Abstract
BACKGROUND
DNA genotyping with mutation-specific TaqMan® probes (Applied Biosystems) is broadly used in detection of single-nucleotide polymorphisms but is less so for somatic mutations because of its limited selectivity for low-level mutations. We recently described coamplification at lower denaturation temperature–PCR (COLD-PCR), a method that amplifies minority alleles selectively from mixtures of wild-type and mutation-containing sequences during the PCR. We demonstrate that combining COLD-PCR with TaqMan technology provides TaqMan genotyping with the selectivity needed to detect low-level somatic mutations.
METHODS
Minor-groove binder–based or common TaqMan probes were designed to contain a nucleotide that matches the desired mutation approximately in the middle of the probe. The critical denaturation temperature (Tc) of each amplicon was then experimentally determined. COLD-PCR/TaqMan genotyping was performed in 2 steps: denaturation at the Tc, followed by annealing and extension at a single temperature (fast COLD-PCR). The threshold cycle was used to identify mutations on the basis of serial dilutions of mutant DNA into wild-type DNA and to identify TP53 (tumor protein p53) and EGFR [epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian)] mutations in tumors.
RESULTS
COLD-PCR/TaqMan genotyping identified G>A mutations within TP53 exon 8 (codon 273 mutation hot spot) and C>T mutations within the EGFR gene (drug-resistance mutation T790M) with a selectivity improvement of 15- to 30-fold over regular PCR/TaqMan genotyping. A second round of COLD-PCR/TaqMan genotyping improved the selectivity by another 15- to 30-fold and enabled detection of 1 mutant in 2000 wild-type alleles. Use of COLD-PCR/TaqMan genotyping allowed quantitative identification of low-level TP53 and T790 mutations in colon tumor samples and in non–small-cell lung cancer cell lines treated with kinase inhibitors.
CONCLUSIONS
The major improvement in selectivity provided by COLD-PCR enables the popular TaqMan genotyping method to become a powerful tool for detecting low-level mutations in clinical samples.
Mutation detection plays a key role in the diagnosis, treatment, and prognosis assessment of cancer patients (1). Methods used for mutation detection include sequencing (2, 3), RFLP analysis (4), MALDI-TOF analysis (5), denaturing HPLC/Surveyor™ (6, 7), ligation-mediated PCR (8), high-resolution melting (9, 10), peptide nucleic acid (PNA)3-locked nucleic acids (11), antiprimer quenching real-time PCR (12, 13), Scorpion primers (14), molecular beacons, and methods based on TaqMan® probes (Applied Biosystems) (15, 16). Because of its simplicity and speed, TaqMan genotyping is frequently used as an end-point approach (17). The reaction consists of 2 primers and 2 probes that match to either the wild-type or mutant allele. The polymorphic nucleotide is usually designed to be in the middle third of the probe, which is labeled with a reporter molecule at the 5′ end and with a non-fluorescent quencher at the 3′ end. Modifications of the TaqMan probe with minor-groove binders (MGBs) (18) or locked nucleic acids (19) increase the probe’s Tm (temperature at which 50% of the probe is denatured from the template) to allow the design of shorter probes and better discrimination between mutant and wild-type alleles. The selectivity limit of Taq-Man genotyping is the detection of mutant alleles present at an abundance of approximately 10%–20% of that of the wild-type allele (17, 20). Because the frequencies of somatic mutations can often be lower (6, 21), this limit poses problems for the use of Taq-Man genotyping in screening for somatic mutations in tumor surgical samples or bodily fluids that are often contaminated with wild-type alleles.
We recently described a new form of PCR, coamplification at lower denaturation temperature–PCR (COLD-PCR), which preferentially enriches “minority alleles” from mixtures of wild-type and mutation-containing sequences, irrespective of where a mutation lies in the sequence (22). COLD-PCR is based on the observations that (a) for each DNA sequence there is a critical denaturation temperature (Tc) that is lower than the Tm of the target sequence and below which PCR efficiency drops abruptly, and (b) Tc is dependent on the DNA sequence. DNA amplicons differing by a single nucleotide have substantially different and reproducible amplification efficiencies when the PCR denaturation temperature is set to the Tc. These features are exploited during PCR amplification to selectively enrich minority alleles that differ by one or more nucleotides at any position in a given sequence. Consequently, COLD-PCR amplification of genomic DNA yields PCR products that contain high percentages of variant alleles, thus permitting their detection. We have demonstrated that COLD-PCR improves the selectivity of RFLP analysis, denaturing HPLC/Surveyor, Sanger sequencing, pyrosequencing, and MALDI-TOF–based mutation detection by one to two orders of magnitude (22). We demonstrate that combining COLD-PCR with the TaqMan genotyping method provides a major improvement in the latter’s ability to quantitatively detect low-level somatic mutations in tumor samples in a real-time format.
Materials and Methods
SOURCE OF GENOMIC DNA
Reference human male genomic DNA was purchased from Promega and used as wild-type DNA in dilution experiments with mutation-containing DNA. Genomic DNA from SW480 and 4 lung adenocarcinoma cell lines (H1975, H820, PC9GR, and H3255GR) were purchased from the ATCC. The H3255GR cell line was developed by exposing H3255 cells to serially increasing concentrations of gefitinib for 6 months until the cells were able to proliferate in 100 nmol/L gefitinib with growth kinetics similar to those of untreated cells (23). Similarly, the PC9GR cell line was derived by gefitinib treatment of PC9 cells (24). Snap-frozen colon tumor samples were obtained from the Massachusetts General Hospital Tumor Bank following Internal Review Board approval. DNA was extracted from cell lines and tumor samples with the DNeasy Blood & Tissue Kit (Qiagen). Primers were synthesized by Integrated DNA Technologies.
SINGLE-ROUND COLD-PCR/TaqMan GENOTYPING
COLD-PCR/TaqMan real-time genotyping for the T790M mutation encoded by EGFR exon 20
See (25, 26) for further details. Real-time PCR reactions were performed directly with 70 ng genomic DNA in the presence of 0.2 μmol/L regular TaqMan probe (5′–6-FAM-CAT GAG CTG CAT GAT GAG CTG-BHQ-1–3′) or 0.1 μmol/L MGB TaqMan probe (5′–6-FAM-TGA GCT GCA TGA TGA GC-MGBNFQ–3′) that fully matches the mutation-containing sequence on DNA from H1975 cells that encodes the T790M mutation (mutation is underlined). The final concentrations of the other reagents were as follows: 1× GoTaq Flexi Buffer (Promega), 1× GoTaq Flexi DNA Polymerase (Promega), 0.2 mmol/L of each deoxynucleoside triphosphate, 0.2 μmol/L forward primer (5′–TGATGGCCAGCGTGGAC–3′), 0.2 μmol/L reverse primer (5′–CAGGAGGCAGCCGAAGG–3′), and 2.5 mmol/L MgCl2. The size of the PCR amplicon is 104 bp. Fast COLD-PCR cycling was performed on a Cepheid SmartCycler™ machine as follows: 95 °C for 120 s; 20 cycles of 95 °C for 15 s and 60 °C (fluorescence reading on) for 30 s; and 30 cycles of 88 °C for 15 s and 60 °C (fluorescence reading on) for 30 s. The 88 °C Tc for this amplicon was determined experimentally, as described previously (22). In brief, a set of PCR reactions were performed at gradually decreasing denaturation temperatures (0.3 °C steps starting from the Tm), and the lowest denaturation temperature that reproducibly yielded a PCR product was chosen.
Quantification of T790M mutations in lung adenocarcinoma cell lines with COLD-PCR/TaqMan genotyping
We first used regular PCR with an intercalating dye on a 104-bp EGFR4 [epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian)] amplicon to quantify the copy number of EGFR exon 20; the PCR was carried out independent of the presence or absence of a mutation. We used 0.1× LCGreendye(Idaho Technology)in this reaction without a TaqMan probe. The PCR cycling conditions were 95 °C for 120 s, and 40 cycles of 95 °C for 15 s and 60 °C (fluorescence reading on) for 60 s. We also used serial dilutions of known concentrations of reference DNA as a calibration reference to quantify the copy numbers of DNA from non–small-cell lung cancer (NSCLC) cell lines.
After determining EGFR allele copy numbers, we tested cell line DNA containing equivalent numbers of EGFR exon 20 copies with COLD-PCR/TaqMan genotyping to quantify the relative amounts of T790M mutations. Our assessment of the amount of mutant T790M allele as a percentage of the wild-type allele with COLD-PCR/TaqMan genotyping was based on a calibration curve of serial dilutions of known amounts of mutation-containing DNA added to wild-type DNA.
COLD-PCR/TaqMan real-time genotyping of the G3A mutation in codon 273 of the TP53 exon 8 fragment
See (27) for further details. Real-time PCR reactions were performed directly from 20 ng genomic DNA in the presence of 0.2 μmol/L of a TaqMan probe (5′–6-FAM-TTT GAG GTG CAT GTT TGT GCC-BHQ-1–3′) that fully matches the mutation-containing sequence in DNA from SW480 cells (mutation is underlined). The final concentrations of the other reagents were as follows: 1× GoTaq Flexi Buffer, 1× GoTaq Flexi DNA Polymerase, 0.2 mmol/L of each deoxynucleoside triphosphate, 0.2 μmol/L forward primer (5′–TGG TAA TCT ACT GGG ACG–3′), 0.2 μmol/L reverse primer (5′–CGG AGA TTC TCT TCC TCT–3′), and 3 mmol/L MgCl2. The size of the COLD- PCR amplicon was 87 bp, with a Tc of 83.5 °C defined experimentally as described above. The fast COLD-PCR cycling conditions were as follows: 95 °C for 120 s; 25 cycles of 95 °C for 15 s and 58 °C (fluorescence reading on) for 60 s; and 25 cycles of 83.5 °C for 15 s and 58 °C (fluorescence reading on) for 60 s. Experiments were repeated at least 5 times in independent experiments.
TWO-ROUND COLD-PCR/TaqMan GENOTYPING FOR T790M MUTATIONS IN EGFR EXON 20
To examine whether a second round of COLD-PCR increases the overall selectivity of the method, we performed a nested second round of COLD-PCR/TaqMan genotyping after completing the first round. After performing the first PCR round with a 104-bp PCR amplicon as described above, we carried out a nested PCR with a 10 000–fold dilution of the first PCR product, a nested forward primer (5′–CTGGGCATCTGC CTCA–3′; amplicon size, 67 bp; Tc, 83.5 °C, defined experimentally as described above), and the same reverse primer used in the first PCR. The fast COLD-PCR cycling conditions were as follows: 95 °C for 120 s; 5 cycles of 95 °C for 15 s and 60 °C (fluorescence reading on) for 30 s; and 35 cycles of 83.5 °C for 15 s and 60 °C (fluorescence reading on) for 30 s. COLD-PCR/Taq-Man genotyping experiments were repeated independently at least 5 times.
Results
PRINCIPLE OF COLD-PCR/TaqMan GENOTYPING
COLD-PCR can be carried out in 2 formats, full COLD-PCR and fast COLD-PCR, depending on whether it is necessary to detect all mutations comprehensively or to detect specific Tm-reducing mutations in a rapid and highly selective fashion (22). The combination of full COLD-PCR with TaqMan genotyping can be applied for Tm-increasing mutations such as A:T>G:C or T:A>G:C or for Tm-decreasing mutations; however, the Tm of a DNA sequence is reduced for the great majority of mutations encountered in cancer samples (28), including the T790M mutation (i.e., C>T, EGFR exon 20) and the codon 273 mutation (G>A, TP53 exon 8) examined in this investigation. In view of the simplicity, speed, and high mutation enrichment achieved via fast COLD-PCR, we focused on developing the combination of fast COLD-PCR with TaqMan genotyping to detect Tm-reducing mutations.
Because the present application is aimed at detecting low-level mutant alleles, the COLD-PCR/TaqMan reaction uses a single TaqMan probe specific for the mutant allele, in which the mutation is placed approximately in the middle of the probe (i.e., there is no need for a second TaqMan probe to detect the wild-type allele, as in conventional TaqMan genotyping). The cycling program includes approximately 20–25 regular PCR cycles to build-up the PCR product, followed by a switch to a 2-step PCR consisting of denaturing at Tc and then lowering to a single temperature for both primer annealing and extension (Fig. 1). At the Tc, the majority of the wild-type amplicons remain double-stranded; however, mutant amplicons are largely denatured at the Tc and function as template for primer and probe binding. Lowering the temperature from the Tc to the annealing and extension temperature allows the probe to bind with the complementary mutant strand. Accordingly, COLD-PCR not only enriches the mutant but also reduces the chance that the probe will mismatch-bind to the wild-type strand by keeping the wild type double-stranded. During the annealing and extension step, the 5′→3′ exonuclease activity of Taq polymerase digests the probe to release the reporter from the quencher, allowing the fluorescence signal to be read at this step (29). The presence and quantity of mutations are detected by recording the threshold cycle of the real-time reaction relative to that of reference samples containing known amounts of the same mutation.
Fig. 1. Outline of COLD-PCR/TaqMan genotyping.

COLD-PCR/TaqMan genotyping is a 2-step amplification that uses denaturation at a Tc and annealing/extension at a single temperature. A TaqMan probe with a centrally located nucleotide matching the mutation is used for realtime detection. WT, wild type; MUT, mutant.
IMPROVEMENT OF TaqMan GENOTYPING VIA REAL-TIME QUANTITATIVE COLD-PCR
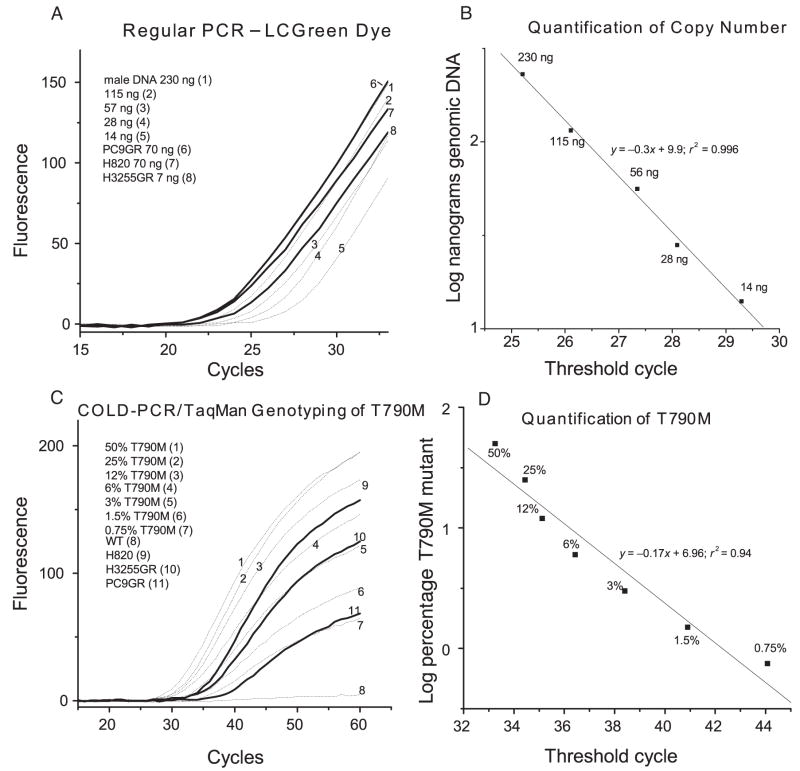
Validation of COLD-PCR/TaqMan genotyping was done by means of serial dilutions of DNA from tumor-derived cell lines containing the gefitinib-resistance mutation [C>T at codon 790 of EGFR exon 20 (cell line H1975)] (30) or TP53 hot-spot mutation G>A at codon 273 of TP53 exon 8 (cell line SW480) (31). These Tm-reducing mutants are suitable for enrichment via fast COLD-PCR. Fig. 2 depicts representative results comparing the selectivities of regular PCR/TaqMan genotyping with COLD-PCR/TaqMan genotyping for a 104-bp amplicon from EGFR exon 20. The selectivity limit of regular PCR/TaqMan genotyping is about 12% mutant allele (Fig. 2A); in contrast, COLD-PCR improves the selectivity to 0.8% (Fig. 2B). Next, we tested whether COLD-PCR/TaqMan genotyping of EGFR exon 20 encoding the T790M variant can quantify the low population of T790M mutations in NSCLC cell lines. Because the EGFR gene is frequently amplified in NSCLC cells, the potential variation in copy number for EGFR exon 20 needs to be considered before T790M quantification. We applied regular PCR in the presence of the LCGreen dye to quantify the copy numbers for the 3 NSCLC lines (H820, H3255GR, and PC9GR; Fig. 3, A and B). H3255GR exhibits 16-fold amplification, and H820 and PC9GR exhibit 4-fold amplification. On the basis of this quantification, we diluted the genomic DNA from these 3 cell lines to obtain equal copy numbers of exon 20 and tested for T790M mutants and known dilutions of T790M mutants added to wild-type DNA (Fig. 3, C and D). The percentage of T790M was calculated by interpolation to be 5% for H820, 2.85% for H3255GR, and 0.4% for PC9GR.
Fig. 2. Comparison of regular PCR/TaqMan genotyping with COLD-PCR/TaqMan genotyping for the T790M C>T mutation in EGFR exon 20.
Serial dilutions into wild-type DNA of DNA from the H1975 cell line containing the T790M mutation encoded by EGFR exon 20 were screened for the T790M mutation with regular PCR/TaqMan genotyping (A) and with COLD-PCR/TaqMan genotyping (MGB probe) (B).
Fig. 3. Quantification of the T790M mutation in NSCLC cell lines.
(A), Primary amplification curve for EGFR exon 20 with the LCGreen dye. DNA from NSCLC cell lines (PC9GR, H820, and H3255GR) was serially diluted into human male DNA and amplified. (B), Plot of the log concentration of input genomic DNA vs threshold cycle to quantify the copy number of EGFR xon 20. (C), Primary amplification curve for the T790M mutation from EGFR exon 20 with COLD-PCR/Taqman genotyping (MGB probe). Serial dilutions of T790M mutant DNA and DNA from NSCLC cell lines were tested. (D), Plot of the log concentration (percent) of T790M mutant DNA vs threshold cycle to quantify the percentage of T790M in NSCLC cell lines. WT, wild type.
Fig. 4. Comparison of regular PCR/TaqMan genotyping with COLD-PCR/TaqMan genotyping for the G>A mutation in codon 273 of TP53 exon 8.
Serial dilutions into wild-type DNA of SW480 cell line DNA containing the G_A mutation in codon 273 of TP53 exon 8 were evaluated for the codon 273 mutation with regular PCR/TaqMan genotyping (A) and COLD-PCR/TaqMan genotyping (B). Clinical tumor samples from 3 NSCLC cancer patients (TL6, TL8, and TL18) and a colon cancer sample (CT20) were evaluated for the codon 273 mutation with regular PCR/TaqMan genotyping (C) and COLD-PCR/TaqMan genotyping (D).
Fig. 4, A and B, presents the results of applying real-time COLD-PCR to TaqMan genotyping of the hot-spot mutations in codon 273 of TP53 exon 8. Whereas the limit of selectivity for regular PCR/Taq-Man genotyping is about 10% mutant allele, COLD-PCR/TaqMan genotyping can detect as little as 0.33% mutant alleles among wild-type alleles, an improvement of approximately 30-fold. Examination of 4 cancer samples, one of which (CT20) is known to contain a codon 273 G>A mutation at a low level (approximately 5%) (6, 7), with COLD-PCR/TaqMan genotyping clearly identified the mutation-containing sample, but regular PCR/TaqMan genotyping did not (Fig. 4, C and D). Thus, our data demonstrate that COLD-PCR improves TaqMan-genotyping selectivity by 15- to 30-fold.
To understand further the improvement in mutation selectivity produced by the application of COLD-PCR, we subjected the PCR product used in TaqMan genotyping of the T790 EGFR exon 20 mutation to an RFLP assay with a restriction enzyme, NlaIII, that selectively recognizes mutation-containing DNA. The digested products were then examined via denaturing HPLC, as reported previously (22). For comparison to COLD-PCR/TaqMan genotyping, we conducted identical experiments after regular PCR/TaqMan genotyping. Fig. 5 demonstrates that the product produced by regular PCR/TaqMan genotyping and digested with NlaIII barely shows the mutant peak (12% mutant relative to wild type), in agreement with the real-time PCR results (Fig. 2). In contrast, NlaIII-digested products produced by COLD-PCR/TaqMan genotyping depict mutant peaks down to 0.8% mutant alleles. The data in Fig. 5 are additional verification that the improved real-time PCR quantification of T790M mutations indeed reflects the anticipated mutation-specific products and not false-positive signals.
Fig. 5. RFLP confirmation of enrichment of the T790M mutation by COLD-PCR/TaqMan genotyping.
DNA from the H1975 cell line DNA serially diluted into wild-type (WT) DNA were screened for the T790M mutation with regular PCR/TaqMan genotyping and with COLD-PCR/TaqMan genotyping. The PCR product was digested with NlaIII, and the digest products were separated by denaturing HPLC to discriminate the mutant peak from the wild-type peak. Cycles
FURTHER IMPROVEMENT OF TaqMan GENOTYPING VIA 2 ROUNDS OF COLD-PCR AMPLIFICATION
Given that a single round of COLD-PCR/TaqMan genotyping can detect as little as 0.8% mutant alleles, we tested whether nested COLD-PCR/TaqMan genotyping can further improve the selectivity of T790M mutant detection. The nested PCR generates a 67-bp product from EGFR exon 20. When applied directly to genomic DNA (i.e., not in a nested format), COLD-PCR/TaqMan genotyping of the 67-bp region had a selectivity of approximately 0.8% T790M mutant alleles (see Fig. 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol55/issue4). When 2 COLD-PCR TaqMan reactions are applied in series (the first COLD-PCR for a 104-bp amplicon and the second a nested PCR for the 67-bp amplicon), the combined selectivity for T790M detection is far superior to the selectivity of a single reaction. Fig. 6A shows that a single round of COLD-PCR/TaqMan genotyping fails to detect 0.1% T790M mutant alleles. In contrast, 2 rounds of COLD-PCR/TaqMan genotyping improved the selectivity to better than 0.05% mutant alleles, whereas 11 replicates of the wild-type DNA remained at the baseline. Thus, 2 rounds of COLD-PCR combined with TaqMan genotyping improve the mutation detection over that obtained with a single round.
Fig. 6. Two-round COLD-PCR/TaqMan genotyping of the T790M mutation.
DNA from the H1975 cell line was serially diluted into human male DNA. Duplicate samples containing 0.1% and 0.05% T790M mutant and 11 replicates of reference wild-type (WT) DNA were tested with COLD-PCR/TaqMan genotyping. (A), Single-round COLD-PCR/TaqMan genotyping of the T790M mutation (104-bp amplicon). (B), Second-round (nested) COLD-PCR/TaqMan genotyping of the T790M mutation (67-bp amplicon).
Discussion
We have described COLD-PCR/TaqMan genotyping, a real-time mutation-detection methodology that combines COLD-PCR and TaqMan genotyping for detecting the EGFR-encoded T790M mutant and TP53 codon 273 mutations in serial dilutions of mutant DNA, in cell lines, and in biological samples. The clinical relevance of these mutations is well established. T790M, an acquired mutation in the EGFR protein that renders NSCLC patients resistant to gefitinib or erlotinib, is found in approximately 50% of tumors from patients who have acquired resistance to these kinase inhibitors (32). The presence of hot-spot mutation at codon 273 of TP53 is a factor for a poor prognosis in NSCLC patients (27). The new method is based on the ability of fast COLD-PCR to enrich Tm-reducing mutations and the ability of the Taq-Man probe to detect mutations in a real-time, quantitative format. Consequently, a single round of COLD-PCR/TaqMan genotyping quantitatively detects as little as 0.8% mutant alleles with a 15- to 30-fold better selectivity than regular PCR/TaqMan genotyping. The addition of a second round of COLD-PCR/TaqMan genotyping further improves the selectivity and reproducibly identifies 1 mutant allele among 2000 wild-type alleles.
Alternative TaqMan-based approaches that detect low amounts of mutant alleles have been described. Allele-specific PCR-based TaqMan genotyping, TaqMAMA, uses a mutant-matched nucleotide at the 3′ end of a primer and a penultimate 3′ mismatch to achieve specific allele discrimination in the PCR (33); however, the optimization of TaqMAMA conditions can be tedious (33). PNA-based TaqMan genotyping uses a PNA to inhibit wild-type DNA and a mutant-specific TaqMan probe to detect mutations (34). The necessity to define experimental conditions such as probe concentration while retaining not only the compatibility between the PNA probe and the TaqMan probe but also the ability of the PNA to inhibit the wild type increases the complexity of assay development. Scorpion assays (35) provide a good alternative to TaqMan genotyping in that the probe and primer are combined on a single oligonucleotide. DxS Ltd. offers a commercially available combination of Scorpion and ARMS® (amplification refractory mutation system) technologies that can detect low-level mutations such as T790Min EGFR with a sensitivity similar to that of the single-round COLD-PCR/TaqMan assay; however, the Scorpion assay is relatively more complex, expensive, and slow (1 h for the COLD-PCR/Taqman assay vs 2–3 h for the Scorpion assay) (35). COLD-PCR achieves real-time mutation detection without tedious optimization or the use of costly PNA probes or Scorpion primers, because COLD-PCR/TaqMan genotyping uses only temperature to inhibit amplification of the wild type. Another potential advantage of the COLD-PCR/TaqMan approach is in the multiplex detection of mutations. Multiplexing would be more difficult to achieve with combinations of PNA and TaqMan probes because of the number of oligonucleotides used in the reaction.
In summary, without relying on special probes and reagents, COLD-PCR/TaqMan genotyping is simple, fast, easy to use, and low in cost compared with other TaqMan-based mutation-detection methods. The major improvement in selectivity obtained with COLD-PCR enables the popular TaqMan genotyping method to become a powerful tool for detecting low-level mutations in clinical samples.
Supplementary Material
Acknowledgments
Research Funding: P.A. Jänne, Pfizer; G.M. Makrigiorgos, NIH grants CA-115439 and CA-111994; J. Li, NIH training grant 5 T32 CA09078.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: P.A. Jänne, AstraZeneca, Roche, Boehringer Ingelheim, and AVEO Pharmaceuticals.
Stock Ownership: None declared.
Expert Testimony: None declared.
Honoraria: None declared.
Nonstandard abbreviations: PNA, peptide nucleic acid; MGB, minor-groove binder; Tm, temperature at which 50% of the probe is denatured from the complementary strand; COLD-PCR, coamplification at lower denaturation temperature–PCR; Tc, critical denaturation temperature; NSCLC, non–small-cell lung cancer; TaqMAMA, allele-specific PCR-based TaqMan genotyping.
Human genes: EGFR, epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian); TP53, tumor protein p53.
References
- 1.Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502–11. doi: 10.1056/NEJMra072367. [DOI] [PubMed] [Google Scholar]
- 2.Bayley H. Sequencing single molecules of DNA. Curr Opin Chem Biol. 2006;10:628 –37. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 3.Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007;373:15–24. doi: 10.1385/1-59745-377-3:15. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins GJ, Chaleshtori MH, Song H, Parry JM. Mutation analysis using the restriction site mutation (RSM) assay. Mutat Res. 1998;405:209 –20. doi: 10.1016/s0027-5107(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 5.Ragoussis J, Elvidge GP, Kaur K, Colella S. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics research. PLoS Genet. 2006;2:e100. doi: 10.1371/journal.pgen.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Berbeco R, Distel RJ, Jänne PA, Wang L, Makrigiorgos GM. s-RT-MELT for rapid mutation scanning using enzymatic selection and real time DNA-melting: new potential for multiplex genetic analysis. Nucleic Acids Res. 2007;35:e84. doi: 10.1093/nar/gkm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeung AT, Hattangadi D, Blakesley L, Nicolas E. Enzymatic mutation detection technologies. Biotechniques. 2005;38:749 –58. doi: 10.2144/05385RV01. [DOI] [PubMed] [Google Scholar]
- 8.Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, Parker AR, et al. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141–7. doi: 10.1038/nmeth713. [DOI] [PubMed] [Google Scholar]
- 9.Lipsky RH, Mazzanti CM, Rudolph JG, Xu K, Vyas G, Bozak D, et al. DNA melting analysis for detection of single nucleotide polymorphisms. Clin Chem. 2001;47:635– 44. [PubMed] [Google Scholar]
- 10.Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem. 2004;50:1156–64. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- 11.Orum H. PCR clamping. Curr Issues Mol Biol. 2000;2:27–30. [PubMed] [Google Scholar]
- 12.Li J, Makrigiorgos GM. Anti-primer quenching-based real-time PCR for simplex or multiplex DNA quantification and single-nucleotide polymorphism genotyping. Nat Protoc. 2007;2:50 – 8. doi: 10.1038/nprot.2007.11. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Wang F, Mamon H, Kulke MH, Harris L, Maher E, et al. Antiprimer quenching-based real-time PCR and its application to the analysis of clinical cancer samples. Clin Chem. 2006;52:624–33. doi: 10.1373/clinchem.2005.063321. [DOI] [PubMed] [Google Scholar]
- 14.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804 –7. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 15.Bernard PS, Wittwer CT. Real-time PCR technology for cancer diagnostics. Clin Chem. 2002;48:1178 – 85. [PubMed] [Google Scholar]
- 16.Makrigiorgos GM. PCR-based detection of minority point mutations. Hum Mutat. 2004;23:406 –12. doi: 10.1002/humu.20024. [DOI] [PubMed] [Google Scholar]
- 17.De la Vega FM, Lazaruk KD, Rhodes MD, Wenz MH. Assessment of two flexible and compatible SNP genotyping platforms: TaqMan SNP Genotyping Assays and the SNPlex Genotyping System. Mutat Res. 2005;573:111–35. doi: 10.1016/j.mrfmmm.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655– 61. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore P. Simplifying the probe set. Nature. 2005;435:238. doi: 10.1038/435238a. [DOI] [PubMed] [Google Scholar]
- 20.Wilkening S, Hemminki K, Thirumaran RK, Bermejo JL, Bonn S, Forsti A, Kumar R. Determination of allele frequency in pooled DNA: comparison of three PCR-based methods. Biotechniques. 2005;39:853– 8. doi: 10.2144/000112027. [DOI] [PubMed] [Google Scholar]
- 21.Jänne PA, Borras AM, Kuang Y, Rogers AM, Joshi VA, Liyanage H, et al. A rapid and sensitive enzymatic method for epidermal growth factor receptor mutation screening. Clin Cancer Res. 2006;12:751– 8. doi: 10.1158/1078-0432.CCR-05-2047. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med. 2008;14:579 – 84. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 23.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale CM, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–14. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 25.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102:7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Taki T, Adachi M, Konishi T, Higashiyama M, Miyake M. Mutations in exon 7 and 8 of p53 as poor prognostic factors in patients with non-small cell lung cancer. Oncogene. 1998;16:2469 –77. doi: 10.1038/sj.onc.1201776. [DOI] [PubMed] [Google Scholar]
- 28.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153– 8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–94. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 30.de La Motte Rouge T, Valent A, Ambrosetti D, Vielh P, Lacroix L. [Clinical and molecular predictors of response to EGFR tyrosine kinase inhibitors in non-small cell lung cancer] Ann Pathol. 2007;27:353– 63. doi: 10.1016/s0242-6498(07)78274-3. [French] [DOI] [PubMed] [Google Scholar]
- 31.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 32.Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 33.Glaab WE, Skopek TR. A novel assay for allelic discrimination that combines the fluorogenic 5′ nuclease polymerase chain reaction (TaqMan) and mismatch amplification mutation assay. Mutat Res. 1999;430:1–12. doi: 10.1016/s0027-5107(99)00147-5. [DOI] [PubMed] [Google Scholar]
- 34.Nagai Y, Miyazawa H, Huqun, Tanaka T, Udagawa K, Kato M, et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276 – 82. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]
- 35.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366 –77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.