Abstract
The pathophysiological mechanisms that drive the development and progression of epithelial ovarian cancer remain obscure. Recently, we identified TCEAL7 as a transcriptional regulatory protein often downregulated in epithelial ovarian cancer. However, the biological significance of such downregulation in cancer is not currently known. Here, we show that TCEAL7 is downregulated frequently in many human cancers and that in immortalized human ovarian epithelial cells this event promotes anchorage-independent cell growth. Mechanistic investigations revealed that TCEAL7 associates with cyclin D1 promoter containing Myc E-box sequence and transcriptionally represses cyclin D1 expression. Moreover, downregulation of TCEAL7 promotes DNA-binding activity of Myc-Max, and upregulates the promoter activity of c-Myc-target gene, ornithine decarboxylase (ODC), whereas enhanced expression of TCEAL7 inhibits Myc-induced promoter activity of ODC. Our findings suggest that TCEAL7 may restrict ovarian epithelial cell transformation by limiting Myc activity. These results also suggest a potential, alternative mechanism by which c-Myc activity may be deregulated in cancer by the downregulation of TCEAL7.
Keywords: TCEAL7, Myc, cyclin D1, tumor suppressor, transformation
Introduction
Genetic alterations that lead to activation of oncogenes and inactivation of tumor suppressors are the underlying causes of cancer. Sequential gain of oncogenes and loss of tumor suppressors provide the necessary foundation for step-wise progression of solid tumors from initiation to transformation and tumor progression. In epithelial ovarian cancer, loss of function of tumor suppressors, such as p53, BRCA1, BRCA2 and Pten, and gain of function of oncogenes such as c-Myc, Ras, Braf, Src and β-catenin are associated with initiation, transformation and progression of epithelial ovarian cancer (Bell, 2005; Fukumoto and Nakayama, 2006). However, additional genetic alterations that contribute to transformation or tumor progression remain to be elucidated.
We have recently identified a putative transcription regulatory factor TCEAL7 as a downregulated protein in epithelial ovarian cancer by suppression subtraction hybridization and by transcriptional profiling analyses (Shridhar et al., 2001, 2002; Chien et al., 2005). This gene encodes a member of the transcription elongation factor A (SII)-like (TCEAL) gene family. Members of this family contain TFA domains and may function as nuclear phosphoproteins that modulate transcription in a promoter context-dependent manner. Multiple family members are located on the X chromosome. The closely related protein transcription elongation factor SII (TFIIS/TCEA1) is involved in transcription elongation and transcript fidelity (Jeon and Agarwal, 1996; Thomas et al., 1998). TFIIS/TCEA1 promotes 3′ endoribonu-clease activity of RNA polymerase II (pol II) and allows pol II to bypass transcript pause or ‘arrest’ during elongation process (Thomas et al., 1998). On the other hand, TCEAL1/SIIR/p21 is a nuclear phosphoprotein implicated in repression of Rous sarcoma virus long-terminal repeat transcription activity and suppression of transformation mediated by Rous sarcoma virus long-terminal repeat in a promoter context-dependent manner (Yeh and Shatkin, 1995).
To better understand the function of TCEAL7 and the consequences of its downregulation in cancer, we analysed the transformation potential of an immortalized, non-transformed ovarian surface epithelial cell line (OSEtsT/hTERT) following the RNAi-mediated downregulation of TCEAL7. Moreover, we analysed the effect of TCEAL7 expression on the DNA-binding activity of 54 transcription factors. In this study, we show that TCEAL7 regulates the transformation potential of OSEtsT/hTERT and modulates the trans-activation activity of Myc by interacting with E-box sequences of a Myc-target gene, cyclin D1. Our observations thus uncover a novel component of the negative regulatory pathway that acts to restrict Myc activity and provide an original mechanistic insight into how TCEAL7 may regulate Myc and cyclin D1 and modulate cell proliferation and malignant transformation.
Results
TCEAL7 is frequently downregulated in cancers of diverse origins
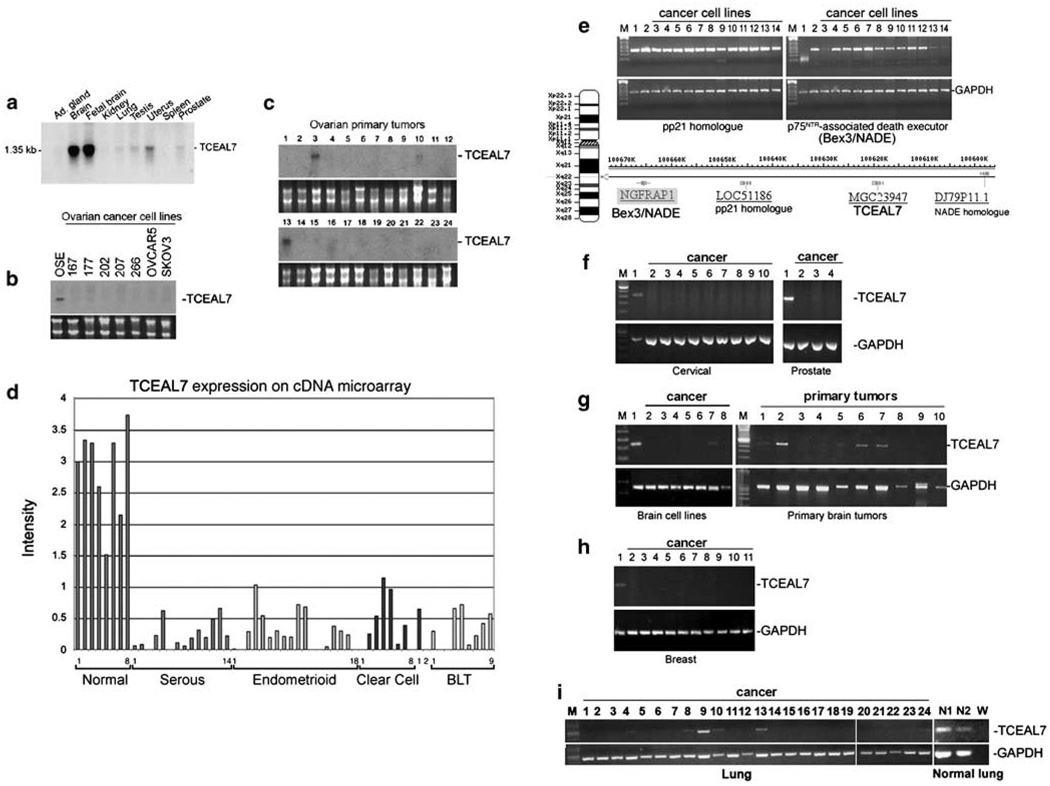
TCEAL7 was originally identified as an epigenetically silenced gene in epithelial ovarian cancer (Chien et al., 2005). To further characterize the expression of TCEAL7 in normal tissues and to validate its down-regulation in epithelial ovarian cancer, TCEAL7 expression was determined by northern analyses in normal tissues, 7 epithelial ovarian cancer cell lines and 24 primary epithelial ovarian tumors. Furthermore, TCEAL7 expression was determined by custom cDNA microarray analysis in 51 primary ovarian tumors. Finally, to characterize the expression of TCEAL7 in human cancers of diverse origins, reverse transcription (RT)-PCR was performed in various cancer cell lines and primary brain tumors. TCEAL7 is highly expressed in normal and fetal brain tissues, and weakly expressed in specific tissues including uterus and ovary (Figure 1a and Supplementary Figure S1). Consistent with our previous report, TCEAL7 is downregulated in 7 epithelial ovarian cancer cell lines (Figure 1b) and in 21 of 24 primary ovarian tumors (Figure 1c), but expressed in short-term cultures of ovarian surface epithelial cells (OSE) (Figure 1b). Microarray analysis of additional 51 ovarian tumors of distinct histologies also indicates downregulation of TCEAL7 transcripts in primary tumors (Figure 1d). To determine whether TCEAL7 downregulation is specific, expression of two other genes mapping close to TCEAL7 was tested in various cell lines by RT-PCR. pp21 homolog and p75NTR-associated death executor (NADE) map 21 and 45 kb centromeric to TCEAL7 (Figure 1e). RT-PCR analysis indicates minimal loss of expression of pp21 homolog and NADE in cancer cell lines (Figure 1e), demonstrating that downregulation of TCEAL7 in cancer is specific. RT-PCR analysis also indicates that TCEAL7 is downregulated in several cervical, prostate, breast, brain and lung cancer cell lines tested and in 6 of 10 primary brain tumors (Figures 1f–i). Furthermore, differential expression analyses of TCEAL7 in human cancers, which are available through the Oncomine database (http://www.oncomine.org), indicate that TCEAL7 is downregulated in cancers and its downregulation is associated with high grade, metastatic or aggressive phenotype in various cancers (Supplementary Table 1).
Figure 1.
TCEAL7 expression in normal and cancer tissues. (a) Northern blot analysis of multiple tissues (Clontech) indicating the predominant expression of TCEAL7 in adult and fetal brains and a modest expression in uterus. See Supplementary Information for TCEAL7 expression in other normal tissues. (b and c) Northern blot analysis of ovarian cancer cell lines (b) and primary epithelial ovarian tumors (c) indicating the downregulation of TCEAL7 in cancer. The lower panels are ethidium bromide stained agarose gels showing 18s and 28s RNA as loading controls. (d) Analysis of TCEAL7 expression in 59 ovarian samples (8 normal ovarian brushings, 42 epithelial ovarian tumors of different histologies and 9 borderline tumors) using custom cDNA microarray. BLT denotes borderline tumors. (e) Reverse transcription (RT)-PCR expression analyses of neighboring genes, pp21 homolog and Bex3/NADE, indicating infrequent downregulation in cancer cell lines. Loading order: 1, OSE; 2, OSEtsT/hTERT; 3, OV167; 4, OV177; 5, OV202; 6, OV207; 7, OV266; 8, OVCAR3; 9, OVCAR5; 10, SKOV3; 11, HMEC; 12, MCF-7; 13, MCF-10A and 14, MDA-MB-157. The relative positions of Bex3/NADE, pp21 homolog, TCEAL7 and NADE homolog in band Xq22.1–22.2 along with the map of X chromosome is shown below. The lower panel with GAPDH represent loading controls. (f–i) The RT-PCR expression analyses of TCEAL7 in various cancers. Loading order is as follow: cervical cell lines (1, normal keratinocytes; 2, SiHa; 3, CaSki; 4, HT-3; 5, HeLa; 6, SW756; 7, MS751; 8, C-33-A; 9, C-4-A and 10, ME-180); prostate cell lines (1, BPH: benign prostate epithelium; 2, DU145; 3, LnCAP; and 4, PC3); brain cell lines (1, Total brain; 2, D32; 3, D37; 4, M067; 5, T989; 6, U148; 7, U251; 8, U373); primary brain tumor samples 1–10; breast cell lines (1, HMEC: normal human mammary epithelial cells; 2, MCF-7; 3, MCF-10A; 4, MDA-MB-157; 5, MDA-MB-361; 6, MDA-MB-435; 7, MDA-MB 468; 8, UACC812; 9, UACC893; 10, BT474; 11, T47D); lung cell lines (1, A549, 2, CL1.0; 3, EKVX; 4, H23; 5, HOP62; 6, H69; 7, H69AR; 8, H82; 9, H146; 10, H157; 11, H827; 12, H209; 13, H226; 14, H358; 15, H441; 16, H460; 17, H526; 18, H650; 19, H1648; 20, H1703; 21, H2122; 22, H2935; N1 and N2, normal lung samples; W, water). M: 100 bp ladder.
Multiple sequence alignment using CLUSTALW analysis indicates that TCEAL7 shares amino-acid sequence homology with other members of transcription elongation factor A-like (TCEAL) family, all mapping to X chromosome (Supplementary Figure S2). Consistent with its putative nuclear function, both endogenous as well as exogenously expressed TCEAL7 localize to the nuclei (Supplementary Figure S3).
TCEAL7 downregulation promotes soft-agar growth and proliferation of nonmalignant OSE cells
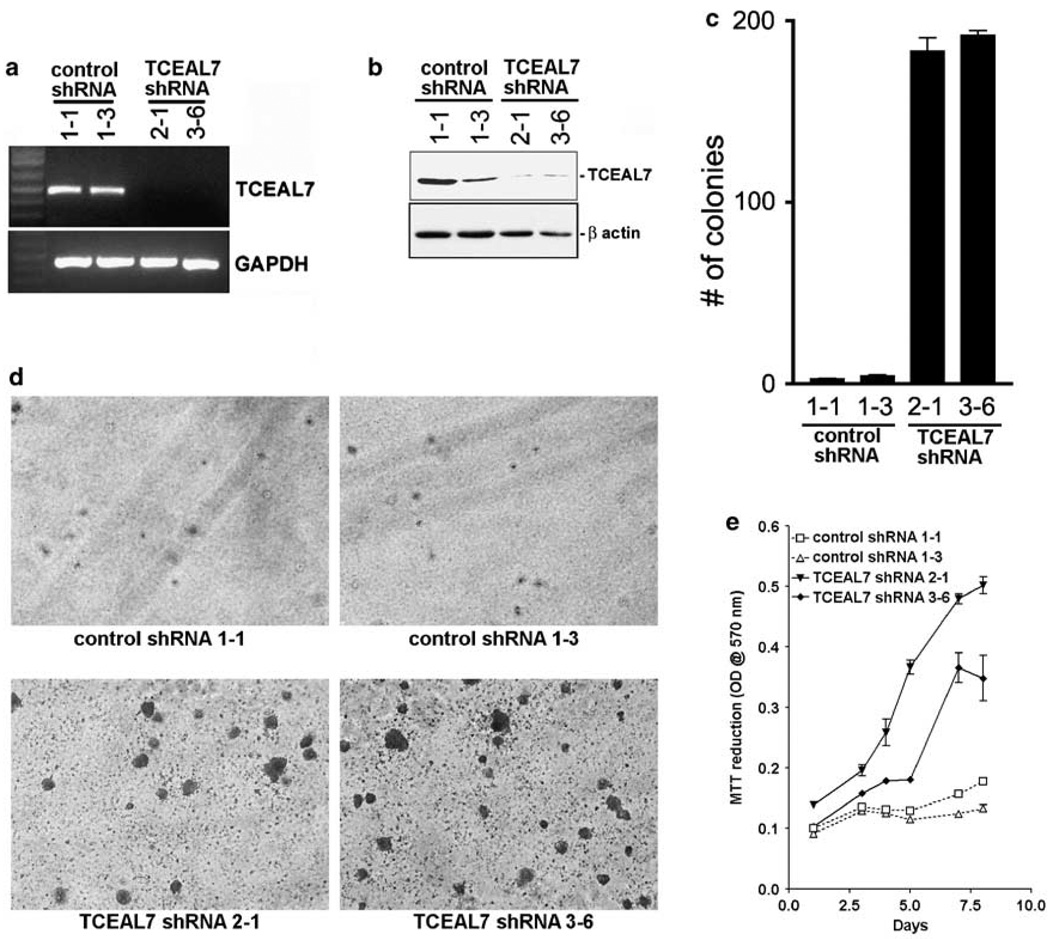
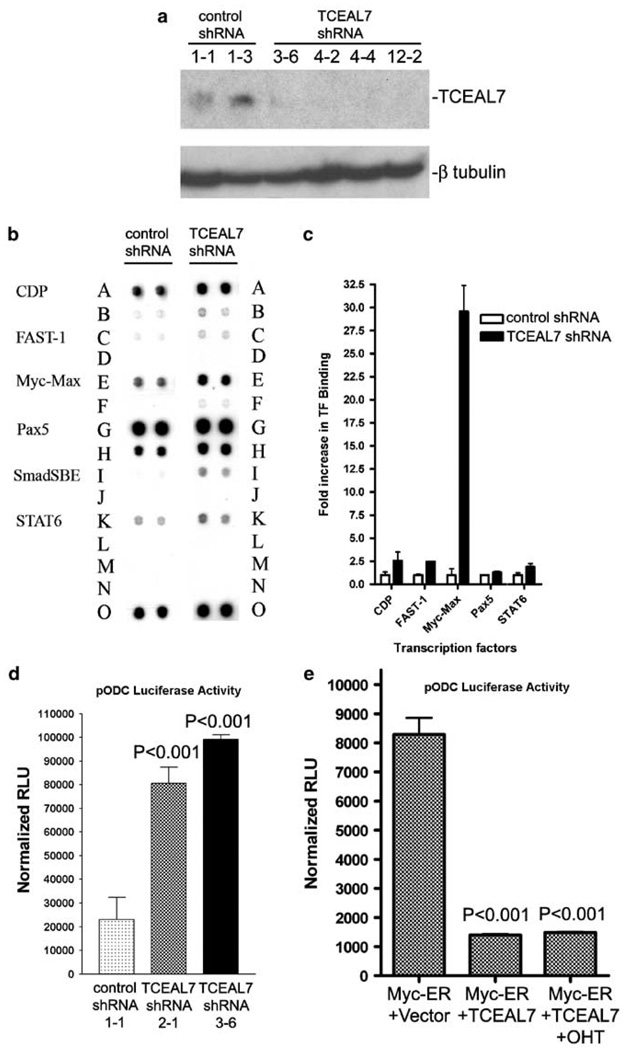
As TCEAL7 is frequently downregulated in cancers of diverse origins, its downregulation may have a function in oncogenesis. To test whether downregulation of TCEAL7 contributes to oncogenesis of epithelial ovarian cancer, we selected an immortalized ovarian epithelial cell line with endogenous TCEAL7 expression (OSEtsT/hTERT) and generated several stable clones expressing short hairpin RNA (shRNA) targeted against TCEAL7 (Figures 2a and b). Transfection with shRNA targeted to TCEAL7 resulted in downregulation of TCEAL7 in clonal lines (Figures 2a and b). No such efficient downregulation was observed in clonal lines expressing empty (control) shRNA construct.
Figure 2.
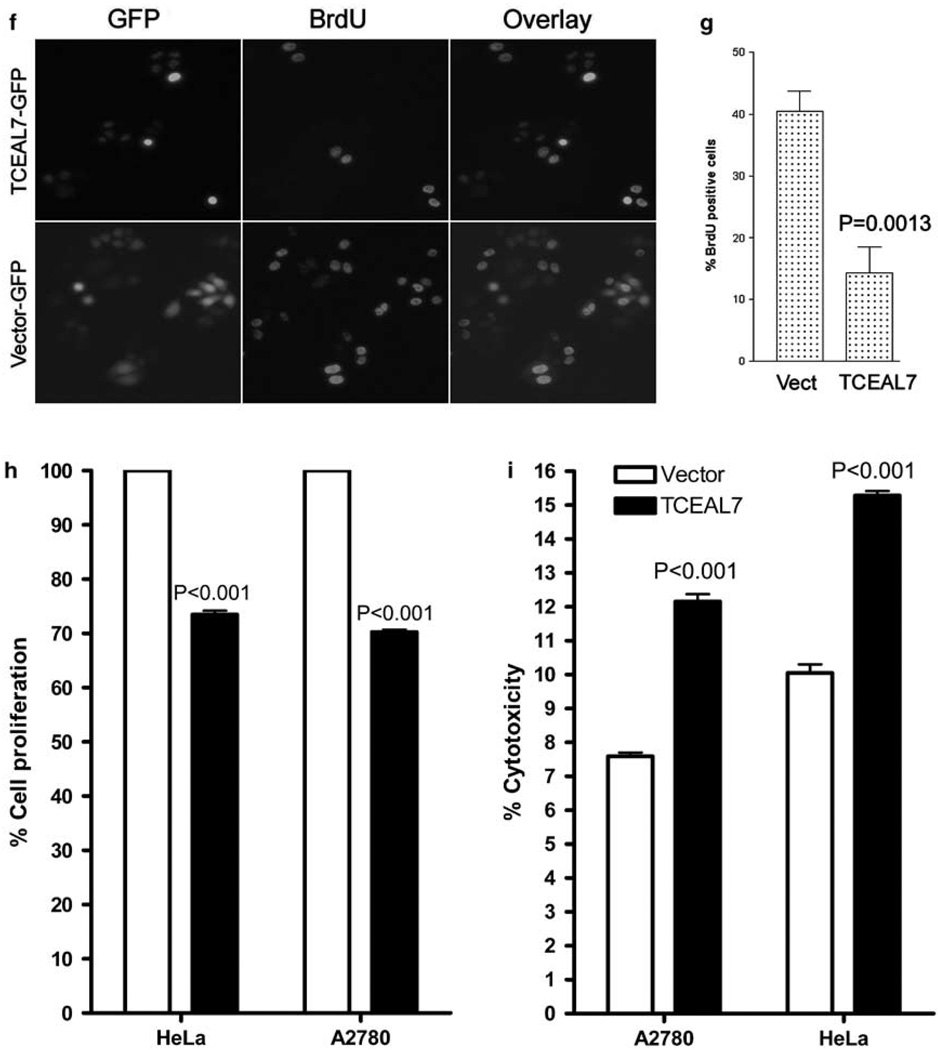
Downregulation of TCEAL7 promotes soft-agar growth of OSEtsT/hTERT. (a) Reverse transcription (RT)-PCR analysis indicates expression of TCEAL7 in two stable clones expressing control short hairpin RNA (shRNA) and downregulation of TCEAL7 in two stable clones expressing TCEAL7 shRNA. 1 kb ladder is shown in the first lane. The lower panel shows control GAPDH levels in the clones. (b) Immunoblot analysis of TCEAL7 in these cells indicates downregulation of TCEAL7 in stable clones expressing TCEAL7 shRNA. The lower panel shows β-actin as protein loading controls. (c) Downregulation of TCEAL7 promotes soft-agar growth of immortalized non-transformed OSEtsT/hTERT. (d) Representative photomicrographs of OSEtsT/hTERT cell lines grown on soft-agar. (e) Downregulation of TCEAL7 also promotes proliferation of OSEtsT/hTERT cells. Data represent the mean ± s.e.m. for two independent experiments. (f) Enhanced TCEAL7 expression in HeLa cells results in reduction of BrdU-positive cells. BrdU labeling was performed as described in the methods section. The red fluorescence, indicating BrdU staining, and TCEAL7-GFP-positive cells were documented using a digital camera. The total numbers of GFP-positive cells as well as BrdU-labeled cells were counted in 5–10 different fields of each well. (g) Quantification of BrdU-positive cells in vector-transfected (control) and TCEAL7-transfected cell populations. (h) Analysis of cell proliferation by CyQuant assay in HeLa cells and A2780 cells transiently transfected with either vector or TCEAL7. (i) Analysis of cytotoxicity by CytoTox-One homogeneous membrane integrity assay in HeLa cells and A2780 cells transiently transfected with either vector or TCEAL7.
Previous studies by Westbrook et al. (2005) indicates that primary epithelial cells immortalized with SV40 T-antigen and human catalytic subunit of telomerase (hTERT) provide a model to test for genes associated with malignant transformation as assessed by soft-agar growth. Therefore, we tested the malignant transformation potential of TCEAL7 downregulation in OSEtsT/hTERT by soft-agar growth assay. These analyses indicate that downregulation of TCEAL7 promotes a significant increase in soft-agar growth of OSEtsT/hTERT cells (Figures 2c and d) and higher rate of proliferation (Figure 2e) compared to control shRNA-transduced clones. Similar results were observed with other clonal lines with downregulated TCEAL7 expression (Supplementary Figure S4). All five shRNA clones showed efficient downregulation of TCEAL7, and there were no phenotypic differences among them. These results indicate that loss of TCEAL7 in cancer cells promote cell proliferation and suggest that endogenous TCEAL7 may regulate cell proliferation. Consistent with its regulation on cell proliferation, enhanced expression of TCEAL7 in cervical cancer cell line HeLa resulted in reduction of cells in S phase as determined by BrdU labeling (Figures 2f and g). In addition, CyQuant cell proliferation analysis indicates a decrease in cell proliferation in both HeLa and ovarian cancer cell line A2780 (Figure 2h), indicating that the decrease in cell proliferation following transient expression of TCEAL7 was not cell line specific. To take into account the contribution of cell death, cytotoxicity assay analysing the release of lactate dehydrogenase into medium by dead cells was performed in the medium collected from HeLa and A2780 cells following transient transfection of TCEAL7. These analyses indicate a statistically significant increase in cell death following TCEAL7 expression (Figure 2i). However, the increased cell death (4–5% over vector transfection) alone could not account for the decrease in cell growth (> 25%), suggesting that inhibition of cell proliferation may also contribute to the decrease in cell growth. Taken together, these observations offered the first evidence that TCEAL7 regulates cell proliferation and oncogenic transformation.
TCEAL7 downregulates expression of cyclins
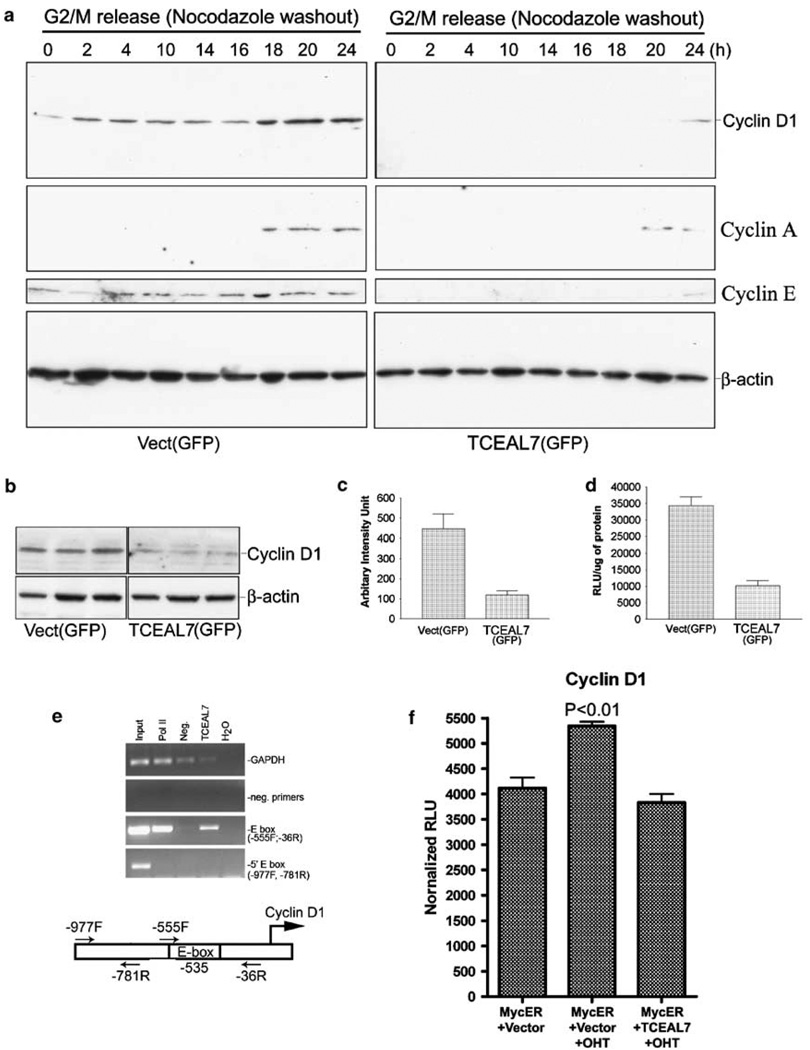
Cyclins have essential functions in cell proliferation and are frequently deregulated in cancer. As exogenous expression of TCEAL7 in HeLa cells attenuated cell proliferation, we tested whether TCEAL7 expression alters the expression of cyclins. Transient re-expression of TCEAL7 in HeLa cells resulted in downregulation of cyclin D1, cyclin A and cyclin E in cell cycle-synchronized HeLa cells (Figure 3a). Moreover, TCEAL7 expression also delayed the cell cycle-regulated expression of these cyclins by at least 6h (Figure 3a). To test whether enhanced expression of TCEAL7 also affected cyclin expression in asynchronous cells, cyclin D1 expression was determined in asynchronous HeLa cells. Enhanced expression of TCEAL7 in a synchronous HeLa cells resulted in downregulation of cyclin D1 expression in triplicate transfections (Figure 3b). Densitometric analysis of cyclin D1 expression indicates that enhanced expression of TCEAL7 resulted in approximately fourfold decrease in cyclin D1 expression (Figure 3c). As TCEAL7 is a nuclear protein that shares sequence homology with the transcription factors TFIIS/TCEA1 and SIIR/TCEAL1, which modulate transcription of target genes, there is a possibility that TCEAL7 may modulate cyclin D1 expression at transcriptional level. To test this possibility, cyclin D1 promoter activity was determined by luciferase promoter assay using full-length cyclin D1 promoter. Cyclin D1 promoter activity was attenuated by approximately 3.5-fold following TCEAL7 expression compared to GFP expression (Figure 3d). These results therefore suggest that TCEAL7 modulates transcription of cyclin D1.
Figure 3.
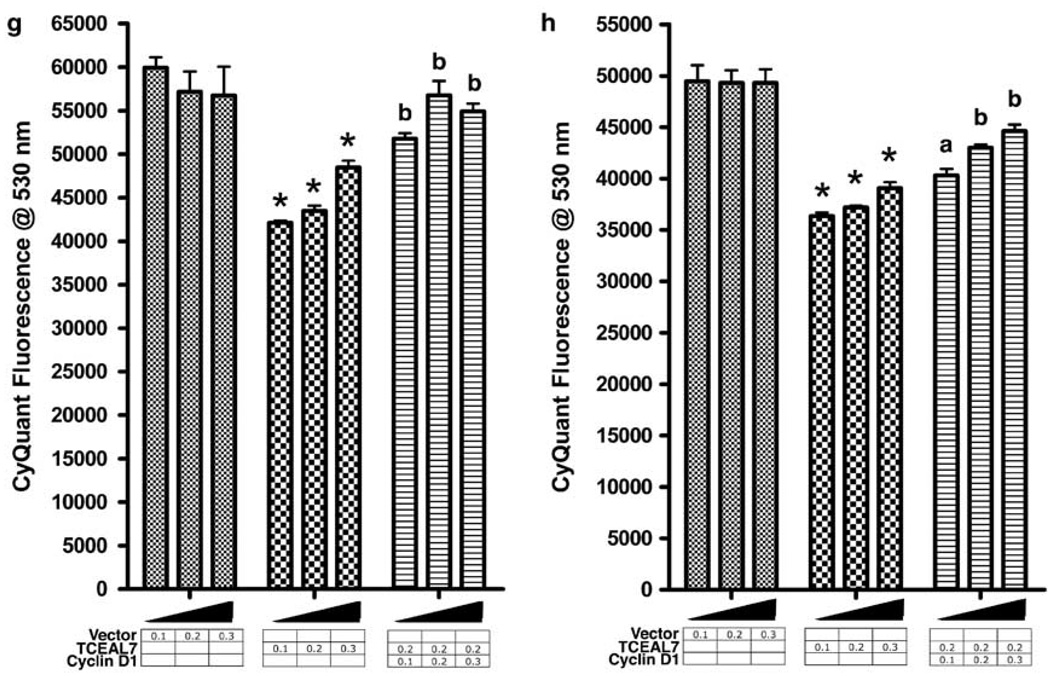
TCEAL7 downregulates expression of cyclins. (a) Suppression of cyclin D1, cyclin A and cyclin E expression by TCEAL7 in cell-cycle synchronized HeLa cells. Cells lysates were collected at different time points after nocodazole washout, and immunoblotting was performed using anti-cyclin D1, A and E antibodies. Please note the basal levels of cyclin D1 in vector-transfected controls and upregulation of cylcin D1 in these cells 18 h after the cell-cycle release. In contrast, TCEAL7-transfected cells showed induction of cyclin D1 only after 24 h following cell-cycle release. (b) TCEAL7 downregulates cyclin D1 expression in asynchronous HeLa cells. Following 48 h of TCEAL7 transfection into HeLa cells, cell lysates were resolved on SDS-polyacrylamide gel electrophoresis, and expression of cylcin D1 was determined with anti-cyclin D1 antibody. Three lanes represent three independent transfections. (c) Quantification of cyclin D1 expression by densitomeric analysis indicates downregulation of cyclin D1 by TCEAL7. (d) TCEAL7 suppresses cyclin D1 promoter activity. (e) OSEtsT/hTERT cells expressing endogenous TCEAL7 were subjected to ChIP analysis with anti-TCEAL7 antibody and PCR amplified with specific cyclin D1 promoter primers as described in the methods section. ChIP analysis indicates specific PCR amplification of cyclin D1 promoter containing E-box (with −36R and −555F primers) in samples immunoprecipitated with either RNA polymerase II or TCEAL7 antibodies. The lower panels show absence of cyclin D1 promoter PCR product outside the region of E-box in ChIP samples (−781R with −997F primer set). Positions of primers and E-box are indicated in the schematic representation of cyclin D1 promoter relative to transcriptional start site. (f) TCEAL7 expression attenuates cyclin D1 promoter activity induced by ectopic activation of MycER by tamoxifen (OHT). (g) Analysis of cell proliferation following rescue of cyclin D1 expression in A2780 cells. (h) Analysis of cell proliferation following rescue of cyclin D1 expression in HeLa cells. Increasing concentrations represent 0.1, 0.2 and 0.3 µg of plasmid. *P<0.001 compared to vector controls; a P<0.05; b P<0.001 compared to TCEAL7 transfection, analysis of variance and Newman-Keuls test.
TCEAL7 binds to cyclin D1 promoter containing Myc E-box sequence
To test whether transcriptional modulation of cyclin D1 expression by TCEAL7 is mediated through its binding to cyclin D1 promoter, endogenous TCEAL7 in OSEtsT/hTERT cells was immunoprecipitated with affinity-purified TCEAL7 antibody. Immunoprecipitation of chromatin with either RNA polymerase II or TCEAL7 antibodies resulted in specific enrichment of cyclin D1 promoter containing Myc-binding E-box sequence (−555F to −36R) (Figure 3e), suggesting that TCEAL7 interacts with E-box region and may transcriptionally modulate cyclin D1 expression induced by c-Myc. Consistent with this model, TCEAL7 expression attenuated cyclin D1 expression induced by ectopic activation of c-Myc by 4-hydroxytamoxifen (OHT) in HeLa cells transfected with c-Myc-estrogen receptor chimeric gene (MycER) (Figure 3f). These results therefore suggest that transient re-expression of TCEAL7 suppresses Myc-induced expression of cyclin D1.
To determine whether rescue of cyclin D1 expression reverses suppression of cell proliferation by TCEAL7, A2780 and HeLa cells were transfected with increasing concentrations of control empty plasmid, TCEAL7 expression plasmid or TCEAL7 plus cyclin D1 expression plasmid. We routinely achieve > 50 transfection efficiency in these cells using standard lipofectamine reagents (Supplementary Figure S5). Twenty-four hours after transfection, cell proliferation was determined by CyQuant Cell Proliferation kit. Consistent with its antiproliferative activity, enhanced expression of TCEAL7 reduces cell proliferation (Figures 3g and h). However, this growth suppression by TCEAL7 was partially rescued by co-expression of cyclin D1, indicating that cyclin D1 downregulation by TCEAL7 is important in the regulation of cancer cell proliferation (Figures 3g and h).
TCEAL7 modulates transactivation activity of c-Myc
To evaluate the specific function of TCEAL7 as modulator of c-Myc transcription factor, we performed protein/DNA-binding assay for 54 transcription factors using nuclear lysates from OSEtsT/hTERT clonal lines with (control shRNA 1–1) or without TCEAL7 (TCEAL shRNA 4–2) (Figure 4a). Analysis of protein/DNA array (TranSignal Array I, Panomics) indicates approximately 30-fold increase in Myc-Max binding to its target DNA sequence in the clonal line without TCEAL7 (TCEAL7 shRNA) compared to the clonal line with TCEAL7 (control shRNA) (Figures 4b and c). These results therefore suggest that endogenous TCEAL7 attenuates transcriptional activity of c-Myc. Among 54 transcription factors tested, TCEAL7 downregulation promoted DNA-binding activities of four transcription factors by more than 20-fold (see Supplementary Table 2 and Supplementary Figure S6). These results further support the function of TCEAL7 as a promoter context-specific transcription modulator.
Figure 4.
TCEAL7 modulates DNA-binding activities of specific transcription factors. (a) Immunoblot analysis of TCEAL7 expression in OSEtsT/hTERT stable clones expressing empty vector or short hairpin RNA (shRNA) against TCEAL7. (b and c) Downregulation of endogenous TCEAL7 in OSEtsT/hTERT (TCEAL7 shRNA, 4–2) resulted in increased Myc-Max transcription factors binding to target DNA when compared to stable clone expressing endogenous TCEAL7 (control shRNA, 1–1). Oligos for each transcription factor are spotted in duplicate (top rows, indicated by label for each transcription factor) as well as the diluted oligos (1:10 dilution) (bottom rows). The last row, designated by the letter ‘O’, indicates biotinylated DNA spotted for the purpose of positive control and alignment. (d) Downregulation of TCEAL7 augments the basal c-Myc transcriptional activity as determined by Ornithine decarboxylase (pODC) promoter luciferase activity. The relative light units (RLUs) are expressed after normalizing with Renilla luciferase and expressed as mean ± s.e.m. from triplicate experiments. (e) TCEAL7 suppresses promoter activity of ornithine decarboxylase induced by endogenous c-Myc or inducible c-Myc. Luciferase activity was measured 6h after OHT treatment and 24 h post-transfection in HeLa cells. All luciferase activities were normalized with Renilla luciferase activity to account for variability in transfection efficiency, and expressed as mean RLU ± s.e.m. from triplicate experiments.
To test whether increased Myc-Max DNA-binding activity, associated with TCEAL7 knockdown, correlates with increased Myc-target promoter activities, we determined the promoter activity of a canonical c-Myc-target gene, ornithine decarboxylase (ODC). Following stable knockdown of TCEAL7 by shRNA, the promoter activity of ODC is increased by approximately fourfold in OSEtsT/hTERT cell lines without TCEAL7 (TCEAL7 shRNA 2–1 and 3–6) compared to the cell line with endogenous TCEAL7 (control shRNA 1–1) (Figure 4d). In addition, to determine whether enhanced expression of TCEAL7 could attenuate c-Myc transactivation activity, HeLa cells were co-transfected with TCEAL7 (or empty vector as control), c-Myc-estrogen receptor (Myc-ER) chimeric gene and luciferase reporter plasmids containing ODC. Enhanced expression of TCEAL7 resulted in attenuation of c-Myc transactivation activity on ODC promoter (Figure 4e). In addition, attenuation of c-Myc activity was observed in cells treated with or without OHT suggesting that TCEAL7 suppressed the activity of both endogenous c-Myc and ectopic activation of c-Myc by OHT. Collectively, these results suggest that TCEAL7 modulates c-Myc activity and affect transcriptional activity of the Myc-target gene ODC.
Discussion
This study provides the first functional characterization of TCEAL7 as a negative regulator of c-Myc. We found that TCEAL7 is specifically downregulated in several cancer cell lines and primary tumors. Furthermore, differential expression analysis of previously published transcriptome studies independently confirmed that TCEAL7 is downregulated in breast carcinoma compared to normal breast, in ER-negative breast cancer compared to ER-positive breast cancer, in grade 3 compared to grade 2 or grade 1 breast cancer, in breast cancer with lymphocytic infiltration compared to those without infiltration, in glioblastoma multiforme, oligodendroglioma and astrocytoma compared to brain from epilepsy patients, and in metastases compared to primary prostate carcinoma or benign prostate tissues (see Supplementary Table 1). These results suggest a functional significance of the downregulation of TCEAL7 in oncogenesis of various cancers.
Consistent with its function in the regulation of oncogenic transformation, downregulation of TCEAL7 in the immortalized, non-transformed ovarian epithelial cell line OSEtsT/hTERT resulted in anchorage-independent growth of OSEtsT/hTERT cells, suggesting that endogenous TCEAL7 may act as a cellular repressor of transformation. To test the effect of TCEAL7 downregulation in malignant transformation in vivo, we performed in vivo tumorigenic assay using OSEtsT/hTERT stably expressing shRNA targeted against TCEAL7. Unfortunately, we observed no tumorigenesis following the knockdown of TCEAL7 in OSEtsT/hTERT cells (data not shown). These data suggest that additional genetic alterations are apparently needed to induce tumorigenesis of OSEtsT/hTERT cells in vivo. However, it should be noted that several ovarian cancer cell lines (such as OV167, OV202 and OV266 established at Mayo Clinic) do not form tumor in mice, although these cell lines were established from primary ovarian tumors. Therefore, lack of tumorigenicity in vivo should not always be equated with absence of neoplastic transformation. OSEtsT/hTERT cells are immortalized by stable transfection of temperature-sensitive large T-antigen (tsT) and catalytic subunit of human telomerase (hTERT). It does not harbor Ras mutation or constitutive activation of Ras as a result of TCEAL7 downregulation in this cell line (data not shown). Therefore, cellular transformation associated with TCEAL7 downregulation is not dependent on Ras activity. Interestingly, the related protein TCEAL1 represses the transformation mediated by Rous sarcoma virus long-terminal repeat (Yeh and Shatkin, 1995). Therefore, this family of proteins may represent cellular repressors of transformation.
Moreover, enhanced expression of TCEAL7 suppresses the expression of cyclin proteins. This repression is most likely mediated through the transcriptional modulation of cyclin D1 gene because TCEAL7 associates with the promoter region of cyclin D1 and suppresses the promoter activity of cyclin D1. Consistent with its repression on cyclin proteins, transient re-expression of TCEAL7 resulted in decreased proliferation. Given the fact that cyclin D1 is deregulated in various cancers including breast, ovarian and brain tumors (Hall and Peters, 1996), our finding that TCEAL7 represses cyclin D1 expression suggests a potential pathway by which cyclin D1 may be deregulated in tumors with loss of TCEAL7 expression. Moreover, although cyclin D1 is frequently overexpressed in various cancers (Hall and Peters, 1996), aberrant expression of cyclin D1 is not always concordant with gene amplification (Gillett et al., 1994). These results suggest that alternative mechanisms, in addition to gene amplification, may account for aberrant expression of cyclin D1 in cancers. Therefore, our observation that TCEAL7 inhibits cyclin D1 promoter activity is highly significant because loss of TCEAL7 in various cancers may provide an alternative mechanism by which cyclin D1 expression is deregulated in cancer. Further correlation studies involving cyclin D1 and TCEAL7 would provide important insights into the function of cyclin D1 in oncogenic transformation mediated by functional loss of TCEAL7.
We also demonstrated that downregulation of TCEAL7 by RNAi resulted in the dramatic increase in Myc-Max DNA-binding activity. Consistent with these results, enhanced expression of TCEAL7 potently inhibited promoter activity of a canonical c-Myc-target gene, ODC, induced by endogenous c-Myc or inducible c-Myc-ER following tamoxifen treatment. Most importantly, our results indicate that downregulation of TCEAL7 expression resulted in increased c-Myc transactivation activity on ODC promoter. Given the fact that ODC function is essential for tumorigenesis (Nilsson et al., 2005), it is most likely that transcriptional repression of ODC by TCEAL7 may also have a central function in the negative regulation of cellular transformation by TCEAL7. These results, therefore, suggest that deregulation of Myc activity, as a result of TCEAL7 downregulation, may also have a function in oncogenesis of human tumors given the fact that the expression of TCEAL7 is downregulated in more than 90% of primary ovarian tumors and cancer cell lines of different origins. More importantly, downregulation of TCEAL7 in cancer is specific as two other genes mapping close to TCEAL7, pp21 homolog and NADE showed minimal loss in the cell lines tested (Figure 2e).
c-Myc is a basic helix-loop-helix-zipper (bHLH-ZIP) transcription factor (Grandori et al., 2000) that is frequently deregulated in tumors of diverse origins due to chromosomal translocations, rearrangements, amplifications, retroviral transductions and viral insertions (Dalla-Favera et al., 1982; Taub et al., 1982; Nesbit et al., 1999; Grandori et al., 2000; Oster et al., 2002). Deregulated Myc expression affects proliferation, differentiation, and apoptosis of cancer cells and contributes to multistep carcinogenesis (Baudino and Cleveland, 2001). Recent studies indicate that as many as 10–15% of all genes in mice and flies may be influenced by Myc activity (Fernandez et al., 2003; Orian et al., 2003). Therefore, future studies investigating global changes in genes expression in the absence of TCEAL7, and whether these changes in gene expression correlate with c-Myc-induced gene expression, are paramount to fully understand the transcriptional modulation mediated by TCEAL7.
Downregulation of TCEAL7 also promoted transcription factor activity of C/EBP, SmadSBE and Brn-3. Interestingly, C/EBPβ is an indispensable effector of cyclin D1 (Lamb et al., 2003). In addition, C/EBPβ also has a critical function in resistance to the growth-inhibitory action of transforming growth factor-β (Gomis et al., 2006). Cytostatic action of transforming growth factor-β in non-transformed epithelial cells is mediated by Smad family of proteins that bind to SmadSBE sequence in the promoter regions of Smad-target genes and requires C/EBPβ (Massague et al., 2005; Gomis et al., 2006). Brn-3b has an important function in cancer progression as it is elevated in breast cancers and in neuroblastoma tumors, where it is associated with increased proliferation, anchorage-independent growth, tumorigenicity, resistance to growth inhibitory stimuli and increased migratory potential (Budhram-Mahadeo and Latchman, 2006). Therefore, additional experiments investigating the modulatory role of TCEAL7 on these transcriptional factors are needed to define the functional significance of these interactions in cell proliferation, inhibition of apoptosis and oncogenesis associated with TCEAL7 downregulation. These additional studies may provide important insights into the function of TCEAL7 as a promoter context-dependent repressor and the functional consequences of the loss of these functions in malignant transformation and oncogenesis.
Materials and methods
Cell culture
Five of eight epithelial ovarian carcinoma cell lines (OV 167, OV 177, OV 202, OV 207 and OV 266) were low passage primary lines established at the Mayo Clinic (Conover et al., 1998), whereas other cell lines were purchased from American Type Culture Collection (Manassas, VA, USA). OSEtsT/hTERT is an ovarian epithelial cell line that was initially immortalized with temperature-sensitive SV40 T-antigen (OSEtsT) (Kalli et al., 2002) and subsequently with catalytic subunit of human telomerase (OSEtsT/hTERT). The primary culture of normal human mammary epithelial cells was purchased from Clonetics Corp. (San Diego, CA, USA). All cells were grown according to the providers’ recommendations.
Northern blot analysis
The probe (PCR amplified open-reading frame) was labeled with 32P using the random primer-labeling system (Invitrogen, Carlsbad, CA, USA) and purified using spin columns (100TE) from Clontech (Palo Alto, CA, USA). Northern blotting was performed on Multiple Tissue Expression Array (Clontech) and primary tumor blots as previously described (Chien et al., 2004).
Semiquantitative RT-PCR
A total of 50–100 ng of reverse-transcribed cDNA was used in PCR reactions with specific primers for TCEAL7 (TCEAL7-99F and TCEAL7-523R), pp21homolog, Bex3/NADE and GAPDH primers (see Supplementary Table 4 for primer sequences and PCR conditions). The products of the reaction were resolved on a 1.6% agarose gel. Band intensities were quantified using the Gel Doc 1000 photo documentation system (Bio-Rad, Hercules, CA, USA) and its associated software and normalized with GAPDH controls.
Plasmid constructs
The coding region of TCEAL7 was amplified by PCR using open-reading frame primers and cloned into pcDNA3/GFP-CT TOPO cloning vector from Invitrogen, following the manufacturer’s recommendation. HA-tagged cyclin D1 expression plasmid was purchased from Addgene (Cambridge, MA, USA).
cDNA microarray analysis
The cDNA microarray data analysis was performed as previously described (Shridhar et al., 2001). Characteristics of tumor samples are listed in Supplementary Table 3.
Transcription factor activity array
For high throughput analysis of transcription factors, we used Panomic’s TranSignal Protein/DNA Array (cat no.; MA1210) containing an array membrane of 54 transcription factors. Vector control (V1–1) and RNAi downregulated TCEAL7 clone (C4–2) in OSEtsT/hTERT were grown in 100 cm dishes for 24 h and harvested for nuclear extract. Nuclear extract was prepared using the Panomic’s Nuclear Extraction kit (cat no.; AY2002). Equal amounts of nuclear extract (15 µg) of both vector and clone were used to perform the array according to the manufacturer’s instructions. The biotin-labeled probes were visualized by streptavidin-HRP and signal intensities were quantitated by Quantity One software program (Bio-Rad).
BrdU labeling
Twenty-four hours after transfection with vector-GFP or TCEAL7 GFP, cells were incubated in growth media containing 10 µM BrdU for 2h. At 2h, the cells were fixed in 4% paraformaldehyde for 10min, permeabilized in 0.2% Triton X-100 for 5min and incubated with anti-BrdU antibody from Becton Dickenson (Mountain View, CA, USA) (1:10 in phosphate-buffered saline, 5mM MgSO4 100U/ml DNAse) for 1 h. Incorporated BrdU was detected by TRITC-conjugated anti-BrdU antibody (Becton Dickinson). The red immunofluorescence, indicating BrdU staining, and TCEAL7-GFP-positive cells were documented using a Spot camera. The total numbers of GFP-positive cells, as well as BrdU-labeled cells, were counted in 5–10 different fields of each well. Two researchers independently made the measurements on all slides. The data were expressed as % BrdU-positive cells.
Cyclin D1 rescue and proliferation assay
Ovarian cancer cell line A2780 was transfected with increasing concentrations (0.1, 0.2, 0.3 µg) of either control plasmid, TCEAL7 or cyclin D1 plus TCEAL7 (0.2 µg) plasmids for 24 h, and cell proliferation was determined by the CyQuant Cell Proliferation Assay Kit (Invitrogen) as recommended by the manufacturer. Each group contains two independent experiments with at least four replicates.
Cytotoxicity assay
CytoTox-One Homogeneous Membrane Integrity Assay (Promega, Madison, WI, USA) was performed according to the manufacturer’s protocol. Medium was collected from cells transfected with either vector or TCEAL7 for 24 h in 96-well plate and subjected to lactate dehydrogenase activity assay. Untransfected cells, lysed in 1% Triton X-100, served as 100% cytotoxicity. Each experiment contains at least triplicates, and was repeated twice.
Luciferase reporter assay
The luciferase reporter gene under the control of the human cyclin D1 promoter (D1-973LUC) was provided by Janknecht (Mayo Clinic, Rochester, MN, USA). ODC promoter was previously described (Elliott et al., 1999). Myc-ER construct was obtained from William Tansey. HeLa cells, seeded at 4 × 104 cells per well in 24-well plates (in triplicates) were transfected with 0.15 µg cyclin D1 promoter luciferase reporter and various plasmids. After 18 h, OHT at 2 nM final concentration was added into wells to induce Myc activity. Luciferase activity was measured 24 h post-transfection with Promega’s Dual-Luciferase Reporter assay system according to the manufacturer’s instructions. The relative light units are expressed after normalizing with Renilla luciferase to account for variability in transfection efficiency, and are representative of at least two independent experiments.
Cell-cycle synchronization
HeLa cells grown to 60–80% confluence in 10 cm dishes were treated with 2.5 µM nocodazole for 16h. The plates were tapped a few times to release the mitotic cells to the medium, and nonadherent cells were collected. Cells were centrifuged, washed two times in phosphate-buffered saline and replated in regular growth medium. Cell lysates were collected at different time points and immunoblotting was performed using anti-cyclin antibodies.
Immunoblotting
Equal amounts of protein (20 µg per lane) were used in immunoblots as previously described (Chien et al., 2006). Blots were first probed with rabbit anti-cyclin D1 (Cell Signaling Inc., Beverly, MA, USA). The blots were stripped and reprobed with antisera that recognize cyclin E (Upstate, Lake Placid, NY, USA) and cyclin A (Upstate) or mouse monoclonal anti-β actin (Sigma, St Louis, MO, USA).
ChIP
The ChIP assays were performed in OSEtsT/hTERT cells with endogenous TCEAL7 expression as previously described (Staub et al., 2007). Immunoprecipitation was performed overnight at 4°C with anti-TCEAL7 antibody, anti-pol II antibody or control IgG (1 µg/100 µl). For PCR, 10 ng of extracted DNA was used in 30 cycles of amplification at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s with cyclin D1-specific promoter primers. The following primers were used: −36R with −555F and −781R with −977F. Primer pairs −36R and −555F span the Myc E-box sequence of the cyclin D1 promoter.
Generation of shRNA downregulated TCEAL7 clones in OSEtsT/hTERT
For these experiments, OSEtsT/hTERT cells were infected with retroviral supernatants in the presence of 4 µg/ml polybrene to transduce pSUPER.retro constructs expressing shRNA targeting TCEAL7 mRNA (TCEAL7 shRNA). For control experiments, empty pSUPER.retro vector (control shRNA) was used. Two shRNAs targeting position 88–106 (T1) and 191–209 (T2) of TCEAL7 mRNA within the open-reading frame were selected and generated as described previously (Berns et al., 2004). Several control shRNA and TCEAL7 shRNA-transduced clones were selected in the presence of both geneticin and puromycin (for maintaining the SV40 T antigen and pSUPER.retro vector) and hygromycin B (for the hTert construct). The efficacy of shRNA-mediated downregulation of TCEAL7 was evaluated by western blot using anti-TCEAL7 antibody.
Soft-agar assay
Soft-agar assay was performed as previously described (Chien et al., 1999). OSEts/hTERT clonal lines (20 000 per well) were plated, and live colonies after 3 weeks of incubation were documented and counted with × 10 objective using a Spot II-RT digital camera (Nikon, Millburn, NJ, USA).
Proliferation assay
OSEtsT/hTERT clonal lines were plated at 2000 cells/well in 96-well plates in triplicates in RPMI medium with 10% fetal calf serum (75 µl per well). Every 24 h for 8 days, MTS solution (5 mg/ml) was added to each well (20 µl per well). Following 1-h incubation at 37 °C, the media was removed and 100 µl of dimethylsulfoxide was added to each well and the plates were read in a microtiter plate reader at 570 nm. The experiment was repeated twice. Data were analysed with GraphPad Prism software (San Diego).
In vivo tumorigenesis assay
OSEtsT/hTERT clonal lines were injected subcutaneously into 5- to 6-week-old nude mice (National Institute of Health, Washington, DC, USA). Mouse studies were carried out following procedures approved by the Institutional Animal Care and Use Committee at the Mayo Clinic College of Medicine. For measurement of tumor growth in vivo, OSEtsT/hTERT clonal lines (V1, vector pool and C4–2 and C4–4, 107 cells per mouse) were mixed with matrigel and injected subcutaneously in the left flanks of mice, 10 mice per clone. The mice were monitored for the appearance of tumors at the site of injection every 3 days for 20 weeks.
Statistical analysis
All values are expressed as the mean ± s.e.m. Differences between two groups were compared using an unpaired two-tailed t-test. When comparing multiple groups, one-way analysis of variance followed by Newman-Keuls test was used. Results were considered significant when P<0.05 at α = 0.05.
Acknowledgements
This study was funded by Fraternal Order of Eagle to VS, by Department of Defense OCRP W81XWH-04-1-0085 to VS, and by Edith and Bernie Waterman Foundation and the Mayo Foundation to VS.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Baudino TA, Cleveland JL. The Max network gone mad. Mol Cell Biol. 2001;21:691–702. doi: 10.1128/MCB.21.3.691-702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DA. Origins and molecular pathology of ovarian cancer. Mod Pathol. 2005;18 Suppl 2:S19–S32. doi: 10.1038/modpathol.3800306. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- Budhram-Mahadeo VS, Latchman DS. Targeting Brn-3b in breast cancer therapy. Expert Opin Ther Targets. 2006;10:15–25. doi: 10.1517/14728222.10.1.15. [DOI] [PubMed] [Google Scholar]
- Chien J, Aletti G, Baldi A, Catalano V, Muretto P, Keeney GL, et al. Serine protease HtrA1 modulates chemotherapy-induced cytotoxicity. J Clin Invest. 2006;116:1994–2004. doi: 10.1172/JCI27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J, Staub J, Avula R, Zhang H, Liu W, Hartmann LC, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- Chien J, Staub J, Hu SI, Erickson-Johnson MR, Couch FJ, Smith DI, et al. A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene. 2004;23:1636–1644. doi: 10.1038/sj.onc.1207271. [DOI] [PubMed] [Google Scholar]
- Chien J, Wong E, Nikes E, Noble MJ, Pantazis CG, Shah GV. Constitutive activation of stimulatory guanine nucleotide binding protein (G(S)alphaQL)-mediated signaling increases invasiveness and tumorigenicity of PC-3M prostate cancer cells. Oncogene. 1999;18:3376–3382. doi: 10.1038/sj.onc.1202690. [DOI] [PubMed] [Google Scholar]
- Conover CA, Hartmann LC, Bradley S, Stalboerger P, Klee GG, Kalli KR, et al. Biological characterization of human epithelial ovarian carcinoma cells in primary culture: the insulinlike growth factor system. Exp Cell Res. 1998;238:439–149. doi: 10.1006/excr.1997.3861. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott K, Sakamuro D, Basu A, Du W, Wunner W, Staller P, et al. Bin1 functionally interacts with Myc and inhibits cell proliferation via multiple mechanisms. Oncogene. 1999;18:3564–3573. doi: 10.1038/sj.onc.1202670. [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto M, Nakayama K. Ovarian epithelial tumors of low malignant potential: are they precursors of ovarian carcinoma? Pathol Int. 2006;56:233–239. doi: 10.1111/j.1440-1827.2006.01960.x. [DOI] [PubMed] [Google Scholar]
- Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, et al. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Aram Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- Jeon C, Agarwal K. Fidelity of RNA polymerase II transcription controlled by elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalli KR, Falowo OI, Bale LK, Zschunke MA, Roche PC, Conover CA. Functional insulin receptors on human epithelial ovarian carcinoma cells: implications for IGF-II mitogenic signaling. Endocrinology. 2002;143:3259–3267. doi: 10.1210/en.2001-211408. [DOI] [PubMed] [Google Scholar]
- Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes & Development. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, et al. Targeting ornithine decarboxylase in Myc-induced lympho-magenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- Shridhar V, Lee J, Pandita A, Iturria S, Avula R, Staub J, et al. Genetic analysis of early- versus late-stage ovarian tumors. Cancer Res. 2001;61:5895–5904. [PubMed] [Google Scholar]
- Shridhar V, Sen A, Chien J, Staub J, Avula R, Kovats S, et al. Identification of underexpressed genes in early- and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002;62:262–270. [PubMed] [Google Scholar]
- Staub J, Chien J, Pan Y, Qian X, Narita K, Aletti G, et al. Epigenetic silencing of HSulf-1 in ovarian cancer: implications in chemoresistance. Oncogene. 2007;26:4969–4978. doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Yeh CH, Shatkin AJ. A cis-acting element in Rous sarcoma virus long terminal repeat required for promoter repression by HeLa nuclear protein p21. J Biol Chem. 1995;270:15815–15820. doi: 10.1074/jbc.270.26.15815. [DOI] [PubMed] [Google Scholar]