576R) did not (Fig. 3E, Fig S6H–I).
Since transrepression requires the tethering of NRs to other transcription factors, we tested whether Nurr1 could bind to transcription factors involved in inflammation, such as NF-κB. Co-immunoprecipitation (Co-IP) assays of Nurr1 in BV2 cells showed interaction with NF-κB-p65 that was significantly enhanced by LPS treatment and independent of changes in Nurr1 protein levels (Fig. 3F and Fig. S6A). Phosphorylation of Serine-468 (S468) in p65 is associated with negative regulation of NF-κB signaling (Buss et al., 2004) and can be mediated by GSK3β, which is activated following TLR4 stimulation in human monocytes (Martin et al., 2005). Furthermore, inactivation of GSK3β results in increased NF-κB-dependent transcription of TNFα without changing the kinase activity of the IKK complex or the nuclear translocation of p65 (Buss et al., 2004). Therefore, we hypothesized that S468 phosphorylation of p65 by GSK3β might provide the docking site for tethering of Nurr1. Consistent with this possibility, the GSK3β-specific inhibitor SB216763 (SB21) inhibited the interaction of Nurr1 and p65 in BV2 cells in a dose-dependent manner (Fig. 3G and Fig. S7D) and prevented the recruitment of Nurr1 to the TNFα-promoter, as determined by ChIP assay (Fig. 3A). To further confirm GSK3β involvement, we performed TNFα-luciferase reporter assays in RAW264.7 cells cotransfected with a kinase-dead mutant of GSK3β (GSK3β-K85R mutant, GSK3β-KD). GSK3β-KD expression abolished the Nurr1-mediated transrepression of the TNFα-promoter in a dose-dependent manner (Fig. S7B). Furthermore, knockdown of GSK3β completely prevented Nurr1-mediated iNOS repression (Fig. 3H). We further validated the contribution of phospho-S468 in p65 by exchanging S468 for alanine (S468A). The p65 S468A mutant, but not wild-type p65, reversed Nurr1-mediated iNOS repression in RAW264.7 cells in a dose-dependent manner (Fig. 3I and Fig. S7E). Finally, GSK3β stimulated the in vitro interaction of Nurr1 with wild-type p65 but not with p65-S486A (Fig. S7C).
The CoREST-repressor complex is required for Nurr1-mediated transcriptional repression
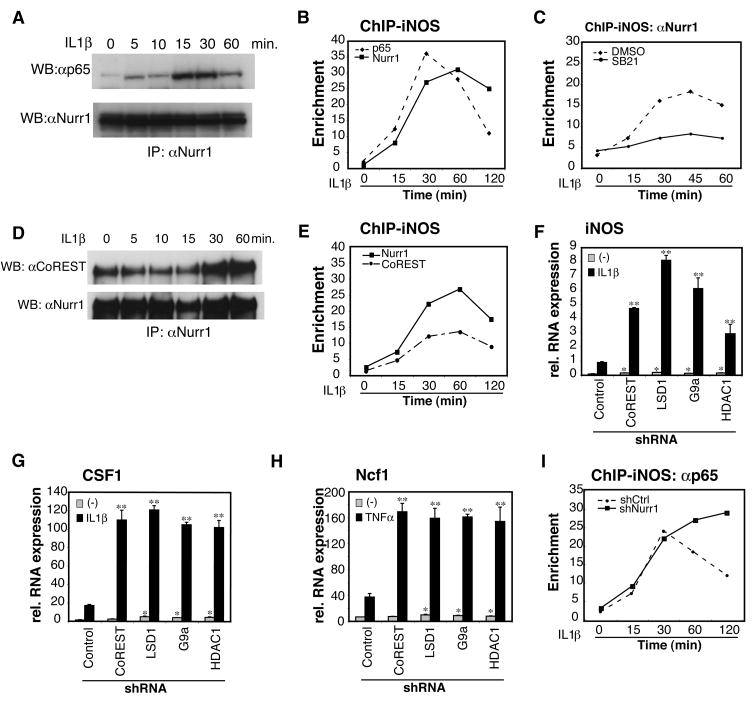
Transcriptional repression requires the recruitment of multiprotein complexes assembled on central scaffolding proteins referred to as co-repressors. We used siRNAs against various candidate corepressors in the iNOS-luciferase reporter assay and identified CoREST as being essential for Nurr1-mediated repression (Fig. 4A and Fig. S8A). CoREST has been considered to be dedicated to repression of neuronal genes in non-neuronal cells or early precursors by binding to neuron-restrictive silencer factor (NRSF)/RE1-silencing transcription factor (REST) (Ballas et al., 2005). CoREST assembles many chromatin-modifying enzymes, including histone methyltransferase G9a, histone demethylase, lysine-specific demethylase (LSD1) and histone deacetylase (HDAC) 1 and 2 (Shi et al., 2003). Using Nurr1-mediated repression of iNOS-luciferase as an assay, we observed that G9a, LSD1 and HDAC1 were also required for Nurr1-CoREST-mediated repression (Fig. S8B). Using co-IP, we also found that LPS stimulated interaction of Nurr1 and CoREST in BV2 cells (Fig. 4B and Fig. S8C). Although the CoREST complex consists of many proteins, the interaction between Nurr1 and CoREST seemed to be direct, as indicated by in vitro GST-pull down assay (Fig. S8D). This interaction was mediated by the DNA-binding domain of Nurr1 (Nurr1-DBD) (Fig. S8E). When Nurr1-DBD was overexpressed in Hela cells, the interaction between Nurr1 and CoREST was inhibited in a dose-dependent manner (Fig. S8F). Furthermore, overexpression of the Nurr1-DBD in RAW264.7 cells altered Nurr1-mediated repression of iNOS-promoter activity (Fig. S8G).
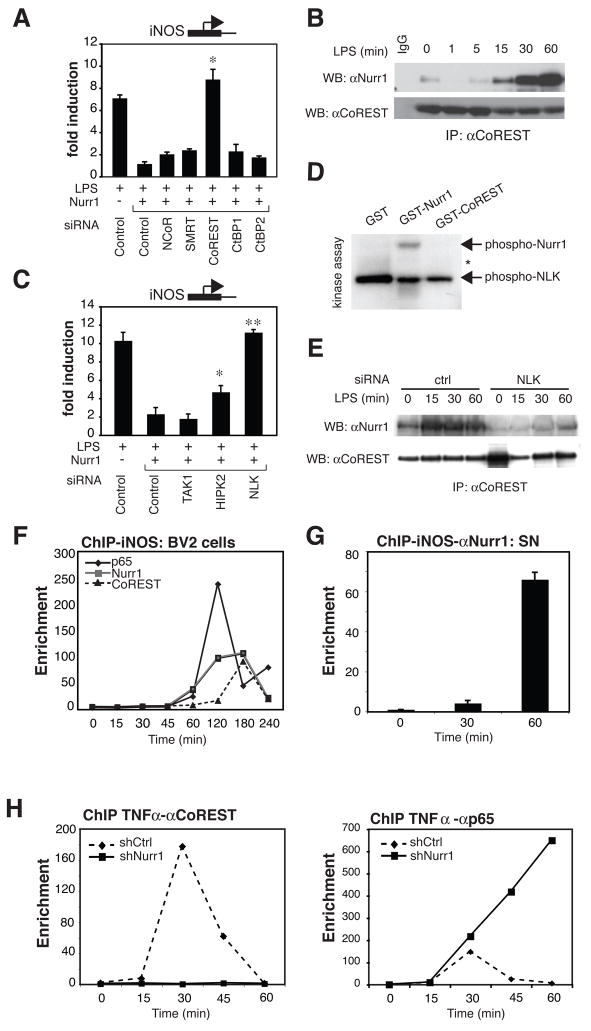
Fig. 4. CoREST repressor complex is required for Nurr1-mediated repression.
A. Corepressor requirements in Nurr1-mediated repression. iNOS-luciferase and Nurr1-expression or control vector as well as siRNAs against the indicated corepressors were transfected into RAW264.7 cells and iNOS-promoter activity was assayed. *, p<0.01 compared to Nurr1 with control siRNA. B. Interaction of Nurr1 and CoREST in BV2 cells. Co-IP was performed with anti-CoREST antibody and Western blots were developed with anti-Nurr1 antibody. C. Effect of siRNAs against the indicated targets on Nurr1-mediated repression of iNOS-promoter activity. *, p<0.01, **, p<0.001 compared to Nurr1 with control siRNA. D. NLK in vitro kinase assay using GST-Nurr1 and GST-CoREST as substrates. Arrows indicate phosphorylated GST-Nurr1 and autophosphorylation of NLK. The migration position of GST-CoREST is indicated by an asterisk. GST substrates and methods are provided in Supplemental Experimental Procedures and Figure S7D. E. Effect of NLK knockdown on interaction of Nurr1 and CoREST. BV2 cells were transfected with siRNA against NLK or control siRNA. Co-IP of Nurr1 and CoREST was performed as described in B. F. Recruitment of Nurr1, CoREST and p65 to the iNOS promoter in BV2 cells shown by ChIP assay. Data represent fold enrichment of iNOS-promoter precipitated by the indicated antibodies compared to control IgG as determined by qPCR. G. ChIP analysis of Nurr1 on the iNOS promoter in the SN before and after LPS stimulation. Data are shown as averages of fold enrichment against control IgG and SD. H. Effect of Nurr1 knockdown on recruitment of CoREST (left) and p65 (right) to the TNFα promoter in BV2 cells. ChIP data are shown as fold enrichment over control IgG.
Since Nurr1 can be phosphorylated by serine/threonine kinases (Nordzell et al., 2004), we speculated that signal-dependent phosphorylation might contribute to the Nurr1-CoREST interaction. Nemo-like kinase (NLK) received our attention because NLK is known to be involved in the repression of various transcription factors (Yasuda et al., 2004). NLK cooperates with TGFβ-activating kinase 1 (TAK1) and homeodomain-interacting kinase 2 (HIPK2) in Wnt signaling (Kanei-Ishii et al., 2004). Knockdown of NLK abolished Nurr1 repression of iNOS-promoter activity, whereas HIPK2 knockdown was much less effective and TAK1 had no effect (Fig. 4C and Fig. S9A). Furthermore, overexpression of kinase-dead NLK (NLKK155M, NLK-KD) in RAW264.7 cells inhibited Nurr1-mediated repression of iNOS in a dose-dependent manner (Fig. S9B). Kinase assays showed that Nurr1, but not CoREST, could be phosphorylated by active NLK in vitro (Fig. 4D). Finally, Nurr1-CoREST interaction was significantly reduced by NLK-knockdown in BV2 cells (Fig. 4E).
To confirm whether CoREST was indeed localized to NF-κB target gene promoters in association with p65 and Nurr1, we performed ChIP assays of the iNOS- and TNFα-promoters in BV2 cells. The occupancy of NF-κB-p65, Nurr1 and CoREST on both the TNFα- and iNOS-promoters by all three proteins was strongly increased upon LPS stimulation (Fig. 4F, Fig. S9C). On the iNOS-promoter, which exhibits relatively slower activation kinetics, p65 binding preceded the binding of Nurr1, which in turn preceded recruitment of CoREST (Fig. 4F). To verify whether this system is indeed functional in vivo, we performed ChIP assays from microdissected SN after the stereotaxic injection of LPS into the SN. Consistent with in vitro data, Nurr1 is recruited to the iNOS- and TNFα–promoters after LPS stimulation (Fig. 4G, Fig. S9D). Finally, we asked whether Nurr1 was indeed essential for the recruitment of the CoREST complex to target gene promoters. ChIP experiments were performed using shNurr1- or shCtrl-BV2 cells. In the absence of Nurr1, CoREST was not recruited to the TNFα-promoter (Fig. 4H, left panel). Interestingly, under these conditions, p65 was present at the TNFα-promoter for extended times (Fig. 4H, right panel). Regulation of p65 acetylation by HDACs, including HDAC1, is known to determine the duration of transcription (Ashburner et al., 2001). HDAC1 is recruited to TNFα or iNOS-promoter in an LPS-dependent manner; however, this recruitment is severely impaired in the absence of Nurr1 (Fig. S9E). Finally, to verify an in vivo role for the genes identified to be involved in Nurr1/CoREST transrepression pathway, BV2 cells were transfected with siRNAs targeting their corresponding mRNAs and were tested for the ability to increase the production of neurotoxic factors. As shown in Figure S10A, knockdown of each of the molecules engaged in this Nurr1/CoREST-mediated transrepression pathway induced significantly higher death of Neuro2A cells compared to control siRNA, as detected by TUNEL ELISA assay
Nurr1 represses the production of neurotoxic factors in astrocytes
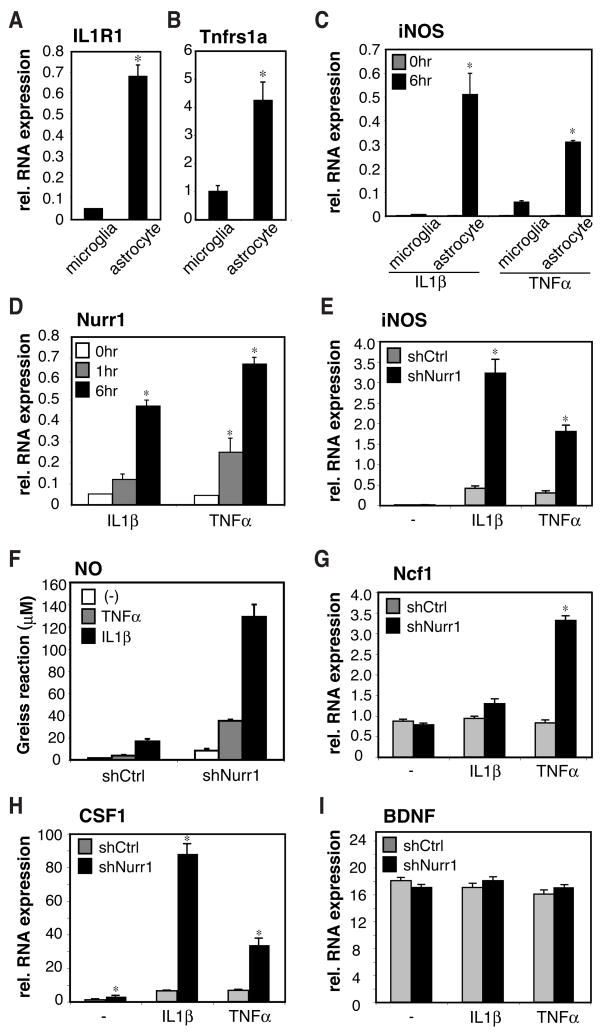
The observation that astrocytes could amplify the neurotoxic effects initiated by microglia (Fig. 2J) suggested that pro-inflammatory cytokines secreted by activated microglia such as TNFα and IL1β could activate the astrocytes and induce the transcription of inflammatory neurotoxic mediators (Fig. 2D–F and Fig. S6A–B). Consistent with this possibility, the receptors for TNFα and IL1β are highly expressed in primary mouse and human astrocytes, but not microglia (Fig. 5A–B, Fig. S11A–B). Nurr1 protein was expressed in resting human and mouse astrocytes (Fig. S1E and F) and Nurr1 mRNA was induced by IL1β or TNFα (Fig. 5C, D and Fig. S11C,D). To test the possibility that Nurr1 also participates in a signal-dependent negative feedback mechanism in astrocytes, primary mouse and human astrocytes were infected with shCtrl- and shNurr1-lentivirues. Activated astrocytes can up-regulate many pro- inflammatory genes, including the iNOS and Ncf1 genes upon IL1β and TNFα stimulation, which are essential enzymes for NO and reactive oxygen species (ROS) production, respectively. Knockdown of Nurr1 in astrocytes drastically increased mRNA expression of both iNOS and Ncf1 in response to IL1β and TNFα and up-regulated NO production (Fig. 5E–G and S11E-F). Furthermore, activated astrocytes can produce macrophage colony stimulating factor (CSF1), which supports the proliferation of microglia (Thery et al., 1992), and knockdown of Nurr1 significantly up-regulated the transcription of CSF1 gene upon both TNFα and IL1β stimulation (Fig. 5H and S11G). In contrast, transcription of brain-derived neurotrophic factor (BDNF), a known neurotrophin for dopaminergic neurons, was not affected by knockdown of Nurr1 (Fig. 5I and S11H). These data indicate that Nurr1 also acts as a transcriptional repressor for inflammatory neurotoxic mediators in astrocytes.
Fig. 5. Nurr1 suppresses inflammatory mediators in murine astrocytes.
A and B. Expression of IL1R1 (A) and p55TNFR (B) mRNA in primary astrocytes and microglia as determined by qPCR assay. *, p<0.01. C. Responses of primary mouse microglia or astrocytes to TNFα or IL1β for 6h. iNOS mRNA level was determined by qPCR. D. Regulation of Nurr1 mRNA in mouse primary astrocytes stimulated with TNFα or IL1β for the indicated times. E–I. Effect of knockdown of Nurr1 in astrocytes on induction of NO (F – measured by Greiss reaction) and mRNAs encoding neurotoxic mediators (E, G–I). Mouse primary astrocytes were infected with shCtrl- or shNurr1-lentivirus and cells were stimulated with TNFα or IL1β for 6h.
The Nurr1/CoREST transrepression pathway functions in astrocytes
Finally, we asked whether the mechanism of transcriptional repression by Nurr1 in astrocytes is similar to that in microglia. Treatment of primary mouse astrocytes with IL1β induced the interaction of Nurr1 with p65 (Fig. 6A) and induced recruitment of both Nurr1 and p65 to the iNOS-promoter (Fig. 6B). Recruitment of Nurr1 to the iNOS promoter was blocked by inhibition of GSK3β by SB21 (Fig. 6C), consistent with TLR4 and IL1β receptors sharing the MyD88 signaling pathway (Verstrepen et al., 2008). Nurr1 also interacted with CoREST in astrocytes in a manner that was stimulated by IL1β (Fig. 6D), and both molecules were recruited to the iNOS-promoter, as was observed in microglia (Fig. 6E). Knockdown of the components of CoREST repressor complex such as LSD1, G9a and HDAC1 also up-regulated iNOS, CSF1 and Ncf1 genes, suggesting that the CoREST-complex is required for Nurr1-mediated transcriptional repression in astrocytes (Fig. 6F–H). Finally, knockdown of Nurr1 in astrocytes resulted in prolonged occupancy of p65 on the iNOS promoter (Fig. 6I), similar to results obtained in microglia (Fig. 4H).
Fig. 6. The CoREST complex is required for Nurr1-mediated repression in astrocytes.
A. Effect of IL1β stimulation on association of Nurr1 and p65 in mouse primary astrocytes. Lysates of astrocytes stimulated with IL1β for the indicated times were immunoprecipitated with anti-Nurr1 antibody and Western blots were developed with anti-p65 antibody. B. Recruitment of Nurr1 and p65 to iNOS-promoter in mouse primary astrocyte shown by ChIP assay. Data represent fold enrichment of iNOS- promoter precipitated with the indicated antibodies compared to control IgG as determined by qPCR. C. Effect of GSK3β-specific inhibitor SB21 on recruitment of Nurr1 to iNOS-promoter. Data represent fold enrichment of iNOS-promoter precipitated with antibody against Nurr1 compared to control IgG as determined by qPCR. D. Interaction of Nurr1 and CoREST in mouse primary astrocytes. Co-IP was performed with anti-Nurr1 antibody and Western blots developed with anti-CoREST or anti-Nurr1 antibodies. E. Recruitment of Nurr1 and CoREST to iNOS-promoter in mouse primary astrocytes shown by ChIP assay. Data represent fold enrichment of iNOS-promoter precipitated with the indicated antibodies compared to control IgG as determined by qPCR. F–H. Effect of knockdown of the components of CoREST-repressor complex on mRNAs encoding inflammatory mediators. Mouse primary astrocytes were infected with lentivirus carrying shRNA against CoREST, LSD1, G9a, HDAC1 or control. Cells were stimulated with IL1β for 6h and mRNA expression of iNOS (F), CSF1 (G) and Ncf1 (H) was determined by qPCR. I. Nurr1-dependent clearance of p65 from iNOS promoter. ChIP assay was performed in shNurr1- or shCtrl-astrocyte and data shown as fold enrichment over control IgG of iNOS promoter precipitated with antibody against p65.
DISCUSSION
Nurr1 exerts anti-inflammatory and neuroprotective effects in glia
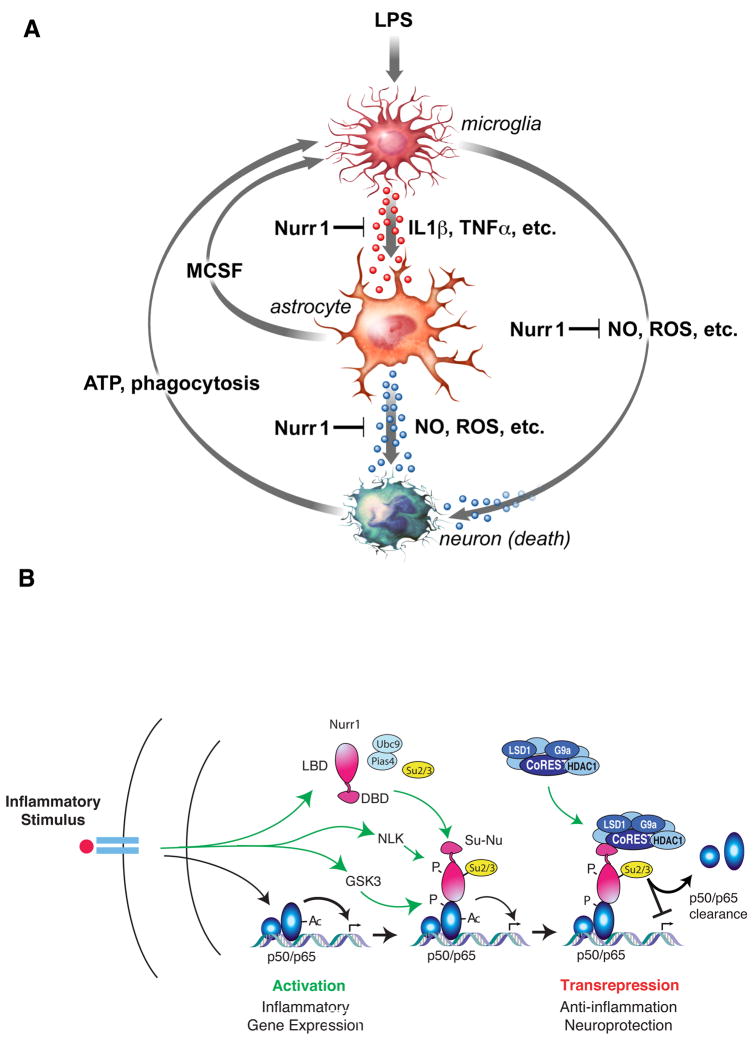
Although a number of genes have been identified as causes of familial PD, the majority of cases are sporadic and of unknown etiology (Farrer, 2006; Moore et al., 2005). An improved understanding of the causes of the more common forms of the disease will therefore be essential in developing broadly applicable treatment strategies. Here, we demonstrate that in addition to its essential roles in the development and maintenance of dopaminergic neurons, Nurr1 plays a previously unexpected role in protecting these neurons from inflammation-induced neurotoxicity. Several lines of evidence suggest that this role is due to its function as an inhibitor of inflammatory gene expression in microglia and astrocytes (Fig. 7A). First, these studies utilized a model system in which neurotoxicity was induced by LPS, which is not effectively sensed by neurons and does not directly cause neuronal death. Second, reduction of Nurr1 expression in the SN (primarily in microglia and astrocytes) did not in itself lead to reduction of TH+ neurons but did result in enhanced expression of inflammatory mediators and accelerated loss of TH+ neurons in response to LPS. Finally, reduction of Nurr1 expression in isolated microglia and astrocytes resulted in their exaggerated production of neurotoxic factors in response to inflammatory stimuli.
Fig. 7. Nurr1 functions to inhibit neurotoxic gene expression in microglia and astrocytes via a CoREST-dependent transrepression pathway.
A. Model for communication among microglia, astrocyte and neurons. B. Model for Nurr1/CoREST-mediated repression. See main text for details.
Experiments employing sequential transfer of cell culture media from microglia to astrocytes or vice versa indicate that astrocytes can act as amplifiers of microglia-derived mediators in the production of neurotoxic factors. Collectively, our data are consistent with a model in which LPS-induced expression of factors such as IL1β and TNFα by microglia results in paracrine activation of astrocytes. This activation in turn is predicted to enhance production of toxic mediators by astrocytes that include NO and ROS. These factors are suggested to act additively or synergistically with neurotoxic factors produced by microglia (Fig. 7A). Experiments using mixed neuronal cultures are of particular interest in this regard because they suggest that activated microglia and astrocytes produce factors that exhibit relative specificity for TH+ neurons (Fig. 2H and I). Conversely, distinct neuronal cell types might exhibit different sensitivities to neurotoxic factors based on protective systems, such as those conferred by genes under the control of the PGC1α coactivitor (St-Pierre et al., 2006). Defining the specific identities of the pathologically important factors produced by microglia and astrocytes and their relative ability to exert toxic effects on different types of neurons will be important future goals.
There is currently no evidence that LPS or bacterial infection contribute directly to the pathogenesis of PD in humans. However, the emerging recognition of the roles of TLRs in a number of inflammatory diseases raises the possibility that endogenous TLR ligands are generated during aging that contribute to disease initiation or progression. For example, components of dying cells, such as Hsp60, have been shown to trigger TLR4 activity (Lehnardt et al., 2008). Other factors, such as ATP, can also induce inflammatory responses through other signaling pathways (Di Virgilio, 2007). A transient event leading to neuronal injury could thus lead to activation of microglia, with subsequent amplification of inflammation by astrocytes. The present findings suggest that Nurr1 protects the CNS from amplification of inflammatory signaling by microglia-astrocyte communication. Strategies to suppress expression of neurotoxins either directly or by interfering with microglia/astrocyte communication may thus have therapeutic utility. Furthermore, it may be necessary to suppress the production of neurotoxic mediators by microglia and astrocytes in order to obtain reconstitution of functional neuronal circuits using cell-based therapies (Brundin et al., 2008).
A Nurr1/CoREST transrepression pathway mediates feedback regulation of inflammatory responses
Members of the NR4A family have been reported to both positively and negatively regulate pro-inflammatory genes (Bonta et al., 2006; Doi et al., 2008; Pei et al., 2006). The present studies demonstrate a potent anti-inflammatory activity of Nurr1 in microglia and astrocytes. We propose that this anti-inflammatory activity is mediated by a Nurr1/CoREST transrepression pathway that operates in a feedback manner to restore transcription of NF-κB target genes to a basal state (Fig. 7B). In this pathway, Nurr1 is recruited to NF-κB on inflammatory gene promoters dependent on GSK3β-mediated phosphorylation of S468 of p65. Nurr1 subsequently recruits the CoREST co-repressor complex in an NLK-dependent manner. Since HDAC1-mediated deacetylation is known to regulate the duration of p65 transcriptional activity, and HDAC1 is a component of the CoREST complex, the Nurr1-CoREST axis might have essential roles in terminating inflammatory responses by p65 clearance from the target promoters. These studies thus establish an unexpected biological role for the CoREST complex, previously considered to mainly be involved in the repression of neuronal genes in NSCs or non-neuronal cells (Ballas et al., 2005). We also find that overexpression of Nurr77 and Nor1 in a macrophage cell line can suppress iNOS activation in response to LPS (K.S., unpublished), suggesting that the CoREST transrepression pathway may be widely used by members of the NR4A family. Of interest, reduction of most of the well-established components of the Co-REST complex severely compromises the anti-inflammatory activity of Nurr1. Quantitative defects in the expression or activities of these proteins could thus predispose certain organ systems to inflammation-sensitive pathologies, such as PD.
Experimental Procedures
Mice and isolation of primary cells
C57BL/6 mice were purchased from Charles River or Harlan and housed according to UCSD or Salk Institute protocols, respectively. Mouse primary microglia cells and astrocytes from the cerebrum of P0 pups. After 10–14 days of culture, microglia cells were isolated from astrocytes by the magnetic sorting using anti-mouse CD11b beads (Miltenyi). Purity of each population was over 98%, as determined by FACS. Murine microglial BV2 cells (kindly provided by Katerina Akassoglou) and macrophage RAW264.7 cells were maintained with DMEM supplemented with 10% FBS (low endotoxin, Hyclone) and penicillin/streptomycin. For other cells, details appear in Supplemental Experimental Procedures.
Reagents
All smart-pool siRNAs and GIPZ lentivirus shRNAmir were purchased from Dharmacon and Open Biosystems, respectively. LPS E. coli 0111:B4 used at 0.1 μg/ml final, SB216763 as indicated, and Cycloheximide used at 10 μg/ml final were obtained from Sigma. Human and mouse IL1β used at10 ng/ml final and TNFα used at 50 ng/ml final were from R&D system.
Stereotaxic Injection of lentivirus and LPS
LPS and/or lentiviruses were delivered to the right SN by stereotaxic injection at AP−3.3 mm, ML−1.2 mm, DV −4.6 mm from bregma (Franklin and Paxinos, 2008). See Supplemental Experimental procedures for full methods.
Immunohistochemistry (IHC)
Experimental animals were anesthetized and perfused transcardially with 0.9% saline followed by 4% paraformaldehyde. Brain samples were postfixed with 4% paraformaldehyde overnight and equilibrated in 30% sucrose. Coronal sections of 40 μm were prepared with a sliding microtome. IHC was performed using mouse anti-tyrosine hydroxylase (Chemicon). Unbiased quantification of TH-immunoreactive neurons in the SN was performed according to the optical fractionator principle (Gundersen et al., 1988). See Supplemental Experimental procedures for full methods.
Transfection assays
The RAW264.7 mouse macrophage cell line was transiently transfected with iNOS- or TNFα-promoters directing luciferase expression (Pascual et al., 2005) using SuperFect (Qiagen). Double transfections with plasmid DNA and siRNAs used Transmessenger reagent (Qiagen). Flag-tagged full-length (FL) mouse Nurr1 was cloned into p3XFLAG-CMV-7.1 vector (Sigma). Mutant constructs of Nurr1 were generated with the Quick-change site-direct mutagenesis kit (Stratagene). Retrovirus production (Openbiosystems) and infection into BV2 cells were performed according to the manufacturer’s protocol. Lentivirus p156RRLsinPPTCMV-GFP-PREU3Nhe and pHAGE vector packaging was done using Virapower (Invitrogen). GIPZ lentivirus shRNAmir (Openbiosystems) packaging was performed according to the manufacturer’s protocols. See Supplemental Experimental procedures for full methods.
Biochemical assays
Cell death detection kit (Roche) was used for TUNEL assay. SUMOylation assays were performed as described before (Pascual et al., 2005). For endogenous co-IP experiments, anti-Nurr1 (E-20, Santa Cruz), anti-CoREST (E-15, Santa Cruz and Millipore) and anti-p65 (C-20, Santa Cruz) were used for IP and Western blotting. For immunoprecipitation of tagged proteins, M2 anti-Flag-agarose (Sigma) beads were used. NLK (Millipore) in vitro kinase reaction (Kanei-Ishii et al., 2004) was performed using GST-fusion substrates.
Chromatin immunoprecipitation (ChIP) assays
For each experimental condition, 2 × 107 BV2 cells or 6 × 106 mouse primary astrocytes were used. Cells were stimulated with LPS for BV2 cells and IL1β for astrocytes for the indicated time before crosslinking for 10 minutes with 1% formaldehyde. Anti-Nurr1 (E- 20, Santa Cruz Biotechnology) anti-p65 (C-20, Santa Cruz Biotechnology), anti-CoREST (Millipore) or control rabbit IgG (Santa Cruz Biotechnology) were used for IP. See details in Supplemental Experimental Procedures.
RNA isolation and quantitative PCR
Total RNA was isolated by RNAeasy kit (Qiagen) from cells or SN samples microdissected from the brain. One microgram of total RNA was used for cDNA synthesis using Superscript III (Invitrogen), and quantitative PCR was performed with SYBR-GreenER (Invitrogen) detected by 7300 Real Time PCR System (ABI). The sequences of qPCR primers used for mRNA quantification in this study were obtained from PrimerBank (Wang and Seed, 2003).
Statistical analyses
Standard deviation, Chi-square and two-tail Student’s t-test were performed with the Prism 4 program. p<0.01 was considered significant. For IHC and IF analyses, Bonferroni was used for post hoc analysis when a significant difference was found with ANOVA. Unpaired two-tailed t test was used for other comparisons, including comparisons between control and injected sides within one group. All data are presented as mean ± SD.
Supplementary Material
Acknowledgments
The authors are grateful to Christian Schmedt, Amy Sullivan and Mary Lynn Gage for critical reading of the manuscript. We thank Katerina Akassoglou for BV2 cells, Alexander Mata de Urquiza for the NBRE-luciferase construct, Inder Verma for the lentivirus vector, Amir Gamliel for the Myc-SUMO1, 2 and 3 constructs, Roberto Jappelli and Roland Riek for the lentivirus encoding A30P α-Synuclein, Richard Mulligan and Jeng-Shin Lee for pHAGE vector. We thank Chris Benner for statistical analysis, Eunice Meija, Robert Aigner and Rosa Luna for technical assistance and Lynn Bautista for help with preparing figures. KS, JGC and CKG are supported by grants from the NIH (CA52599). FHG, BW, LB and CC are supported by grants from CIRM and the Picower Foundation. BW is a Feodor-Lynen fellow of the Alexander von Humboldt Foundation. MGR is an investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarnisalo P, Kim CH, Lee JW, Perlmann T. Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J Biol Chem. 2002;277:35118–35123. doi: 10.1074/jbc.M201707200. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson TJ. Toll-like receptors, transduction-effector pathways, and disease diversity: evidence of an immunobiological paradigm explaining all human illness? Int Rev Immunol. 2008;27:255–281. doi: 10.1080/08830180801959072. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005;19:2466–2477. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci. 2007;8:57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- Bonta PI, van Tiel CM, Vos M, Pols TW, van Thienen JV, Ferreira V, Arkenbout EK, Seppen J, Spek CA, van der Poll T, et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol. 2006;26:2288–2294. doi: 10.1161/01.ATV.0000238346.84458.5d. [DOI] [PubMed] [Google Scholar]
- Brown GC. Mechanisms of inflammatory neurodegeneration: iNOS and NADPH oxidase. Biochem Soc Trans. 2007;35:1119–1121. doi: 10.1042/BST0351119. [DOI] [PubMed] [Google Scholar]
- Brundin P, Li JY, Holton JL, Lindvall O, Revesz T. Research in motion: the enigma of Parkinson’s disease pathology spread. Nat Rev Neurosci. 2008;9:741–745. doi: 10.1038/nrn2477. [DOI] [PubMed] [Google Scholar]
- Buss H, Dorrie A, Schmitz ML, Frank R, Livingstone M, Resch K, Kracht M. Phosphorylation of serine 468 by GSK-3β negatively regulates basal p65 NF-κB activity. J Biol Chem. 2004;279:49571–49574. doi: 10.1074/jbc.C400442200. [DOI] [PubMed] [Google Scholar]
- Di Virgilio F. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol Sci. 2007;28:465–472. doi: 10.1016/j.tips.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Doi Y, Oki S, Ozawa T, Hohjoh H, Miyake S, Yamamura T. Orphan nuclear receptor NR4A2 expressed in T cells from multiple sclerosis mediates production of inflammatory cytokines. Proc Natl Acad Sci U S A. 2008;105:8381–8386. doi: 10.1073/pnas.0803454105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- Fearnley JM, Lees AJ. Ageing and Parkinson’s disease: substantia nigra regional selectivity. Brain . 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- Frank-Cannon TC, Tran T, Ruhn KA, Martinez TN, Hong J, Marvin M, Hartley M, Trevino I, O’Brien DE, Casey B, et al. Parkin deficiency increases vulnerability to inflammation-related nigral degeneration. J Neurosci. 2008;28:10825–10834. doi: 10.1523/JNEUROSCI.3001-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. 3. Academic Press; 2008. [Google Scholar]
- Galleguillos D, Vecchiola A, Fuentealba JA, Ojeda V, Alvarez K, Gomez A, Andres ME. PIASγ represses the transcriptional activation induced by the nuclear receptor Nurr1. J Biol Chem. 2004;279:2005–2011. doi: 10.1074/jbc.M308113200. [DOI] [PubMed] [Google Scholar]
- Gao HM, Kotzbauer PT, Uryu K, Leight S, Trojanowski JQ, Lee VM. Neuroinflammation and oxidation/nitration of α-synuclein linked to dopaminergic neurodegeneration. J Neurosci. 2008;28:7687–7698. doi: 10.1523/JNEUROSCI.0143-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- Gundersen HJ, Bagger P, Bendtsen TF, Evans SM, Korbo L, Marcussen N, Moller A, Nielsen K, Nyengaard JR, Pakkenberg B, et al. The new stereological tools: disector, fractionator, nucleator and point sampled intercepts and their use in pathological research and diagnosis. Apmis. 1988;96:857–881. doi: 10.1111/j.1699-0463.1988.tb00954.x. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Hunot S, Hirsch EC. Inflammation and dopaminergic neuronal loss in Parkinson’s disease: a complex matter. Exp Neurol. 2003;184:561–564. doi: 10.1016/j.expneurol.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- Le WD, Xu P, Jankovic J, Jiang H, Appel SH, Smith RG, Vassilatis DK. Mutations in NR4A2 associated with familial Parkinson disease. Nat Genet. 2003;33:85–89. doi: 10.1038/ng1066. [DOI] [PubMed] [Google Scholar]
- Lehnardt S, Massillon L, Follett P, Jensen FE, Ratan R, Rosenberg PA, Volpe JJ, Vartanian T. Activation of innate immunity in the CNS triggers neurodegeneration through a Toll-like receptor 4-dependent pathway. Proc Natl Acad Sci U S A. 2003;100:8514–8519. doi: 10.1073/pnas.1432609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnardt S, Schott E, Trimbuch T, Laubisch D, Krueger C, Wulczyn G, Nitsch R, Weber JR. A vicious cycle involving release of heat shock protein 60 from injured cells and activation of toll-like receptor 4 mediates neurodegeneration in the CNS. J Neurosci. 2008;28:2320–2331. doi: 10.1523/JNEUROSCI.4760-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira M, Martens C, Philips A, Drouin J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol. 1999;19:7549–7557. doi: 10.1128/mcb.19.11.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol. 2005;6:777–784. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeer PL, McGeer EG. Glial reactions in Parkinson’s disease. Mov Disord. 2008;23:474–483. doi: 10.1002/mds.21751. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Meredith GE, Sonsalla PK, Chesselet MF. Animal models of Parkinson’s disease progression. Acta Neuropathol. 2008;115:385–398. doi: 10.1007/s00401-008-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson’s disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Nagatsu T, Sawada M. Inflammatory process in Parkinson’s disease: role for cytokines. Curr Pharm Des. 2005;11:999–1016. doi: 10.2174/1381612053381620. [DOI] [PubMed] [Google Scholar]
- Nordzell M, Aarnisalo P, Benoit G, Castro DS, Perlmann T. Defining an N-terminal activation domain of the orphan nuclear receptor Nurr1. Biochem Biophys Res Commun. 2004;313:205–211. doi: 10.1016/j.bbrc.2003.11.079. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280:29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol. 2006;20:786–794. doi: 10.1210/me.2005-0331. [DOI] [PubMed] [Google Scholar]
- Sakurada K, Ohshima-Sakurada M, Palmer TD, Gage FH. Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development. 1999;126:4017–4026. doi: 10.1242/dev.126.18.4017. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Tansey MG, McCoy MK, Frank-Cannon TC. Neuroinflammatory mechanisms in Parkinson’s disease: potential environmental triggers, pathways, and targets for early therapeutic intervention. Exp Neurol. 2007;208:1–25. doi: 10.1016/j.expneurol.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teismann P, Schulz JB. Cellular pathology of Parkinson’s disease: astrocytes, microglia and inflammation. Cell Tissue Res. 2004;318:149–161. doi: 10.1007/s00441-004-0944-0. [DOI] [PubMed] [Google Scholar]
- Thery C, Stanley ER, Mallat M. Interleukin 1 and tumor necrosis factor-α stimulate the production of colony-stimulating factor 1 by murine astrocytes. J Neurochem. 1992;59:1183–1186. doi: 10.1111/j.1471-4159.1992.tb08366.x. [DOI] [PubMed] [Google Scholar]
- Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R. TLR-4, IL-1R and TNF-R signaling to NF-κB: variations on a common theme. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- Whitton PS. Inflammation as a causative factor in the aetiology of Parkinson’s disease. Br J Pharmacol. 2007;150:963–976. doi: 10.1038/sj.bjp.0707167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J, Yokoo H, Yamada T, Kitabayashi I, Sekiya T, Ichikawa H. Nemo-like kinase suppresses a wide range of transcription factors, including nuclear factor-κB. Cancer Sci. 2004;95:52–57. doi: 10.1111/j.1349-7006.2004.tb03170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.