Abstract
The genome of Saccharomyces cerevisiae contains 35 members of the mitochondrial carrier protein family, most of which have not yet been functionally identified. Here the identification of the mitochondrial carrier for S-adenosylmethionine (SAM) Sam5p is described. The corresponding gene has been overexpressed in bacteria and the protein has been reconstituted into phospholipid vesicles and identified by its transport properties. In confirmation of its identity, (i) the Sam5p–GFP protein was found to be targeted to mitochondria; (ii) the cells lacking the gene for this carrier showed auxotrophy for biotin (which is synthesized in the mitochondria by the SAM-requiring Bio2p) on fermentable carbon sources and a petite phenotype on non-fermentable substrates; and (iii) both phenotypes of the knock-out mutant were overcome by expressing the cytosolic SAM synthetase (Sam1p) inside the mitochondria.
Keywords: mitochondria/proteomics/Saccharomyces cerevisiae/S-adenosylmethionine carrier/YNL003c
Introduction
In Saccharomyces cerevisiae, S-adenosylmethionine (SAM) is synthesized from ATP and methionine by two synthetases, Sam1p (Thomas and Surdin-Kerjan, 1987) and Sam2p (Thomas and Surdin-Kerjan, 1991), that are both localized exclusively in the cytosol (Kumar et al., 2002). SAM has therefore to be imported into the mitochondria, where it is required as a methyl group donor for DNA, RNA and protein methylation (Gantt and Schneider, 1979; Dubin et al., 1982; Pintard et al., 2002; Santamaria et al., 2003), for sterol methylation (McCammon et al., 1984) and as essential cofactor in the last step of biotin and lipoic acid biosyntheses catalyzed by biotin synthetase (Bio2p) and lipoate synthetase (Lip5p), respectively (Sulo and Martin, 1993; Zhang et al., 1994; Marquet et al., 2001; Kumar et al., 2002).
Despite the importance of SAM in mitochondrial metabolism, its transport across the inner mitochondrial membrane has been investigated only by Horne et al. (1997) in rat liver mitochondria. This study led to the hypothesis that SAM is transported into mitochondria by a carrier-mediated system. However, the protein responsible for this transport has not been identified hitherto.
Here we report the identification and functional characterization of the mitochondrial SAM carrier (Sam5p) encoded by YNL003c, also known as PET8 (Mortimer et al., 1992; Lalo et al., 1994; el Moualij et al., 1997; Roussel et al., 2002; Steinmetz et al., 2002). It is 284 amino acids long and has the characteristic features of the mitochondrial carrier family (Palmieri et al., 2000; Palmieri, 2003). Sam5p was overexpressed in bacteria, reconstituted into phospholipid vesicles and identified from its transport properties as a SAM carrier. The green fluorescent protein (GFP) fused to Sam5p was found to be targeted to mitochondria. sam5Δ cells exhibited auxotrophy for biotin on fermentative carbon sources and a petite phenotype on non-fermentative substrates. Furthermore, both phenotypes of the knock-out yeast strain were restored by directing the cytosolic SAM synthetase (Sam1p) into the mitochondria.
Results
Bacterial expression of Sam5p protein
Sam5p was expressed at high levels in Escherichia coli C0214(DE3) (Figure 1, lane 4). It accumulated as inclusion bodies and was purified by centrifugation and washing (Figure 1, lane 5). The apparent molecular mass of the recombinant protein was 30.9 kDa (calculated value with initiator methionine, 31 027 Da). The protein was not detected in bacteria harvested immediately before induction of expression (Figure 1, lane 2), nor in cells harvested after induction but lacking the coding sequence in the expression vector (Figure 1, lane 3). The identity of the purified protein was confirmed by N-terminal sequencing. Approximately 60 mg of Sam5p were obtained per liter of culture.
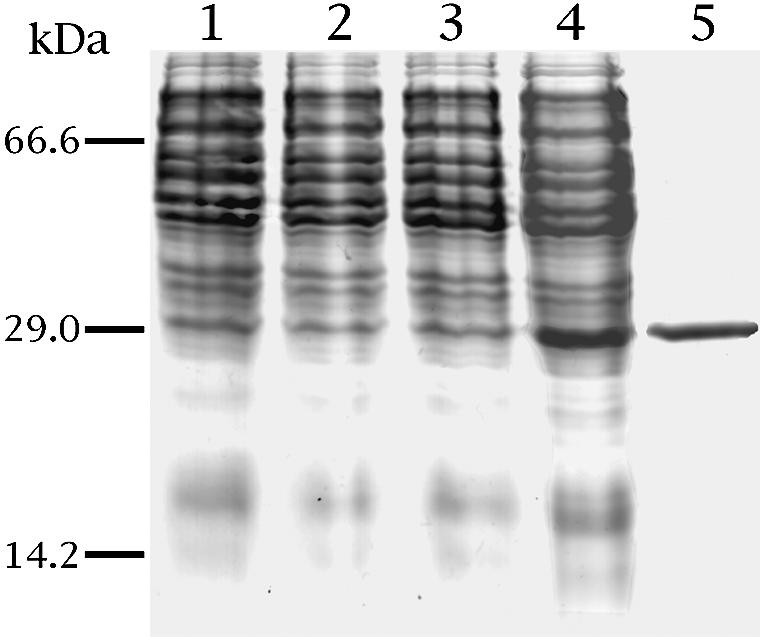
Fig. 1. Expression of yeast Sam5p in E.coli and its purification. Proteins were separated by SDS–PAGE and stained with Coomassie Blue dye. The positions of the markers (bovine serum albumin, carbonic anhydrase and cytochrome c) are shown on the left. Lanes 1–4, E.coli CO214(DE3) containing the expression vector with (lanes 2 and 4) and without (lanes 1 and 3) the coding sequence of Sam5p. Samples were taken at the time of induction (lanes 1 and 2) and 5 h later (lanes 3 and 4). The same number of bacteria was analyzed in each sample. Lane 5, purified Sam5p (3 µg) originating from bacteria shown in lane 4.
Functional characterization of recombinant Sam5p
Sam5p was reconstituted into liposomes, and its transport activities for a large variety of potential substrates were tested in homo-exchange experiments (i.e. with the same substrate inside and outside). Using external and internal substrate concentrations of 1 and 10 mM, respectively, we found by chance that the recombinant protein catalyzed an active [3H]SAM/SAM exchange, that was completely inhibited by 20 mM pyridoxal 5′-phosphate and 0.4 mM p-hydroxymercuribenzoate. No such activity was observed with Sam5p that had been boiled before incorporation into liposomes nor by reconstitution of sarkosyl-solubilized material from bacterial cells either lacking the expression vector for Sam5p or harvested immediately before induction of expression. Likewise, no [3H]SAM/SAM exchange activity was detected in liposomes reconstituted with three unrelated mitochondrial carriers, Odc1p, Ant1p and Tpc1p (Palmieri et al., 2001a,b; Marobbio et al., 2002), which had been expressed and purified from E.coli using the same expression vector. Furthermore, reconstituted Sam5p did not catalyze homo-exchanges for phosphate, pyruvate, ADP, ATP, malonate, malate, oxoglutarate, ketoisocaproate, citrate, carnitine, biotin, aspartate, ornithine, lysine, arginine, histidine, glutathione, choline, spermine, proline and threonine.
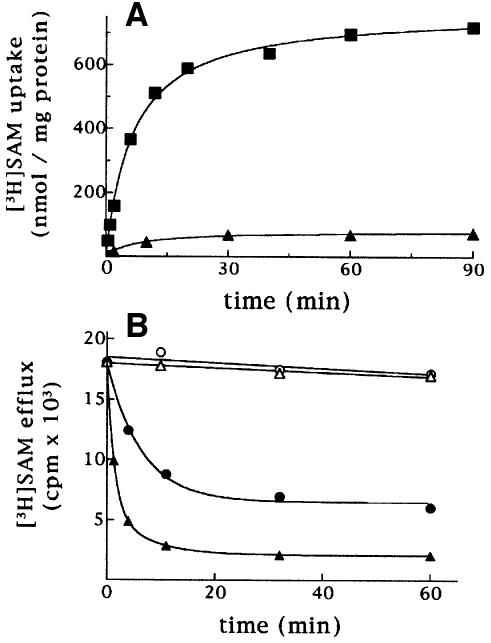
In Figure 2A, the kinetics are compared for the uptake by proteoliposomes of 1 mM [3H]SAM measured either as uniport (in the absence of internal SAM), or as exchange (in the presence of 10 mM SAM). Both the exchange and the uniport reactions catalyzed by Sam5p followed first-order kinetics, isotopic equilibrium being approached exponentially. The ratio of maximal substrate uptake by the exchange and by the uniport was 10.1, in agreement with the expected value of 10 from the intraliposomal concentrations at equilibrium (1 and 10 mM for uniport and exchange, respectively). The initial rates of SAM uptake deduced from the time-courses (Palmieri et al., 1995) were 82.2 and 7.6 µmol/min per g protein for the exchange and the uniport reaction, respectively. Sam5p is therefore able to catalyze a unidirectional transport of SAM, besides the exchange reaction. A more convenient assay for unidirectional transport is to measure the efflux of [3H]SAM from pre-labeled proteoliposomes (Palmieri et al., 1995). As shown in Figure 2B, a substantial efflux of [3H]SAM from proteoliposomes occurred when adding buffer alone. The addition of 10 mM SAM induced a greater efflux of SAM, and the efflux either with or without added substrate was prevented by inhibitors of SAM transport.
Fig. 2. Sam5p catalyses the transport of SAM. (A) Kinetics of [3H]SAM uniport and [3H]SAM/SAM exchange by Sam5p. 1 mM [3H]SAM was added to proteoliposomes containing 10 mM SAM (exchange, filled square) or 10 mM NaCl and no substrate (uniport, filled triangle). Similar results were obtained in three independent experiments. (B) Efflux of [3H]SAM from proteoliposomes reconstituted with Sam5p. Proteoliposomes were reconstituted in the presence of 1 mM SAM. The internal substrate pool was labeled by carrier-mediated exchange equilibration. Then the proteoliposomes were passed through Sephadex G-75. The efflux of [3H]SAM was started by adding buffer A alone (filled circle), 20 mM pyridoxal 5′-phosphate and 0.4 mM p-hydroxymercuribenzoate in buffer A (open circle), 10 mM SAM in buffer A (filled triangle) or 10 mM SAM, 20 mM pyridoxal 5′-phosphate and 0.4 mM p-hydroxymercuribenzoate in buffer A (open triangle). Similar results were obtained in three independent experiments.
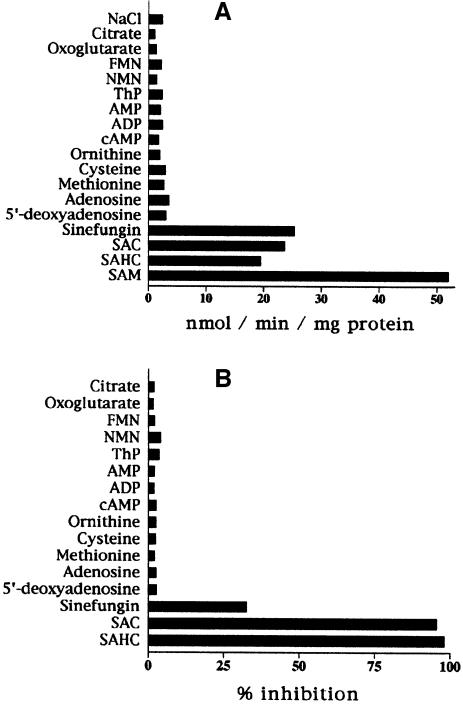
The substrate specificity of reconstituted Sam5p was examined in detail by measuring the uptake of [3H]SAM into proteoliposomes that had been pre-loaded with various potential substrates (Figure 3A). The highest rate of [3H]SAM uptake into proteoliposomes was with internal SAM. High activities were also found with adenosylornithine (sinefungin), S-adenosylcysteine (SAC) and S-adenosylhomocysteine (SAHC). In contrast, the uptake of [3H]SAM was low in the presence of internal 5′-deoxyadenosine, adenosine, methionine, cysteine, ornithine, cAMP, ADP, AMP, thiamine pyrophosphate (ThP), NMN, FMN, oxoglutarate, citrate and (not shown) thiamine, NAD, FAD and coenzyme A. The residual activity in the presence of these substrates was approximately the same as the activity observed in the presence of sodium chloride, in agreement with the uniport mode of transport of [3H]SAM.
Fig. 3. Substrate specificity of yeast Sam5p. (A) Dependence of Sam5p activity on internal substrate. Proteoliposomes were pre-loaded internally with various substrates (concentration, 10 mM). Transport was started by addition of 75 µM [3H]SAM and terminated after 45 s. Similar results were obtained in at least three independent experiments. (B) Inhibition of the rate of [3H]SAM uptake by external substrates. Transport was started by adding 75 µM [3H]SAM to proteoliposomes containing 10 mM SAM, and stopped after 45 s. External substrates (concentration, 0.75 mM) were added together with [3H]SAM. The extents of inhibition (%) from a representative experiment are reported.
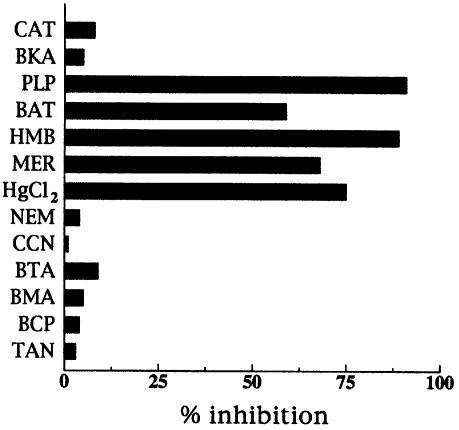
The [3H]SAM/SAM exchange was inhibited markedly by mercurials (p-hydroxymercuribenzoate, mersalyl and mercuric chloride), pyridoxal 5′-phosphate and bathophenanthroline (which are all inhibitors of several mitochondrial carriers) (Figure 4). In contrast, carboxyatractyloside, bongkrekate, N-ethylmaleimide, α-cyano-4-hydroxycinnamate, 1,2,3-benzenetricarboxylate, butylmalonate, bromocresol purple and tannic acid, inhibitors of other characterized mitochondrial carriers, had little or no effect on the activity of reconstituted Sam5p.
Fig. 4. Effect of inhibitors on the [3H]SAM/SAM exchange by Sam5p. Proteoliposomes were pre-loaded internally with 10 mM SAM and transport was initiated by adding 75 µM [3H]SAM. The incubation time was 45 s. Thiol reagents and α-cyanocinnamate were added 2 min before the labeled substrate; the other inhibitors were added together with [3H]SAM. The final concentrations of the inhibitors were 0.2 mM (HMB, p-hydroxymercuribenzoate; MER, mersalyl; HgCl2, mercuric chloride), 20 mM (PLP, pyridoxal 5′ phosphate; BAT, bathophenanthroline), 1 mM (NEM, N-ethylmaleimide; CCN, α-cyanocinnamate), 2.0 mM (BTA, benzene-1,2,3-tricarboxylate; BMA, butylmalonate), 0.3 mM (BCP, bromocresol purple), 0.2% (TAN, tannic acid) and 10 µM (CAT, carboxyatractyloside; BKA, bongkrekic acid). The extents of inhibition (%) from a representative experiment are given.
In addition, [3H]SAM uptake was inhibited strongly by external addition of SAC or SAHC (Figure 3B). Sinefungin was much less effective. In contrast, no significant inhibition was detected with 5′-deoxyadenosine, adenosine, methionine, cysteine, ornithine, cAMP, ADP, AMP, ThP, NMN, FMN, oxoglutarate, citrate and (not shown) substrates of other known mitochondrial carriers.
Kinetic characteristics of recombinant Sam5p
The kinetic constants of the recombinant purified Sam5p were determined by measuring the initial transport rate at various external [3H]SAM concentrations, in the presence of a constant saturating internal concentration of 10 mM SAM. The Km and Vmax values for SAM uptake were 75 ± 8.8 µM and 105.5 ± 30.2 µmol/min per g protein, respectively (mean values from 18 experiments). SAC, SAHC and sinefungin inhibited SAM uptake competitively (not shown), and their Ki values were 14.5 ± 3.2 µM, 6.8 ± 0.9 µM and 1.14 ± 0.12 mM, respectively (means of four experiments for each inhibitor).
Subcellular localization of Sam5p and Sah1p
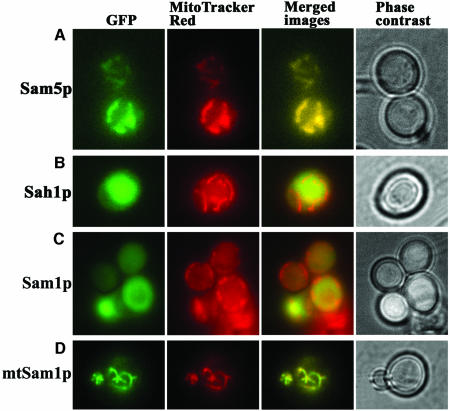
To test the subcellular localization of Sam5p and S-adenosylhomocysteine hydrolase (Sah1p) (Knudsen and Yall, 1972; Kanehisa et al., 2002), we constructed an in-frame SAM5 or SAH1 fusion with GFP cDNA in the vector pUG35 under the control of the MET25 promoter. This vector allows the expression of the GFP fusion proteins in yeast when methionine is absent from the medium. We chose to transform the yeast strain YPH499 with our constructs since it is not auxotrophic for methionine. Sam5p–GFP expressing cells showed a green fluorescence of the mitochondrial network (Figure 5A). With the Sah1p–GFP fusion the fluorescence was largely diffuse (Figure 5B). With the mitochondrial-specific dye, MitoTracker Red, the same cells showed a pattern of fluorescence that coincides with the mitochondria (control). From the overlapping images it is clear that Sam5p–GFP was localized to mitochondria, whereas Sah1p–GFP was localized to the cytoplasm. The structural integrity of the cells was documented by phase contrast microscopy.
Fig. 5. Subcellular localization of Sam5p–GFP, Sah1p–GFP, Sam1p–GFP and mtSam1p–GFP fusion proteins after expression in S.cerevisiae cells. Wild-type YPH499 cells expressing (A) Sam5p–GFP (SAM carrier fused to GFP from the SAM5-GFP-pUG35 vector); (B) Sah1p–GFP (S-adenosylhomocysteine hydrolase fused to GFP from the SAH1-GFP-pUG35 vector); (C) Sam1p-GFP (SAM synthetase1 fused to GFP from the SAM1-GFP-pYES2 vector); and (D) mtSam1p–GFP (intramitochondrially directed SAM synthetase1 fused to GFP from the Su9(1–69)-SAM1-GFP-pYES2 vector) were grown as described in Materials and methods. MitoTracker Red was used to locate mitochondria in the cells (MitoTracker panel), and phase contrast microscopy to monitor the integrity of the cells. The same cells were photographed first with a GFP filter set and then with the MitoTracker filter set. Identical fields are presented.
sam5Δ yeast cells are auxotrophic for biotin on fermentative carbon sources
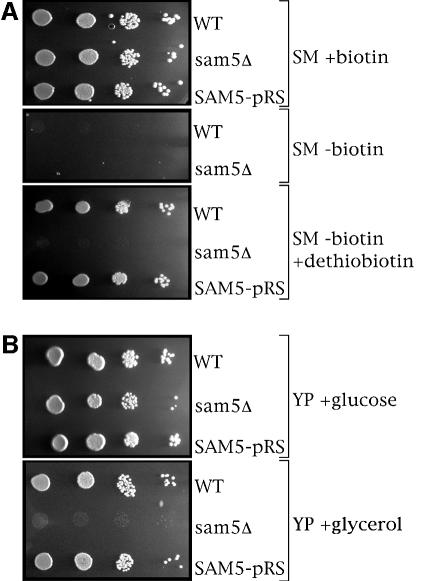
Having established the transport function of Sam5p by in vitro assays and its subcellular localization, the effect of deleting its gene on yeast cells was investigated. The sam5Δ strain was tested for its ability to grow in the absence of different cofactors. On synthetic minimal (SM) medium supplemented with glucose or (not shown) galactose yeast cells lacking sam5 did not grow in the absence of biotin, similarly to the wild-type cells (Figure 6A). On this medium the growth of wild-type cells was fully restored by the addition of dethiobiotin (Figure 6A), a precursor of biotin synthesis that is converted to biotin by Bio2p, a SAM-requiring enzyme (Zhang et al., 1994; Marquet et al., 2001) localized in the mitochondria (Kumar et al, 2002). In contrast, sam5Δ cells did not grow after the addition of dethiobiotin (Figure 6A). The growth phenotype of sam5Δ cells in glucose or galactose-supplemented SM medium containing dethiobiotin was restored by complementing the deletion strain with the SAM5-pRS416 plasmid, containing the gene for the mitochondrial SAM carrier (Figure 6A), demonstrating that the difference in phenotype is the result of the absence of Sam5p protein and not a secondary effect. In other experiments it was found that none of the other cofactors tested, i.e. pyridoxine, panthotenate, riboflavin, folate, thiamine, p-aminobenzoate and inositol, was necessary for the growth of yeast cells lacking Sam5p when removed singly from the SM medium (not shown). Taken together, these results suggest that the deletion of sam5 causes a defect in the reaction catalyzed by biotin synthetase, probably due to a shortage of SAM in the mitochondria.
Fig. 6. sam5Δ mutant is auxotrophic for biotin. Growth behavior of sam5Δ mutant in various conditions. Four-fold serial dilutions of wild-type BY4741 cells, sam5Δ cells (BY4741 cells lacking sam5) and SAM5-pRS cells (sam5Δ cells transformed with the SAM5-pRS416 plasmid) were plated on (A) solid SM medium supplemented with glucose or (B) solid YP medium supplemented with glucose or glycerol. Where indicated, 20 µg/l biotin or 20 µg/l dethiobiotin was also present.
Growth of sam5Δ cells on yeast extract (YP) medium containing fermentative carbon sources was virtually indistinguishable from wild-type cells (Figure 6B). However, sam5Δ cells did not grow on the same medium containing the non-fermentative carbon sources glycerol (Figure 6B) and (not shown) lactate, ethanol, acetate and pyruvate. The latter findings are in agreement with the previous observations that the knock-out of YNL003c resulted in a petite ρ+ (PET8) (Mortimer et al., 1992; Lalo et al., 1994; el Moualij et al., 1997; Roussel et al., 2002; Steinmetz et al., 2002). Growth on YP medium containing glycerol was fully restored by complementing the deletion strain with the SAM5-pRS416 plasmid (Figure 6B), demonstrating that under these conditions too the difference in phenotype is the result of the absence of Sam5p protein only.
The growth phenotype of sam5Δ cells is restored by expression of Sam1p in the mitochondria
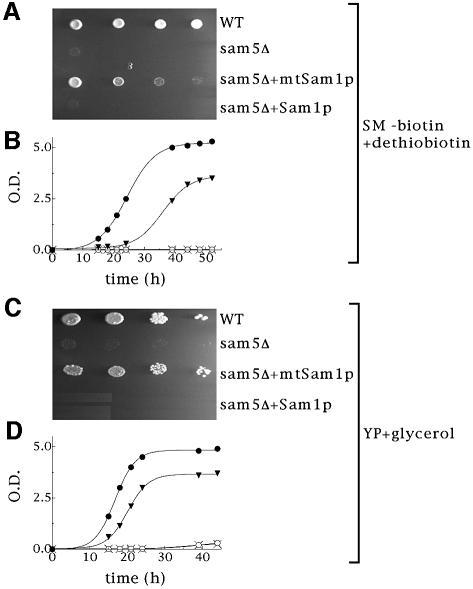
In S.cerevisiae SAM is synthesized by Sam1p and Sam2p exclusively in the cytosol (Kumar et al., 2002). Assuming that the function of Sam5p is to import SAM into the mitochondria, we checked whether the lack of growth of sam5Δ cells on fermentable carbon sources in biotin-less SM and on non-fermentable carbon sources in YP media was restored by expressing Sam1p in the mitochondria and hence providing SAM inside the mitochondria. For this reason, we transformed the sam5Δ yeast strain with the plasmid SAM1-pYES2 or Su9(1–69)-SAM1-pYES2 in order to direct Sam1p in the cytosol or in the mitochondria. Figure 5C and D show that Sam1p and Su9(1–69)-Sam1p (i.e. mtSam1p) fused with GFP are indeed localized to the cytosol and to the mitochondria, respectively. The lack of growth of sam5Δ cells on biotin-less galactose-supplemented SM and on glycerol-supplemented YP was largely restored in cells expressing Sam1p in the mitochondria, but not in cells expressing Sam1p in the cytosol (Figure 7A and C). The growth of sam5Δ cells, transformed with Su9(1–69)-SAM1-pYES2 or SAM1-pYES2 and inoculated in liquid media, are shown as a function of the time of culture in Figure 7B and D. Similar results were obtained using ethanol and pyruvate as non-fermentative carbon sources, instead of glycerol, in both solid and liquid media (not shown). These results indicate that the defects of sam5Δ cells to grow on fermentative and non-fermentative carbon sources are overcome if SAM is synthesized inside the mitochondria.
Fig. 7. Effect of intramitochondrially directed Sam1p (mtSam1p) or cytosolic Sam1p (Sam1p) on the growth of sam5Δ cells. Wild-type BY4741 cells, sam5Δ cells (BY4741 cells lacking sam5), sam5Δ + mtSam1p cells (sam5Δ cells transformed with the Su9(1–69)-SAM1-pYES2 vector), sam5Δ + Sam1p cells (sam5Δ cells transformed with the SAM1-pYES2 vector) were pre-incubated in YP medium supplemented with 0.4% galactose for 6 h, washed three times with biotin-less SM medium and then (A) spotted on solid or (B) inoculated in liquid SM medium supplemented with 2% galactose plus 20 µg/l dethiobiotin, (C) spotted on solid or (D) inoculated in liquid YP medium supplemented with 3% glycerol. The values of optical density at 600 nm given in (B) and (D) refer to cell cultures after the indicated times of growth. Filled circle, wild-type; crosses, sam5Δ; filled triangle, sam5Δ + mtSam1p; and open circles, sam5Δ + Sam1p.
Discussion
The transport characteristics and kinetic parameters of the recombinant and reconstituted Sam5p from S.cerevisiae and its targeting to mitochondria, reported here, show that it is the mitochondrial transporter for SAM and SAHC. Sam5p also transports, to a lesser extent, the non-physiological structurally-related compounds SAC and sinefungin, but none of the many other compounds tested. This is the first time that a mitochondrial carrier for SAM has been identified from any organism at the molecular level. Sam5p (encoded by YNL003c) does not show significant sequence homology with any other mitochondrial carrier functionally identified until now in yeast, mammals and plants (Fiermonte et al., 2002; Marobbio et al., 2002; Hoyos et al., 2003 and references therein). In a phylogenetic tree of the S.cerevisiae members of the mitochondrial carrier family (el Moualij et al., 1997; Nelson et al., 1998; Palmieri et al., 2000), Sam5p clusters with YMR166c, YJL133w and YKR052c (amino acid identity between 25 and 30%), none of which has been identified hitherto. However, several protein sequences available in data bases show a significant homology (38–44% amino acid identity) with Sam5p and may be orthologs of this transporter in other organisms. These sequences include AX349357 from Homo sapiens, BC037142 from Mus musculus, AL355930 from Neurospora crassa, AE003753 from Drosophila melanogaster and Z68160 from Caenorhabditis elegans.
Since SAM is produced in the cytosol by Sam1p and Sam2p (Kumar et al., 2002), a primary function of Sam5p is to catalyze the uniport uptake of SAM into the mitochondria, where it is required for the activity of Bio2p, Lip5p and for methylation of DNA, RNA, proteins and other compounds. In addition, since SAHC, which is produced from SAM in the methylation reactions, is hydrolyzed in the cytosol by Sah1p, a further important physiological function of Sam5p is probably to catalyze the exchange between cytosolic SAM and intramitochondrial SAHC. The other metabolites produced from SAM within the mitochondria by the action of Bio2p and Lip5p, i.e. 5′-deoxyadenosine and methionine, cannot be exported in exchange for SAM by Sam5p, because they are not substrates of this carrier. The rate of the exchange is much higher than that of the uniport. However, the uniport reaction is necessary in special conditions (when the intramitochondrial level of SAM has to be increased), for example, when cells divide or when cells are transferred to a medium containing non-fermentative carbon sources.
Saccharomyces cerevisiae cells lacking sam5 show auxotrophy for biotin on fermentable carbon sources and a petite phenotype on non-fermentable substrates. Both of these phenotypes of the knock-out mutant are restored by expression of the SAM synthetase, Sam1p, enzyme in the mitochondria making SAM available inside the organelles. These findings are consistent with and strongly support Sam5p controlling the uptake of SAM into mitochondria.
The sam5Δ strain does not grow on fermentable carbon sources even in the presence of the biotin precursor dethiobiotin. Dethiobiotin is directly converted to biotin in the last step of biotin synthesis catalyzed by the intramitochondrial, SAM-requiring biotin synthetase Bio2p (Zhang et al., 1994; Marquet et al., 2001; Kumar et al., 2002). We first thought that Sam5p was required for the uptake of dethiobiotin or for the export of biotin. However, direct transport assays showed that the recombinant Sam5p was unable to transport biotin or dethiobiotin in reconstituted liposomes. Thus, the reconstituted protein catalyzed neither the [14C]biotin/biotin homoexchange nor the heterologous exchanges [14C]biotin/dethiobiotin, [3H]SAM/biotin and [3H]SAM/dethiobiotin. Furthermore, studies with intact mitochondria have suggested that biotin diffuses across the mitochondrial membrane (Said et al., 1992). The most likely explanation for the sam5Δ mutant biotin auxotrophy, in dethiobiotin-containing glucose- or galactose-supplemented SM medium, is that it is caused by insufficient supply of SAM to the mitochondrial matrix, where it is indispensable for the conversion of dethiobiotin to biotin by Bio2p. This explanation is in line with the finding that recombinant Sam5p is able to transport SAM in the reconstituted system and with the ability of mitochondrially directed SAM synthetase to cure the auxotrophy for biotin. Our results further indicate that the limiting factor for the sam5Δ strain growing on fermentative carbon sources is the intramitochondrial SAM-dependent synthesis of biotin.
The lack of growth of sam5Δ cells on non-fermentable carbon sources even in YP medium suggests that the non-supply of SAM to the mitochondria causes additional defects (apart from the synthesis of biotin which is present in the medium) that prevent cells from growing under respiratory conditions. The molecular bases of these alterations that involve the mitochondria are emerging from recent studies. It should be mentioned that S.cerevisiae mitochondria contain two mitochondrial proteins, Pet56p and Mrm2p, which are essential for the growth on non-fermentable carbon sources (Struhl, 1985; Pintard et al., 2002; Steinmetz et al., 2002). They have methyltransferase activity, possess a SAM-binding site and methylate two nucleotides located at the peptidyl transferase center of 21S rRNA (Sirum-Connolly and Mason, 1993; Pintard et al., 2002). Studies in knock-out mice deficient in hepatic SAM synthetase (MAT1A–/–), and in cultured rat hepatocytes using a medium without methionine or in the presence of cycloleucine, a known inhibitor of MAT1 activity, have shown a post-translational down-regulation of prohibitin (PHB1) and of subunits I and II of cytochrome c oxidase without any decreases in their mRNA transcripts (Santamaria et al., 2003). Two members of the prohibitin family (encoded by YGR132c and YGR231c) have been identified in S.cerevisiae (Berger and Yaffe, 1998; Steglich et al., 1999; Nijtmans et al., 2000; Back et al., 2002). The two yeast prohibitins are mitochondrial, chaperone-like proteins involved in the correct folding and assembly of some of the components of the respiratory chain (Nijtmans et al., 2000, 2002). It is therefore likely that the reduced methylation of prohibitins causes alterations in the respiratory chain complexes compromising mitochondrial function with no loss of mitochondrial DNA (Santamaria et al., 2003). Finally, we can recall that the yeast knock-out for LIP5 (lipoate synthetase) shows a growth defect on non-fermentable substrates (Steinmetz et al., 2002), because the SAM-dependent intramitochondrial synthesis of lipoate is indispensable for the activities of pyruvate dehydrogenase and oxoglutarate dehydrogenase complexes. The multiple defects, responsible for the lack of sam5Δ cell growth on non-fermentable carbon sources in YP medium, are all caused by the shortage of SAM in the mitochondria, as demonstrated by the finding that the petite phenotype is restored by the expression of SAM synthetase in the mitochondria. Taken together, our results on the phenotypic analysis of the sam5Δ mutant on fermentative and non-fermentative substrates suggest that in mitochondria SAM plays a role essential for cell life (i.e. the synthesis of biotin) and roles of importance for respiratory growth only (energy conservation and related functions).
Materials and methods
Yeast strains, media and growth conditions
Wild-type BY4741 and sam5Δ yeast strains were provided by the EUROFAN resource centre EUROSCARF (Frankfurt). The SAM5 (YNL003c) locus of S.cerevisiae strain BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) (Brachmann et al., 1998) was replaced by kanMX4. The deletion of SAM5 was verified by PCR. Wild-type fungi and the deletion strain were grown in rich medium containing 2% bactopeptone and 1% YP or SM medium (Sherman, 1991). All media were supplemented with either fermentable or non-fermentable carbon sources (2% glucose or 2% galactose, and 3% glycerol, 2% ethanol, 3% acetate, 2% lactate or 2% pyruvate). The final pH was adjusted to 4.5 or, with pyruvate or acetate, to 6.5.
Construction of expression plasmids
The coding sequence of SAM5 (corresponding to ORF YNL003c) was cloned into the pMW7 vector for expression in E.coli. The SAM5 ORF was amplified from S.cerevisiae genomic DNA by PCR using primers corresponding to the extremities of the coding sequence with additional NdeI and HindIII sites. The plasmid SAM5-pRS416 was constructed by cloning a DNA fragment consisting of the SAM5 ORF, of 412 bp upstream and of 322 bp downstream the ORF (amplified from S.cerevisiae genomic DNA by PCR using primers with additional HindIII and BamHI sites) into the pRS416 vector (Sikorski and Hieter, 1989). The plasmids SAM1-pYES2 and Su9(1–69)-SAM1-pYES2 were constructed by cloning the SAM1 ORF (YLR180w) and a DNA fragment encoding the first 69 amino acids of the Neurospora crassa Fo ATPase subunit 9 in frame with the SAM1 ORF, respectively, into the pYES vector (Invitrogen) behind the inducible Gal-Cyc promoter. The SAM1 ORF was amplified from S.cerevisiae genomic DNA with additional BamHI and EcoRI sites. The Fo ATPase fragment was amplified from the pVT100U-GFP vector (Westermann and Neupert, 2000) kindly provided by Dr Hanspeter Rottensteiner, with additional HindIII and BamHI sites. Transformants of E.coli DH5α cells were selected on ampicillin (100 µg/ml), and screened by direct colony PCR and by restriction digestion of purified plasmids. The sequences of the inserts were verified.
Bacterial expression and purification of Sam5p
Sam5p was overexpressed at 30°C in E.coli CO214(DE3) (Palmieri et al., 2001c). Control cultures with the empty vector were processed in parallel. Inclusion bodies were purified on a sucrose density gradient (Fiermonte et al., 1993), washed at 4°C with TE buffer (10 mM Tris–HCl, 1 mM EDTA pH 6.5), then twice with a buffer containing Triton X-114 (3%, w/v), 1 mM EDTA and 10 mM PIPES–NaOH pH 7.0, and once again with TE buffer. Sam5p was solubilized in 1.8% sarkosyl (w/v), and a small residue was removed by centrifugation (258 000 g, 1 h). Proteins were analysed by SDS–PAGE in 17.5% and stained with Coomassie Blue. The N-terminus was sequenced, and the yield of purified Sam5p was estimated by laser densitometry of stained samples (Fiermonte et al., 1998).
Transport assays
The recombinant protein in sarkosyl was reconstituted into liposomes in the presence of 10 mM PIPES at pH 7.0 and with or without substrate (Palmieri et al., 1995). External substrate was removed from proteoliposomes on a Sephadex G-75 columns, pre-equilibrated with 50 mM NaCl and 10 mM PIPES at pH 7.0 (buffer A). The amount of protein incorporated into liposomes was measured as described previously (Fiermonte et al., 1998); in all cases, it was ∼20%. Transport at 25°C was started by adding S-[methyl-3H]-adenosyl-l-methionine (NEN Life Science Products) to substrate-loaded proteoliposomes (exchange) or to unloaded proteoliposomes (uniport), and terminated by addition of 20 mM pyridoxal 5′-phosphate and 0.4 mM p-hydroxymercuribenzoate. In controls, inhibitors were added with the labeled substrate. Entrapped radioactivity was counted (Palmieri et al., 1995). The experimental values were corrected by subtracting control values. The initial transport rate was calculated from the radioactivity taken up by proteoliposomes after 45 s (in the initial linear range of substrate uptake). Various other transport activities were also assayed by the inhibitor-stop method. For efflux measurements, proteoliposomes containing 1 mM SAM were labeled with 5 µM [3H]SAM by carrier-mediated exchange equilibration (Palmieri et al., 1995). After 60 min, the external radioactivity was removed by passing the proteoliposomes through Sephadex G-75. Efflux was started by adding unlabeled external substrate or buffer A alone, and terminated by adding the inhibitors indicated above.
Intracellular localization of Sam5p, Sah1p, Sam1p and Su9(1–69)-Sam1p (mtSam1p)
The coding sequences for Sam5p (YNL003c) and Sah1p (YER043c) were amplified by PCR from S.cerevisiae genomic DNA without termination codons and with additional BamHI and EcoRI (Sam5p) and BamHI and HindIII (Sah1p) sites. The products were cloned into the pUG35 vector in frame with the yEGFP (yeast enhanced green fluorescence protein) coding sequence (amplified from the pVT100U-GFP vector) under the control of the MET25 promoter. Sam1p and Su9(1–69)-Sam1p (mtSam1p), amplified as described above but without termination codons, were cloned in frame with the yEGFP coding sequence into the pYES2 vector. The yeast strain YPH499 (MATa ade2-101 his3-Δ200 leu 2-Δ1 ura3-52 trp1-Δ63 lys2-801) was transformed with each of the GFP fusion constructs. Cells were grown in glycerol-supplemented SM medium (SMGly) to mid-logarithmic phase, transferred in methionine-less SMGly (for the expression of Sam5p and Sah1p) or 0.4% galactose-supplemented SMGly (for the expression of Sam1p and mtSam1p). Then, the cells were incubated for 1 h at 30°C in the presence of 50 nM MitoTracker Red CMXRos (Molecular Probes, Leiden, The Netherlands), washed twice with fresh medium and imaged at the microscope according to published protocols (Westermann and Neupert, 2000). We employed a Zeiss Axiovert 200 inverted epifluorescence microscope equipped with a Plan-Neofluar 100×/1.30 Ph3 oil objective (Carl Zeiss Jena, Germany), and with a CoolSNAP HQ CCD camera (Roper Scientific, Trenton, NJ, USA) using the Metamorph software (Universal Imaging Corporation, Downington, PA, USA).
Acknowledgments
Acknowledgements
This paper is dedicated to Professor Eraldo Antonini. This work was supported by grants from MIUR-PRIN, MIUR-FIRB, MIUR L.488/92 and L.95/95, CNR-MIUR project ‘Genomica funzionale’, CEGBA and by the European Social Fund.
References
- Back J.W., Sanz,M.A., De Jong,L., De Koning,L.J., Nijtmans,L.G., De Koster,C.G., Grivell,L.A., Van Der Spek,H. and Muijsers,A.O. (2002) A structure for the yeast prohibitin complex: structure prediction and evidence from chemical crosslinking and mass spectrometry. Protein Sci., 11, 2471–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger K.H. and Yaffe,M.P. (1998) Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 4043–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J.D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Dubin D.T., Green,C.M. and Prince,D.L. (1982) Effects of cycloleucine on mitochondrial RNA. In Usdin,E., Borchardt,R.T. and Creveling,C.R. (eds), Biochemistry of S-Adenosylmethionine and Related Compounds. McMillan Press, London, pp. 289–296. [Google Scholar]
- el Moualij B., Duyckaerts,C., Lamotte-Brasseur,J. and Sluse,F.E. (1997) Phylogenetic classification of the mitochondrial carrier family of Saccharomyces cerevisiae. Yeast, 13, 573–581. [DOI] [PubMed] [Google Scholar]
- Fiermonte G., Walker,J.E. and Palmieri,F. (1993) Abundant bacterial expression and reconstitution of an intrinsic membrane transport protein from bovine mitochondria. Biochem. J., 294, 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiermonte G., Dolce,V. and Palmieri,F. (1998) Expression in Escherichia coli, functional characterization and tissue distribution of isoforms A and B of the phosphate carrier from bovine mitochondria. J. Biol. Chem., 273, 22782–22787. [DOI] [PubMed] [Google Scholar]
- Fiermonte G., Palmieri,L., Todisco,S., Agrimi,G., Palmieri,F. and Walker,J.E. (2002) Identification of the mitochondrial glutamate transporter: bacterial expression, reconstitution, functional characterization and tissue distribution of two human isoforms. J. Biol. Chem., 277, 19289–19294. [DOI] [PubMed] [Google Scholar]
- Gantt R. and Schneider,W.C. (1979) Presence of 5-methylcytosine in rat liver mitochondrial and microsomal DNA. In Usdin,E., Borchardt,R.T. and Creveling,C.R. (eds), Transmethylation. Elsevier/North-Holland, New York, NY, pp. 465–471. [Google Scholar]
- Horne D.W., Holloway,R.S. and Wagner,C. (1997) Transport of S-adenosylmethionine in isolated rat liver mitochondria. Arch. Biochem. Biophys., 343, 201–206. [DOI] [PubMed] [Google Scholar]
- Hoyos M.E., Palmieri,L., Wertin,T., Arrigoni,R., Polacco,J.C. and Palmieri,F. (2003) Identification of a mitochondrial transporter for basic amino acids in Arabidopsis thaliana by functional reconstitution into liposomes and complementation in yeast. Plant J., 33, 1027–1035. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto,S., Kawashima,S. and Nakaya,A. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res., 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen R.C. and Yall,I. (1972) Partial purification and characterization of S-adenosylhomocysteine hydrolase isolated from Saccharomyces cerevisiae. J. Bacteriol., 112, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. (2002) Subcellular localization of the yeast proteome. Genes Dev., 16, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo D., Stettler,S., Mariotte,S., Gendreau,E. and Thuriaux,P. (1994) Organization of the centromeric region of chromosome XIV in Saccharomyces cerevisiae. Yeast, 10, 523–533. [DOI] [PubMed] [Google Scholar]
- Marobbio C.M.T., Vozza,A., Harding,M., Bisaccia,F., Palmieri,F. and Walker,J.E. (2002) Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J., 21, 5653–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet A., Bui,B.T. and Florentin,D. (2001) Biosynthesis of biotin and lipoic acid. Vitam. Horm., 61, 51–101. [DOI] [PubMed] [Google Scholar]
- McCammon M.T., Hartmann,M.A., Bottema,C.D. and Parks,L.W. (1984) Sterol methylation in Saccharomyces cerevisiae. J. Bacteriol., 157, 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R.K., Contopoulou,C.R. and King,J.S. (1992) Genetic and physical maps of Saccharomyces cerevisiae. Yeast, 8, 817–902. [DOI] [PubMed] [Google Scholar]
- Nelson D.R., Felix,C.M. and Swanson,J.M. (1998) Highly conserved charge-pair networks in the mitochondrial carrier family. J. Mol. Biol., 277, 285–308. [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., de Jong,L., Artal Sanz,M., Coates,P.J., Berden,J.A., Back,J.W., Muijsers,A.O., van der Spek,H. and Grivell,L.A. (2000) Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J., 19, 2444–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans L.G., Artal,S.M., Grivell,L.A. and Coates,P.J. (2002) The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life Sci., 59, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F. (2003) The mitochondrial transporter family (SLC25): physiological and pathological implications. In Hediger,M.A. (ed.), The ABC of Solute Carriers: Physiological, Pathological And Therapeutic Implications Of Human Membrane Transport Proteins. Eur. J. Physiol., in press. [Google Scholar]
- Palmieri F., Indiveri,C., Bisaccia,F. and Iacobazzi,V. (1995) Mitochondrial metabolite carrier proteins: purification, reconstitution and transport studies. Methods Enzymol., 260, 349–369. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Runswick,M.J., Fiermonte,G., Walker,J.E. and Palmieri,F. (2000) Yeast mitochondrial carriers: bacterial expression, biochemical identification and metabolic significance. J. Bioenerg. Biomembr., 32, 67–77. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Agrimi,G., Runswick,M.J., Fearnley,I.M., Palmieri,F. and Walker,J.E. (2001a) Identification in Saccharomyces cerevisiae of two isoforms of a novel mitochondrial transporter for 2-oxoadipate and 2-oxoglutarate. J. Biol. Chem., 276, 1916–1922. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Rottensteiner,H., Girzalsky,W., Scarcia,P., Palmieri,F. and Erdmann,R. (2001b) Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J., 20, 5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L. et al. (2001c) Citrin and aralar1 are Ca2+-stimulated aspartate/glutamate transporters in mitochondria. EMBO J., 20, 5060–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L., Bujnicki,J.M., Lapeyre,B. and Bonnerot,C. (2002) MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J., 21, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel D., Harding,M., Runswick,M.J., Walker,J.E. and Brand,M.D. (2002) Does any yeast mitochondrial carrier have a native uncoupling protein function? J. Bioenerg. Biomembr., 34, 165–176. [DOI] [PubMed] [Google Scholar]
- Said H.M., McAlister-Henn,L., Mohammadkhani,R. and Horne,D.W. (1992) Uptake of biotin by isolated rat liver mitochondria. Am. J. Physiol., 263, G81–G86. [DOI] [PubMed] [Google Scholar]
- Santamaria E., Avila,M.A., Latasa,M.U., Rubio,A., Martin-Duce,A., Lu,S.C., Mato,J.M. and Corrales,F.J. (2003) Functional proteomics of nonalcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc. Natl Acad. Sci. USA, 100, 3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirum-Connolly K. and Mason,T.L. (1993) Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science, 262, 1886–1889. [DOI] [PubMed] [Google Scholar]
- Steglich G., Neupert,W. and Langer,T. (1999) Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol., 19, 3435–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L.M. et al. (2002) Systematic screen for human disease genes in yeast. Nature Genet., 31, 400–404. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1985) Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res., 13, 8587–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulo P. and Martin,N.C. (1993) Isolation and characterization of LIP5. A lipoate biosynthetic locus of Saccharomyces cerevisiae. J. Biol. Chem., 268, 17634–17639. [PubMed] [Google Scholar]
- Thomas D. and Surdin-Kerjan,Y. (1987) SAM1, the structural gene for one of the S-adenosylmethionine synthetases in Saccharomyces cerevisiae. Sequence and expression. J. Biol. Chem., 262, 16704–16709. [PubMed] [Google Scholar]
- Thomas D. and Surdin-Kerjan,Y. (1991) The synthesis of the two S-adenosyl-methionine synthetases is differently regulated in Saccharomyces cerevisiae. Mol. Gen. Genet., 226, 224–232. [DOI] [PubMed] [Google Scholar]
- Westermann B. and Neupert,W. (2000) Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast, 16, 1421–1427. [DOI] [PubMed] [Google Scholar]
- Zhang S., Sanyal,I., Bulboaca,G.H., Rich,A. and Flint,D.H. (1994) The gene for biotin synthase from Saccharomyces cerevisiae: cloning, sequencing and complementation of Escherichia coli strains lacking biotin synthase. Arch. Biochem. Biophys., 309, 29–35. [DOI] [PubMed] [Google Scholar]