Abstract
Constitutive heterochromatin in Arabidopsis thaliana is marked by repressive chromatin modifications including DNA methylation, histone H3 dimethylation at lysine 9 (H3K9me2), and monomethylation at lysine 27 (H3K27me1). The enzymes catalyzing DNA methylation and H3K9me2 have been identified and mutations in these proteins lead to the reactivation of silenced heterochromatic elements. The enzymes responsible for heterochromatic H3K27me1, in contrast, remain unknown. Here we show that the divergent SET-domain proteins ARABIDOPSIS TRITHORAX-RELATED PROTEIN5 (ATXR5) and ATXR6 exhibit H3K27 monomethyltransferase activity and double mutants have reduced H3K27me1 in vivo and show partial heterochromatin decondensation. Mutations in atxr5 and atxr6 also lead to transcriptional activation of repressed heterochromatic elements. Interestingly, H3K9me2 and DNA methylation are unaffected in the double mutant. These results indicate that ATXR5 and ATXR6 form a novel class of H3K27 methyltransferases and that H3K27me1 represents a new pathway required for transcriptional repression in Arabidopsis.
In eukaryotes, chromatin modifications such as the methylation of DNA and specific histone residues are associated with epigenetic gene silencing and heterochromatin formation1. Nuclei of the model plant Arabidopsis contain highly condensed regions of constitutive heterochromatin referred to as chromocenters, which are primarily composed of pericentromeric repeats, transposons, and rDNA genes2. Chromocenters are enriched with several epigenetic marks, including DNA methylation at CG, CHG, and CHH (where H=A, T, C) sites, as well as histone modifications such as dimethylation at H3K9 (H3K9me2) and monomethylation at H3K27 (H3K27me1)3–7. The enzymes responsible for the establishment and the maintenance of DNA methylation include DOMAINS REARRANGED METHYLASE 2 (DRM2), which is responsible for the establishment of DNA methylation in all three sequence contexts 8,9. In addition, METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) are required for proper maintenance of CG and CHG methylation, respectively 10–12. Dimethylation at H3K9 (H3K9me2) is mediated by Su(var)3–9 homologs (SUVH), such as SUVH2, KRYPTONITE (KYP)/SUVH4, SUVH5 and SUVH6 11,13–15. The reduction of H3K9me2 in a kyp/suvh4 mutant leads to the decrease of DNA methylation levels at some loci, suggesting a link between DNA and histone methylation 11,16. Removal of DNA methylation and/or H3K9me2 leads to the transcriptional activation of transposable and repeat elements 17. In contrast to H3K9me2, the role of H3K27me1 at chromocenters is less well understood. Data suggests, however, that H3K27me1 is likely to be present independent of DNA methylation/H3K9me2, as H3K27 methylation is unaffected in kyp and met1 mutants 18,19.
With respect to the chromatin modifications associated with Arabidopsis chromocenters, a major unanswered question is the role of H3K27me1. Studies addressing the significance of this mark have been hampered by the inability to identify the enzymes responsible for H3K27 monomethylation. The eukaryotic enzymes that have been demonstrated to methylate H3K27 in vivo are all homologs of the Drosophila melanogaster SET-domain protein Enhancer of zeste (E(Z))20. E(Z) acts as part of the Polycomb repressive complex 2 (PRC2) and requires the WD-40 protein Extra Sex Combs (ESC) for activity in Drosophila21. Arabidopsis contains three E(Z) homologs: MEDEA (MEA), CURLY LEAF (CLF), and SWINGER (SWN) 22. Both CLF and SWN are expressed during postembryonic development and are likely to have redundant functions 23, whereas MEA expression is limited to the female gametophyte and embryo development 24. Although CLF and SWN are the only known H3K27 methyltransferases to be expressed in adult plants, H3K27me1 at chromocenters is unaffected in clf swn double mutants 18. Furthermore, a mutation in FERTILIZATION INDEPENDENT ENDOSPERM (FIE), the sole Arabidopsis homolog of ESC, also shows no effect on H3K27me1 18. These results strongly suggest that H3K27 methylation at chromocenters is catalyzed by unknown proteins that are not homologous to E(Z).
Here we demonstrate that the Arabidopsis proteins ATXR5 and ATXR6 act as H3K27 monomethyltransferases in vitro and in vivo. Moreover, atxr5 atxr6 double mutants show reduced H3K27me1 at chromocenters and partial heterochromatin decondensation. In addition, our results clarify the relationship between different epigenetic marks present in heterochromatin and their roles in gene silencing. Transcriptional activation of repressed elements is observed in atxr5 atxr6 mutant plants, however, DNA methylation and H3K9me2 levels remain unchanged. Thus, DNA methylation and H3K9me2 occur independently of H3K27me1 and gene silencing at constitutive heterochromatin requires the presence of both H3K27me1 and DNA methylation/H3K9me2.
RESULTS
ATXR5 and ATXR6 function as H3K27 monomethyltransferases
Histone lysine methylation is primarily catalyzed by SET-domain proteins25. The substrate specificity of most SET-domain proteins can be predicted by sequence comparison to biochemically-characterized proteins. Phylogenetic analysis of 32 SET-domain proteins from Arabidopsis, however, shows that the homologous proteins ATXR5 and ATXR6 (Supplementary Fig. 1) belong to a divergent, functionally uncharacterized group22,26. Therefore, it is not possible to predict which histone lysine residue might be methylated by these two proteins. ATXR5 and ATXR6 appear to be plant specific and their origin likely coincides with the emergence of land plants, as homologues of these proteins are present in the moss Physcomitrella patens (Supplementary Fig. 1), but not in the unicellular green algae Chlamydomonas reinhardtii.
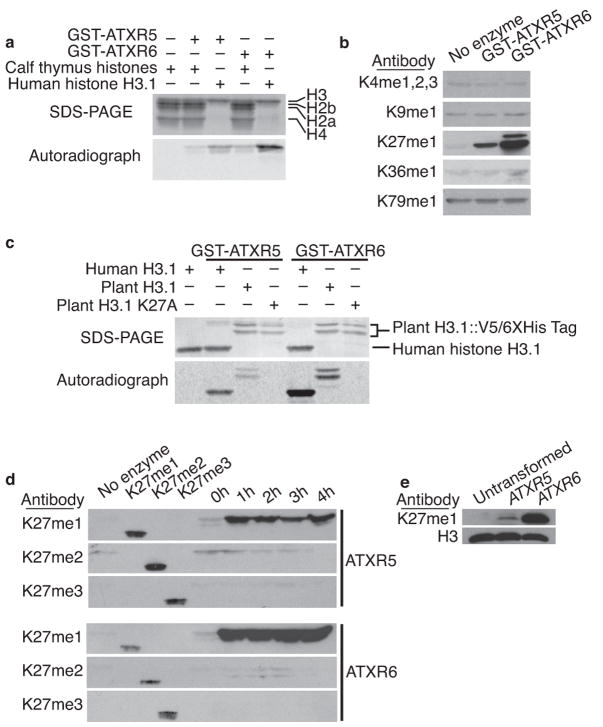
To determine if ATXR5 and ATXR6 are involved in histone methylation, we performed in vitro histone methyltransferase (HMT) assays. We mixed purified GST-tagged ATXR5 and ATXR6 and radiolabeled S-adenosyl methionine (SAM) with calf thymus histones or recombinant human H3.1. Both ATXR5 and ATXR6 methylated H3, but not other histone proteins (Fig. 1a). H3 contains four lysine residues that have been reported to be methylated in Arabidopsis: K4, K9, K27, and K3627,28. In addition to these four H3 lysine residues, K79 is also methylated by the non-SET domain protein Dot1 in yeasts and animals 29–32. To identify which site is methylated by ATXR5 and ATXR6, we repeated the HMT assay with non-radiolabeled SAM and used antibodies that recognize the five different monomethylated lysines of H3. Of these, only the H3K27me1 antibody detected a strong difference between control and enzyme-treated samples (Fig. 1b and Supplementary Fig. 2). To confirm that K27 is indeed the residue methylated by ATXR5 and ATXR6, we created a plant H3 variant in which K27 is changed to alanine (H3K27A) and repeated the HMT assay. ATXR5 and ATXR6 efficiently methylated the wild-type plant H3 protein, but not H3K27A (Fig. 1c).
Figure 1.
ATXR5 and ATXR6 monomethylate H3K27. (a) In vitro histone methyl transferase assay using radiolabeled SAM; only H3 is methylated by ATXR5 and ATXR6. (b) The products of non-radioactive HMT assays were analyzed by Western blot using antibodies against different methylated lysines of H3. (c) ATXR5 and ATXR6 cannot methylate a mutant plant H3 protein where lysine 27 has been replaced with alanine. (d) ATXR5 and ATXR6 HMT time-course assay products were analyzed by Western blot using antibodies specific for mono-, di-, and trimethylated forms of H3K27. H3K27 methylated peptides were included to confirm the specificity of the antibodies and a no-enzyme negative control was included. (e) Western blot analysis of histones extracted from S. cerevisiae expressing ATXR5 and ATXR6.
We then investigated the possibility that ATXR5 and ATXR6 may add more than one methyl group to H3K27. The level of H3K27 methylation (mono-, di- or trimethylated) has important functional implications. For example, immunofluorescence experiments in Arabidopsis have shown that H3K27me1 is enriched in the constitutive heterochromatin, whereas H3K27me3 is restricted to euchromatin18,19. Therefore, we repeated the HMT assay with antibodies that recognize H3K27me1, H3K27me2 and H3K27me3. Even after prolonged incubation, only monomethylation at H3K27 was detected for both ATXR5 and ATXR6 (Fig. 1d), suggesting that these proteins function only as H3K27 monomethyltransferases. To obtain additional evidence that ATXR5 and ATXR6 monomethylate H3K27, we expressed ATXR5 and ATXR6 in the budding yeast Saccharomyces cerevisiae. This model system is particularly well suited for this experiment as methylation of H3K27 is either absent or very minimal33. Our results show that expression of ATXR5 or ATXR6 in yeast does indeed induce monomethylation at lysine 27 (Fig. 1e). These results demonstrate that ATXR5 and ATXR6 are the first examples of eukaryotic H3K27 methyltransferases that are not related to the Drosophila E(Z) protein.
ATXR5 and ATXR6 have redundant functions in development
To investigate the function of ATXR5 and ATXR6 in vivo, we obtained T-DNA insertional mutants (atxr5, SALK_130607; atxr6, SAIL_240_0134). The T-DNA insertion in atxr5 is located in the coding sequence upstream of the SET domain (Fig. 2a and Supplementary Fig. 1). For atxr6, however, the closest available T-DNA insertion is located 147 bp upstream of the predicted start codon. Semi-quantitative RT-PCR analysis shows that the mutants have reduced atrx5 and atrx6 mRNA levels relative to wild-type Columbia (Col) plants (Fig. 2b). No alterations in growth or development were observed in the single mutants. Given the similarities in sequence (Supplementary Fig. 1), biochemical activity (Fig. 1), and partially overlapping expression patterns (Fig. 2c), we suspected that ATXR5 and ATXR6 might have redundant functions. Therefore, we created an atxr5 atxr6 double mutant. atxr5 atxr6 plants were smaller than wild type (Fig. 2d), especially when grown under short days. This phenotype appears to be primarily due to reduced leaf size, as the rate of leaf initiation was similar to wild type (Fig. 2e).
Figure 2.

ATXR5 and ATXR6 play redundant roles in leaf development.
(a) Schematic drawings indicate the genomic structure of ATXR5 and ATXR6; thin horizontal lines represent the chromosome, thick lines represent exons. Horizontal lines below the drawings represent the regions amplified by each primer pair. (b, c) Semi-quantitative RT-PCR analysis of ATXR5 and ATXR6 expression in (b) fully expanded leaves and (c) various tissues. UBQ was used as a constitutively expressed control. (d) Single and double mutants of atxr5 and atxr6 grown under short-day conditions. (e) Rate of leaf initiation of plants were grown under short days.
Chromatin condensation and gene silencing require H3K27me1
Because H3K27me1 is associated with constitutive heterochromatin in Arabidopsis18,19, we examined the effect of atxr5 and atxr6 mutations on chromatin structure. Several DAPI-stained chromocenters characteristic of constitutive heterochromatin are visible in nuclei isolated from fully expanded Arabidopsis leaves (Fig. 3a, b, also Figs. 4 and 5). atxr5 and atxr6 single mutants displayed no visible defect in the organization of the heterochromatin. In contrast, approximately 65% of atxr5 atxr6 nuclei showed partial decondensation of the chromocenters (Fig. 3a, b, also Figs. 4 and 5). Mutations in a number of chromatin-modifying and chromatin-associated proteins have been shown to lead to heterochromatin decondensation35. These mutations, however, can have different effects on chromatin structure. For example, met1 and decrease in DNA methylation1 (ddm1) mutants show decondensation of pericentromeric sequences, but not the 180bp centromeric repeats35. In contrast, variant in methylation1 (vim1) mutants have been reported to show decondensation of centromeric sequences36. Analysis of 180bp-repeat sequences by fluorescence in situ hybridization (FISH) indicates that the centromeric sequences are not strongly decondensed in atxr5 atxr6 double mutants (Fig. 3b). Thus the disruptions in heterochromatin structure observed in atxr5 atxr6 plants appear to be more similar to those observed in met1/ddm1 mutants.
Figure 3.
atxr5 atxr6 mutations lead to disruption of constitutive heterochromatin, reduced H3K27 monomethylation, and reactivation of silenced elements.
(a) Leaf interphase nuclei were stained with DAPI and analyzed for immunofluorescence with an anti-H3K27me1 antibody. Approximately 65% of atxr5 atxr6 nuclei show severe (a, upper panel) or moderate (a, lower panel) chromocenter decondensation and reduced H3K27me1 staining. Scale bar = 5 μm. (b) FISH analysis of leaf interphase nuclei using a 180-bp centromeric repeat probe. The DNA was counterstained with DAPI. Scale bar = 5 μm. (c) Semi-quantitative RT-PCR analysis of heterochromatic elements in Col, atxr5, atxr6, and atxr5 atxr6. UBQ was used as a constitutively expressed control. (d) ChIP analysis of repetitive elements using H3K27me1 antibodies. Black and white bars indicate relative levels of immunoprecipitated DNA normalized to ACTIN, as determined by real-time PCR, from wild type and atxr5 atxr6 leaves, respectively. Grey bars represent no antibody controls. The data are presented as mean ± SEM for three individual experiments.
Figure 4.
Di- and trimethylation of H3K27 are not altered in atxr5 atxr6 mutants
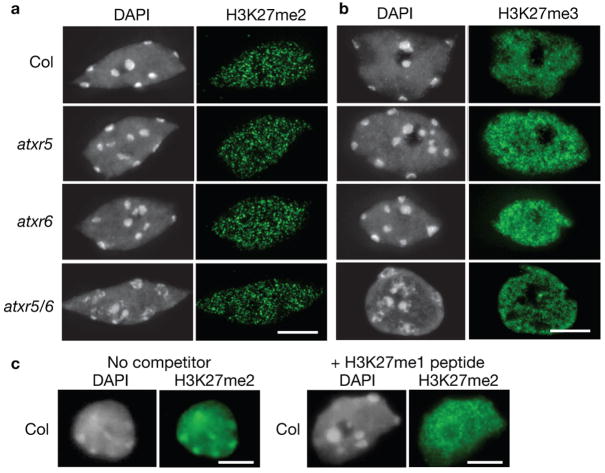
(a, b) Leaf interphase nuclei were stained with DAPI and analyzed for immunoflorescence with (a) anti-H3K27me2 and (b) anti-H3K27me3 antibodies. c, Leaf interphase nuclei were stained with DAPI and analyzed for immunofluorescence with anti-H3K27me238 in the presence or absence of an H3K27me1 peptide. Scale bars = 5 μm.
Figure 5.

Mutations in atxr5 and atxr6 do not affect H3K9 dimethylation or DNA methylation.
(a) Leaf interphase nuclei were stained with DAPI and analyzed for immunoflorescence with anti-H3K9me2 antibodies. Scale bar = 5 μm. (b, d) DNA methylation analysis by (b) locus-specific (Ta3 and CACTA) and (d) genome-wide BS-Seq. Black and white bars represent wild type and atxr5 atxr6, respectively. (c) ChIP analysis of repetitive elements using H3K9me2 antibodies. Black and white bars indicate relative levels of immunoprecipitated DNA normalized to ACTIN, as determined by real-time PCR, from wild type and atxr5 atxr6, respectively. Grey bars represent no antibody controls. The data are presented as mean ± SEM for three individual experiments. (e) Distribution of methylation along the five Arabidopsis chromosomes. (f) Average methylation levels within protein coding genes. (g) Average methylation levels within pseudogenes and transposons. (e–g) A horizontal blue line indicates zero percent methylation. Panels on the left correspond to wild type and panels on the right correspond to atxr5 atxr6 double mutant. CG methylation is indicated by green lines, CHG methylation is indicated by yellow lines, and CHH methylation is indicated by red lines. (e) Vertical blue lines are used to separate different chromosomes. (f, g) Vertical blue lines mark the boundaries between upstream regions and gene bodies and between gene bodies and downstream regions. (h) Leaf interphase nuclei of wild-type plants and kyp suvh5 suvh6 triple mutants were stained with DAPI and analyzed for immunofluorescence with anti-H3K27me1. Scale bar = 5 μm.
Mutations that affect chromatin structure often lead to transcriptional reactivation of repressed heterochromatic elements37. In atxr5 atxr6 plants, we observed that chromatin decondensation is also accompanied by the release of silencing at a DNA repeat (TRANSCRIPTIONALLY SILENT INFORMATION-TSI) and transposons (Ta3, AtMuI, and CACTA) localized in chromocenters (Fig. 3c). To determine if the loss of silencing is associated with reduced H3K27me1, chromatin immunoprecipitation (ChIP) was performed on TSI, Ta3, and CACTA. These experiments indicate that H3K27me1 is reduced at these loci (Fig. 3d), suggesting that ATXR5 and ATXR6 may act directly at these elements. Taken together, these results suggest that ATXR5 and ATXR6 play functionally redundant roles in the formation of constitutive heterochromatin and the silencing of heterochromatic elements in Arabidopsis.
ATXR5 and ATXR6 are required for heterochromatic H3K27me1
Previous experiments have shown that the enrichment of H3K27me1 at chromocenters is not due to the activities of the known E(Z) homologs, MEA, CLF, and SWN18. To determine if ATXR5 and/or ATXR6 play a role in H3K27 methylation, we performed immunocytochemistry using antibodies specific for the three different methylated forms of H3K27. The specificity of each antibody was confirmed by Western blot analysis using methylated H3 peptides (Fig. 1d). Di- and trimethylation are not affected in single or double mutants (Fig. 4a, b). Although previous studies have suggested that H3K27me2 is enriched in chromocenters18,19, chromocenter staining was not observed with the H3K27me2 antibody (Abcam) used in this study (Fig. 4a). The antibody used in previous work shows substantial cross reactivity with H3K27me138. This, together with the observation that H3K27me1 is roughly four times more abundant than H3K27me2 in Arabidopsis27, led us to suspect that the reported H3K27me2 localization in chromocenters may be a result of the antibody cross reacting with H3K27me1. Consistent with this model, immunostaining with this antibody38 in the presence of an H3K27me1 peptide competitor eliminates chromocenter staining (Fig. 4c). These results suggest that the previously reported chromocenter enrichment of H3K27me2 is likely due to antibody cross reactivity with H3K27me1.
In contrast to di- and tri-methylation, H3K27 monomethylation is affected in atxr5 atxr5 mutants. In wild type (Col), fluorescence is observed throughout the nucleus with the strongest staining occurring at chromocenters (Fig. 3a). This pattern is unchanged in atxr5 or atxr6 single mutants. In the atxr5 atxr6 double mutant, however, staining is strongly reduced in chromocenters (Fig. 3a) and total H3K27me1 levels are decreased by an average of 22% (Supplemental Fig. S3). It should be noted that, although reduced, H3K27me1 was not always totally eliminated from the chromocenters of atxr5 atxr6 nuclei (Fig. 3a, bottom panels). This may be explained by residual ATXR6 activity, as our mutant allele is likely not a null (Fig. 2b), and/or the remaining H3K27me1 may be catalyzed by other H3K27 methyltransferases (i.e., MEA/CLF/SWN). Because H3K27me2 and H3K27me3 are unaffected in atxr5 atxr6 mutants (Fig. 4a, b) and ChIP coupled with shotgun sequencing (ChIP-seq) shows that H3K27me3 is not found at Ta3, AtMuI, and CACTA (data not shown), this suggests that the loss of gene silencing and chromatin deconsensation in atxr5 atxr6 mutants is due to reduced H3K27me1.
H3K27me1 does not affect DNA methylation and H3K9me2
In addition to H3K27me1, H3K9me2 and DNA methylation at symmetric (CG and CHG) and asymmetric (CHH) sites are also enriched in constitutive heterochromatin and are tightly associated with transcriptional silencing in Arabidopsis17. The relationship between different epigenetic marks is complex. For example, it is well documented that mutations in the enzymes responsible for DNA methylation can affect H3K9 dimethylation and vice versa17,37. Given the observed reactivation of heterochromatic elements, we predicted that the reduction in H3K27me1 in atxr5 atxr6 double mutants might also be accompanied by reductions in DNA methylation and/or H3K9me2. Interestingly, the global distribution of H3K9me2 did not appear altered in atxr5 atxr6 (Fig. 5a). Also, no changes in DNA methylation (Fig. 5b) or H3K9me2 (Fig. 5c) were detected at loci that showed reactivation in the atxr5 atxr6 double mutant, suggesting that the release of silencing in atxr5 atxr6 double mutants is independent of H3K9me2/DNA methylation. Because this result was unexpected, we used bisulfite conversion coupled with Illumina/Solexa shotgun sequencing (BS-Seq)3 to examine DNA methylation at a global level. Consistent with our locus-specific analysis (Fig. 5b), DNA methylation patterns were indeed similar between wild type and atxr5 atxr6 double mutants genome wide (Fig. 5d–g). Likewise,
These results indicate that the reduced heterochromatic H3K27me1 in atxr5 atxr6 mutants does not affect DNA methylation or H3K9me2. The independence between H3K27me1 and the other epigenetic modifications found at chromocenters seems to be bi-directional, as H3K27me1 levels are not changed in met1 or kyp mutants18,19. Recent work has shown that KYP is partially redundant with SUVH5 and SUVH6 in catalyzing the dimethylation of H3K9 (ref. 14). To provide additional evidence that H3K27me1 levels are independent of H3K9me2, we assessed the distribution of H3K27me1 in a kyp suvh5 suvh6 triple mutant. Our results demonstrate that H3K27me1 at chromocenters is not affected in the triple mutant (Fig. 5h). Thus, H3K27me1 and DNA methylation/H3K9me2 occur via independent pathways and both sets of modifications are required for gene silencing.
DISCUSSION
In this work, we have shown that the divergent SET-domain proteins ATXR5 and ATXR6 are H3K27 methyltransferases that are not homologous to the Drosophila protein E(Z). ATXR5 and ATXR6 are the only enzymes that have been shown biochemically to catalyze the methylation of H3K27 in Arabidopsis. Despite the fact that MEA, SWN, and CLF are also thought to methylate H3K27 based on their similarity to E(Z) and reduced H3K27me2/3 levels in mutants 18,22,26,39–41, these proteins remain biochemically uncharacterized.
The post-translational processing of H3K27 is complex, both in terms of the types of modifications and the spatial distribution of modified forms. H3K27 in Arabidopsis has been shown to exist in H3K27me1, H3K27me2, H3K27me3, as well as unmethylated (H3K27me0) forms 27,28. In contrast to some eukaryotes, H3K27 is not acetylated in Arabidopsis28. Although all methylated forms of H3K27 are associated with transcriptional repression (this work; ref. 42), distinct nuclear localization patterns are observed by immunostaining; H3K27me1 is enriched in constitutive heterochromatin, whereas H3K27me2 and H3K27me3 are primarily localized to euchromatin (this work; 18,19). Thus, three interesting questions regarding H3K27 methylation are the mechanisms by which the various methylation states are generated, how they are localized to particular regions of the genome, and the functional differences between the different methylated forms.
The data presented here indicate that ATXR5 and ATXR6 play a role only in H3K27 monomethylation. H3K27me1 is reduced at heterochromatin in atxr5 atxr6 double mutants. Consistent with this result, ATXR5 and ATXR6 show only H3K27 monomethyltransferase activity in vitro. This strongly suggests that the primary role of ATXR5 and ATXR6 is H3K27 monomethylation and that other enzymes are responsible for H3K27me2 and H3K27me3. Di- and trimethylation of H3K27 is likely carried out by MEA, SWN, and CLF. Although the enzymatic activity of MEA, SWN, and CLF has not been established biochemically, these proteins have been shown to be present in Polycomb Repressor Complex2 (PRC2)-like complexes in Arabidopsis 43,44. The PRC2 complex is highly conserved and has been shown to be responsible for H3K27me2/3 in many eukaryotes 20,45,46. Our results indicate that monomethylation of H3K27 by ATXR5 and ATXR6 is not required prior to di- and trimethylation of H3K27, as H3K27me2 and H3K27me3 levels are not affected in atxr5 atxr6 mutants. This suggests that MEA, SWN and CLF are able to catalyze the formation of H3K27me2/3 directly from H3K27me0.
Although ATXR5 and ATXR6 contribute to H3K27me1 levels in chromocenters, we observed that this histone mark is only decreased by approximately 22% overall in atxr5 atxr6 double mutants. The remaining H3K27me1 could be due to residual ATXR6 activity, as the allele used in this study is not a null. Another, non-mutually exclusive, explanation is that some H3K27me1 may be catalyzed by MEA/SWN/CLF, either directly via monomethyltransferase activity or indirectly through the conversion of H3K27me2/3 to H3K27me1 by histone demethylases. While the chromocenter enrichment of H3K27me1 is clearly visible by immunostaining, it remains unclear whether this mark is also abundant in euchromatin. In mammals, H3K27me1 is enriched, as in Arabidopsis, at the pericentromeric heterochromatin38. In addition, it is also broadly distributed in euchromatin, with the exception of regions surrounding the transcription start site of active genes47. Some evidence suggests that H3K27me1 might also be found outside of the constitutive heterochromatin in plants. In the gymnosperms Pinus sylvestris and Picea abies, H3K27me1 was shown to be uniformly distributed along the complete length of all chromosomes 48. In Arabidopsis, it is possible that the heterochromatic enrichment of H3K27me1, as observed by immunostaining, is partly due to the compact structure of the chromocenters. Precise mapping of H3K27me1 in the Arabidopsis genome will be needed to determine the prevalence of this mark outside of the constitutive heterochromatin.
In addition to their SET domains, ATXR5 and ATXR6 each contain a plant homeo-domain (PHD) domain and a PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA)-interacting protein (PIP) box49. Given the role of ATXR5 and ATXR6 in catalyzing H3K27me1 at the chromocenters, an interesting question is how the activity of these enzymes is targeted to heterochromatin. Some PHD domains have been shown to mediate interactions with specific post-translationally modified forms of H3 (ref. 50,51). Thus, the PHD domains of ATXR5 and ATXR6 could potentially direct the activity of these proteins to chromocenters by interacting with specific forms of H3 that are enriched in heterochromatin. Further studies on ATXR5 and ATXR6 may also shed light on how the H3K27me1 mark is maintained through DNA replication. Given that ATXR5 and ATXR6 likely interact directly with PCNA during S phase of the cell cycle49, it is possible that these proteins play a role in modifying newly synthesized histones that are incorporated into heterochromatin during DNA synthesis. This model of action could help to explain the epigenetic inheritance of H3K27me1 in Arabidopsis. SETDB1 and G9A are two examples of histone methyltransferases that have been shown to localize at the replication fork in mammals52,53.
In conclusion, our work demonstrates that ATXR5 and ATXR6 comprise a novel class of H3K27 methyltransferases that play critical roles in H3K27 monomethylation, chromatin condensation, and gene silencing in Arabidopsis. Our results, in combination with previous studies showing that H3K27me1 levels were found to be unchanged in DNA methylation or H3K9 methyltransferase mutants18,19, indicate that H3K27me1 and DNA methylation/H3K9me2 are two essential and independent pathways required for the silencing of heterochromatic elements in Arabidopsis. These two pathways are also required for regulating the structural organization of chromocenters. It will be interesting to understand to which extent the H3K27me1- and DNA methylation/H3K9me2 pathways overlap, and for which biological mechanisms do they provide unique functions.
METHODS
Plant material
atxr5 (SALK_130607) and atxr6 (SAIL_240_H01) are in the Col genetic background and were obtained from the Arabidopsis Biological Resource Center (Columbus, Ohio). Plantswere grown under cool-white fluorescent light (~100 μmol per m−2 s−1 under long-day (16 hours light followedby 8 hours darkness) or short-day conditions (8 hours light followed by 16 hours darkness).
Gene expression analysis
For semi-quantitative RT-PCR analysis, we performed reverse transcription and PCR as described previously54 with slight modifications. 5 μg of RNA was used for cDNA synthesis and GoTaq DNA Polymerase(Promega) was used for amplification. The data shown are representative of at least three independent experiments. The primers and PCR conditions are shown in Supplementary Table 1.
Chromatin immunoprecipitation (ChIP)
We performed ChIP assays as described previously4. For the experiment, 300 mg of fully expanded leaves and 4.5 μg of H3K27me1 or 4 μg of H3K9me2 antibody were used.
Antibodies
Antibodies used in the histone methyltransferase assays, ChIP assays, and immunostaining were from Millipore (H3K4me1/2/3, 05-791; H3K9me1, 07-450; H3K27me1, 07-448; H3K27me3, 07-449; H3, 07-690) and Abcam (H3K9me2, ab1220; H3K27me2, ab24684; H3K36me1, ab9048; H3K79me1, ab2886). Antibodies for Western blots were used according to the manufacturer’s recommendations.
Locus-specific bisulfite sequencing
We bisulfite treated 350–550 ng of Col and atxr5 atxr6 DNA with EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA) according to manufacturer’s instructions. One microliter of bisulfite-treated DNA was used for PCR with Ta3- and CACTA-specific primers (#1274 5′CCACTRATTCCTRAAACACAACATTTCTRCTRATA3′; #1269 5′GAGAATYAGGTTAATAAGAAAGTGAAGTGTT3′; #6059 5′AACATCCTTTCCTTCAACTTAACATCACACTC3′; #6076 5′ATAGATGAGTTATTTGTTAGTTTTGGTYGTGTTGGTT3′). PCR products were gel purified and cloned into pCR4 Blunt TOPO vector (Invitrogen, Carlsbad, CA). Total of 19–27 clones were sequenced for each genotype and locus.
Genomic bisulfite sequencing and methylation data analysis
We generated whole genome shotgun bisulfite-treated DNA libraries following a published protocol 3, except that the Plant DNeasy Maxi Kit (Qiagen) was used to isolate Arabidopsis genomic DNA. Ultra-high-throughput sequencing of libraries on an Illumina/Solexa 1G Genome Analyzer was carried out according to manufacturer’s instructions. Sequencing data was first processed using initial steps of version 0.2.2.4 of the Solexa Analysis Pipeline, and then by previously published customized software that performs basecalling, bisulfite alignment, and non-conversion filtration3. In total, 1,858,393 and 1,534,127 aligned 31 bp reads were obtained for WT (Col) Arabidopsis and atxr5 atxr6 double mutant, respectively. Methylated cytosines were identified as cytosines (or guanines as appropriate) in reads aligned to genomic cytosines, while unmethylated cytosines were identified as thymines (or adenines as appropriate) in reads aligned to genomic cytosines. Chromosomal views and metaplots of Figure 4E–G were produced as described in a previous report3.
Constructs
We used pGEX-6P to clone ATXR5 (a.a. 57-379, PHD-SET; a.a. 57-133, PHD only) and ATXR6 (a.a. 25-349, PHD-SET; a.a. 25-103, PHD only) for histone methyltransferase assays. For the plant histone 3 constructs, At1g09200 was used as a template to amplify a plant histone 3.1 gene. The resulting PCR fragment was cloned into pENTR/D (Invitrogen) and subcloned into pET-DEST42 (Invitrogen). The lysine to alanine mutation at position 27 was engineered by overlapping PCR. The primers used for construction of the plasmids are available upon request.
Histone methyltransferase assay
We incubated10 μg of calf thymus histones (Roche), 2.5 μg of recombinant human histone 3.1 (New England Biolabs), or 2.5 μg of plant histone 3.1 (wild-type or mutant) with 5 μg of GST-ATXR5 or GST-ATXR6 in 40 μl methylation buffer (50 mM Tris-HCl pH 8.5, 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 250 mM sucrose) supplemented with 250 nCi of S-adenosyl-L-(methyl-14C) methionine (14C-SAM) (GE Life Sciences) or 5 nmol of unlabeled SAM (Sigma) for 3 hr at 30°C. Reactions were stopped by adding 5X SDS-PAGE sample buffer followed by heating to 95°C for 5 min. After SDS-PAGE on 12 or 15% gels and transfer to PVDF membrane, detection was performed using a PhosphorImager or by Western blot using specific histone 3 antibodies.
Detection of histone modifications in yeast
We transformed pYES-DEST52-ATXR5 and pYES-DEST52-ATXR6 into an INVSc1 yeast strain (Invitrogen) and expressed according to manufacturer’s recommendations. Yeast cells were harvested 18 hours after induction. Nuclear proteins were extracted from yeast cells as described previously55 and analyzed by Western blot.
Protein expression, Immunofluorescence, and FISH analysis are described in supplemental methods.
Supplementary Material
Acknowledgments
We thank C.E. Walczak for assistance with microscopy. This work was supported by grants to S.D.M. from the NSF (grant no. IOB-0447583) and NIH (GM075060) and to Y.J. from NATEQ (Fonds québécois de recherche sur la nature et les technologies). Y. V. B. was supported by USPHS National Research Sevice Award GM07104. Jacobsen lab research was supported by NIH grant GM60398. S.E.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
Y.J. performed the genetic and biochemical characterization of ATXR5 and ATXR6. Y.J. and C.A.L. generated gene expression and ChIP data. S.F. generated and sequenced BS-Seq libraries. Y.V.B. performed locus-specific bisulfite sequencing analyses. H.S. validated ChIP and RT-PCR results. C.A.L., Y.V.B., and L.M.J. performed the immunofluorescence studies. S.C., M.P., S.F., and S.E.J. analyzed BS-Seq data. S.D.M., M.P., and S.E.J. participated in the design of experiments.
COMPETING INTEREST STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Maluszynska J, Heslop-Harrison JS. Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1991;1:159–166. [Google Scholar]
- 3.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–7. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 5.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–9. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 6.Tariq M, et al. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A. 2003;100:8823–7. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci U S A. 2002;99 (Suppl 4):16491–8. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–44. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 10.Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–7. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 11.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–60. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 12.Finnegan EJ, Dennis ES. Isolation and Identification by Sequence Homology of a Putative Cytosine Methyltransferase from Arabidopsis-Thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol. 2005;25:10507–15. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–76. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naumann K, et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005;24:1418–29. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–52. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaillant I, Paszkowski J. Role of histone and DNA methylation in gene regulation. Curr Opin Plant Biol. 2007;10:528–33. doi: 10.1016/j.pbi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Lindroth AM, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–96. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–91. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 21.Ketel CS, et al. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol Cell Biol. 2005;25:6857–68. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumbusch LO, et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–33. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanvivattana Y, et al. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–76. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 24.Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–50. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 25.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 26.Springer NM, et al. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–25. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson L, et al. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–8. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K, Sridhar VV, Zhu J, Kapoor A, Zhu JK. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE. 2007;2:e1210. doi: 10.1371/journal.pone.0001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–56. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 30.Ng HH, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–27. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–4. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 32.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 33.Garcia BA, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–55. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 34.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–7. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 35.Soppe WJ, et al. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–59. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–77. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–24. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 38.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 39.Makarevich G, et al. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–52. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schonrock N, et al. Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 2006;20:1667–78. doi: 10.1101/gad.377206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubert D, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–49. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohler C, Grossniklaus U. Epigenetic inheritance of expression states in plant development: the role of Polycomb group proteins. Curr Opin Cell Biol. 2002;14:773–9. doi: 10.1016/s0955-0674(02)00394-0. [DOI] [PubMed] [Google Scholar]
- 43.De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci U S A. 2008;105:16831–6. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood CC, et al. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A. 2006;103:14631–6. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyes JC, Grossniklaus U. Diverse functions of Polycomb group proteins during plant development. Semin Cell Dev Biol. 2003;14:77–84. doi: 10.1016/s1084-9521(02)00139-8. [DOI] [PubMed] [Google Scholar]
- 46.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuchs J, Jovtchev G, Schubert I. The chromosomal distribution of histone methylation marks in gymnosperms differs from that of angiosperms. Chromosome Res. 2008;16:891–8. doi: 10.1007/s10577-008-1252-4. [DOI] [PubMed] [Google Scholar]
- 49.Raynaud C, et al. Two cell-cycle regulated SET-domain proteins interact with proliferating cell nuclear antigen (PCNA) in Arabidopsis. Plant J. 2006;47:395–407. doi: 10.1111/j.1365-313X.2006.02799.x. [DOI] [PubMed] [Google Scholar]
- 50.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 52.Esteve PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 54.Michaels SD, Bezerra IC, Amasino RM. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:3281–5. doi: 10.1073/pnas.0306778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kizer KO, Xiao T, Strahl BD. Accelerated nuclei preparation and methods for analysis of histone modifications in yeast. Methods. 2006;40:296–302. doi: 10.1016/j.ymeth.2006.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.