Abstract
The energetic contributions of hydrogen bonding to protein folding are still unclear, despite over 70 years of study. This is due partly to the difficulty of extracting thermodynamic information about specific interactions from protein mutagenesis data, and partly to the context dependence of hydrogen bond strengths. Herein, we test the hypothesis that hydrogen bond strengths depend on the polarity of their microenvironment, with stronger hydrogen bonds forming in non-polar surroundings. Double mutant thermodynamic cycle analysis using a combination of amide-to-ester backbone mutagenesis and traditional side chain mutagenesis revealed that hydrogen bonds can be stronger by up to 1.2 kcal mol−1 when they are sequestered in hydrophobic surroundings than when they are solvent exposed. Such large coupling energies between hydrogen bond strengths and local polarity suggest that the context dependence of hydrogen bond strengths must be accounted for in any comprehensive account of the forces responsible for protein folding.
Introduction
A quantitative understanding of the forces that enable and oppose protein folding is required to fully appreciate this complex process. Conformational entropy, the hydrophobic effect, and hydrogen bonding terms dominate the thermodynamics of protein folding1. Amongst these, the contribution of hydrogen bonding to protein folding thermodynamics may be the least clear and most contentious. Hydrogen bonding was first believed to be the primary source of protein native state stability2. However, this hypothesis fell out of favor when the hydrophobic effect was introduced3. It is now clear that both the hydrophobic effect and hydrogen bonding contribute to the stability of protein native states, and we are beginning to better understand the latter, especially in light of recent findings on the solvation of the protein backbone4–6.
Hydrogen bonds form between a hydrogen atom attached to an electronegative atom (the hydrogen bond donor; e.g., a peptide backbone N-H) and another electronegative atom (the hydrogen bond acceptor; e.g., a peptide backbone carbonyl oxygen). Hydrogen bonding per se is an unambiguously thermodynamically favorable process. For example, hydrogen bonds formed in non-polar solvents between molecules of N-methyl acetamide (a model for the protein backbone) have enthalpies of formation of around −4 kcal mol−1 (refs. 7,8). The dispute over the contribution of hydrogen bonding to protein folding thermodynamics has two origins. First, hydrogen bond formation between protein donors and acceptors does not yield a net gain of hydrogen bonds upon protein folding, because the donors and acceptors were initially hydrogen bonded to water7. Thus, intermolecular protein donor–water and protein acceptor–water hydrogen bonds are merely exchanged for intramolecular protein donor–protein acceptor hydrogen bonds. The net contribution of backbone-backbone, backbone–side chain and side chain–side chain hydrogen bonding to protein folding thermodynamics therefore depends on the difference in strengths between protein-water and protein-protein hydrogen bonds. Second, hydrogen bond strengths are context dependent; i.e., they appear to depend on the polarity of the microenvironment in which they are formed, as expected given the largely electrostatic nature of hydrogen bonds9. A connection between hydrogen bond strength and microenvironment polarity was first proposed by Nemethy et al.10, and the theoretical grounds for this connection has since been explored extensively11,12. A recent survey of the lengths of surface vs. buried hydrogen bonds in soluble proteins and integral membrane proteins has recently provided experimental evidence for the polarity dependence of hydrogen bond strengths13. Hydrogen bonds at the surfaces of soluble proteins were found to be the longest, averaging 2.08 Å. Buried hydrogen bonds in both soluble and integral membrane proteins were shorter (2.02 Å), and interfacial hydrogen bonds in integral membrane proteins, which are exposed to the interiors of lipid membranes, were the shortest (1.98 Å). Thus, hydrogen bond lengths appear to decrease as the environment becomes more non-polar, suggesting that hydrogen bonds are stronger in non-polar environments. Further anecdotal evidence for hydrogen bond strengths increasing as local polarity decreases comes from studies employing mutagenesis approaches14–21. For example, amide-to-ester mutations (in which an amide in the protein backbone is replaced by an ester, eliminating the donor NH and weakening the acceptor CO) in the Pin WW domain, a β-sheet rich, 34-residue domain from the human Pin 1 protein, were most destabilizing when an amide that formed a buried hydrogen bond was perturbed19,20.
The magnitude of the context dependence of hydrogen bond strengths has important ramifications for quantifying the forces that drive protein folding. Herein, we report a study designed to prospectively scrutinize the hypothesis that hydrogen bond strengths depend on the polarity of their microenvironment, and to quantify the extent of this dependence, if observed. By combining protein backbone and side chain mutagenesis to enable double mutant thermodynamic cycle analysis, we demonstrate that decreasing the polarity of the microenvironment of a hydrogen bond can increase its strength by up to 1.2 kcal mol−1 in both helix- (the B domain of protein A from Staphylococcus aureus) and sheet- (the WW domain from the human Pin1 protein) rich contexts. This result indicates that the context dependence of hydrogen bond strengths in protein tertiary structures could be critical for protein structure prediction, protein engineering, and any comprehensive account of the forces responsible for protein folding.
Results
Design
To quantify the influence of local hydrophobicity on hydrogen bond strengths, we selected backbone amides that formed hydrogen bonds that could be buried or exposed by mutating neighboring side chains. As mentioned above, we employed two well behaved model proteins in this undertaking, namely the human Pin WW domain and the B domain of protein A from Staphylococcus aureus (hereafter protein A). These proteins have been thoroughly characterized19,20,22–31, they are both two-state folders, and are small enough to be synthesized chemically. The Pin WW domain is predominantly a β-sheet protein, whereas protein A is largely α-helical.
Extracting the energy of a specific interaction from the difference in folding free energies between a protein that has the interaction in question and a mutant in which it has been abolished can be difficult32. Corrections have to be applied to the folding free energy difference to account for other effects of the mutation, including changes in desolvation energy, secondary structure propensity, introduction of cavities (when a group is deleted from the interior of a protein), and so on14,21,32. Fortunately, we are interested primarily in the influence of microenvironments on hydrogen bond strengths and not in hydrogen bond strengths themselves, which enables us to take advantage of double mutant thermodynamic cycle analysis32–34.
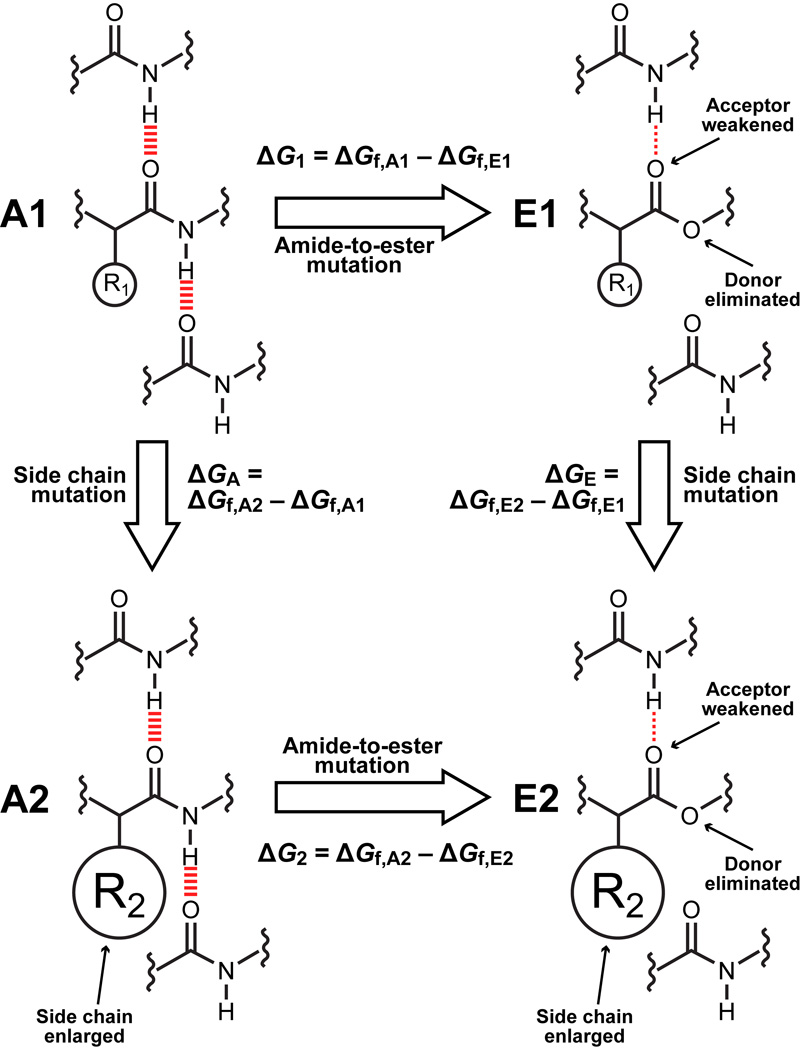
Double mutant cycles are commonly used to assess energetic coupling between two side chains in a protein32–34. The design of a double mutant cycle for extracting the context dependence of hydrogen bonds is depicted in Fig. 1. A1 and A2 represent all-amide proteins; A2 always has a bulkier hydrophobic side chain proximal to the hydrogen bond of interest than A1. E1 and E2 are proteins harboring a mutation of the protein backbone that perturbs the hydrogen bond of interest. Several types of backbone-modified proteins have been studied, including those incorporating β-amino acids35–38, flexible dipeptide mimics like 5-aminopentanoic acid23, E-olefin isosteres39–43, and several others44. For our purposes, however, amide-to-ester mutants of proteins provide the best combination of synthetic ease and hydrogen bond perturbation21,45. Thus, E1 and E2 have a single amide-to-ester backbone mutation that eliminates the hydrogen bond of interest. Note that E1 and E2 bear the same side chains as A1 and A2, respectively. The folding free energy difference between A1 and E1 (ΔG1) primarily reflects the energy of the eliminated hydrogen bond, although, as noted above, it likely is influenced by other factors as well21,42,43. Analogously, the folding energy difference between A2 and E2 (ΔG2) primarily reflects the energy of the same hydrogen bond, but in a less polar microenvironment because of the larger hydrophobic side chain. If ΔG1 ≠ ΔG2, we can conclude that the strength of the hydrogen bond is different in the presence of the different side chains on A1 and A2. We define ΔΔG2–1 = ΔG2 – ΔG1 as the “thermodynamic coupling energy” between the pairs of mutants A1 and E1, and A2 and E2. If negative, ΔΔG2–1 indicates that the hydrogen bond perturbed by the amide-to-ester mutation is stronger in the less polar microenvironment provided by the bulkier side chain in A2 (however, see below for a discussion of an alternative interpretations).
Figure 1. Double mutant cycle design.
Amide-to-ester mutation of the backbone perturbs hydrogen bonding while traditional side chain mutation of a nearby residue perturbs microenvironment polarity. The differences in the free energies of folding of the pairs of mutants A1/E1 and A2/E2, ΔG1 = ΔGf,A1 – ΔGf,E1 and ΔG2 = ΔGf,A2 – ΔGf,E2 respectively, primarily (but not exclusively) reflect the strength of the hydrogen bond lost in the context of a smaller, less hydrophobic side chain (A1/E1) or a larger, more hydrophobic side chain (A2/E2). The differences in the free energies of folding of the pairs of mutants A2/A1 and E2/E1, ΔGA = ΔGf,A2 – ΔGf,A1 and ΔGE = ΔGf,E2 – ΔGf,E1 respectively, primarily reflect the effect of the side chain on folding free energy in the presence or absence of the hydrogen bond(s) formed by the amide of interest. The thermodynamic coupling energy, ΔΔG2–1 = ΔG2 – ΔG1 = ΔGA – ΔGE reflects the effect of the microenvironment on hydrogen bond strength (barring a structural rearrangement in the amide-to-ester mutants): if it is negative, the hydrogen bond is stronger in the more hydrophobic microenvironment; if it is positive, the opposite is true; finally, if it is 0, then either the hydrogen bond is unaffected by its microenvironment or its microenvironment does not change upon folding.
Stronger hydrogen bonds in nonpolar environments: Pin WW domain
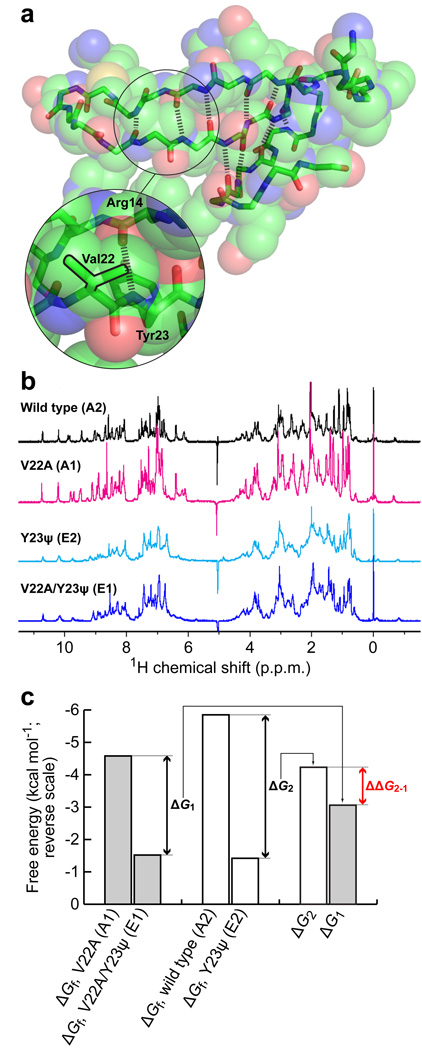
A complete set of scanning backbone mutagenesis data for the Pin WW domain reveals that the NH group of Tyr23 forms a strong hydrogen bond with the CO of Arg14 (Fig. 2a)19,20. This hydrogen bond is partially buried by the adjacent isopropyl side chain of Val22. A double mutant thermodynamic cycle analysis, as outlined in Fig 1, was used to scrutinize the sensitivity of the hydrogen bond between the Tyr23 amide NH and the Arg14 amide CO to its microenvironment. In this double mutant cycle, the hydrogen bond of interest was perturbed using an amide-to-ester mutation at Tyr23 and the microenvironment was perturbed by mutating Val22 to Ala. However, it is known that an amide-to-ester mutation at Tyr23 seriously compromises native state stability of the Pin 1 WW domain19,20. To circumvent this problem, the 6-residue loop 1 sequence (–SRSSGR–) in the Pin WW domain was replaced with the 5-residue loop 1 sequence (–SADGR–) from the Formin-binding protein (FBP) WW domain. This modification enhances the thermodynamic stability of the native state by more than 2 kcal mol−1 (ref. 26), enabling the amide-to-ester mutant to remain folded. Thus, the double mutant cycle employed the loop-1–modified wild-type Pin WW domain with the Val22 side chain as A2, and the V22A side chain mutant as A1. The Y23ψ amide-to-ester mutation, where ψ denotes the α-hydroxy acid equivalent of Tyr, in the context of Val22 served as E2. Finally, the double mutant V22A/Y23ψ served as E1.
Figure 2. Environmental dependence of hydrogen bond strength in the loop-1–modified Pin WW domain at Tyr23.
(a) Structure of the Pin WW domain (pdb ID 1PIN, rendered using PyMol) shown as space filling spheres. The spheres are transparent to reveal the backbone (sticks). The hydrogen bonds defining the 3-stranded β-sheet are shown as thick dashed lines. The region around residues 22 and 23 is enlarged to show the side chains and hydrogen bond that are perturbed in the double mutant cycle. The isopropyl side chain of Val22 is outlined in black. (b) 1D 1H-NMR spectra of the loop-1–modified Pin WW domain variants used in the double mutant cycle. (c) Bar graph displaying the thermodynamic data for the double mutant cycle in kcal mol−1 (note the reverse scale). Shown are the free energies of folding of the four variants in the double mutant cycle, and the free energy differences between A1 and E1 (ΔG1), and A2 and E2 (ΔG2). The thermodynamic coupling energy (ΔΔG2–1 = ΔG2 – ΔG1) is indicated in red.
The four Pin WW domain variants (A1, A2, E1, and E2), synthesized by solid phase peptide synthesis, were monomeric in solution according to analytical ultracentrifugation analysis (data not shown). Retention of the native three-dimensional structure of the Pin WW domain fold upon side chain or combined backbone and side chain mutagenesis is supported by the characteristic far-ultraviolet circular dichroism spectra (Supplementary Fig. 1) and nearly identical one-dimensional 1H NMR spectra (Fig. 2b).
Guanidine hydrochloride (GuHCl) denaturation studies reveal that the Pin WW domain variants described above exhibit two-state behavior, enabling the folding free energies to be determined from their denaturation curves19,20 (Fig. 2c and Supplementary Table 1). The thermodynamic data in Fig. 2c reveal that ΔG2, the difference between the folding free energies of the loop-1–modified wild-type and Y23ψ Pin WW domain variants, is −4.23 ± 0.12 kcal mol−1. This large decrease in folding free energy upon amide-to-ester mutation is consistent with ΔG2 being primarily influenced by the loss of the strong hydrogen bond between the amide NH of Tyr23 and the amide CO of Arg14. ΔG1, the difference between the folding free energies of the loop-1–modified V22A and V22A/Y23ψ Pin WW domain variants, is −3.06 ± 0.03 kcal mol−1, thus, the value of ΔΔG2–1 is −1.17 ± 0.12 −kcal mol−1. As argued above, this large, negative value for the thermodynamic coupling energy indicates that the presence of the larger, more hydrophobic side chain of Val relative to Ala appears to substantially increase the strength of the backbone-backbone hydrogen bond between Tyr23 and Arg14.
Stronger hydrogen bonds in nonpolar environments: protein A*
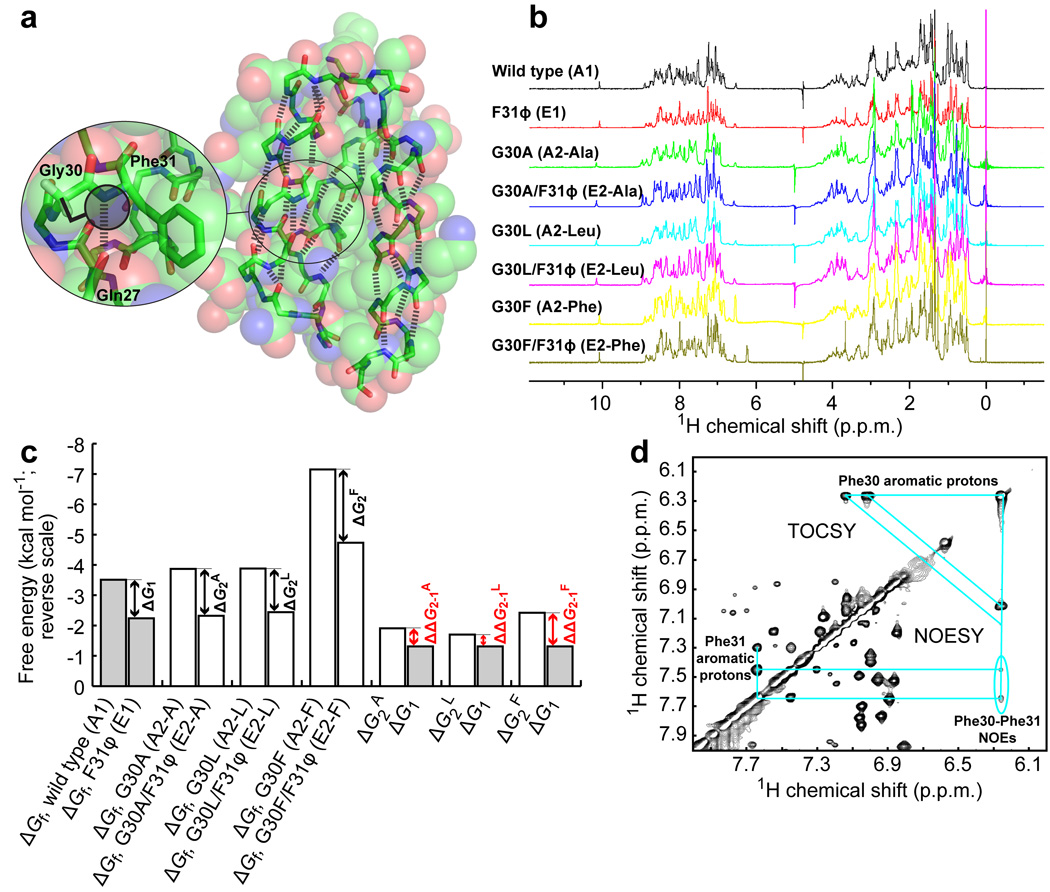
Protein A is a 60-residue three-helix bundle protein30 known to be highly tolerant of side chain mutations31. Like the Pin WW domain, the folding of protein A is two state, enabling its folding free energy to be extracted from chaotrope denaturation curves46. The Gly30-Phe31 amide bond (amide 30–31) forms two backbone hydrogen bonds, one in which the NH of Phe31 hydrogen bonds to the CO of Gln27 and one in which the CO of Gly30 hydrogen bonds to the NH of Cys34 (Fig. 3a). The lack of a side chain on Gly30 leaves the hydrogen bonds formed by amide 30–31 exposed to solvent (Fig. 3a). Mutating Gly30 to an amino acid with a larger side chain, like Phe, should substantially decrease the polarity of the microenvironment surrounding amide 30–31.
Figure 3. Environmental dependence of hydrogen bond strength in protein A* at Phe31.
(a) Structure of the B domain of protein A (pdb ID: 1SS1, rendered using PyMol) shown as space filling spheres. The spheres are transparent to reveal the backbone (sticks). The hydrogen bonds defining the three α-helices are shown as thick dashed lines. The region around residues 30 and 31 is enlarged to show the side chains and hydrogen bond that are perturbed in the double mutant cycle. The potential of a larger side chain on residue 30 to occlude the hydrogen bond formed by the NH of Phe31 is illustrated by the shaded circle. (b) 1D 1H-NMR spectra of the protein A* variants used in the double mutant cycles. (c) Bar graph displaying the thermodynamic data for the double mutant cycles in kcal mol−1 (note the reverse scale). Shown are the free energies of folding of the eight variants in the three double mutant cycles (in which Gly30 is replaced by Ala, Leu, or Phe), and the free energy differences between A1 and E1 (ΔG1), A2-A and E2-A (ΔG2A), A2-L and E2-L (ΔG2L), and A2-F and E2-F (ΔG2F). The thermodynamic coupling energies (ΔΔG2–1X = ΔG2X – ΔG1) are indicated in red. (d) 1H-1H NOESY and TOCSY spectra of G30F showing inter-residue cross peaks between aromatic protons of Phe30 and Phe31. Cyan lines connect the relevant intra- and inter-residue cross peaks. The critical Phe30–Phe31 NOEs are circled.
To enable a double mutant thermodynamic cycle analysis that could be used to test the polarity dependence of the strength of the hydrogen bonds formed by amide 30–31, four variants of protein A were synthesized by native chemical ligation. Note that Ser34 was changed to Cys to enable the ligation reaction; protein A with the S34C mutation will hereafter be denoted protein A*. The wild-type protein A* served as A1, F31ϕ as E1 (ϕ denotes the α-hydroxy acid equivalent of Phe), G30F as A2-F, and G30F/F31ϕ as E2-F. These four protein A* variants fold into native structures based on their one-dimensional 1H NMR spectra (Fig. 3b) and their far-UV circular dichroism spectra (Supplementary Fig. 2). Retention of native-like structure by the G30F/F31ϕ double mutant is further corroborated by 2D 1H-1H NOESY spectra. The fingerprint (NH-NH and NH-Hα) spectral regions of the G30F/F31ϕ variant exhibit resonances that are as well dispersed as those of the all-amide G30F protein A* variant (Supplementary Fig. 3). The cross peaks of the G30F and G30F/F31ϕ spectra display strictly analogous patterns, with the expected chemical shift perturbations. The majority of the NOE cross peaks were assigned for the G30F/F31ϕ double mutant, with the aid of COSY/TOCSY spectra and previously published wild-type protein A chemical shift assignments31. The methyl group of Ala49 in wild-type protein A packs against the benzene ring of Phe31, leading to upfield shifted Ala49 β-protons31. The analogous cross peak between the Phe31 aromatic protons and Ala49 β-protons was observed in G30F/F31ϕ protein A* (Supplementary Fig. 4). Moreover, the chemical shift of Ala49 β-protons in the double mutant G30F/F31ϕ is at 0.523 p.p.m., similar to that of wild-type protein A* (0.497 p.p.m.). Collectively, these data indicate that the double mutant has a native structure.
The folding free energies of the protein A* variants mentioned above were extracted from their GuHCl denaturation curves (Fig. 3c, Supplementary Table 1, and Supplementary Fig. 5). The thermodynamic data in Figure 3c show that an amide-to-ester mutation at Phe31 destabilizes protein A* by a ΔG1 = −1.31 ± 0.05 kcal mol−1 when residue 30 is Gly. When residue 30 is Phe, however, an amide-to-ester mutation at Phe31 causes a larger destabilization, with ΔG2F = −2.42 ± 0.09 kcal mol−1. This yields a thermodynamic coupling energy of ΔΔG2–1F = −1.11 ± 0.10 kcal mol−1, again consistent with the notion that decreasing the polarity of the microenvironment increases the strength of backbone-backbone hydrogen bonds.
The thermodynamic coupling energy reported above suggests that there should be a detectable interaction between residues Phe30 and Phe31 in the G30F mutant. Indeed, such an interaction was confirmed by 2D 1H-1H NOE cross peaks between the aromatic protons of Phe30 and Phe31 (Fig. 3d, circled). This Phe-Phe interaction corroborates the assertion that the side chain of Phe30 substantially shields amide 30–31 from solvent, thereby decreasing the solvent accessibility and the polarity of its microenvironment.
It is reasonable to expect that the magnitude of the thermodynamic coupling energy discussed directly above should correlate with how effectively the side chain at position 30 shields the hydrogen bonds formed by amide 30–31 from water. Thus, side chains that are smaller than that of Phe but larger than that of Gly should yield a value of ΔΔG2–1 between 0 and −1.11 kcal mol−1 when subjected to double mutant thermodynamic cycle analysis. To test this hypothesis, protein A* variants with Ala or Leu at position 30 were synthesized, again by using a native chemical ligation strategy47. G30A (A2-A) and G30A/F31ϕ (E2-A) for Ala, and G30L (A2-L) and G30L/F31ϕ (E2-L) for Leu were used with the wild-type (A1) and F31ϕ (E1) variants of protein A*, which were already studied above, to enable the desired double mutant thermodynamic cycle analyses.
Inserting the resulting free energies of folding (Fig. 3c and Supplementary Table 1) into the double mutant cycle yielded values of ΔΔG2-1 of −0.60 ± 0.08 kcal mol−1 and −0.39 ± 0.08 kcal mol−1 for the Ala and Leu mutations at position 30, respectively. Both of these values are between 0 and −1.11 kcal mol−1, as expected. It is perhaps surprising that the thermodynamic coupling energy between the side chain and amide-to-ester mutations is slightly more negative for Ala (by −0.21 ± 0.11 kcal mol−1) than it is for Leu. However, this result is consistent with the results of Bai et al.48, who found that Ala and Leu have similar abilities to protect amides on their C-terminal side from acid catalyzed hydrogen exchange by steric blockage.
Limits on the polarity dependence of hydrogen bond strength
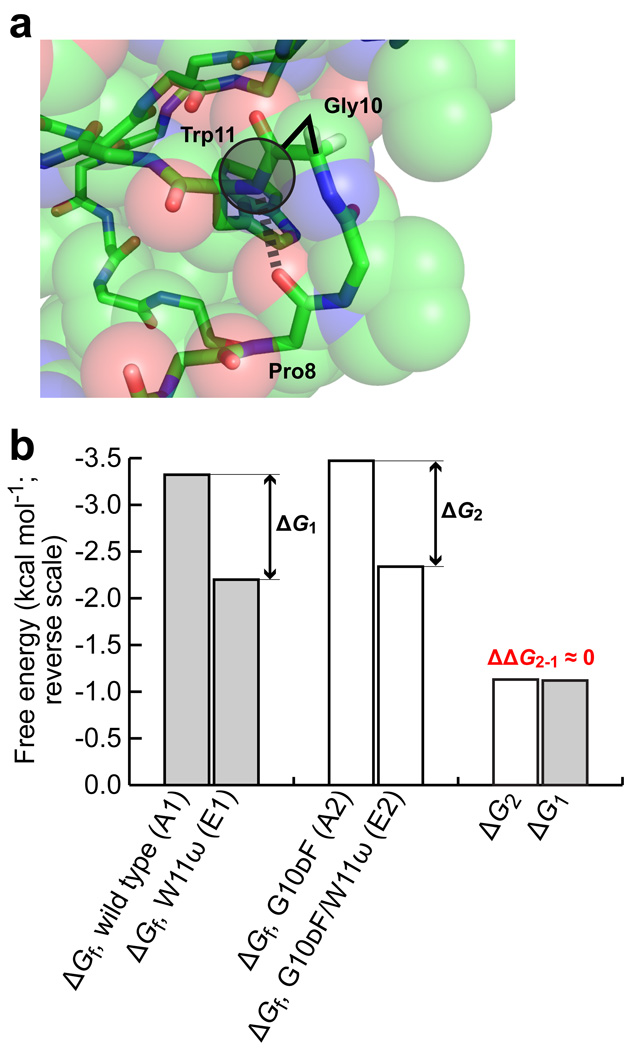
To further explore the influence of hydrophobic microenvironments on hydrogen bond strength, we applied the double mutant cycle analysis to the region around Trp11 of the Pin WW domain. Pro8 and Trp11 are residues i and i+3 of a type II β-turn (Fig. 4a). The amide NH of Trp 11 therefore makes a hydrogen bond to the amide CO of Pro8. This hydrogen bond occurs between residues that are closer in sequence than in any of the cases described above.
Figure 4. Environmental dependence of hydrogen bond strength in the Pin WW domain at Trp11.
(a) Enlargement of the region of the Pin WW domain around residues 10 and 11, showing the side chain and hydrogen bond that are perturbed in the double mutant cycle. The potential of a larger side chain on residue 10 to occlude the hydrogen bond formed by the NH of Trp11 is illustrated by the shaded circle. (b) Bar graph displaying the thermodynamic data for the double mutant cycle in kcal mol−1 (note the reverse scale). Shown are the free energies of folding of the four variants in the double mutant cycle (note that the values for A1 and E1 were determined previously19), and the free energy differences between A1 and E1 (ΔG1), and A2 and E2 (ΔG2). The thermodynamic coupling energy is close to 0, as indicated in red.
To determine whether this hydrogen bond behaves like the others examined in this study upon a decrease in the polarity of its microenvironment, a double mutant thermodynamic cycle was constructed. Mutating residue Gly10 to dPhe (the d configuration was used to avoid a steric clash with Asn26, His27, and/or Ile28 in the native state) places a large, hydrophobic side chain next to amide 10–11. Thus, the following Pin WW domain variants enable the thermodynamic coupling energy to be determined after extracting their folding free energies from GuHCl denaturation curves: wild type (A1), W11ω (E1), G10dF (A2), and G10dF/W11ω (E2). Note that these variants of the Pin WW domain contained the native loop 1 sequence (–SRSSGR–). The folding free energies of these variants are displayed in Figure 4b (the folding free energies of the wild type and W11ω variants of the Pin WW domain were determined previously19,20). Inserting these folding free energies into the double mutant thermodynamic cycle analysis yielded values of −1.12 kcal mol−1 for ΔG1 (from literature data19) and −1.13 ± 0.06 kcal mol−1 for ΔG2. Since ΔG1 and ΔG2 are almost equal, there is no thermodynamic coupling between the G10dF and W11ω mutations.
Clearly, introducing a larger side chain on the N-terminal side of amide 10–11 has a different effect on the folding free energy than the cases examined previously, indicating that even the environmental dependence of hydrogen bond strengths may itself be context dependent. We offer two explanations for this result. Given the sequence proximity of Trp11 and Pro8, perhaps the most likely explanation is that the hydrogen bond between the amide NH of Trp11 and the amide CO of Pro8 is at least partially formed in the denatured state, as has been observed in both small peptide fragments and other proteins49–53. Thus, mutating Gly10 to dPhe would increase the strength of this hydrogen bond in both the native and the denatured states, muting the effect on the folding free energy. A second explanation is that the dPhe side chain at position 10 could have adopted a conformation that left amide 10–11 solvent exposed. In either case, no thermodynamic coupling between the G10dF and W11ω mutations would have been observed.
Discussion
Stronger hydrogen bonds vs. structural rearrangement
In the interpretation of our results, we have reasoned that the thermodynamic coupling energies measured in the Pin WW domain and protein A* resulted from native state hydrogen bonds becoming stronger in the all-amide variants with larger side chains (the A2 variants). Our results could alternatively be explained by structural rearrangements in the native states of the amide-to-ester mutants that allowed water to hydrogen bond to the acceptor carbonyl that is left unsatisfied by the amide-to-ester mutation (the amide CO of Arg14 in the loop-1–modified Pin WW domain and the amide CO of Gln27 in protein A*). Such rearrangements could provide better access to water in the amide-to-ester mutants with small side chains (E1) than those with large side chains (E2), resulting in higher stability of the E1 variants relative to the E2 variants, while the hydrogen bond strength remained the same in the all-amide variants with small (A1) and large (A2) side chains. The difference between the folding free energies of E2 and E1 (ΔGE; see Fig. 1) would then be positive, whereas the difference between the folding free energies of A2 and A1 (ΔGA; see Fig. 1) would be close to 0. In this circumstance, negative values of ΔΔG2-1 (recall that ΔΔG2–1 = ΔG2 – ΔG1 = ΔGA – ΔGE; see Fig. 1) would arise largely from the positive value of ΔGE.
While the results of Blaber et al.54 with T4 lysozyme have demonstrated that structural rearrangements can make it possible for water to compensate for lost hydrogen bonds in the interior of a protein, we nevertheless believe that the dependence of hydrogen bond strengths on microenvironment polarity is primarily responsible for the thermodynamic coupling energies we observe for the following reasons. First, Alber et al.55 have shown, also in T4 lysozyme, that mutational elimination of a hydrogen bond sometimes simply leaves the remaining donor or acceptor unsatisfied. In fact, a survey of protein crystal structures by McDonald and Thornton showed that a small, but substantial, percentage of main chain donors and acceptors, ∼1.3% for NH groups and 1.8% for CO groups, are unsatisfied56 (it should be noted, however, that Fleming and Rose have suggested that unsatisfied hydrogen bond donors and acceptors are even less common57). Second, and more importantly, none of the E1 variants are substantially more stable than the E2 variants in the double mutant cycles examined herein (see Fig. 2c, Fig. 3c, and Supplementary Table 1). In the loop-1–modified Pin WW domain variants, the E1 and E2 variants are nearly equally stable (−1.52 ±0.02 vs. −1.42 ± 0.05 kcal mol−1, respectively; ΔGE = 0.1 ± 0.05 kcal mol−1); in the protein A* variants the E1 variants are actually less stable than the E2 variants (−3.47 ± 0.03 vs. −3.67 ± 0.04 for E2-A, −3.77 ± 0.06 for E2-L, and −4.73 ± 0.07 kcal mol−1 for E2-F; ΔGE = −0.20 ± 0.05, −0.30 ± 0.07, and −1.26 ± 0.08 kcal mol−1, respectively). Thus, it seems unlikely that a structural rearrangement that allowed water to compensate for lost hydrogen bonds could have occurred in the E1 variants. The negative values of ΔΔG2–1 for both the loop-1–modified Pin WW domain variants and the protein A* variants instead seem to be better explained by increased hydrogen bond strengths in the A2 variants, in which the hydrogen bonds in question are embedded in less polar microenvironments.
The above arguments notwithstanding, the possibility of structural rearrangements that compensate for lost hydrogen bonds can be rigorously excluded only by high-resolution structures of E1 and E2. Despite substantial effort, we have been unable to obtain such structures. We therefore suggest that the observed thermodynamic coupling energies should be considered upper limits on the effect of microenvironment polarity on hydrogen bond strengths. If rearrangements that enabled solvent to compensate for lost hydrogen bonds actually took place in the E1 variants, the values of ΔΔG2–1 would overestimate the true increases in hydrogen bond strengths by an amount equal to the stability difference between the normal and rearranged structures of E1 (which is in turn equal to the difference between the energetic cost of losing a hydrogen bond in the normal native state and the energetic cost of the strain induced by the conformational changes in the rearranged state).
Denatured vs. native state effects
Another alternative explanation for our results is that shielding of amide groups from water in the denatured state decreases the desolvation energy of the amides, thereby stabilizing the native state. Denatured state effects certainly could make some contribution to the observed thermodynamic coupling energies. However, it is unlikely that denautured state effects could outweigh native state effects in our double mutant thermodynamic cycles. Bai and Englander58 have suggested that amide shielding in the denatured state should stabilize the native state by no more than 0.2 kcal mol−1. This amount of energy can account for small effects, like the differences among the β-sheet propensities of the amino acids58, but not the large thermodynamic coupling energies that are reported above. Our thermodynamic coupling energies are more in line with the differences in hydrogen bond enthalpies observed by Franzen and Stephens59 for N-methylacetamide in cis-dichloroethylene (dielectric constant = 9.13, ΔH = −1.6 kcal mol−1) and the much less polar trans-dichloroethylene (dielectric constant = 2.25, ΔH = −3.2 kcal mol−1). This observation is consistent with our thermodynamic coupling energies originating in changes in the hydrogen bond’s native state microenvironment. It should be kept in mind, however, that denatured state effects could be important in some instances. For example, detergents like SDS are often used to unfold membrane proteins13. In the environment of SDS micelles, hydrogen bonds formed in the denatured state could be stronger than typical solvent-protein hydrogen bonds.
Energy increment upon hydrogen bond burial
The thermodynamic coupling energies determined from our double mutant thermodynamic cycles are summarized in Table 1. The data in Table 1 show that the average thermodynamic coupling energy measured herein is roughly −0.7 kcal mol−1, but can range from 0 to almost −1.2 kcal mol−1. As discussed above, these energies can be taken to be upper limits on the increase in hydrogen bond strength when the polarity of the microenvironment is decreased. Thus, these results demonstrate not only that the effect of microenvironment polarity on hydrogen bond strength can be quite large, but also that it can vary considerably. This finding is consistent with literature data. The last row in Table 1 shows the difference between the average changes in folding free energies (ΔΔGf values) for buried and exposed amide-to-ester mutants from previous studies (see Supplementary Table 2; note that, for simplicity, only amide-to-ester mutations in which the affected amide donates, but does not accept, a hydrogen bond are used). This difference is −1.1 kcal mol−1, which is consistent with the largest of the thermodynamic coupling energies that we report. In addition, this difference has a large standard deviation, again consistent with our results. It is worth noting that this large standard deviation primarily results from the variability in ΔΔGf values for the amide-to-ester mutations at buried sites; at exposed sites, the ΔΔGf values are more narrowly distributed (see Supplementary Table 2). This observation again emphasizes the context dependence of hydrogen bond strengths.
Table 1.
Summary of the thermodynamic coupling energies determined herein and comparison to literature data (s.d. = standard deviation, s.e. = standard error.)
| Double mutant cycle | ΔΔG2–1 (kcal mol−1) |
|---|---|
| Loop-1–modified Pin WW domain | −1.17 |
| Protein A*, G30A | −0.60 |
| Protein A*, G30L | −0.39 |
| Protein A*, G30F | −1.11 |
| Wild-type loop 1 Pin WW domain | −0.01 |
| Average | −0.66, s.d. = 0.49, s.e. = 0.22 |
| Difference between average literature ΔΔGf values for buried and exposed amide-to-ester mutantsa | −1.08, s.d. = 1.33, s.e. = 0.54 |
Data from Supplementary Table 2. Mutations at sites with fractional backbone exposure ≤ 0.20 were defined as buried.
Implications for studies of protein folding
We have demonstrated that under some circumstances, introducing a hydrophobic side chain at the surface of a protein can stabilize either helix- or sheet-rich proteins by up to 1.2 kcal mol−1, most likely by increasing the strength of a nearby hydrogen bond in the native state. Thus, the effect of context dependence on hydrogen bond strengths can be a substantial fraction of the overall free energies of protein folding, which are typically1,60 between −5 and −20 kcal mol−1. Thus, burial or solvent exposure of a few hydrogen bonds near the surface of a protein can dramatically stabilize or destabilize a protein’s native state, respectively11,12. Understanding this point is likely to be important for a number of practical problems, including protein structure prediction, protein engineering, and the design of foldable, non-protein polymers.
When local hydrophobicity and hydrogen bonding are thermodynamically coupled, it creates an energetic “hot spot” that buttresses the native state against unfolding. It is equally important to recognize, however, that placing hydrophobic side chains close to hydrogen bonded amides favors the native state in some, but not all, cases. Defining the conditions that must be met to observe cooperativity between hydrophobic environments and hydrogen bond strengths is an important future undertaking that will require close collaboration between theory and experiment.
Methods
Protein preparation
We synthesized the all amide sequences of the Pin WW domain on an ABI 433 peptide synthesizer using Fmoc/tBu chemistry. We prepared the ester mutants through a solid-phase Boc strategy as described previously16. To ensure that the double mutant is well-folded, some Pin WW domain variants (V22Y23, A22Y23, V22ψ23 and A22ψ23) incorporated the 5-residue FBP loop 1 sequence –SADGR– instead of the 6-residue Pin1 loop 1 sequence –SRSSGR–. The shortened loop increases the thermodynamic stability of Pin WW domain by 2.3 kcal mol−1 (ref. 26). The sequences of the Pin WW domain variants used in the V22A/Y23ψ double mutant cycle were:
| 6 | 11 | 21 | 31 | |
|---|---|---|---|---|
| A2: | KLPPG | WEKRMSAD-G | RVYYFNHITN | ASQWERPSG |
| A1: | KLPPG | WEKRMSAD-G | RAYYFNHITN | ASQWERPSG |
| E2: | KLPPG | WEKRMSAD-G | RVψYFNHITN | ASQWERPSG |
| E1: | KLPPG | WEKRMSAD-G | RAψYFNHITN | ASQWERPSG |
The sequences of the Pin WW domain variants used in the G10dF/W11ω double mutant cycle were:
| 6 | 11 | 21 | 31 | |
|---|---|---|---|---|
| A1: | KLPPG | WEKRMSRSSG | RVYYFNHITN | ASQWERPSG |
| A2: | KLPPdF | WEKRMSRSSG | RVYYFNHITN | ASQWERPSG |
| E1: | KLPPG | ωEKRMSRSSG | RVYYFNHITN | ASQWERPSG |
| E2: | KLPPdF | ωEKRMSRSSG | RVYYFNHITN | ASQWERPSG |
We carried out the chemical synthesis of 60-residue protein A* through a fragment condensation strategy47 enabled by mutation of solvent exposed Ser34 to Cys. We synthesized N-terminal fragments (T1-Q33-COSR) on S-trityl-β-mercaptopropionyl-Leu-Pam resin through a Boc chemistry strategy. Automated peptide synthesis yielded C-terminal fragments (C34-A60), which we ligated to N-fragments to give intact protein A*. The sequences of the protein A* variants used in the double mutant cycles were:
| 1 | 11 | 21 | 31 | 41 | 51 | |
|---|---|---|---|---|---|---|
| A1: | TADNKFNKEQ | QNAFWEILHL | PNLNEEQRNG | FIQCLKDDPS | QSANLLAEAK | KLNDAQAPKA |
| A2-X: | TADNKFNKEQ | QNAFWEILHL | PNLNEEQRNX | FIQCLKDDPS | QSANLLAEAK | KLNDAQAPKA |
| E1: | TADNKFNKEQ | QNAFWEILHL | PNLNEEQRNG | ϕIQCLKDDPS | QSANLLAEAK | KLNDAQAPKA |
| E2-X: | TADNKFNKEQ | QNAFWEILHL | PNLNEEQRNX | ϕIQCLKDDPS | QSANLLAEAK | KLNDAQAPKA |
For A2-F and E2-F, X = F; for A2-A and E2-Ala, X = A; and for A2-L and E2-L, X = L, We purified all protein variants by RP-HPLC and size exclusion chromatography to give >95% purity and chemical identity confirmed by ESI-MS.
Structure characterization
We subjected chemically synthesized Protein A* and Pin WW domain variants to spectroscopic analysis to ensure that they adopted native-like structures. Analytical ultracentrifugation experiments confirmed that the protein variants are monomeric under the experimental conditions used. We compared circular dichroism spectra, Trp fluorescence emission, and NMR spectra (both 1D 1H NMR and 2D 1H-1H NOESY data) of the mutants to those of the wild type to confirm their structural similarity.
Assessing the thermodynamic stability
We measured the thermodynamic parameters of all protein variants through guanidine hydrochloride denaturation experiments19,20. We monitored the denaturation process by both circular dichroism (ellipticity 222 nm for protein A* and 227 nm for Pin WW domain variants) and Trp fluorescence (fluorescence intensity a 355nm for protein A* and fluorescence intensity ratio 355 nm / 342 nm for Pin WW domain). We obtained essentially identical results from the two different methods. Fitting the denaturation curves to a two-state model, with pre- and post-transition baselines determined by independently fitting the pre- and post-transition regions of the denaturation curves to straight lines, yielded the Cm, m values and folding free energies ΔGf of all protein variants. Examples of the denaturation data are shown in Supplementary Figure 5.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM051105), the Lita Annenberg Hazen foundation, and The Skaggs Institute for Chemical Biology. D.A.B. thanks the National Institutes of Health for a post-doctoral fellowship (NS047024).
References
- 1.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 2.Pauling L, Corey RB, Branson HR. The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. U. S. A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 4.Bolen DW, Rose GD. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu. Rev. Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 5.Avbelj F, Baldwin RL. Limited validity of group additivity for the folding energetics of the peptide group. Proteins. 2006;63:283–289. doi: 10.1002/prot.20756. [DOI] [PubMed] [Google Scholar]
- 6.Avbelj F, Baldwin RL. Origin of the change in solvation enthalpy of the peptide group when neighboring peptide groups are added. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3137–3141. doi: 10.1073/pnas.0813018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klotz IM, Franzen JS. Hydrogen bonds between model peptide groups in solution. J. Am. Chem. Soc. 1962;84:3461–3466. [Google Scholar]
- 8.Kresheck GC, Klotz IM. Thermodynamics of transfer of amides from an apolar to an aqueous solution. Biochemistry. 1969;8:8–12. doi: 10.1021/bi00829a002. [DOI] [PubMed] [Google Scholar]
- 9.Umeyama H, Morokuma K. The origin of hydrogen bonding: an energy decomposition study. J. Am. Chem. Soc. 1977;99:1316–1332. [Google Scholar]
- 10.Nemethy G, Steinberg IZ, Scheraga HA. Influence of water structure and of hydrophobic interactions on the strength of side-chain hydrogen bonds in proteins. Biopolymers. 1963;1:43–69. [Google Scholar]
- 11.Fernandez A, Berry RS. Extent of hydrogen-bond protection in folded proteins: A constraint on packing architectures. Biophys. J. 2002;83:2475–2481. doi: 10.1016/S0006-3495(02)75258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez A, Zhang X, Chen JP. Folding and wrapping soluble proteins: Exploring the molecular basis of cooperativity and aggregation. Prog. Mol. Biol. Trans. Sci. 2008;83:53–87. doi: 10.1016/S0079-6603(08)00602-8. [DOI] [PubMed] [Google Scholar]
- 13.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453:1266–1270. doi: 10.1038/nature06977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers JK, Pace CN. Hydrogen bonding stabilizes globular proteins. Biophys J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albeck S, Unger R, Schreiber G. Evaluation of direct and cooperative contributions towards the strength of buried hydrogen bonds and salt bridges. J. Mol. Biol. 2000;298:503–520. doi: 10.1006/jmbi.2000.3656. [DOI] [PubMed] [Google Scholar]
- 16.Blankenship JW, Balambika R, Dawson PE. Probing backbone hydrogen bonds in the hydrophobic core of GCN4. Biochemistry. 2002;41:15676–15684. doi: 10.1021/bi026862p. [DOI] [PubMed] [Google Scholar]
- 17.Takano K, Scholtz JM, Sacchettini JC, Pace CN. The contribution of polar group burial to protein stability is strongly context-dependent. J. Biol. Chem. 2003;278:31790–31795. doi: 10.1074/jbc.M304177200. [DOI] [PubMed] [Google Scholar]
- 18.Scheike JA, et al. Amide-to-ester substitution in coiled coils: the effect of removing hydrogen bonds on protein structure. Angew. Chem. Int. Ed. Engl. 2007;46:7766–7769. doi: 10.1002/anie.200702218. [DOI] [PubMed] [Google Scholar]
- 19.Deechongkit S, Dawson PE, Kelly JW. Toward assessing the position-dependent contributions of backbone hydrogen bonding to beta-sheet folding thermodynamics employing amide-to-ester perturbations. J. Am. Chem. Soc. 2004;126:16762–16771. doi: 10.1021/ja045934s. [DOI] [PubMed] [Google Scholar]
- 20.Deechongkit S, et al. Context-dependent contributions of backbone hydrogen bonding to beta-sheet folding energetics. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 21.Powers ET, Deechongkit S, Kelly JW. Backbone-backbone H-bonds make context-dependent contributions to protein folding kinetics and thermodynamics: Lessons from amide-to-ester mutations. Adv. Protein Chem. 2006;72:39–78. doi: 10.1016/S0065-3233(05)72002-7. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson N, Johnson CM, Macias M, Oschkinat H, Fersht A. Ultrafast folding of WW domains without structured aromatic clusters in the denatured state. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13002–13007. doi: 10.1073/pnas.221467198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson N, et al. Using flexible loop mimetics to extend phi-value analysis to secondary structure interactions. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13008–13013. doi: 10.1073/pnas.221467398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Socolich M, et al. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 25.Jager M, Nguyen H, Crane JC, Kelly JW, Gruebele M. The folding mechanism of a beta-sheet: the WW domain. J. Mol. Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 26.Jager M, et al. Structure-function-folding relationship in a WW domain. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deechongkit S, et al. Beta-sheet folding mechanisms from perturbation energetics. Curr. Opin. Struct. Biol. 2006;16:94–101. doi: 10.1016/j.sbi.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen H, Jager M, Kelly JW, Gruebele M. Engineering a beta-sheet protein toward the folding speed limit. J. Phys. Chem. B. 2005;109:15182–15186. doi: 10.1021/jp052373y. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen H, Jager M, Moretto A, Gruebele M, Kelly JW. Tuning the free-energy landscape of a WW domain by temperature, mutation, and truncation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3948–3953. doi: 10.1073/pnas.0538054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, Karimi A, Dyson HJ, Wright PE. Absence of a stable intermediate on the folding pathway of protein A. Protein Sci. 1997;6:1449–1457. doi: 10.1002/pro.5560060709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato S, Religa TL, Daggett V, Fersht AR. Testing protein-folding simulations by experiment: B domain of protein A. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6952–6956. doi: 10.1073/pnas.0401396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 33.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 34.Horovitz A, Fersht AR. Co-operative interactions during protein folding. J. Mol. Biol. 1992;224:733–740. doi: 10.1016/0022-2836(92)90557-z. [DOI] [PubMed] [Google Scholar]
- 35.Seebach D, Beck AK, Bierbaum DJ. The world of beta-and gamma-peptides comprised of homologated proteinogenic amino acids and other components. Chem. Biodivers. 2004;1:1111–1239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 36.Horne WS, Price JL, Gellman SH. Interplay among side chain sequence, backbone composition, and residue rigidification in polypeptide folding and assembly. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9151–9156. doi: 10.1073/pnas.0801135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price JL, Horne WS, Gellman SH. Discrete heterogeneous quaternary structure formed by alpha/beta-peptide foldamers and alpha-peptides. J. Am. Chem. Soc. 2007;129:6376–6377. doi: 10.1021/ja071203r. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee S, Roy RS, Balaram P. Expanding the polypeptide backbone: hydrogen-bonded conformations in hybrid polypeptides containing the higher homologues of alpha-amino acids. J. R. Soc. Interface. 2007;4:587–606. doi: 10.1098/rsif.2006.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hann MM, Sammes PG, Kennewell PD, Taylor JB. Double bond isosteres of the peptide bond - Enkephalin analog. Chem. Comm. 1980:234–235. [Google Scholar]
- 40.Xiao JB, Weisblum B, Wipf P. Trisubstituted (E)-alkene dipeptide isosteres as beta-turn promoters in the gramicidin S cyclodecapeptide scaffold. Org. Lett. 2006;8:4731–4734. doi: 10.1021/ol0617704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenkins CL, Vasbinder MM, Miller SJ, Raines RT. Peptide bond isosteres: ester or E-alkene in the backbone of the collagen triple helix. Org. Lett. 2005;7:2619–2622. doi: 10.1021/ol050780m. [DOI] [PubMed] [Google Scholar]
- 42.Fu YW, Gao JM, Bieschke J, Dendle MA, Kelly JW. Amide-to-E-olefin versus amide-to-ester backbone H-bond perturbations: Evaluating the O-O repulsion for extracting H-bond energies. J. Am. Chem. Soc. 2006;128:15948–15949. doi: 10.1021/ja065303t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao JM, Kelly JW. Toward quantification of protein backbone-backbone hydrogen bonding energies: An energetic analysis of an amide-to-ester mutation in an alpha-helix within a protein. Protein Sci. 2008;17:1096–1101. doi: 10.1110/ps.083439708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stigers KD, Soth MJ, Nowick JS. Designed molecules that fold to mimic protein secondary structures. Curr. Opin. Chem. Biol. 1999;3:714–723. doi: 10.1016/s1367-5931(99)00030-7. [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Wang M, Fitzgerald MC. Analysis of protein folding and function using backbone modified proteins. Bioorg. Chem. 2004;32:438–449. doi: 10.1016/j.bioorg.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Myers JK, Oas TG. Preorganized secondary structure as an important determinant of fast protein folding. Nat. Struct. Biol. 2001;8:552–558. doi: 10.1038/88626. [DOI] [PubMed] [Google Scholar]
- 47.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 48.Bai YW, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyson HJ, Rance M, Houghten RA, Lerner RA, Wright PE. Folding of immunogenic peptide fragments of proteins in water solution. I. Sequence requirements for the formation of a reverse turn. J. Mol. Biol. 1988;201:161–200. doi: 10.1016/0022-2836(88)90446-9. [DOI] [PubMed] [Google Scholar]
- 50.Wright PE, Dyson HJ, Lerner RA. Conformation of peptide fragments of proteins in aqueous solution: implications for initiation of protein folding. Biochemistry. 1988;27:7167–7175. doi: 10.1021/bi00419a001. [DOI] [PubMed] [Google Scholar]
- 51.Yi Q, Scalley-Kim ML, Alm EJ, Baker D. NMR characterization of residual structure in the denatured state of protein L. J. Mol. Biol. 2000;299:1341–1351. doi: 10.1006/jmbi.2000.3816. [DOI] [PubMed] [Google Scholar]
- 52.Zhang O, Forman-Kay JD. NMR studies of unfolded states of an SH3 domain in aqueous solution and denaturing conditions. Biochemistry. 1997;36:3959–3970. doi: 10.1021/bi9627626. [DOI] [PubMed] [Google Scholar]
- 53.Anil B, Li Y, Cho JH, Raleigh DP. The unfolded state of NTL9 is compact in the absence of denaturant. Biochemistry. 2006;45:10110–10116. doi: 10.1021/bi060636o. [DOI] [PubMed] [Google Scholar]
- 54.Blaber M, et al. Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala-->Ser and Val-->Thr substitutions in T4 lysozyme. Biochemistry. 1993;32:11363–11373. doi: 10.1021/bi00093a013. [DOI] [PubMed] [Google Scholar]
- 55.Alber T, et al. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987;330:41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- 56.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 57.Fleming PJ, Rose GD. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 2005;14:1911–1917. doi: 10.1110/ps.051454805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai YW, Englander SW. Hydrogen bond strength and beta-sheet propensities - the role of a side chain blocking effect. Proteins. 1994;18:262–266. doi: 10.1002/prot.340180307. [DOI] [PubMed] [Google Scholar]
- 59.Franzen JS, Stephens RE. Effect of a dipolar solvent system on interamide hydrogen bonds. Biochemistry. 1963;2:1321–1327. doi: 10.1021/bi00906a025. [DOI] [PubMed] [Google Scholar]
- 60.Pace CN. The stability of globular proteins. CRC Crit. Rev. Biochem. 1975;3:1–43. doi: 10.3109/10409237509102551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.