Abstract
Purpose of review
Patient response to the asthma drug classes, bronchodilators, inhaled corticosteroids and leukotriene modifiers, are characterized by a large degree of heterogeneity, which is attributable in part to genetic variation. Herein, we review and update the pharmacogenetics and pharmaogenomics of common asthma drugs.
Recent findings
Early studies suggest that bronchodilator reversibility and asthma worsening in patients on continuous short-acting and long-acting β-agonists are related to the Gly16Arg genotype for the ADRB2. More recent studies including genome-wide association studies implicate variants in other genes contribute to bronchodilator response heterogeneity and fail to replicate asthma worsening associated with continuous β-agonist use. Genetic determinants of the safety of long-acting β-agonist require further study. Variants in CRHR1, TBX21, and FCER2 contribute to variability in response for lung function, airways responsiveness, and exacerbations in patients taking inhaled corticosteroids. Variants in ALOX5, LTA4H, LTC4S, ABCC1, CYSLTR2, and SLCO2B1 contribute to variability in response to leukotriene modifiers.
Summary
Identification of novel variants that contribute to response heterogeneity supports future studies of single nucleotide polymorphism discovery and include gene expression and genome-wide association studies. Statistical models that predict the genomics of response to asthma drugs will complement single nucleotide polymorphism discovery in moving toward personalized medicine.
Keywords: asthma, genes, personalized medicine, polymorphisms, response heterogeneity
Introduction
Asthma imposes a serious burden on our society with respect to mortality, morbidity, and healthcare costs. The long-range goal of asthma pharmacogenetics and pharmacogenomics is to personalize asthma pharmacotherapy using genetic information, which is expected to reduce the asthma burden. Novel sequence variants have been recently identified that associate with response to commonly used asthma drug classes [bronchodilators, inhaled corticosteroid (ICS), and leukotriene modifiers) and that support continuing research in asthma pharmacogenetics and pharmacogenomics. Herein, we review the pharmacogenetics and pharmacogenomics of bronchodilators, ICSs, and leukotriene modifiers focusing on recently published work.
Bronchodilators
β-agonists are the most commonly used bronchodilators in the treatment of asthma. Short-acting β-agonists (SABAs) are used by virtually all patients with asthma as rescue bronchodilator medications to treat acute bronchoconstrictive symptoms, whereas long-acting β-agonists (LABAs) are used in combination with ICSs to provide prolonged bronchodilation and control asthma symptoms. The bronchodilator response to SABA, that is, bronchodilator reversibility, is highly variable [1] (Fig. 1). The majority of asthma pharmacogenetic and pharmacogenomic studies have focused mainly on SABA and LABA, particularly on genetic determinants of bronchodilator reversibility and on asthma worsening related to continuous use of SABA or LABA.
Figure 1.
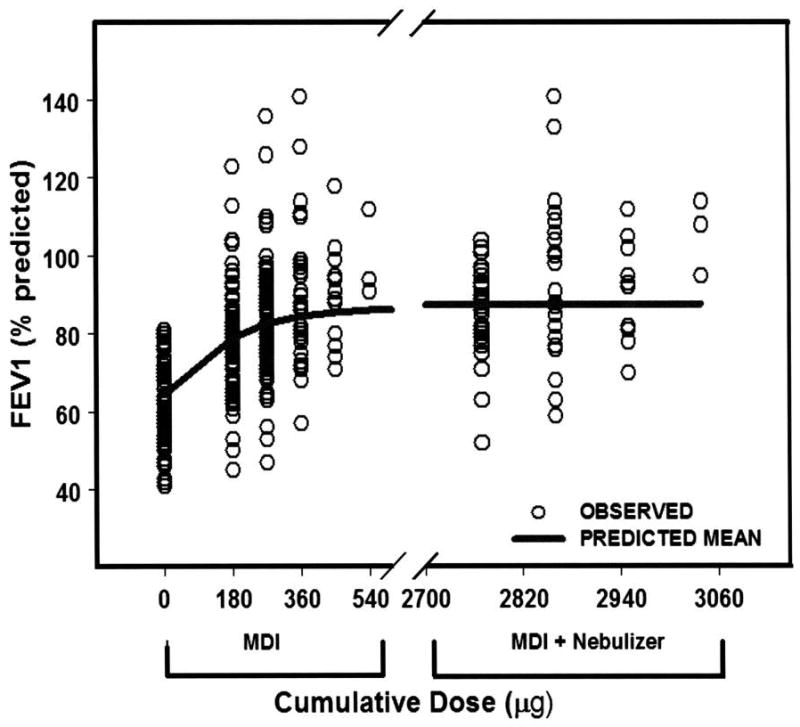
Fitted dose–response after cumulative doses of albuterol (solid line) gr1
Open circles represent individual patient responses. Response is percentage predicted FEV1. The cumulative doses of albuterol administered from the metered dose inhaler (MDI) were 180, 270, 360, 450, and 540 μg, and the cumulative doses from the MDI and nebulizer were 2770 μg (270 μg MDI + 2500 μg nebulized), 2860 μg (360 μg MDI + 2500 μg nebulized), 2950 μg (450 μg MDI + 2500 μg nebulized), and 3040 μg (540 μg MDI + 2500 μg nebulized). A population-based pharmacodynamic model was fitted to the data and predicted maximal bronchodilator effect of 24% (Emax) above baseline (average of percentage predictive FEV1, 62%) with an EC50 (dose of albuterol achieving 50% of Emax) of 141 μg. The baseline ranged between 40 and 80%, whereas the Emax ranged between 50 and 140% predicted. Adapted from [1].
The safety of pressurized β2-agonist aerosols has been controversial since their introduction in the1950s. Two epidemics of asthma mortality occurring in the 1960s and 1970s coincided with increased use of isoproterenol and fenoterol [2–5]. Subsequently, with the discovery and sequencing of the β2-adrenergic receptor gene (ADRB2), interest has focused on pharmacogenetics to explain both the widespread variability in bronchodilator response observed between patients and the occurrence of worsening asthma control noted in small numbers of asthmatics treated with inhaled β2-agonists.
The ADRB2 is a small, intronless gene, which has been recently resequenced in multiple ethnic populations to determine polymorphic variability and haplotype structure [6,7]. Of 80 polymorphisms identified, 45 single nucleotide polymorphisms (SNPs) and two insertion/deletion variants have been validated. Two common nonsynonymous variants at amino acid positions 16 (Gly16Arg) and 27 (Gln27Glu) have functional relevance in vitro [8,9], and most clinical studies have focused on outcomes resulting from the Gly16Arg polymorphism.
Initial studies exploring associations between bronchodilator response to SABA and the Gly16Arg polymorphism in outpatients found that Arg16 homozygotes had a greater bronchodilator response than Gly16 homozygotes [10–12]. Subsequent studies found opposite results or no association [6,13–17]. More recently, no association was found between bronchodilator response and ADRB2 haplotype tagging SNPs in over 500 asthmatic patients [18]. Additionally, both a candidate gene and genome-wide association analysis failed to associate any polymorphism in ADRB2 with bronchodilator response but did identify associations with SNPs in novel genes (ARG1 and ABLIM2) [19,20]. Association studies between ADRB2 polymorphisms and bronchodilator response to high-dose SABA during treatment of severe acute asthma have only begun to be reported [21,22].
In the early 1990s, regularly scheduled fenoterol use was found to worsen asthma control [23], but a similar study with albuterol in mild asthmatic patients [the Beta-Agonist in Mild Asthma (BAGS) Trial] found no harmful effects [24]. However, when the BAGS trial was analyzed by genotype, Arg16 homozygotes had worse asthma control on regularly scheduled albuterol than Gly16 homozygotes on regularly scheduled albuterol and Arg16 homozygotes treated as needed with albuterol [25]. Prospective [Beta-Adrenergic Response by Genotype (BARGE) Study] and retrospective studies have been consistent with these findings [26–28].
The effect of LABA therapy on asthma control by ADRB2 genotype has generated significant interest. The safety of LABAs has been hotly debated [29–31]. A review of 26 trials with over 60,000 patients found that salmeterol (with and without ICSs) was associated with slight but significantly worse asthma control compared with placebo [32••]. The first published pharmacogenetic association analyses of LABA (salmeterol) on asthma control were consistent with studies of SABA [33,34]. However, numerous retrospective pharmacogenetic analyses in all racial groups and in both children and adults have failed to find any association between the Gly16Arg genotype and asthma control in patients treated with salmeterol or formoterol [35–39]. A prospective clinical trial by the Asthma Clinical Research Network is currently underway. This randomized, double-blind, crossover, placebo-controlled trial will examine the effects of regularly scheduled long-acting β-agonist in a group of asthmatic patients harboring the Arg16 homozygous genotype and in a separate group matched for forced expiratory volume in 1s (FEV1) and matched and race-matched (whites versus nonwhites) patients harboring the Gly16 homozygous genotype at the β2-adrenergic receptor. Both groups will receive concurrent ICSs (website: www.acrn.org).
The discrepant findings of coding block polymorphisms on β2-agonist response have prompted investigation into effects of ADRB2 regulatory regions on receptor function. Early study of a variant in the in the ADRB2 promoter region affected receptor translation and density but not transcription [40]. A variable-length poly-C tract polymorphism in the 3′UTR has been found to influence β2-adrenergic receptor expression, mRNA expression, mRNA degradation, and agonist-induced receptor downregulation in vitro [41•]. However, this polymorphism was not found to associate with effects of salmeterol on asthma control [39]. Clearly, coding block variants in ADRB2 do not reliably predict the response to SABA and LABA as was initially expected. Meticulously designed large prospective studies of asthmatic patients with homogenous phenotypes, carefully controlled environmental influences, and analysis of gene–gene interactions are clearly needed to move forward toward personalizing SABA and LABA therapy.
Corticosteroids
ICSs are the most effective and commonly used drugs for the chronic treatment of asthma but may result in serious adverse effects [42]. There is substantial interindividual variability in the response to ICSs [43,44] (Fig. 2), and the intraindividual response to ICSs in patients with asthma is highly repeatable [45]. The combination of wide interindividual variability and high intraindividual repeatability supports a genetic difference for the response to ICSs in asthma [45].
Figure 2.
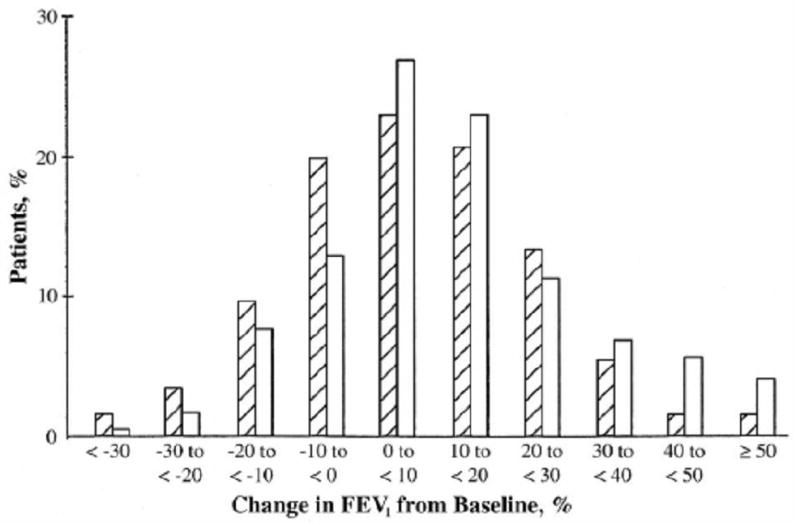
Distribution of treatment responses for FEV1 gr2
The response distributions are shown as histograms for predefined intervals of percentage change in FEV1. Striped bars represent patients receiving monelukast, 10 mg once daily; white bars represent patients receiving inhaled beclomethasone, 200 mg twice daily, Adapted from [43].
In a study of 14 candidate genes selected for their biologic relevance to the corticosteroid pathway, a significant association between 8-week FEV1 response to ICSs and SNPs from the corticotropin-releasing hormone receptor 1 (CRHR1) gene in both adult and pediatric asthmatic patients was noted [46]. Rs242941 (minor allele frequency ∼30%) was associated with about 2½ times the improvement in FEV1 in both the Adult Study and Childhood Asthma Management Program (CAMP) populations (P=0.025 and 0.006, respectively). In CAMP, although ICS usage was associated with improved FEV1, evaluation of the placebo arm revealed no association with change in lung function (interaction P=0.02). One CRHR1 haplotype had even larger improvements in FEV1 on inhaled steroids. However, the overall explained phenotypic variance was small (<5% in both populations). Moreover, the same CRHR1 variants were not associated with protection against lung function decline in a cohort of adult asthmatic patients taking ICSs [47]. Although differences in the studies may be related to the much smaller sample size in the lung function decline cohort, additional factors (including other genetic loci) clearly contribute to FEV1 response variability to ICSs. Interestingly, the initial association may be related to a large structural inversion, rather than to an effect within the CRHR1 gene itself [48••].
TBX21 encodes for T-bet, a transcription factor crucial for naive T-lymphocyte production. The T-bet knockout mouse spontaneously develops airways inflammation and hyperresponsiveness suggestive of asthma [49]. One common nonsynonymous SNP has been identified in the TBX21 gene, encoding for a replacement of histidine by glutamine at amino acid position 33 (H33Q). Four and a half percent of CAMP children are heterozygous for this variant. Each H33Q white individual on ICSs demonstrated a marked improvement in airways responsiveness, as measured by PC20, when compared with either H33H homozygotes or individuals not taking inhaled steroids (interaction P=0.0002) [50]. The average improvement in the level of PC20 in those H33Q individuals taking ICSs was similar to that associated with nonasthmatic individuals.
A novel variant in FCER2 (which encodes for the low-affinity IgE receptor) has recently been associated with asthma exacerbations while on ICSs [51••]. The SNP, rs28364072, was associated with increased risk of exacerbations in asthmatic children taking ICSs despite generally protective effects of this medication class. Relative risk, expressed as hazard ratios, for exacerbations in those homozygous for the variant allele was 3.95 [95% confidence interval (CI), 1.64–9.51] for white children and 3.08 (95% CI, 1.00–9.47) for African–American children. Of interest, this novel variant was also associated with both higher IgE levels and with differential expression of the FCER2 gene, supporting the contention that variation in FCER2 can adversely affect normal negative feedback in the control of IgE synthesis and action.
In addition to studies evaluating genetic associations, genomic association studies, focusing on differential expression of genes influencing treatment response, have commenced. Hakonarson et al. [52] evaluated glucocorticoid-sensitive and resistant asthmatic patients. Using oligonucleotide microarrays (11 812 genes), expression was measured in blood mononuclear cells at baseline and following stimulation with IL-1β/TNF-α with or without dexamethasone. Fifteen genes predicted glucocorticoid response category with a reported accuracy of 84%. This study focused only on phenotypic extremes; additional studies of this type are warranted. Nonetheless, the promise of rapidly identifying a small subset of candidate genes by expression profiling in this manner is clearly appealing.
In summary, pharmacogenetic studies of ICS response in asthma have shown significant associations with lung function, airways responsiveness, and exacerbations. Future studies are needed to identify novel loci that contribute to response heterogeneity and will achieve the goal of personalizing ICS therapy.
Leukotriene modifiers
Leukotriene modifiers act by inhibiting the action of leukotrienes, which are a family of products generated from the metabolism of arachidonic acid in leukocytes [53]. Leukotrienes can be produced in other cells that lack the complete cassette of enzymes required for this production through the process of transcellular biosynthesis [54]. Two classes of leukotriene modifiers are available for use in the treatment of asthma: 5-lipoxygenase (5-LO) inhibitors (zileuton) and leukotriene receptor antagonists (LTRAs), montelukast, zafirlukast and pranlukast. There is little question that leukotriene modifiers are an important addition to asthma controller therapy owing to their unique mechanism of action, safety (especially LTRAs) and efficacy, and convenient once-daily (for LTRAs) oral dosing. However, heterogeneity in response to leukotriene modifiers seriously detracts from their advantages [43,55] (Fig. 2).
An important question is which polymorphisms contribute to the heterogeneity in response to leukotriene modifiers? The addition/deletion variant in the promoter of ALOX5 reported by Drazen et al. [56] for ABT-761, a 5-LO inhibitor similar to zileuton, has not been replicated in asthma. Recently, we [57] reported that montelukast but not placebo was associated with a 73% reduced risk of an asthma exacerbation in carriers of the mutant allele (number of repeats, five) as compared with homozygous wildtype (five repeats on each allele), which was not consistent with the findings of Drazen et al. [56]. Consistent with the study by Drazen et al. [56], Telleria et al. [58] reported that montelukast treatment decreased the number of asthma exacerbations, improved FEV1, and decreased the use of inhaled β2-agonists in patients with 5/5 or 4/5 repeats but not in participants carrying 4/4 homozygotes. However, both reports [57,58] of montelukast studied a small number of white participants (n=61), and the Telleria study was not blinded. Clearly, associations between this polymorphism and response to LTRAs should be studied in larger and more diverse populations with asthma.
Early reports suggested an association between the response to LTRAs and the C allele of the LTC4 synthase gene promoter (A-444C) polymorphism [59–61]. We replicated this association and also identified three novel SNPs in the ALOX5 (rs2115819), ABCC1 (rs119774), and LTA4H (rs2660845) genes for changes in FEV1 or exacerbation rates [57]. Klotsman et al. [62] also identified novel SNPs in ALOX5 and CYSLTR2 that was associated with response to montelukast in asthma.
Montelukast (and probably other LTRAs) undergoes transport-mediated absorption by OATP2B1 and likely other transporters [63]. A nonsynonymous G1199A (rs12422149; R312G) in SLCO2B1 gene associates with the asthma symptom utility index (ASUI) [64], which is a validated tool that assesses patient preferences of asthma-related symptoms and drug effects on a scale from worse (0) to best possible state (1) and with plasma levels of montelukast obtained from individuals who participated in a large asthma clinical trial [65]. Plasma concentrations of montelukast and ASUI were significantly higher in R312 homozygotes [66] than in heterozygotes. Thus, SNPs in transporters expressed in the gut may have an important influence on the pharmacokinetics of montelukast and other LTRAs, which in turn could influence the patient response to these drugs. However, these associations require replication.
Conclusion
During the past 2 years, significant progress has been made toward identifying novel sequence variants that contribute to the heterogeneity in response to drugs commonly used to treat asthma. In order to move forward toward achieving the goal of personalizing asthma pharmacotherapy, we need to continue to identify novel polymorphisms that associate with response focusing on relevant phenotypes, including bronchodilator reversibility and indices of asthma control in large pharmacogenomic studies. Gene expression studies and studies utilizing genome-wide association methods promise to identify novel loci that contribute to heterogeneity in response to asthma pharmacotherapy. Additionally, the development of statistical models that predict the genomics of response to asthma drugs will complement SNP discovery in moving toward personalized medicine. Currently, we are using Bayes Networks to develop multi-SNP predictions of asthma exacerbations and bronchodilator reversibility. This method is promising for performing multi-SNP, multi-gene prediction of drug response.
Acknowledgments
Funding sources: National Institutes of Health (R01 HL71394, R01 HL074755, K23HL081245, U01 HL065899), American Lung Association Asthma Clinical Research Centers.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 000–000).
- 1.Blake KV, Madabushi R, Derendorf H, Lima JJ. Population phamacodynamic model of bronochodilator response to inhaled albuterol in children and adults with asthma. Chest. 2008 doi: 10.1378/chest.07-2991. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Giuntini CG, Paggiaro PL. Present state of the controversy about regular inhaled beta-agonists in asthma. Eur Respir J. 1995;8:673–678. [PubMed] [Google Scholar]
- 3.Inman WH, Adelstein AM. Rise and fall of asthma mortality in England and Wales in relation to use of pressurised aerosols. Lancet. 1969;2:279–285. doi: 10.1016/s0140-6736(69)90051-8. [DOI] [PubMed] [Google Scholar]
- 4.Sears MR. Relationships between asthma mortality and treatment. Ann Allergy. 1993;70:425–426. [PubMed] [Google Scholar]
- 5.Pearce N, Beasley R, Crane J, et al. End of the New Zealand asthma mortality epidemic. Lancet. 1995;345:41–44. doi: 10.1016/s0140-6736(95)91159-6. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins GA, Tantisira K, Meyers DA, et al. Sequence, haplotype, and association analysis of ADRbeta2 in a multiethnic asthma case-control study. Am J Respir Crit Care Med. 2006;174:1101–1109. doi: 10.1164/rccm.200509-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortega VE, Montealegre F, Chardon D, et al. Sequencing of the beta2 adrenergic receptor gene in Puerto Ricans with asthma [abstract] Am J Respir Crit Care Med. 2007;175:A461. [Google Scholar]
- 8.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. (Erratum in Biochemistry 1994 29; 33:14368) [DOI] [PubMed] [Google Scholar]
- 9.Green SA, Turki J, Bejarano P, et al. Influence of beta 2-adrenergic receptor genotypes on signal transduction in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 1995;13:25–33. doi: 10.1165/ajrcmb.13.1.7598936. [DOI] [PubMed] [Google Scholar]
- 10.Lima JJ, Mohamed M, Eberle LV, et al. Impact of genetic polymorphisms of the β2-adrenergic receptor on albuterol bronchodilator pharmacodynamics. Clin Pharmacol Ther. 1999;65:519–525. doi: 10.1016/S0009-9236(99)70071-8. [DOI] [PubMed] [Google Scholar]
- 11.Martinez FD, Graves PE, Baldini M, et al. Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. J Clin Invest. 1997;100:3184–3188. doi: 10.1172/JCI119874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choudhry S, Ung N, Avila PC, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med. 2005;171:563–570. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 13.Drysdale CM, McGraw DW, Stack CB, et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci U S A. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silverman EK, Kwiatkowski DJ, Sylvia JS, et al. Family-based association analysis of beta2-adrenergic receptor polymorphisms in the childhood asthma management program. J Allergy Clin Immunol. 2003;112:870–876. doi: 10.1016/s0091-6749(03)02023-2. [DOI] [PubMed] [Google Scholar]
- 15.Taylor DR, Epton MJ, Kennedy MA, et al. Bronchodilator response in relation to beta2-adrenoceptor haplotype in patients with asthma. Am J Respir Crit Care Med. 2005;172:700–703. doi: 10.1164/rccm.200501-092OC. [DOI] [PubMed] [Google Scholar]
- 16.Hall IP, Blakey JD, Al Balushi KA, et al. Beta2-adrenoceptor polymorphisms and asthma from childhood to middle age in the British 1958 birth cohort: a genetic association study. Lancet. 2006;368:771–779. doi: 10.1016/S0140-6736(06)69287-8. [DOI] [PubMed] [Google Scholar]
- 17.Tsai HJ, Shaikh N, Kho JY, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet. 2006;119:547–557. doi: 10.1007/s00439-006-0169-2. [DOI] [PubMed] [Google Scholar]
- 18.Blake KV, Feng H, Tantisira K, et al. Bronochodilator response and B2-adrenergic receptor ADRB2 haplotype tagging single nucleotide polymorphisms (htSNPs) [abstract] Am J Respir Crit Care Med. 2008;177:A568. [Google Scholar]
- 19.Litonjua AA, Lasky-Su JA, Schneiter K, et al. ARG1 is a novel bronchodilator response gene: screening and replication in four asthma cohorts. Am J Respir Crit Care Med. 2008;178:688–694. doi: 10.1164/rccm.200709-1363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litonjua AA, Tantisira K, Lasky-Su J, et al. Genome-wide association analysis of bronchodilator response in asthma [abstract] Am J Respir Crit Care Med. 2008;177:A776. [Google Scholar]
- 21.Carroll CL, Stoltz P, Schramm CM, Zucker AR. Beta2-adrenergic receptor polymorphisms affect response to treatment during acute asthma exacerbations in children. Am J Respir Crit Care Med. 2008;177:A567. doi: 10.1378/chest.08-2041. abstract. [DOI] [PubMed] [Google Scholar]
- 22.Martin AC, Zhang G, Rueter K, et al. Beta2-adrenoceptor polymorphisms predict response to beta2-agonists in children with acute asthma. J Asthma. 2008;45:383–388. doi: 10.1080/02770900801971792. [DOI] [PubMed] [Google Scholar]
- 23.Sears MR, Taylor DR, Print CG, et al. Regular inhaled beta-agonist treatment in bronchial asthma [see comments] Lancet. 1990;336:1391–1396. doi: 10.1016/0140-6736(90)93098-a. [DOI] [PubMed] [Google Scholar]
- 24.Drazen JM, Israel E, Boushey HA, et al. Comparison of regularly scheduled with as-needed use of albuterol in mild asthma. Asthma Clinical Research Network. N Engl J Med. 1996;335:841–847. doi: 10.1056/NEJM199609193351202. [DOI] [PubMed] [Google Scholar]
- 25.Israel E, Drazen JM, Liggett SB, et al. The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med. 2000;162:75–80. doi: 10.1164/ajrccm.162.1.9907092. [DOI] [PubMed] [Google Scholar]
- 26.Israel E, Chinchilli VM, Ford JG, et al. Use of regularly scheduled albuterol treatment in asthma: genotype-stratified, randomised, placebo-controlled cross-over trial. Lancet. 2004;364:1505–1512. doi: 10.1016/S0140-6736(04)17273-5. [DOI] [PubMed] [Google Scholar]
- 27.Hancox RJ, Sears MR, Taylor DR. Polymorphism of the beta2-adrenoceptor and the response to long-term beta2-agonist therapy in asthma. Eur Respir J. 1998;11:589–593. [PubMed] [Google Scholar]
- 28.Taylor DR, Drazen JM, Herbison GP, et al. Asthma exacerbations during long term beta agonist use: influence of beta(2) adrenoceptor polymorphism. Thorax. 2000;55:762–767. doi: 10.1136/thorax.55.9.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castle W, Fuller R, Hall J, Palmer J. Serevent nationwide surveillance study: comparison of salmeterol with salbutamol in asthmatic patients who require regular bronchodilator treatment. BMJ. 1993;306:1034–1037. doi: 10.1136/bmj.306.6884.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson HS, Weiss ST, Bleecker ER, et al. The Salmeterol Multicenter Asthma Research Trial: a comparison of usual pharmacotherapy for asthma or usual pharmacotherapy plus salmeterol. Chest. 2006;129:15–26. doi: 10.1378/chest.129.1.15. [DOI] [PubMed] [Google Scholar]
- 31.Taylor DR, Town GI, Herbison GP, et al. Asthma control during long-term treatment with regular inhaled salbutamol and salmeterol. Thorax. 1998;53:744–752. doi: 10.1136/thx.53.9.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Cates CJ, Cates MJ. Regular treatment with salmeterol for chronic asthma: serious adverse events. Cochrane Database Syst Rev. 2008:CD006363. doi: 10.1002/14651858.CD006363.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]; This Cochrane review included 26 clinical trials of over 60 000 asthmatic patients to evaluate risks of fatal and nonfatal serious adverse events from exposure to salmeterol. A strength of this review is that the influence of ICSs was minimized by excluding trials that included ICSs as part of the trial design, allowing for a clearer estimate of the intrinsic effects of salmeterol on asthma control. A statistically significant odds ratio (9.52) was observed for the effect of salmeterol on asthma-related death as compared with albuterol or placebo. Thus, despite large numbers of retrospective pharmacogenetic reviews demonstrating no effect by ADRB2 genotype, further research is needed to uncover the mechanisms for LABA toxicity.
- 33.Wechsler ME, Lehman E, Lazarus SC, et al. Beta-adrenergic receptor polymorphisms and response to salmeterol. Am J Respir Crit Care Med. 2006;173:519–526. doi: 10.1164/rccm.200509-1519OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee DK, Currie GP, Hall IP, et al. The arginine-16 beta2-adrenoceptor polymorphism predisposes to bronchoprotective subsensitivity in patients treated with formoterol and salmeterol. Br J Clin Pharmacol. 2004;57:68–75. doi: 10.1046/j.1365-2125.2003.01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleecker ER, Yancey SW, Baitinger LA, et al. Salmeterol response is not affected by beta2-adrenergic receptor genotype in subjects with persistent asthma. J Allergy Clin Immunol. 2006;118:809–816. doi: 10.1016/j.jaci.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 36.Bleecker ER, Postma DS, Lawrance RM, et al. Effect of ADRB2 polymorphisms on response to longacting beta2-agonist therapy: a pharmacogenetic analysis of two randomised studies. Lancet. 2007;370:2118–2125. doi: 10.1016/S0140-6736(07)61906-0. [DOI] [PubMed] [Google Scholar]
- 37.Bleecker ER, Lawrance R, Ambrose H, Goldman M. Beta2-adrenergic receptor Gly16Arg variation: effect on response to budesonide/formoterol (BUD/FM) or budesonide (BUD; postformoterol) in children and adolescents with asthma [abstract] Am J Respir Crit Care Med. 2008;177:A776. [Google Scholar]
- 38.Bleecker ER, Lawrance RM, Ambrose HJ, Goldman M. Beta2-adrenergic receptor gene polymorphisms: is Arg/Arg genotype associated with serious adverse events during treatment with budesonide and formoterol in one pressurized metered-dose inhaler (BUD/FM pMDI) within racial groups? Am J Respir Crit Care Med. 2008;177:A775. Ref Type: Abstract. [Google Scholar]
- 39.Anderson W, Bacanu SA, Bleecker ER, et al. A prospective haplotype analysis of beta2-adrenergic receptor polymorphisms and clinical response to salmeterol and salmeterol/fluticasone propionate [abstract] Am J Respir Crit Care Med. 2008;177:A775. [Google Scholar]
- 40.McGraw DW, Forbes SL, Kramer LA, Liggett SB. Polymorphisms of the 5′ leader cistron of the human beta2-adrenergic receptor regulate receptor expression. J Clin Invest. 1998;102:1927–1932. doi: 10.1172/JCI4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Panebra A, Schwarb MR, Swift SM, et al. Variable-length poly-C tract polymorphisms of the beta2-adrenergic receptor 3′UTR alter expression and agonist regulation. Am J Physiol Lung Cell Mol Physiol. 2008;294:L190–L195. doi: 10.1152/ajplung.00277.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first study to evaluate the regulatory effects of a poly-C tract polymorphism on β2-receptor function in the 3′UTR of ADRB2.
- 42.National Asthma Education and Prevention Program. Expert panel report 2: guidelines for the diagnosis and management of asthma. Bethesda, MD: U.S. Department of Health and Human Services, Public Health Service, National Institutes of Health, National Heart, Lung, and Blood Institute; Apr, 1997. Report No. 97–4051. (Artwork) [Google Scholar]
- 43.Malmstrom K, Rodriguez-Gomez G, Guerra J, et al. Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. A randomized, controlled trial. Montelukast/Beclomethasone Study group. Ann Intern Med. 1999;130:487–495. doi: 10.7326/0003-4819-130-6-199903160-00005. [DOI] [PubMed] [Google Scholar]
- 44.Szefler SJ, Phillips BR, Martinez FD, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115:233–242. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 45.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–1070. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 46.Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13:1353–1359. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- 47.Dijkstra A, Koppelman GH, Vonk JM, et al. Pharmacogenomics and outcome of asthma: no clinical application for long-term steroid effects by CRHR1 polymorphisms. J Allergy Clin Immunol. 2008;121:1510–1513. doi: 10.1016/j.jaci.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 48••.Tantisira KG, Lazarus R, Litonjua AA, et al. Chromosome 17: association of a large inversion polymorphism with corticosteroid response in asthma. Pharmacogenet Genomics. 2008;18:733–737. doi: 10.1097/FPC.0b013e3282fe6ebf. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the association of a structural inversion with corticosteroid response in asthma.
- 49.Finotto S, Neurath MF, Glickman JN, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 50.Tantisira KG, Hwang ES, Raby BA, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A. 2004;101:18099–18104. doi: 10.1073/pnas.0408532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Tantisira KG, Silverman ES, Mariani TJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007;120:1285–1291. doi: 10.1016/j.jaci.2007.09.005. [DOI] [PubMed] [Google Scholar]; This is the first association of a genetic variant with exacerbations in association with inhaled corticosteroid usage in asthma.
- 52.Hakonarson H, Bjornsdottir US, Halapi E, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc Natl Acad Sci U S A. 2005;102:14789–14794. doi: 10.1073/pnas.0409904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters-Golden M, Henderson WR., Jr Leukotrienes. N Engl J Med. 2007;357:1841–1854. doi: 10.1056/NEJMra071371. [DOI] [PubMed] [Google Scholar]
- 54.Folco G, Murphy RC. Eicosanoid transcellular biosynthesis: from cell-cell interactions to in vivo tissue responses. Pharmacol Rev. 2006;58:375–388. doi: 10.1124/pr.58.3.8. [DOI] [PubMed] [Google Scholar]
- 55.Lima JJ. Treatment heterogeneity in asthma: genetics of response to leukotriene modifiers. Mol Diagn Ther. 2007;11:97–104. doi: 10.1007/BF03256228. [DOI] [PubMed] [Google Scholar]
- 56.Drazen JM, Yandava CN, Dube L, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to antiasthma treatment. Nat Genet. 1999;22:168–170. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 57.Lima JJ, Zhang S, Grant A, et al. Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am J Respir Crit Care Med. 2006;173:379–385. doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Telleria JJ, Blanco-Quiros A, Varillas D, et al. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir Med. 2008;102:857–861. doi: 10.1016/j.rmed.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 59.Sampson AP, Siddiqui S, Buchanan D, et al. Variant LTC(4) synthase allele modifies cysteinyl leukotriene synthesis in eosinophils and predicts clinical response to zafirlukast. Thorax. 2000;55(Suppl 2):S28–S31. doi: 10.1136/thorax.55.suppl_2.S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asano K, Shiomi T, Hasegawa N, et al. Leukotriene C4 synthase gene A(-444)C polymorphism and clinical response to a CYS-LT(1) antagonist, pranlukast, in Japanese patients with moderate asthma. Pharmacogenetics. 2002;12:565–570. doi: 10.1097/00008571-200210000-00009. [DOI] [PubMed] [Google Scholar]
- 61.Whelan GJ, Blake K, Kissoon N, et al. Effect of montelukast on time-course of exhaled nitric oxide in asthma: influence of LTC4 synthase A(−444)C polymorphism. Pediatr Pulmonol. 2003;36:413–420. doi: 10.1002/ppul.10385. [DOI] [PubMed] [Google Scholar]
- 62.Klotsman M, York TP, Pillai SG, et al. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics. 2007;17:189–196. doi: 10.1097/FPC.0b013e3280120043. [DOI] [PubMed] [Google Scholar]
- 63.Mougey EB, Feng H, Castro M, et al. Transporter mediated absorption of montelukast in a CACO-2 model system [abstract] Clin Pharmacol Ther. 2008;83:S67. [Google Scholar]
- 64.Revicki DA, Leidy NK, Brennan-Diemer F, et al. Integrating patient preferences into health outcomes assessment: the multiattribute Asthma Symptom Utility Index. Chest. 1998;114:998–1007. doi: 10.1378/chest.114.4.998. [DOI] [PubMed] [Google Scholar]
- 65.ALA, ACRC. Clinical trial of low-dose theophylline and montelukast in patients with poorly controlled asthma. Am J Respir Crit Care Med. 2007;175:235–242. doi: 10.1164/rccm.200603-416OC. [DOI] [PubMed] [Google Scholar]
- 66.Mougey EB, Feng H, Irvin C, Lima J. Montelukast plasma concentration associates with a common SNP on SLCO2B1 [abstract] Clin Pharmacol Ther. 2008;83:S67. [Google Scholar]


