Abstract
Eukaryotes and prokaryotes have developed mutually beneficial relationships over millennia of evolutionary adaptation. Bacteria in our gut rely on our diet and the protected environment of our bodies just as our health depends on byproducts of microbial metabolism. Microorganisms of the gut microbiota ferment carbohydrates into short-chain fatty acids, convert dietary and endogenous nitrogenous compounds into ammonia and microbial protein, and synthesize and activate B vitamins and vitamin K. The benefit from their activity is multiplex and translates into increased energy for the gut epithelial cells, balanced absorption of salt and water, nitrogen recycling, breakdown of complex lipids and cholesterol, and detoxification of waste compounds.
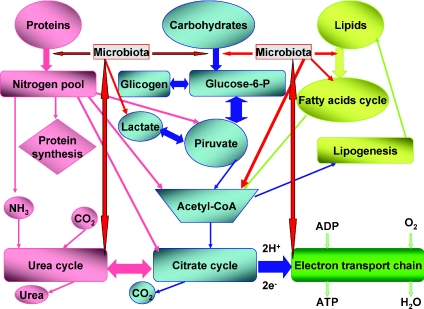
Humans are not the predominant species in our world, as the most numerous organisms are represented by bacteria (Fraser et al. 2009; Large & Lund, 2009; Thane Papke, 2009; Wilmes et al. 2009). Moreover, humans depend greatly for survival from the environment and other organisms. Furthermore, human's adaptability is considerably slower than organisms endowed with a faster and more efficient metabolism, like prokaryotes and unicellular eukaryotes. (de la Cueva-Méndez & Pimentel, 2007; Boles & Singh, 2008; Bayliss, 2009). Among organisms with greater ability to survive a critical change in environment are microorganisms, parasites, bacteria, viruses and elemental forms like plasmids and prions (Laan & Hogeweg, 1995; Raes & Bork, 2008; Crombach & Hogeweg, 2009). Thus, to survive, complex organisms, including humans, have developed symbiosis as a common way to overcome genetic and metabolic deficiencies (Allen et al. 2009; Tylianakis, 2009; Martin et al. 2009). At length, symbiosis has proven beneficial for all partners considered, and has become our way of life. So it happens that Homo sapiens’ largest “organ” is the gut microbiota, comprising two major divisions of bacteria, Bacteroidetes and Firmicutes (Zoetendal et al. 2008; Neish, 2009; Gill et al. 2006; Xu et al. 2003). The colonization of the human gut begins at birth through exposure to other humans, to animals and to the environment, and through our diet (Adlerberth & Wold, 2009). From the very beginning, the gut microbiota plays an important role in our life by modulating cell differentiation and by mediating many of the effects of the diet on our general health. In particular, the microbiota performs functions that are not encoded in the human genome, including the processing of undigestible dietary polysaccharides, metabolism of complex proteins, synthesis of vitamins and production of energy to preserve gut homeostasis (Fig. 1) (Cani & Delzenne, 2009; Martens et al. 2009; Paliy et al. 2009; Wilmes & Bond, 2009). However, considering that more than 60% of the bacteria in our gut cannot be isolated in culture, and thus cannot be characterized in full, we may expect the definition of some other metabolic process relying greatly on the human microbiota in the near future thanks to continuous advances in biotechnology.
Figure 1. Digestion of the main components of an unrestricted mammal diet involves a sequential process of degradation/digestion, absorption and recycling.
The gut microbiota has evolved a parallel processing of foods that complements and in some steps of the process substitutes for the host in the handling of nutrients. This is particularly true for the complete digestion of complex carbohydrates and vitamins.
In this article, we will review the general role of the microbiota in human nutrient handling, particularly, the active cooperation of microorganisms in digestion and production of intermediate compounds that contribute to our well-being. The effect of the microbiota on the expression of digestive enzymes and metabolites by the enterocytes has been described excellently elsewhere (Hooper et al. 2002) and will not be repeated here. This article will comprise four sections corresponding to the main components of our diet: proteins, carbohydrates, lipids, and vitamins and minerals. Particular emphasis will be given to calcium metabolism, as an aspect of host–commensal cooperation that has very recently emerged as one of the many important contribution of the microbiota to mammalian nutrients handling.
Microbiota and human nitrogen balance
The normal digestion of proteins begins in the stomach and duodenum, where proteins are broken down into large peptides, and these into small peptides and amino acids. A good portion of these products are absorbed transiting the small intestine. While bacteria contribute to the digestive process at all these levels, the abundant proteases of pancreatic origin play the greatest role in protein metabolism in the upper gastrointestinal (GI) tract and ileum (Schuchert-Shi & Hauser, 2009). However, in direct relation to the dietary content, about 6–18 g of nitrogenous material daily reaches the large intestinal lumen through the ileocaecal valve. This material is metabolized by the colonic flora and at the end about 15 g of nitrogenous material is excreted in the faeces, 1 g of which is bacterially derived. The microbiota will deaminate amino acids, hydrolyse urea, and recycle ammonia into new microbial cells accumulating metabolites including ammonia, hydrogen sulfide, short and branched-chain fatty acids, amines, phenolic, indolic and N-nitroso compounds (Hooper et al. 2002). Not all of these metabolites are beneficial to the gut mucosa and the organism in general, and their effect on the gut homeostasis is largely dependent on their luminal concentrations, the detoxifying ability of enterocytes, oxidative metabolism, and electrolyte movements through the colonic epithelium. However, enterocytes can utilize ammonia via synthetic pathways involving glutamate, glutamine, citrulline and urea thus maintaining physiologic gut homeostasis (Bergen & Wu, 2009). It is important to remember that colonic micro-organisms such as sulphate-reducing bacteria, bifidobacteria and clostridia respond selectively to specific dietary components, thus conditioning the quality and quantity of byproducts produced. In normal subjects on a standard diet, four-fifths of the urea produced is excreted in urine and the rest is endogenously degraded. Seventy per cent of the nitrogen and 63% of the carbon of the degraded urea are recycled into the urea pool. In healthy volunteers on a nitrogen-free diet or after neomycin treatment with regular diet, degradation of urea is significantly reduced. In septic patients, degradation and recycling of urea is near abolished (Long et al. 1978). In conditions in which nitrogen balance is disrupted, as in hepatic and/or renal failure, or after intestinal injury, new findings have shown that gut microbial metabolism may be improved by treatment with prebiotics and probiotics (Nicaise et al. 2008). These bioactive agents have proved useful in improving the overall nitrogen balance by significantly decreasing toxic secondary nitrogen metabolites (Sharma et al. 2008; Forsyth et al. 2009).
Microbiota and carbohydrate metabolism
As we adapt to changes of the environment, so the gut microbiota adapts to qualitative and quantitative changes of our diet. Particularly, bacteria of the large intestine respond to changes in diet, especially to the type and quantity of dietary carbohydrate. One of the main consequences of increased carbohydrate intake is to decrease the pH of the gut lumen, significantly altering bacterial metabolism and commensal prevalence (Sonnenburg et al. 2006; Martens et al. 2008; Underwood et al. 2009).
Mammals lack the ability to degrade complex polysaccharides, but actively transport simple dietary sugars, like glucose, through the small intestinal enterocytes. The undigested polysaccharides transit to portions of the GI tract where the bacterial colonization is heavier (ileum, colon) and composed of large families of fermentors. Thus undigested polysaccharides such as cellulose, xylan and undigested starch, and host-derived glycans (mucins, glycosphingolipids) have to reach the largest part of the microbiota to be broken down and fully utilized. The microbiota degrades the monosaccharides further, and the byproducts of this fermentation (short chain fatty acids, SCFAs) are absorbed and used by the enterocytes as their main energy source (McFarlane & McFarlane, 2003; Brinkworth et al. 2009; Jacobs et al. 2009).
The recognition of the importance of complex polysaccharides in our diet and the beneficial effects of these nutrients in term of enrichment and selection of fermentors among the microbiota has prompted the application of dietary manipulation with purified oliosaccharides (prebiotics) with or without probiotic bacteria to improve the health of the gut, and the body as a whole, in conditions of chronic inflammation or frailty, like inflammatory bowel disease, autoimmune diseases and cancer, and as a potentially preventative measure against many of these conditions (Fujimori et al. 2009; Salonen et al. 2009).
Microbiota and lipid metabolism
While the findings in germ-free and conventionally colonized mice have shown that the microbiota is essential for full handling of polysaccharides in the diet, the same studies revealed that the surplus of carbohydrates breakdown is channelled by components of the microbiota into energy storage. In fact, certain commensal species (Firmicutes) suppress circulating lipoprotein lipase inhibitors, thus inducing increased enzyme activity and energy accumulation as fat (Wolf, 2006; Backhed et al. 2004). Since obesity results from alterations in the body's regulation of energy intake, expenditure and storage, and the microbiota can regulate energy storage from the diet, the result is that an abnormal microbiota activity associated with an unbalanced diet is a cause of increased fat accumulation in the body (Turnbaugh et al. 2006; Isken et al. 2009; Ley et al. 2005; Wall et al. 2009). More on this subject is amply explained in another review of this series (Turnbaugh & Gordon, 2009). Moreover, recent studies showed that the microbiota metabolism of oligosaccharides not only regulates energy uptake and storage, but suppresses liver triglyceride and VLDL synthesis, resulting in significant decrease of circulating triglyceride, and to a lesser extent cholesterol, levels (Taylor & Williams, 1998). However, the microbiota handling of fat metabolism is not limited to promoting carbohydrate-derived energy storage into fat; commensal bacteria intervene directly in fatty acid and cholesterol metabolism. In particular, the handling of cholesterol by the gut is the result of a balance between absorption, excretion and metabolism by the microbiota. Between 34 and 57% of dietary cholesterol is absorbed from the human intestine. Cholesterol absorption depends on the amount of cholesterol consumed, fibre content of the diet, amount of plant sterols, gut transit time and relative ratio of fatty acids in the diet. Faecal cholesterol (150 mg day−1) results from biliary secretions into the intestine, sloughing of epithelial cells and unabsorbed dietary cholesterol. Abnormal intestinal bacterial metabolism of cholesterol may cause severe colonic disorders (colitis, bacterial overgrowth, malabsorption) (Lichtenstein, 1990). Furthermore, commensal bacteria are essential for the handling of bile acids, promoting their recirculation and regulating their end fate. Bacterial bile salt hydrolases (BSHs) modify bile acid in the gut, and since bile acids regulate their own synthesis, lipid absorption, cholesterol homeostasis, and mucosal defenses, bacterial BSHs promote gut homeostasis (Jones et al. 2008).
Microbiota role in vitamin and mineral metabolism
Vitamins were discovered during the first half of the 20th century, by analysing human and animal diet, and through the identification of ‘growth factors’ for bacterial isolation (Burkholder & McVeigh, 1942; Peterson & Peterson, 1945), but only in the last decades of the last century was the association between bacteria and vitamin synthesis made (Leviton & Hargrove, 1952; Smith, 1952). Most vitamin-producing bacteria use the 2-methyl-d-erythritol 4-phosphate pathway for the synthesis of vitamins and isoprenoids, and for certain vitamins their presence in the microbiota is crucial for our survival (vitamins K and B12). Table 1 summarizes data on bacteria-derived vitamins and their function. It is foreseeable that genetically engineered commensals will become an efficient source of vitamins, essential elements and growth factors in the future, thus overcoming the severe deficiencies associated with malnutrition, radiation- and chemo-therapy, antibiotic therapy, and space travel.
Table 1.
Essential vitamins produced by intestinal bacteria in humans and/or animals
| Vitamin | Source | Function |
|---|---|---|
| Vitamin K | Diet: green leaf vegetables. Synthesized by bacteria in the large intestine. | Synthesis of blood-clotting factors in the liver; deficiency causes bleeding disorders. |
| Vitamin C | Diet: fruits and vegetables. Most animals’ microbiota can synthesize ascorbic acid, with the exception of primates (including humans), guinea pigs and Mongolian fruit bats. | Synthesis of hydroxyproline (collagen, connective tissues, cartilage, bone, teeth, wound healing). Deficiency: scurvy. |
| Niacin | Diet: meats, leafy green vegetables, potatoes and peanuts. Synthesized by intestinal bacteria. | Precursor to the coenzymes nicotinamide adenine dinucleotide (NAD) and nicotinamide adenine dinucleotide phosphate (NADPH) (glycolysis, Krebs cycle and oxidative phosphorylation). |
| Panthotenic acid | Diet: grain, legumes, egg yolk, meat. Synthesized by intestinal bacteria. | Precursor to coenzyme A (oxidation and/or synthesis of carbohydrates and fatty acids). |
| Biotin | Diet: egg yolk, legumes, nuts, liver. Synthesized by intestinal bacteria. | Coenzyme for enzymes that catalyse carboxylation, decarboxylation and deamination. |
| Vitamin B12 | Microbial synthesis is only source in nature. Diet: derived from ingestion of animal products. Not present in vegetables. | Coenzyme involved in many metabolic pathways. Deficiency: pernicious anaemia. |
| Folic acid | Diet: dark-green vegetables, beef, eggs, whole grains. Synthesized by intestinal bacteria. | Coenzyme in the synthesis of amino acids, purines and thymine. Deficiency: growth failure and anaemia. |
Minerals and essential elements are crucial components of enzymes, structural proteins and redox transport chains, and without them life as we know it would not be possible. In this too the gut microbiota acts as a fully fledged partner: copper, cobalt, iron and zinc are elements that can be exchanged in the gut between bacteria and their host. However, it is calcium absorption and metabolism that, owing to its central role in their survival, has induced in both microorganisms and hosts a series of adaptive changes to assure that physiological levels of this element are kept in balance over a lifetime. We and others have found that byproducts of carbohydrate metabolism, i.e. SCFAs, have, among other functions, the ability to induce expression of the vitamin D receptor on eukaryotic cells (Feng et al. 2005; Christakos et al. 2007). Vitamin D, being one of the key regulators of calcium metabolism in mammals, ensures calcium absorption from dietary intake and intracellular storage and deposition in bone and teeth (Ordóñez-Morán & Muñoz, 2009).
We have found that components of the gut microbiota have the ability to enhance expression of the vitamin D receptor (VDR) in intestinal epithelial cells in a SCFA-dependent and -independent manner, and to regulate through the MAPK and PKC mediated signalling pathways calcium transport across the epithelial cells (Fig. 2) and its storage in intracellular compartments (Feng et al. 2005). Moreover, commensal and probiotic bacteria can enhance SCFA transport inside the intestinal epithelium by inducing the expression of the main transporter for these compounds, the monocarboxylate transporter (MCT1) (Lee et al. 2006), further promoting cellular energy increase and enhanced expression of VDR and calcium binding proteins inside the cells (calbindin, Sp100, calretinin). Furthermore, we have shown that the expression of the main calcium transporter in the intestinal epithelial cells, TRPV6, is also markedly induced by commensal and probiotic bacteria in a MAPK-dependent manner (author's unpublished observations). The increase in calcium handling at the gut level translates into more replete stores in the body. Since bones and teeth are the main storage compartments for calcium in mammals, the result of the microbiota activity in calcium handling is stronger bones and teeth. We confirmed this hypothesis by treating animal models of chronic gut and joint inflammation, which showed osteopaenia and/or osteoporosis as a consequence of their disease process, with commensal and probiotic bacteria, and found that the treatment significantly improved calcium transport in the gut, and restored to normal levels bone calcification as measured by bone density index and direct calcium content of isolated bones (Resta-Lenert & Lee, 2006, 2007; Resta-Lenert et al. 2007, 2008).
Figure 2.
While enterocytes have an established pathway that enables calcium absorption and its regulation through hormones such as vitamin D and parathyroid hormone, the microbiota has developed a complex redundancy system that allows for enhanced expression of the vitamin D nuclear receptor and of various channels and transporters (TRPV6, NCX1, TRPC) as well as binding proteins, which are essential for calcium extraction from dietary sources, its recycling inside the body of the host and storage in cells and at the levels of specialized tissues (bone, cartilage).
Conclusions
Do we really need the gut microbiota to survive? The answer is yes, unless we choose to live in a bubble or on a sterile planet. It could be argued that there are examples of human survival in the absence of significant microbial bio-mass (large bowel resection, chronic antibiotic therapy). In these cases, besides the presence of residual bacteria in the remaining GI tract, or, in case of infants, through microbiota conditioning to induce immune priming, special diets and vitamin supplementation can support survival for long times with limited reduction of energy balance. However, these are particular circumstances that are outside what we consider normality: barring a radical change in our environment and our dietary habits, our survival will still depend on a healthy relationship with our gut microbiota.
Acknowledgments
These studies were supported by NIH grant DK062096 and by UCSD-DDRDC DK080506 to S.C.R.
Glossary
Abbreviations
- SCFA
short chin fatty acid
- MAPK
mitogen-activated protein kinase
- VDR
vitamin D receptor
Definitions
- Commensal
microorganism with established mutualistic relationship with the host
- probiotics
commensal bacteria selected for their beneficial effects on the host health
- prebiotics
undigestible polysaccharides which upon fermentation by commensal bacteria generate byproducts with beneficial (SCFA) effects on the host
References
- Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- Allen JM, Light JE, Perotti MA, Braig HR, Reed DL. Mutational meltdown in primary endosymbionts: selection limits Muller's ratchet. PLoS One. 2009;4:e4969. doi: 10.1371/journal.pone.0004969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss CD. Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol Rev. 2009;33:504–520. doi: 10.1111/j.1574-6976.2009.00162.x. [DOI] [PubMed] [Google Scholar]
- Bergen WG, Wu G. Intestinal nitrogen recycling and utilization in health and disease. J Nutr. 2009;139:821–825. doi: 10.3945/jn.109.104497. [DOI] [PubMed] [Google Scholar]
- Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth GD, Noakes M, Clifton PM, Bird AR. Comparative effects of very low-carbohydrate, high-fat and high-carbohydrate, low-fat weight-loss diets on bowel habit and faecal short-chain fatty acids and bacterial populations. Br J Nutr. 2009;101:1493–1502. doi: 10.1017/S0007114508094658. [DOI] [PubMed] [Google Scholar]
- Burkholder PR, McVeigh I. Synthesis of vitamins by intestinal bacteria. Proc Natl Acad Sci U S A. 1942;28:285–289. doi: 10.1073/pnas.28.7.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15:1546–1558. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- Christakos S, Dhawan P, Benn B, Porta A, Hediger M, Oh GT, Jeung EB, Zhong Y, Ajibade D, Dhawan K, Joshi S. Vitamin D: molecular mechanism of action. Ann N Y Acad Sci. 2007;1116:340–348. doi: 10.1196/annals.1402.070. [DOI] [PubMed] [Google Scholar]
- Crombach A, Hogeweg P. Evolution of resource cycling in ecosystems and individuals. BMC Evol Biol. 2009;9:122–126. doi: 10.1186/1471-2148-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cueva-Méndez G, Pimentel B. Gene and cell survival: lessons from prokaryotic plasmid R1. EMBO Rep. 2007;8:458–464. doi: 10.1038/sj.embor.7400957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Lee SJ, Resta-Lenert S. Effect of probiotic and commensal bacteria on vitamin D-receptor and calcium transport protein expression in intestinal epithelial cells in vitro. Gastroenterology. 2005;128:A–47204. [Google Scholar]
- Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, Keshavarzian A. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol. 2009;43:163–172. doi: 10.1016/j.alcohol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- Fujimori S, Gudis K, Mitsui K, Seo T, Yonezawa M, Tanaka S, Tatsuguchi A, Sakamoto C. A randomized controlled trial on the efficacy of synbiotic versus probiotic or prebiotic treatment to improve the quality of life in patients with ulcerative colitis. Nutrition. 2009;25:520–525. doi: 10.1016/j.nut.2008.11.017. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Isken F, Klaus S, Osterhoff M, Pfeiffer AF, Weickert MO. Effects of long-term soluble vs. insoluble dietary fibre intake on high-fat diet-induced obesity in C57BL/6J mice. J Nutr Biochem. 2009 doi: 10.1016/j.jnutbio.2008.12.012. in press. [DOI] [PubMed] [Google Scholar]
- Jacobs DM, Gaudier E, van Duynhoven J, Vaughan EE. Non-digestible food ingredients, colonic microbiota and the impact on gut health and immunity: a role for metabolomics. Curr Drug Metab. 2009;10:41–54. doi: 10.2174/138920009787048383. [DOI] [PubMed] [Google Scholar]
- Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan JD, Hogeweg P. Predator-prey coevolution: interactions across different timescales. Proc Biol Sci. 1995;259:35–42. [Google Scholar]
- Large AT, Lund PA. Archaeal chaperonins. Front Biosci. 2009;14:1304–1324. doi: 10.2741/3310. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Feng J, Resta-Lenert S. A synbiotic upregulates MCT1 expression and butyrate transport in intestinal epithelial cells. FASEB J. 2006;20:A1271. [Google Scholar]
- Leviton A, Hargrove RE. Microbiological synthesis of vitamin B12 by propionic acid bacteria. Ind Eng Chem. 1952;44:2651–2655. [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein AH. Intestinal cholesterol metabolism. Ann Med. 1990;22:49–52. doi: 10.3109/07853899009147241. [DOI] [PubMed] [Google Scholar]
- Long CL, Jeevanandam M, Kinney JM. Metabolism and recycling of urea in man. Am J Clin Nutr. 1978;31:1367–1382. doi: 10.1093/ajcn/31.8.1367. [DOI] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: The bacteroidetes Sus-like paradigm. J Biol Chem. 2009 doi: 10.1074/jbc.R109.022848. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FP, Sprenger N, Yap IK, Wang Y, Bibiloni R, Rochat F, Rezzi S, Cherbut C, Kochhar S, Lindon JC, Holmes E, Nicholson JK. Panorganismal gut microbiome-host metabolic crosstalk. J Proteome Res. 2009;8:2090–2105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise C, Prozzi D, Viaene E, Moreno C, Gustot T, Quertinmont E, Demetter P, Suain V, Goffin P, Devière J, Hols P. Control of acute, chronic, and constitutive hyperammonemia by wild-type and genetically engineered Lactobacillus plantarum in rodents. Hepatology. 2008;48:1184–1192. doi: 10.1002/hep.22445. [DOI] [PubMed] [Google Scholar]
- Ordóñez-Morán P, Muñoz A. Nuclear receptors: genomic and non-genomic effects converge. Cell Cycle. 2009;8:1675–1680. doi: 10.4161/cc.8.11.8579. [DOI] [PubMed] [Google Scholar]
- Paliy O, Kenche H, Abernathy F, Michail S. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl Environ Microbiol. 2009;75:3572–3579. doi: 10.1128/AEM.02764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson EH, Peterson MS. Relation of bacteria to vitamins and other growth factors. MMBR. 1945;9:49–109. doi: 10.1128/br.9.2.49-109.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Resta-Lenert S, Lee SJ, Maruggi M. Aging: synbiotics protect colitic mice from developing altered BMD. Nat Med. 2007:P13, 267. [Google Scholar]
- Resta-Lenert S, Lee SJ. Expression of receptor activator of nuclear factor-κB ligand (RANKL) and osteoprotegerin (OPG) in IL-10 -/- and mdr1a -/- mouse models of colitis. Gastroenterology. 2006;130:A548. [Google Scholar]
- Resta-Lenert S, Lee SJ. Probiotics and synbiotics protect mdr1a -/- colitic mice from developing altered bone mineral density. Gastroenterology. 2007;132:A1804. [Google Scholar]
- Resta-Lenert S, Lee SJ, Maruggi M. Synbiotics prevent development of osteoporosis in chronic inflammation of young and old mdr1a -/- mice. FASEB J. 2008;22:883.10. [Google Scholar]
- Schuchert-Shi A, Hauser PC. Peptic and tryptic digestion of peptides and proteins monitored by capillary electrophoresis with contactless conductivity detection. Anal Biochem. 2009;387:202–207. doi: 10.1016/j.ab.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Salonen A, Palva A, de Vos WM. Microbial functionality in the human intestinal tract. Front Biosci. 2009;14:3074–3084. doi: 10.2741/3436. [DOI] [PubMed] [Google Scholar]
- Sharma P, Sharma BC, Puri V, Sarin SK. An open-label randomized controlled trial of lactulose and probiotics in the treatment of minimal hepatic encephalopathy. Eur J Gastroenterol Hepatol. 2008;20:506–511. doi: 10.1097/MEG.0b013e3282f3e6f5. [DOI] [PubMed] [Google Scholar]
- Smith EL. The discovery and identification of vitamin B12. Proc Nutr Soc. 1952:295–299. doi: 10.1079/bjn19520031. Jan 5th. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GR, Williams CM. Effects of probiotics and prebiotics on blood lipids. Br J Nutr. 1998;80:225–230. [PubMed] [Google Scholar]
- Thane Papke R. A critique of prokaryotic species concepts. Methods Mol Biol. 2009;532:379–395. doi: 10.1007/978-1-60327-853-9_22. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;17:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Tylianakis JM. Ecology. Warming up food webs. Science. 2009;323:1300–1301. doi: 10.1126/science.1170909. [DOI] [PubMed] [Google Scholar]
- Underwood MA, Salzman NH, Bennett SH, Barman M, Mills DA, Marcobal A, Tancredi DJ, Bevins CL, Sherman MP. A randomized placebo-controlled comparison of 2 prebiotic/probiotic combinations in preterm infants: impact on weight gain, intestinal microbiota, and fecal short-chain fatty acids. J Pediatr Gastroenterol Nutr. 2009;48:216–225. doi: 10.1097/MPG.0b013e31818de195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall R, Ross RP, Shanahan F, O’Mahony L, O’Mahony C, Coakley M, Hart O, Lawlor P, Quigley EM, Kiely B, Fitzgerald GF, Stanton C. Metabolic activity of the enteric microbiota influences the fatty acid composition of murine and porcine liver and adipose tissues. Am J Clin Nutr. 2009;89:1393–1401. doi: 10.3945/ajcn.2008.27023. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Simmons SL, Denef VJ, Banfield JF. The dynamic genetic repertoire of microbial communities. FEMS Microbiol Rev. 2009;33:109–132. doi: 10.1111/j.1574-6976.2008.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P, Bond PL. Microbial community proteomics: elucidating the catalysts and metabolic mechanisms that drive the Earth's biogeochemical cycles. Curr Opin Microbiol. 2009;12:310–317. doi: 10.1016/j.mib.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Wolf G. Gut microbiota: a factor in energy regulation. Nutr Rev. 2006;64:47–50. doi: 10.1111/j.1753-4887.2006.tb00173.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]