Abstract
p53 protects against cancer through its capacity to induce cell cycle arrest or apoptosis under a large variety of cellular stresses. It is not known how such diversity of signals can be integrated by a single molecule. However, the literature reveals that a common denominator in all p53-inducing stresses is nucleolar disruption. We thus postulated that the impairment of nucleolar function might stabilize p53 by preventing its degradation. Using micropore irradiation, we demonstrate that large amounts of nuclear DNA damage fail to stabilize p53 unless the nucleolus is also disrupted. Forcing nucleolar disruption by anti-upstream binding factor (UBF) microinjection (in the absence of DNA damage) also causes p53 stabilization. We propose that the nucleolus is a stress sensor responsible for maintenance of low levels of p53, which are automatically elevated as soon as nucleolar function is impaired in response to stress. Our model integrates all known p53-inducing agents and also explains cell cycle-related variations in p53 levels which correlate with established phases of nucleolar assembly/disassembly through the cell cycle.
Keywords: apoptosis/micropore irradiation/NPM/nucleolus/p53
Introduction
p53 acts as a tumour suppressor through its capacity to induce cell cycle arrest and apoptosis in response to a variety of cellular stresses (reviewed in Levine, 1997). These properties are exploited in anti-cancer therapy, mainly by triggering a p53 response through genotoxic stress (Weinstein et al., 1997). However, more than 20 years after the discovery of the protein, the mechanism of induction of a p53 response still remains unresolved. A p53 response typically involves stabilizing the short-lived protein and unlocking its capacity to transactivate cell cycle arrest and apoptosis genes (Levine, 1997). In normal cell growth conditions, p53 protein levels are kept low by the action of the Mdm2 protein which targets p53 for proteasomal degradation (Ljungman, 2000). Activation of p53 usually involves some form of disruption of its interaction with Mdm2, and it has been shown that co-compartmentalization of both proteins is essential for p53 degradation (Xirodimas et al., 2001). The main puzzle, however, resides in the variety of cellular stresses that can stabilize p53, which include DNA damage in the form of both adducts and strand breaks, transcription inhibition, depletion of nucleotide pools, oncogene expression, viral infection and heat shock (Ljungman, 2000; Pluquet and Hainaut, 2001). Many of these stresses induce covalent modifications of the p53 protein, which have been proposed as induction mechanisms, but these vary from one stimulus to another, making it difficult to understand how p53 can integrate such a wide range of stimuli. Importantly, recent studies strongly argue against allosteric modifications as activators of p53 as a transcription factor and suggest that it is the elevation of nuclear p53 levels that causes transcriptional activation (Blattner et al., 1999; Meek, 1999; Kaeser and Iggo, 2002). Neither is it clear what the stress sensors are or whether p53 senses stresses directly. There is strong evidence that the p53 response to UV irradiation and DNA adducts is mediated by inhibition of RNA polymerase II (Pol II) transcription, and in fact it can be induced by RNA Pol II inhibitors (Ljungman et al., 1999), but it is not clear how the transcription inhibition signal is translated into a p53 response. Finally, no p53 stabilization model can explain the cell cycle-dependent variation of p53 levels which are raised in early G1 but remain low for the rest of the cell cycle (see for example David-Pfeuty, 1999).
Mdm2 mediates proteasomal degradation of p53. While initially thought to provide both the leucine-rich nuclear export signal (NES) and the ubiquitylation responsible for p53 export and degradation, respectively (Tao and Levine, 1999), later evidence indicated that a more likely mechanism is an initial C-terminal monoubiquitylation of p53 by Mdm2 which in turn exposes the C-terminal leucine-rich NES of p53 (Stommel et al., 1999; Boyd et al., 2000). Probably the best characterized p53 activation pathway is that employed by oncogene expression and some viral infections, which involves expression of the ARF gene product (Sherr and Weber, 2000). When induced, p14ARF in humans (p19ARF in mouse) disrupts the interaction between p53 and Mdm2 and sequesters the latter to the nucleolus (Tao and Levine, 1999; Sherr and Weber, 2000). Two non-mutually exclusive mechanisms have been proposed for this effect: nucleolar sequestration of Mdm2, and blockage of a postulated nucleolar route of p53 export for cytoplasmic degradation (Tao and Levine, 1999; Sherr and Weber, 2000). This latter mechanism, based on the participation of the nucleolus in the export of macromolecular complexes, was described by Sherr and Weber (2000) as p53–Mdm2 complexes ‘riding the ribosome’.
Several lines of evidence support a nucleolar export model for p53 degradation. Marechal et al. (1994) demonstrated the simultaneous association of the ribosomal protein L5 with both Mdm2 and Mdm2–p53. The same group went on to demonstrate that nucleo-cytoplasmic shuttling of Mdm2 uses the same pathway as the human immunodeficiency virus (HIV) Rev protein (Roth et al., 1998) which also displays nucleolar localization, and suggested that Mdm2 export may rely on its capacity to bind to the L5 protein, since L5 participates in the export of the HIV Rev protein (see Schatz et al., 1998; and references therein). Importantly, both p53 and Rev utilize CRM1 for their export, which is also employed by 40S and 60S ribosomal subunits and is specifically inhibited by leptomycin B (LMB; Freedman and Levine, 1998; Thomas and Kutay, 2003). In addition, p53 can be covalently linked to 5.8S rRNA, and it has been shown that cytoplasmic p53 is associated with a subset of ribosomes in which 5.8S rRNA is covalently linked to protein (Samad and Carroll, 1991; Fontoura et al., 1992, 1997). In spite of all these observations, lack of evidence for p53 in the nucleolus prevented acceptance of the model of Mdm2–p53 complexes ‘riding the ribosome’, even though nucleolar levels of p53 in transit should be expected to be very low. However, for cells grown under normal conditions, we have reported recently that following permeabilization, where most soluble nucleoplasmic p53 is eliminated, nuclear-bound p53 is readily detectable in the nucleoli (Rubbi and Milner, 2003). Klibanov et al. (2001) subsequently have demonstrated accumulation of nucleolar p53 after proteasome inhibition.
From these findings, we explored whether the nucleolar export model of p53 could be extended to explain the induction of a p53 response under a variety of cell stresses. The strongest evidence yet for this extension of the model comes from the observations of Pestov et al. (2001) on the nucleolar protein Bop1, implicated in pre-rRNA processing and assembly of ribosomal subunits. When a dominant-negative Bop1 mutant was introduced into cells, it interfered with nucleolar function, inducing p53 stabilization and a p53-dependent cell cycle arrest in G1 (Pestov et al., 2001). While the authors proposed that some sort of stress sensor might monitor nucleolar function and respond to its impairment by inducing a p53 response, the links between p53 and nucleolar components mentioned above suggest to us that a simpler explanation is that it is nucleolar function itself that is required for proper degradation of p53, without the need to invoke an intermediary stress sensor. We base our proposition on the fact that a common denominator of all p53-inducing stresses, some of which are summarized in Table I, is that they all cause nucleolar disruption and compromise nucleolar function. The nucleolus is thus a sensor responsive to a wide range of cellular stresses. Under these nucleolus-disrupting stresses, p14ARF appears to be dispensable since ARF-null cells can mount a normal p53 response (Stott et al., 1998). Therefore, we now postulate that the model of nucleolar export of Mdm2–p53 complexes proposed for the explanation of p14ARF induction of p53 (see above) can be extended to explain p53 induction by a wide range of cellular stresses, all of which cause disruption of nucleolar organization. In Table I, two agents, LMB and the proteasome inhibitor MG132, have been listed separately since they can induce p53 without apparent nucleolar disruption. In the nucleolar export model, these two agents act precisely downstream of nucleolar disruption and are thus the only p53-stabilizing agents which are not expected to compromise nucleolar function.
Table I. Comparison of nucleolar-disrupting activity of various agents and their p53 activation capability.
| Agent | Nucleolar disruption | Reference | p53 stabilization | Reference |
|---|---|---|---|---|
| UV | ✓ | Zatsepina et al. (1989) | ✓ | Pluquet and Hainaut (2001) |
| Cis-Pt | ✓ | Jordan and Carmo-Fonseca (1998) | ✓ | Pluquet and Hainaut (2001) |
| 5-FU | ✓ | Ghoshal and Jacob (1994) | ✓ | Pritchard et al. (1997) |
| DRB | ✓ | David-Pfeuty et al. (2001) | ✓ | David-Pfeuty et al. (2001) |
| Actinomycin D | ✓ | Haaf and Ward (1996) | ✓ | Andera and Wasylyk (1997) |
| α-Amanitin | ✓ | Haaf and Ward (1996) | ✓ | Andera and Wasylyk (1997) |
| Camptothecin | ✓ | Buckwalter et al. (1996) | ✓ | Ljungman (2000) |
| NTP depletion | ✓ | Grummt and Grummt (1976) | ✓ | Linke et al. (1996) |
| Bleomycin | ✓ | Vazquez-Nin et al. (1979) | ✓ | Pluquet and Hainaut (2001) |
| Heat shock | ✓ | Y.Liu et al. (1996) | ✓ | Ljungman (2000) |
| Hypoxia | ✓ | Yung et al. (1991) | ✓ | Graeber et al. (1994) |
| LMB | – | – | ✓ | Freedman and Levine (1998) |
| MG132 | – | – | ✓ | Klibanov et al. (2001) |
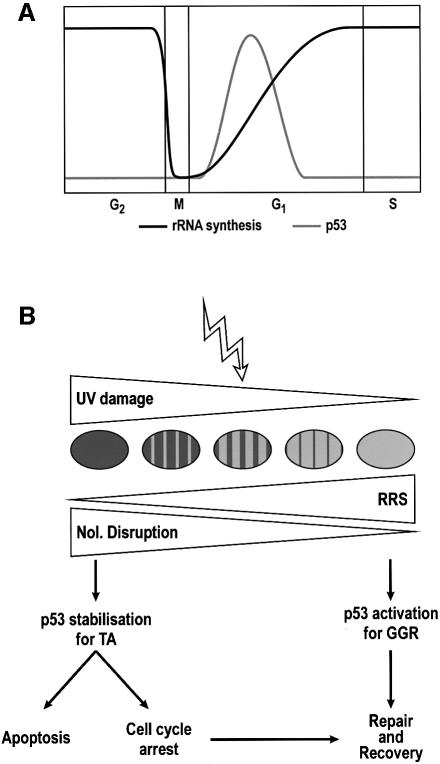
The correlation between impairment of nucleolar function and p53 stabilization can be extended further: mammalian cells lose their nucleoli during mitosis, and full nucleolar functionality, in the form of the maximum level of rRNA synthesis, is not achieved until late in G1 phase (Klein and Grummt, 1999). This period of recovery of nucleolar functionality is precisely the window in which p53 levels are increased during the cell cycle (David-Pfeuty, 1999; see scheme in Figure 5A). Thus, nucleolar disruption is a unifying model that can explain the cell cycle-dependent variation in p53 levels. Moreover, agents that arrest cells in mitosis (e.g. nocodazole) induce a p53 response (Pluquet and Hainaut, 2001) and at the same time prevent nucleolar reformation.
Fig. 5. (A) Schematic representation of the variations of rRNA synthesis rates and p53 levels along the normal cell cycle. (B) Scheme of the p53 response to high and low level DNA damage following UV irradiation. High level DNA damage implies compromise of the majority of the nucleolar population, impairing the total nucleolar function of the nucleus; low level refers to either localized high density damage or low whole area damage, such that the overall nucleolar function is not compromised. Recovery of RNA synthesis (RRS) will be faster the lower the DNA damage is. High level DNA damage stabilizes p53 and induces transactivation of downstream effector genes. Low level damage elicits a p53 response, with low (resting) levels consisting of chromatin relaxation for global NER.
A series of elegant experiments by David-Pfeuty (1999) and David-Pfeuty et al. (2001) demonstrated a correlation between inhibition of cyclin-dependent kinases, nucleolar fragmentation and p53 accumulation. Intriguingly, nucleolar disruption by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; a casein kinase II inhibitor) could be impaired by overexpression of p21 (David-Pfeuty et al., 2001). Transfectants with higher p21 expression levels were more resistant to DRB-induced nucleolar disruption and, for the majority of cells reported (as is apparent in figure 8 in David-Pfeuty et al., 2001), it is clearly noticeable that cells with low p21 levels (high nucleolar disruption) showed high levels of p53 induction by DRB treatment, and vice versa. In the absence of p21 overexpression, DRB induces a rapid and massive p53 stabilization in all cells (David-Pfeuty et al., 2001; and see Figure 1H, this work). Since high p21 cells were refractory to both nucleolar disruption and p53 induction by DRB, the nucleolar disruption model appears to offer a better explanation for p53 induction by DRB than RNA Pol II inhibition (Ljungman et al., 1999).
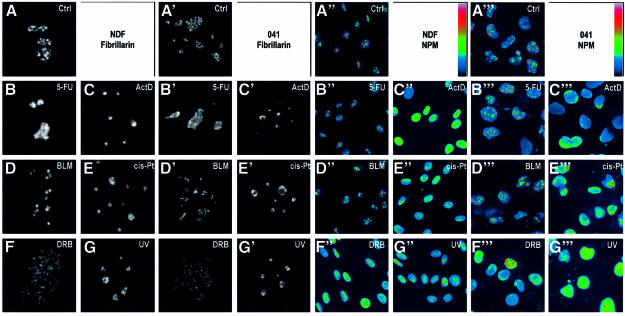
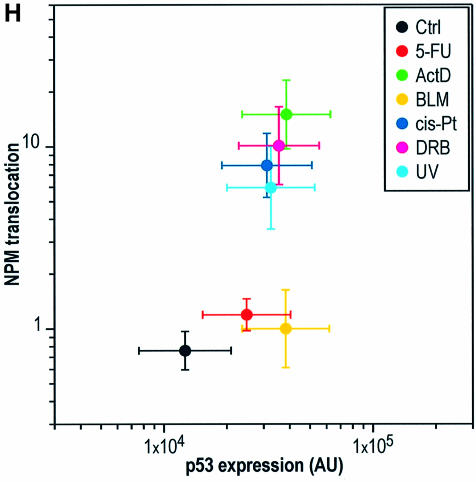
Fig. 1. Nucleolar disruption is independent of p53. (A–G′) Single confocal sections of nuclei of NDFs (A–G) and 041 cells (A′–G′) stained for fibrillarin, 6 h after treatment with each indicated agent. Each image corresponds to a single nucleus. (A′′–G′′′) wide field images of NDFs (A′′–G′′) and 041 cells (A′′′–G′′′) stained for NPM after the same treatments. A pseudo-colour scale (indicated) was applied to each image to highlight all intensity ranges. (H) Dual plot of nuclear p53 expression level (DO-1 staining) and NPM translocation index for NDFs treated for 6 h with the indicated agents.
Thus, a solid bibliographic background supports the nucleolar export model of p53 degradation. In the present work, we aimed to test the validity of the model under three conditions. First, we ensured, for a representative group of p53-inducing stresses, that nucleolar disruption occurs in the absence of p53, thus rejecting the possibility that the correlation might be due to p53 blocking, for example, rRNA synthesis (Cairns and White, 1998; Budde and Grummt, 1999; Zhai and Comai, 2000) and causing the nucleolar disruption. Secondly, using localized UV irradiation of cell nuclei with micropore filters, we demonstrate that cells can tolerate a large amount of DNA damage without inducing a p53 response provided it is localized and the nucleoli are not disrupted. Thus, DNA damage by itself does not cause p53 stabilization. Moreover, we show that the lower minimum response dose (MRD) for p53 induction by UV irradiation displayed by Cockayne syndrome (CS) cells (the basis for the model of p53 stabilization by transcription inhibition; Yamaizumi and Sugano, 1994; Ljungman and Zhang, 1996) also correlates with a lower minimum UV response dose for nucleolar disruption, further confirming that the intermediary between DNA damage and p53 stabilization is nucleolar disruption. Finally, since the model predicts that p53 should be stabilized whenever nucleolar disruption occurs, even in the absence of DNA damage, of phosphorylation inhibition or of metabolic stresses, we tested this prediction by microinjecting antibodies against the nucleolar protein upstream binding factor (UBF). We observed nucleolar disruption, induction of p53 and a low but noticeable induction of p21. We conclude that the nucleolar disruption model of p53 stabilization offers a unifying explanation for the induction of p53 under a wide range of cellular stresses. Our model predicts that a p53 response, rather that being induced, has to be constantly prevented by a fully functional nucleolus.
Results
Nucleolar disruption is independent of p53
We first had to define a criterion for nucleolar disruption. The cellular stresses summarized in Table I have all been shown to disrupt nucleolar function in one form or another. The confocal sections of fibrillarin staining of human normal diploid fibroblasts (NDFs) shown in Figure 1A–G exemplify the dispersion of nucleolar structures produced by some common p53-inducing agents. Figure 1A′–G′ indicates that these nucleolar modifications also occur in cells not expressing p53 (041 fibroblasts derived from Li–Fraumeni cells harbouring a deletion in one p53 allele, which subsequently have lost the remaining wtp53 allele; P.K.Liu et al., 1996). Since different treatments cause different alterations of nucleolar morphology, we decided to employ a more quantifiable marker of nucleolar disruption. For this, a criterion often used is translocation of the nucleolar protein nucleophosmin/B23 (NPM), which has been found to correlate with the cytotoxicity of a number of agents (Chan et al., 1996). NPM translocation from the nucleolus to the nucleoplasm following cell stress is evident in both NDFs and 041 cells, as shown in Figure 1A′′–G′′′. In these images, a pseudo-colour scale (indicated) is used in order to evidence the changes in nucleoplasmic staining, usually much weaker than the nucleolus since translocated NPM is diluted into a larger volume. For some agents such as 5-fluorouracil (5-FU) and bleomycin (BLM), we found that NPM translocation is not a good marker of nucleolar disruption. This is also evidenced by analysing the correlation between NPM translocation and p53 stabilization (fluorescent staining with DO-1 antibody) for different agents (Figure 1H).
NPM translocation has been proposed to retain p53 in the nucleoplasm, thus mediating p53 stabilization (Colombo et al., 2002). While this may occur in a number of stresses, we find that it cannot explain all situations of p53 stabilization, as we have found at least two common agents (5-FU and BLM) which stabilize p53 without extensive NPM translocation, while disrupting the nucleoli (compare Figure 1B, B′, B′′, B′′′, D, D′, D′′, D′′′ and H).
DNA damage does not directly cause p53 stabilization
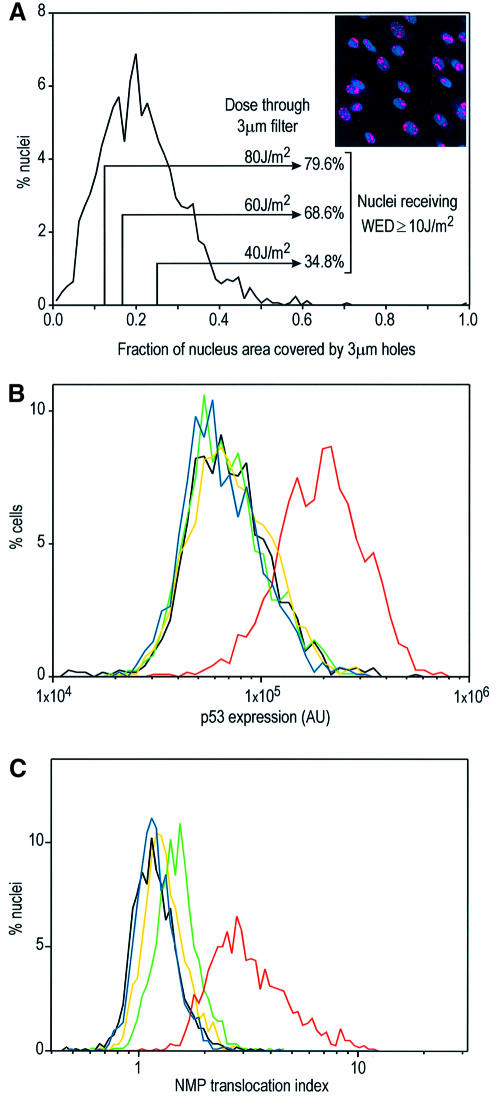
When an individual nucleolus is inactivated by localized UV irradiation, the remaining intact nucleoli within the same nucleus expand such that the whole nucleolar function is not compromised (Zatsepina et al., 1989). UV irradiation can be expected to cause nucleolar disruption in two ways: by direct blockage of rDNA transcription and by global inhibition of RNA Pol II transcription. When UV irradiation is localized, the overall nucleoplasmic transcription is not affected (Mone et al., 2001). We reasoned that if our hypothesis that p53 is stabilized by nucleolar disruption is correct, then localized UV irradiation would not cause p53 stabilization, since few of the nucleoli, if any, would be inactivated. To test this, we UV irradiated NDFs through Isopore filters with 3 µm pores in conditions that ensured that cells received a UV dose equal to or larger than the minimum required for p53 stabilization (10 J/m2, see below). We defined whole-nucleus irradiation equivalent density (WED) as the UV density at which a whole nucleus has to be irradiated in order to receive the same amount of DNA damage as that received by localized irradiation through micropores. To determine the fraction of nuclei that would receive a WED of ≥10 J/m2, we irradiated NDFs through 3 µm filters, immediately fixed them and labelled the areas of damaged DNA with an anti-cyclobutane pyrimidine dimer (CPD) antibody (see Materials and methods), and built a distribution of the fraction of nuclear projected area harbouring DNA damage (Figure 2A). It can be seen in Figure 2A that in these conditions practically all of the nuclei are irradiated on <50% of their area. Since irradiation through Isopore filters consists of 100% transmission through holes and 0% outside, multiplication of the irradiating density by the fraction of exposed nuclear area yields the WED for each nucleus. The percentages of cells receiving WED ≥10 J/m2 are indicated in Figure 2A. Quantitation of p53 levels in cells fixed 6 h after whole-nucleus irradiation at 10 J/m2 indicates a strong p53 stabilization (Figure 2B). UV irradiation through 3 µm Isopore filters at 40, 60 and 80 J/m2 (WED ≥10 J/m2 in 34.8, 68.6 and 79.6% of nuclei, respectively) caused no p53 stabilization (Figure 2B). Non-parametric Kolmogorov–Smirnov (K–S) analysis of the immunofluorescence distributions (Young, 1977) showed no statistical difference between the unirradiated control and all micropore irradiations, even at P = 0.1 (800–1200 nuclei analysed). At this level, histogram simulations indicated that K–S analysis for these sample sizes could detect as low as 7% positives, 5–11 times less than the percentages of cells irradiated at WED ≥10 J/m2. This result indicates that nuclei can withstand high levels of UV DNA damage without p53 stabilization provided irradiation is restricted to a fraction of the nuclear area. Likewise, Figure 2C shows that nucleolar disruption by UV irradiation (NPM translocation) only occurs when the whole nucleus is irradiated, as levels of NPM translocation in nuclei irradiated through micropores are similar to the unirradiated control.
Fig. 2. Effects of micropore irradiation on p53 expression and NPM translocation in NDFs. (A) Distribution of the fraction of irradiated areas on NDF nuclei (n = 1699) observed by the ratio between the area exposed to UV irradiation through 3 µm Isopore filters (irradiated area detected by antibody labelling of photolesions) and the total nuclear projected area (Hoechst 33324). The insert shows an example field of micropore-irradiated nuclei, with CPDs labelled red and nuclei blue (Hoechst). (B) p53 expression levels in NDFs whole-nucleus irradiated at 10 J/m2 (red), micropore irradiated at 40 (green), 60 (yellow) and 80 J/m2 (blue), and non-irradiated (black). The fractions of nuclei receiving WED ≥10 J/m2 (see text) under each micropore irradiation condition are indicated in (A). (C) NPM translocation indext for NDFs irradiated in the same conditions as in (B). Cells in (A) were fixed immediately after irradiation, while cells in (B) and (C) were fixed 6 h after irradiation.
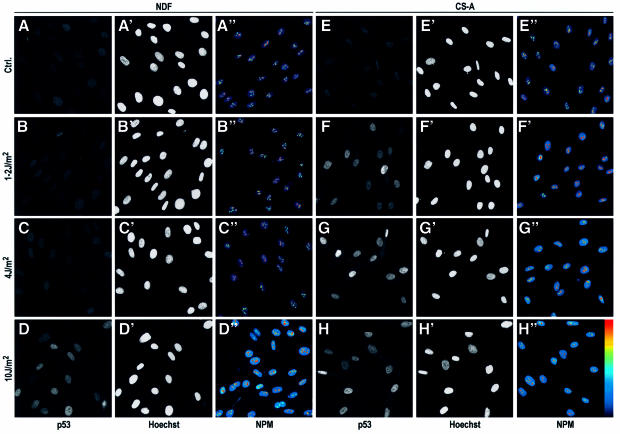
Thus, we conclude that DNA damage per se does not induce p53 stabilization and that nucleolar disruption appears to be the mediator of p53 stabilization in this DNA damage situation. UV photolesions cause RNA Pol II transcription inhibition, and it has been proposed that this inhibition is in turn the cause of p53 stabilization. This concept was mainly derived from the observation that CS cells, which cannot remove photolesions from transcribed genes, have a lower MRD for UV induction of p53 (Yamaizumi and Sugano, 1994; Ljungman and Zhang, 1996). In order for our model of p53 stabilization to hold, a lower MRD for p53 induction should correlate with a lower MRD for NPM translocation. We observed exactly this by irradiation of normal and CS skin fibroblasts at different UV doses (Figure 3). In our system, no p53 induction was observed in NDFs 6 h after UV doses of ≤4 J/m2 (Figure 3), with a few positives appearing at 6 J/m2 (data not shown) and a clear strong response at 10 J/m2, in agreement with previous reports (Yamaizumi and Sugano, 1994; Ljungman and Zhang, 1996). CS cells, on the other hand, showed p53 stabilization at doses as low as 1–2 J/m2, as expected (Figure 3). These MRDs for p53 induction completely correlated with the MRD for NPM translocation (Figure 3). In these experiments, nucleoli of both NDFs and CS cells obviously received the same irradiation doses and in both cell types photolesions in rDNA are expected to be resolved with equal efficiency, since they are repaired by global genomic nucleotide excision repair (NER; Christians and Hanawalt, 1993). This indicates that RNA Pol II inhibition is the major cause of nucleolar disruption by UV irradiation.
Fig. 3. Correlation of MRD for p53 expression and NPM translocation in NDFs and Cockayne syndrome complementation group A (CS-A) fibroblasts. NDFs (A–D) and CS-A cells (E–H) were irradiated at the indicated UV densities, fixed 6 h later and stained for p53 expression (A–D and E–H, with positions of nuclei indicated by Hoechst staining in A′–D′ and E′–H′) or NPM (A′′–D′′ and E′′–H′′). All images are wide-field. Pseudo-colour is used in NPM images.
Direct interference with nucleolar structure/function stabilizes p53
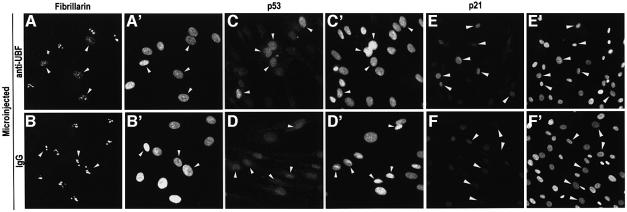
Our model predicts that DNA damage is dispensable for p53 stabilization and can be by-passed by interfering with nucleolar structure/function. To test this prediction, we chose to block nucleolar function by nuclear microinjection of an antibody against UBF, which is required for transcription of rDNA genes (Klein and Grummt, 1999). In these conditions, no obvious DNA damage is introduced. Microinjection of a non-specific antibody causes neither nucleolar disruption nor p53 stabilization (Figure 4B and D). However, anti-UBF caused nucleolar disruption and, as predicted, p53 stabilization (Figure 4A and C). This p53 stabilization was sufficient to cause a noticeable induction of p21, a p53 downstream gene that mediates cell cycle arrest (compare Figure 4E and F). No p21 induction was observed in p53-null cells (data not shown). We conclude that a p53 response can be induced by interfering with nucleolar function in the absence of any genotoxic insult.
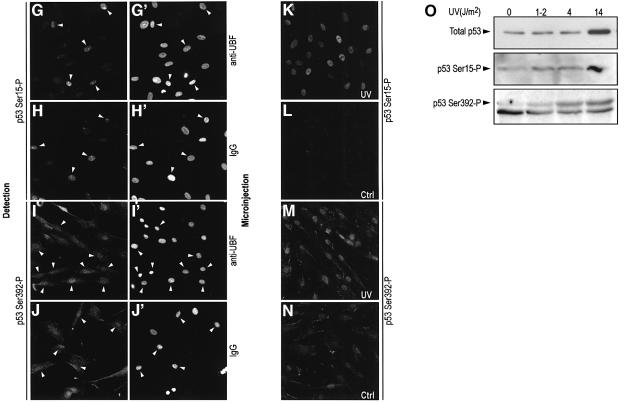
Fig. 4. Effect of nucleolar disruption by microinjection of an anti-UBF antibody in NDFs. (A, C and E) Microinjection of anti-UBF (IgG1) with fibrillarin distribution (A) and p53 expression (C) assayed 6 h post-injection, and p21 expression (E) 18 h post-injection. (B, D and F) Microinjection of control purified mouse IgG1. (A′–F′) Reference Hoechst images. Arrows indicate microinjected cells. Lower magnification was used in p21 images in order to incorporate more cells into the field of view. (G–J) p53 phosphorylation in response to microinjection of anti-UBF (G and I) and non-specific IgG (H and J). p53 phosphorylation was detected for Ser15 (G and H) and Ser392 (I and J). (G′–J′) reference Hoechst images. (K–N) Phosphorylation at Ser15 and Ser392 in control and UV-irradiated NDFs (14 J/m2) as indicated. (O) Western blot analysis of p53 expression and phosphorylation (Ser15 and Ser392) in control and UV-irradiated NDFs (1–2, 4 and 14 J/m2). Cells were harvested 6 h post-irradiation.
To resolve further the issue of p53 stabilization by nucleolar disruption as opposed to post-translational modification of p53, we analysed whether anti-UBF microinjection might specifically induce p53 phosphorylation at Ser15. Figure 4G shows that anti-UBF microinjection does indeed induce Ser15 phosphorylation. However, as shown in Figure 4H, microinjection of non-specific purified IgG also causes the same phosphorylation. Since non-specific IgG does not cause p53 stabilization (Figure 4D), we conclude that the observed phosphorylation at Ser15 of p53 is a response to the microinjection stress and does not cause p53 stabilization. We also looked at p53 phosphorylation at Ser392, a residue specifically phosphorylated in response to UV irradiation (Meek, 1999). Figure 4I and J shows that microinjection of anti-UBF or non-specific IgG, respectively, does not cause any detectable change in the phosphorylation status of Ser392, which is nevertheless phosphorylated in response to UV irradiation (Figure 4M and N). Thus, for Ser392, phosphorylation is uncoupled from p53 stabilization induced by anti-UBF disruption of the nucleolus.
Lack of participation of phosphorylation in p53 stabilization was demonstrated further by western blot analysis of total cell extracts from NDFs untreated or UV irradiated at 1–2, 4 and 14 J/m2. In agreement with Figure 3 and previous reports (Yamaizumi and Sugano, 1994; Ljungman and Zhang, 1996), levels of p53 are only increased by UV doses >10 J/m2 (Figure 4O, upper panel). However, irradiation at lower UV doses (1–2 and 4 J/m2) causes detectable phosphorylation of Ser15 and Ser392 (Figure 4O, middle and lower panels). Taken together, these data indicate that p53 phosphorylation and stabilization are separable.
Discussion
The model for p53 stabilization proposed in the present work, namely prevention of p53 degradation by nucleolar disruption, is, to our knowledge, the only model that can provide a unifying and coherent explanation for the action of all known p53-stabilizing agents. The model is robust in that all p53 inducers also cause nucleolar disruption, and no known p53-stabilizing agent contradicts it. Importantly, cell cycle-dependent variations in p53 levels can be accounted for by the model (see Introduction). As discussed in the Introduction, current knowledge of the biology of p53 fully agrees with a model where the nucleolus or nucleolar components participate in the Mdm2-mediated export and degradation of p53.
Maintenance of p53 levels in the nucleolar disruption model
The model proposed here presents a radically new view of the p53 response: rather than being actively induced from a ‘default’ resting level, a p53 response has to be constantly prevented from occurring, and the ‘default’ condition is p53 stabilization unless a fully functional nucleolus promotes its degradation. The nucleolar disruption mechanism is intrinsically safer than any inductive mechanism which, if failing by mutation, etc., would leave the cell incapable of reacting to stresses. This way of using the nucleolus as a stress sensor is analogous to the ‘dead man’s foot’ safety system: a p53 response will occur unless the nucleolus is constantly capable of promoting its degradation.
An important implication of the model is that nucleolar disruption or interference with nucleolar function frequently associated with viral infections (Hiscox, 2002) will cause p53 stabilization. This explains why it is important for many viruses to inactivate p53, even when a downstream effector of cell cycle arrest such as Rb is also inactivated or, alternatively, in the absence of ARF.
If the loss of the p53 degradation capacity is an intrinsic property of a stressed cell nucleus, then there is no need to postulate post-translational modifications of p53 for its stabilization. This agrees with the number of observations that question the role of covalent modifications for p53 stabilization and indicate that the main player is the Mdm2 protein (Bottger et al., 1997; Meek, 1999; Blattner et al., 1999; Kaeser and Iggo, 2002). Likewise, unlocking the transactivation capability of stabilized p53 may also not require covalent modification, with the rise in p53 levels being sufficient (Kristjuhan and Maimets, 1995; Ljungman, 2000; and references therein). In agreement with this, our data on p53 stabilization by direct nucleolar disruption (Figure 4) indicate that p53 phosphorylation and stabilization are separable, with the latter only occurring when nucleoli are disrupted.
Post-translational modifications, however, may still have a role in fine tuning the p53 response and perhaps in the decision between cell cycle arrest and apoptosis. However, the recent findings of Yin et al. (2002) indicate that an alternative p53 translation product lacking the first transactivation domain (called p53/47) may be produced under Mdm2 control. In this model, the balance between synthesis of full-length p53 and p53/47 would decide the set of genes to be transactivated. This model requires p53 binding to ribosomes and its own mRNA in the cytoplasm, which has in fact been reported to happen (Fontoura et al., 1997). It is thus possible that two p53 export pathways may exist: one for p53 degradation with p53 ubiquitylated by Mdm2, exposing its NES, and another with p53 bound to its own mRNA, in agreement with the similarities with HIV Rev (Roth et al., 1998).
While we found nucleolar disruption to be independent of p53 (Figure 1), it is still possible that certain p53 effects such as inhibition of rRNA synthesis (Cairns and White, 1998; Budde and Grummt, 1999; Zhai and Comai, 2000) or mobilization of nucleolar proteins (Daniely et al., 2002) can prevent recovery of nucleolar functionality. How then can the p53 response be limited? The Mdm2 protein, whose transcription is promoted by p53 can interact with p53, inhibiting its transactivation capacity (Blaydes et al., 1997; Meek, 1999; Ljungman, 2000). However, it is possible that in a situation of nucleolar disruption, p53 degradation might be performed by nuclear proteasomes, as Shirangi et al. (2002) have shown to occur in the downregulation of a p53 response.
Stabilization-dependent and -independent functions of p53
We recently have reported that p53 participates in NER by promoting the chromatin relaxation required for lesion detection in the global genome (Rubbi and Milner, 2003). We reported that the chromatin relaxation function of p53 in NDFs could be triggered at UV densities of 4 J/m2 and also by micropore UV irradiation. Here we show that in those conditions, the p53 protein is not stabilized (Figures 2, 3 and 4O), yet these low UV irradiation densities induce p53 phosphorylation (Figure 4O). We conclude that the role of p53 in repair of DNA damage does not require p53 stabilization, in contrast to induction of cell cycle arrest and apoptosis for which p53 levels have to be elevated. This implies that the DNA repair and the cell cycle arrest/apoptosis functions of p53 are separable and take two different pathways: low level DNA damage will activate the NER function of p53, while only a DNA damage extensive enough to compromise nucleolar function will result in cell cycle arrest or apoptosis (Figure 5B). Thus, the p53 stabilization model presented here, in conjunction with our earlier report of p53-dependent DNA repair in non-stabilizing conditions for p53, offers an explanation for the cellular decision between repair or death.
Tumours developed in p53 heterozygous individuals (both Li–Fraumeni patients and p53+/– mice) have a surprisingly low frequency of loss of heterozygosity (LOH) for a gene that is expected to behave as a tumour suppressor (Varley et al., 1997; Venkatachalam and Donehower, 1998). This suggests that tumour development in haploinsufficient individuals may be related to a DNA repair deficiency, rather than to an impairment of cell cycle arrest/apoptosis function. In fact, there is evidence that NER efficiency can be affected by p53 haploinsufficiency (Ford and Hanawalt, 1995; Abrahams et al., 1998). Our demonstration that the DNA repair function of p53 can operate with normal ‘resting’ levels of p53 suggests that NER could be highly sensitive to p53 haploinsufficiency, while functions dependent on p53 stabilization may appear normal.
Induction of a p53 response without DNA damage
The model for p53 stabilization proposed here opens up the possibility of identifying a number of potential targets for nucleolar disruption and thus for p53 induction. A p53 response can be induced by nucleolar disruption without resorting to DNA-damaging agents, as indicated in Figure 4. This is a tempting avenue for development of anti-tumour therapies, since it avoids the principal problem of DNA damage-based therapies of inducing secondary tumours due to their intrinsic carcinogenic nature. We show that microinjection of monoclonal antibodies is a viable means of identifying potential targets for impairment of nucleolar function and p53 stabilization. As demonstrated by David-Pfeuty et al. (2001), nucleolar disruption and p53 response can be triggered by kinase inhibitors, which implies potential pharmacological exploitation of the model. In addition, a recent proteomic analysis of isolated nucleoli revealed ∼250 proteins, 30% of which were previously unknown, with a composition varying depending on cell growth conditions (Andersen et al., 2002). It may thus be possible that a number of tumour-specific nucleolar proteins could be found that can be used for triggering therapeutically useful p53 responses by nucleolar disruption.
Materials and methods
Cells
GM00038 human skin NDFs were obtained from Coriell Repositories (Camden, NJ). The MDAH041 cell line and CS complementation group A (CS-A) fibroblasts were gifts from Drs Michael Tainsky and Alan Lehmann, respectively. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) + 15% fetal calf serum (FCS).
Micropore irradiation
A range of Isopore filters (Millipore) were tested and 3 µm filters were chosen as they deposit micropores on practically all of the nuclei and the fraction of the projected nuclear area exposed rarely exceeds 0.5 (see Figure 2A). Irradiations at densities of ≥60–80 J/m2 followed by staining with anti-photoproduct antibodies revealed a low but significant UV transmittance through the membrane. For this reason, we coated the filters with aluminium prior to use and the problem was eliminated. UV fluencies were measured using a radiometer (UVP).
Immunofluorescence
Cells grown in coverslips were washed with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde and blocked with PBS + 10% of serum of the same origin as the secondary antibodies. Anti-p53 (DO-1) and -p21 antibodies (from Santa Cruz), anti-NPM (Upstate) and anti-fibrillarin antibodies (Cytoskeleton) were all used at a 1:100 dilution. Secondary antibodies were labelled with either Cy3 (Jackson ImmunoResearch) or fluorescein isothiocyanate (FITC; Sigma). Samples were mounted in Mowiol supplemented with DABCO and Hoechst 33342.
Microscopy and image analysis
Confocal imaging was performed with a Carl Zeiss LSM410 confocal system using a ×63 1.4 NA planapochromatic objective. Wide field imaging was performed with a Carl Zeiss Axiovert 200 equipped with an AxioCam HR, using either ×63 or ×40 oil immersion objectives. For quantitation of total nuclear fluorescence, Hoechst images were binarized by semi-automated thresholding and morphing, and expression levels were integrated inside those nuclear masks. For NPM translocation, total NPM was quantitated as before and, from the NPM images, nucleoli were semi-automatically thresholded in order to generate nucleolar masks. Translocation indices were expressed as total nucleoplasmic fluorescence/total nucleolar fluorescence. In all cases, images were corrected for shading and glare before analysis. All operations were performed using programs written in-house using the LSM410 software (Carl Zeiss).
Microinjection
Microinjections were performed as described (Rubbi and Milner, 2003) using a purified anti-UBF mouse monoclonal IgG1 antibody (Santa Cruz) and purified mouse IgG1 (Sigma). All immunoglobulins were injected at a concentration of 2–3 mg/ml. Injected cells and expression of the protein of interest (fibrillarin, p53, p53Ser15-P, p53Ser392-P or p21) were identified by dual labelling of the injected and staining antibodies.
Western blot
NDFs were cultured in 10 cm Petri dishes, irradiated and harvested 6 h later. Total cell lysates were loaded on 10% SDS–polyacrylamide gels at equal protein concentrations (DC Protein Assay, BioRad, Hemel Hempstead, UK), run and transferred to nitrocellulose membranes. Membranes were probed with either DO-1 antibody, anti-p53Ser15-P rabbit polyclonal (Santa Cruz) or anti-p53Ser392-P mouse monoclonal antibodies (AutogenBioclear, Calne, UK), followed by horseradish peroxidase (HRP)-labelled secondary antibody (Dako, Ely, UK). Blocking and chemiluminescence solutions (Roche Diagnostics, Lewes, UK) were used as indicated by the manufacturer.
Acknowledgments
Acknowledgements
We are grateful to Meg Stark for aluminium coating the micropore filters, to Drs Michael Tainsky and Alan Lehmann for providing the 041 and CS-A cells, respectively, and to Andrei Okorokov for valuable discussions. This work was supported by a Yorkshire Cancer Research core funding to J.M.
References
- Abrahams P.J. et al. (1998) Impaired DNA repair capacity in skin fibroblasts from various hereditary cancer-prone syndromes. Mutat. Res., 407, 189–201. [DOI] [PubMed] [Google Scholar]
- Andera L. and Wasylyk,B. (1997) Transcription abnormalities potentiate apoptosis of normal human fibroblasts. Mol. Med., 3, 852–863. [PMC free article] [PubMed] [Google Scholar]
- Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Blattner C., Tobiasch,E., Litfen,M., Rahmsdorf,H.J. and Herrlich,P. (1999) DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene, 18, 1723–1732. [DOI] [PubMed] [Google Scholar]
- Blaydes J.P., Gire,V., Rowson,J.M. and Wynford-Thomas,D. (1997) Tolerance of high levels of wild-type p53 in transformed epithelial cells dependent on auto-regulation by mdm-2. Oncogene, 14, 1859–1868. [DOI] [PubMed] [Google Scholar]
- Bottger A., Bottger,V., Sparks,A., Liu,W.L., Howard,S.F. and Lane,D.P. (1997) Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol., 7, 860–869. [DOI] [PubMed] [Google Scholar]
- Boyd S.D., Tsai,K.Y. and Jacks,T. (2000) An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nature Cell Biol., 2, 563–568. [DOI] [PubMed] [Google Scholar]
- Buckwalter C.A., Lin,A.H., Tanizawa,A., Pommier,Y.G., Cheng,Y.C. and Kaufmann,S.H. (1996) RNA synthesis inhibitors alter the subnuclear distribution of DNA topoisomerase I. Cancer Res., 56, 1674–1681. [PubMed] [Google Scholar]
- Budde A. and Grummt,I. (1999) p53 represses ribosomal gene expression. Oncogene, 19, 1119–1124. [DOI] [PubMed] [Google Scholar]
- Cairns C.A. and White,R.J. (1998) p53 is a general repressor of RNA polymerase III transcription. EMBO J., 17, 3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.K., Qi,Y., Amley,J. and Koller,C.A. (1996) Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett., 100, 191–197. [DOI] [PubMed] [Google Scholar]
- Christians F.C. and Hanawalt,P.C. (1993) Lack of transcription-coupled repair in mammalian ribosomal RNA genes. Biochemistry, 32, 10512–10518. [DOI] [PubMed] [Google Scholar]
- Colombo E., Marine,J.C., Danovi,D., Falini,B. and Pelicci,P.G. (2002) Nucleophosmin regulates the stability and transcriptional activity of p53. Nature Cell Biol., 4, 529–533. [DOI] [PubMed] [Google Scholar]
- Daniely Y., Dimitrova,D.D. and Borowiec,J.A. (2002) Stress-dependent nucleolin mobilization mediated by p53–nucleolin complex formation. Mol. Cell. Biol., 22, 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Pfeuty T. (1999) Potent inhibitors of cyclin-dependent kinase 2 induce nuclear accumulation of wild-type p53 and nucleolar fragmentation in human untransformed and tumor-derived cells. Oncogene, 18, 7409–7422. [DOI] [PubMed] [Google Scholar]
- David-Pfeuty T., Nouvian-Dooghe,Y., Sirri,V., Roussel,P. and Hernandez-Verdun,D. (2001) Common and reversible regulation of wild-type p53 function and of ribosomal biogenesis by protein kinases in human cells. Oncogene, 20, 5951–5963. [DOI] [PubMed] [Google Scholar]
- Fontoura B.M., Sorokina,E.A., David,E. and Carroll,R.B. (1992) p53 is covalently linked to 5.8S rRNA. Mol. Cell. Biol., 12, 5145–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura B.M., Atienza,C.A., Sorokina,E.A., Morimoto,T. and Carroll,R.B. (1997) Cytoplasmic p53 polypeptide is associated with ribosomes. Mol. Cell. Biol., 17, 3146–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J.M. and Hanawalt,P.C. (1995) Li–Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV resistance. Proc. Natl Acad. Sci. USA, 92, 8876–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman D.A. and Levine,A.J. (1998) Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol., 18, 7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K. and Jacob,S.T. (1994) Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res., 54, 632–636. [PubMed] [Google Scholar]
- Graeber T.G., Peterson,J.F., Tsai,M., Monica,K., Fornace,A.J. and Giaccia,A.J. (1994) Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol. Cell. Biol., 14, 6264–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. and Grummt,F. (1976) Control of nucleolar RNA synthesis by the intracellular pool sizes of ATP and GTP. Cell, 7, 447–453. [DOI] [PubMed] [Google Scholar]
- Haaf T. and Ward,D.C. (1996) Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure and dispersion of chromosomal domains. Exp. Cell Res., 224, 163–173. [DOI] [PubMed] [Google Scholar]
- Hiscox J.A. (2002) The nucleolus—a gateway to viral infection? Arch. Virol., 147, 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. and Carmo-Fonseca,M. (1998) Cisplatin inhibits synthesis of ribosomal RNA in vivo. Nucleic Acids Res., 26, 2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser M.D. and Iggo,R.D. (2002) Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc. Natl Acad. Sci. USA, 99, 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J. and Grummt,I. (1999) Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl Acad. Sci. USA, 96, 6096–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klibanov S.A., O’Hagan,H.M. and Ljungman,M. (2001) Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J. Cell Sci., 114, 1867–1873. [DOI] [PubMed] [Google Scholar]
- Kristjuhan A. and Maimets,T. (1995) Protein p53 modulates transcription from a promoter containing its binding site in a concentration-dependent manner. Eur. J. Biochem., 234, 827–831. [DOI] [PubMed] [Google Scholar]
- Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Linke S.P., Clarkin,K.C., Di Leonardo,A., Tsou,A. and Wahl,G.M. (1996) A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev., 10, 934–947. [DOI] [PubMed] [Google Scholar]
- Liu P.K., Kraus,E., Wu,T.A., Strong,L.C. and Tainsky,M.A. (1996) Analysis of genomic instability in Li–Fraumeni fibroblasts with germline p53 mutations. Oncogene, 12, 2267–2278. [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liang,S. and Tartakoff,A.M. (1996) Heat shock disassembles the nucleolus and inhibits nuclear protein import and poly(A)+ RNA export. EMBO J., 15, 6750–6757. [PMC free article] [PubMed] [Google Scholar]
- Ljungman M. (2000) Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia, 2, 208–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M. and Zhang,F. (1996) Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene, 13, 823–831. [PubMed] [Google Scholar]
- Ljungman M., Zhang,F., Chen,F., Rainbow,A.J. and McKay,B.C. (1999) Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene, 18, 583–592. [DOI] [PubMed] [Google Scholar]
- Marechal V., Elenbaas,B., Piette,J., Nicolas,J.C. and Levine,A.J. (1994) The ribosomal L5 protein is associated with mdm-2 and mdm-2–p53 complexes. Mol. Cell. Biol., 14, 7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W. (1999) Mechanisms of switching on p53: a role for covalent modification? Oncogene, 18, 7666–7675. [DOI] [PubMed] [Google Scholar]
- Mone M.J., Volker,M., Nikaido,O., Mullenders,L.H., van Zeeland,A.A., Verschure,P.J., Manders,E.M. and van Driel,R. (2001) Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep., 2, 1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestov D.G., Strezoska,Z. and Lau,L.F. (2001) Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol., 21, 4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluquet O. and Hainaut,P. (2001) Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett., 174, 1–15. [DOI] [PubMed] [Google Scholar]
- Pritchard D.M., Watson,A.J., Potten,C.S., Jackman,A.L. and Hickman,J.A. (1997) Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc. Natl Acad. Sci. USA, 94, 1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Dobbelstein,M., Freedman,D.A., Shenk,T. and Levine,A.J. (1998) Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J., 17, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner,J. (2003) p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J., 22, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A. and Carroll,R.B. (1991) The tumor suppressor p53 is bound to RNA by a stable covalent linkage. Mol. Cell. Biol., 11, 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Oft,M., Dascher,C., Schebesta,M., Rosorius,O., Jaksche,H., Dobrovnik,M., Bevec,D. and Hauber,J. (1998) Interaction of the HIV-1 rev cofactor eukaryotic initiation factor 5A with ribosomal protein L5. Proc. Natl Acad. Sci. USA, 95, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. and Weber,J.D. (2000) The ARF/p53 pathway. Curr. Opin. Genet. Dev., 10, 94–99. [DOI] [PubMed] [Google Scholar]
- Shirangi T.R., Zaika,A. and Moll,U.M. (2002) Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J., 16, 420–422. [DOI] [PubMed] [Google Scholar]
- Stommel J.M., Marchenko,N.D., Jimenez,G.S., Moll,U.M., Hope,T.J. and Wahl,G.M. (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J., 18, 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott F.J. et al. (1998) The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J., 17, 5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W. and Levine,A.J. (1999) p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl Acad. Sci. USA, 96, 6937–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F. and Kutay,U. (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci., 116, 2409–2419. [DOI] [PubMed] [Google Scholar]
- Varley J.M., Evans,D.G. and Birch,J.M. (1997) Li–Fraumeni syndrome—a molecular and clinical review. Br. J. Cancer, 76, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Nin G.H., Echeverria,O.M. and Pedron,J. (1979) Ultrastructural and autoradiographic study of the effects of bleomycin on the interphase nucleus of cultured normal cells. Cancer Res., 39, 4218–4223. [PubMed] [Google Scholar]
- Venkatachalam S. and Donehower,L.A. (1998) Murine tumor suppressor models. Mutat. Res., 400, 391–407. [DOI] [PubMed] [Google Scholar]
- Weinstein J.N. et al. (1997) An information-intensive approach to the molecular pharmacology of cancer. Science, 275, 343–349. [DOI] [PubMed] [Google Scholar]
- Xirodimas D.P., Stephen,C.W. and Lane,D.P. (2001) Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res., 270, 66–77. [DOI] [PubMed] [Google Scholar]
- Yamaizumi M. and Sugano,T. (1994) U.V.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene, 9, 2775–2784. [PubMed] [Google Scholar]
- Yin Y., Stephen,C.W., Luciani,M.G. and Fahraeus,R. (2002) p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nature Cell Biol., 4, 462–467. [DOI] [PubMed] [Google Scholar]
- Young I.T. (1977) Proof without prejudice: use of the Kolmogorov–Smirnov test for the analysis of histograms from flow systems and other sources. J. Histochem. Cytochem., 25, 935–941. [DOI] [PubMed] [Google Scholar]
- Yung B.Y., Yang,Y.H. and Bor,A.M. (1991) Nucleolar protein B23 translocation after deferoxamine treatment in a human leukemia cell line. Int. J. Cancer., 48, 779–784. [DOI] [PubMed] [Google Scholar]
- Zatsepina O.V., Voronkova,L.N., Sakharov,V.N. and Chentsov,Y.S. (1989) Ultrastructural changes in nucleoli and fibrillar centers under the effect of local ultraviolet microbeam irradiation of interphase culture cells. Exp. Cell Res., 181, 94–104. [DOI] [PubMed] [Google Scholar]
- Zhai W. and Comai,L. (2000) Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol., 20, 5930–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]