Summary
Human neutrophil α-defensins (HNPs) are cationic antimicrobial peptides that are synthesized in vivo as inactive precursors (proHNPs). Activation requires proteolytic excision of their anionic N-terminal inhibitory pro peptide. The pro peptide of proHNP1 also specifically interacts with and inhibits the antimicrobial activity of HNP1 intermolecularly. In the light of the opposite net charges segregated in proHNP1, functional inhibition of the C-terminal defensin domain by its pro peptide is generally thought to be of electrostatic nature. Using a battery of analogs of the pro peptide and of proHNP1, we identified residues in the pro peptide region important for HNP1 binding and inhibition. Only three anionic residues in the pro peptide, Glu15, Asp20 and Glu23, were modestly important for interactions with HNP1. By contrast, the hydrophobic residues in the central part of the pro peptide, and the conserved hydrophobic motif Val24Val25Val26Leu28 in particular, were critical for HNP1 binding and inhibition. Neutralization of all negative charges in the pro peptide only partially activated the bactericidal activity of proHNP1. Our data indicate that hydrophobic forces play a dominant role in mediating the interactions between HNP1 and its pro peptide – a finding largely contrasting the commonly held view that the interactions are of electrostatic nature.
Keywords: defensin, HNP, antimicrobial peptide, pro peptide, electrostatic interaction, hydrophobic interaction
Introduction
Defensins are small cationic antimicrobial peptides commonly found in leukocytes and epithelial cells of mammals1; 2; 3; 4. These molecules directly kill microbes through microbial membrane disruption, constituting the first line of innate immune defense against foreign pathogens. Defensins also act as immunomodulators by inducing expression of cytokines as well as chemoattracting and activating immune cells, presumably through receptor-mediated signaling events5. Based on the pattern of three disulfide linkages, defensins are categorized intoα, β and θ families. Alpha- and β-defensins form a three-stranded beta-sheet structure stabilized by disulfide bridges, whereas θ-defensins adopt a close-end, two-stranded beta-sheet conformation with a ladder pattern of disulfides.
To date, six human α-defensins have been identified. The first four, also known as human neutrophil peptides 1–4 or HNPs 1–4, are abundant in the azurophilic granules of neutrophils6; 7, while human α defensins 5 and 6, also known as HD5 and HD6, are mainly expressed in intestinal Paneth cells8; 9. Human α defensins are synthesized in vivo as inactive precursors and activated in a sequence of posttranslational proteolytic events10. Unlike HD5 and HD6, which are released as prodefensins and processed extracellularly4; 11; 12; 13, HNPs 1–4 are processed intracellularly14.
PreproHNP1, consisting of a 19-residue N-terminal signal peptide, a 45-residue anionic pro sequence and a 30-residue C-terminal mature domain10, is synthesized in neutrophil precursor cells in bone marrow14, and proteolytically processed in ER, Golgi complex, and azurophilic granules before its final maturation and storage10. Liu and Ganz (1995) first discovered that the pro peptide facilitates sub-cellular trafficking and sorting of proHNP114. Deletion of the N-terminal two fifths of the propiece – a stretch of 18 amino acid residues – had little effect on defensin biosynthesis. However, deletion of the succeeding 13 amino acid residues, encompassing a conserved hydrophobic region, was detrimental to correct defensin trafficking and sorting. This hydrophobic region was therefore considered essential for the biosynthesis and transport of HNP114.
ProHNP1 itself is functionally inactive against bacteria15. Further, an exogenously added anionic pro peptide can specifically interact with and inhibit the antimicrobial function of HNP1 in a dose-dependent manner15; 16. Ouellette and colleagues demonstrated in a recent study that replacement of the anionic residues (Asp/Glu) in the pro peptide region of a mouse α-defensin precursor, proCryptdin-4, by either Gly or Asn/Gln activated proCryptdin-4 with respect to its membrane and bactericidal activity17. In the light of opposing net charges generally observed in pro-region and mature defensin, electrostatic forces are thought to be critical for inter-molecular interactions between cationic α-defensins and their anionic pro peptides, and for the intra-molecular functional inhibition seen in proHNPs and proCryptdins as well15; 18.
In studying the impact of the pro region on the folding and function of HNP1, however, we found that a charge-reversing (cationic) variant of the proHNP1 pro peptide, where Arg/Lys residues were changed to Asp and Asp/Glu residues to Lys, was still capable of binding HNP1 effectively19. This finding raises an intriguing question about the precise nature of the molecular determinants for the recognition of HNP1 by its pro peptide. Here we report on the binding of mature HNP1 to a series of synthetic analogs of its pro peptide using surface plasmon resonance. In addition, we determined the inhibition of the bactericidal activity of HNP1 by these pro peptide analogs using a virtual colony counting (vCC) assay. Selected analogs of full-length proHNP1 were also prepared and functionally characterized.
Results
Design of pro peptide analogs
To evaluate the effects of length, charge and hydrophobicity of the pro peptide on its binding to HNP1, we designed three different types of modifications: N-terminal truncation, substitution of acidic residues, and replacement of hydrophobic residues. First, we truncated the N-terminal 9 and 19 residues in the 45-residue pro peptide, generating two analogs designated as Δ9 and Δ19. Second, we replaced with Ala each of eight acidic residues (Asp and Glu) individually, obtaining eight single-mutation pro peptides. In addition, all Asp and Glu residues were simultaneously substituted for Asn and Gln, respectively, yielding one octal-mutation pro peptide termed DE/NQ. Third, we changed the aromatic residue Trp30 to Ala, and an adjacent hydrophobic motif (Val24Val25Val26Leu28) conserved in many mammalian alpha-defensins to Ala24Ala25Ala26Ala28, resulting in two analogs W30A and VVVL/4A, respectively. The wild type pro peptide, all 13 pro peptide analogs, and folded HNP1 were purified by HPLC to homogeneity, and their molecular masses were ascertained by ESI-MS (data not shown). The amino acid sequence of the proHNP1 pro peptide aligned with 33 unique pro peptide sequences of other mammalian species is listed in Figure 1.
Figure 1.
Sequence alignment of 34 unique pro peptides of known mammalian alpha-defensins (human, mouse, rat, rhesus macaque – MACMU, rabbit – RABIT, chimpanzee – PANTR, and guinea pig – CAVPO). The acidic residues in the proHNP1 pro peptide are in bold typeface and underlined. The central hydrophobic regions are shaded in grey (residues 10–19) and black (residues 24–28). Also shaded in black are hydrophobic motifs conserved in the pro peptides of most mammalian alpha-defensins.
To study the mutational effects of length, charge and hydrophobicity in the context of proHNP1, we chemically synthesized and folded wild type proHNP1 and a selected group of proHNP1 analogs using a published protocol16; 20, yielding Δ9-, DE/NQ-,VVVL/4A-, and W30A-proHNP1. Immediately following a Met residue in the amino acid sequence, the C-terminal defensin domain was conveniently released by CNBr cleavage of proHNP1 and its folded analogs. Comparison of CNBr-released HNP1 with a previously characterized synthetic HNP1 using analytic RP-HPLC, ESI-MS and antibacterial activity assay indicated that the four proHNP1 analogs all folded correctly (data not shown). We failed, however, to produce correctly folded Δ19-proHNP1 due to massive precipitation of the polypeptide during the oxidative folding process.
Binding versus inhibition
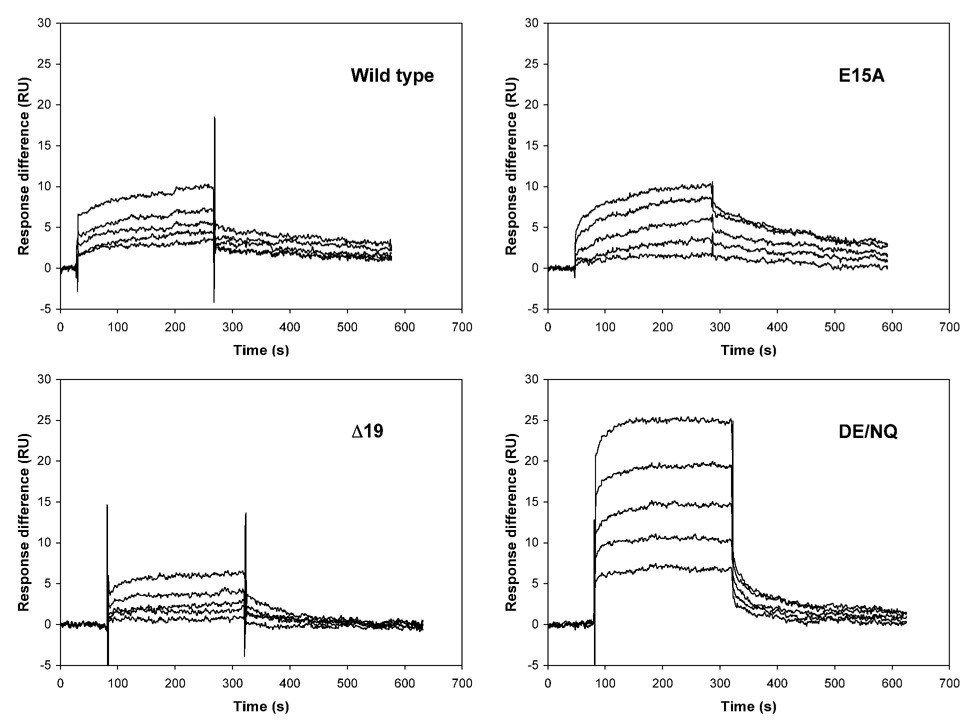
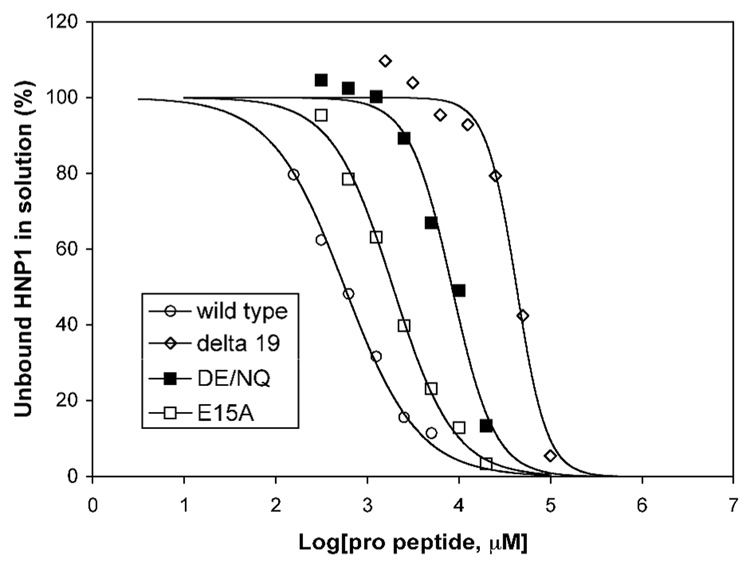
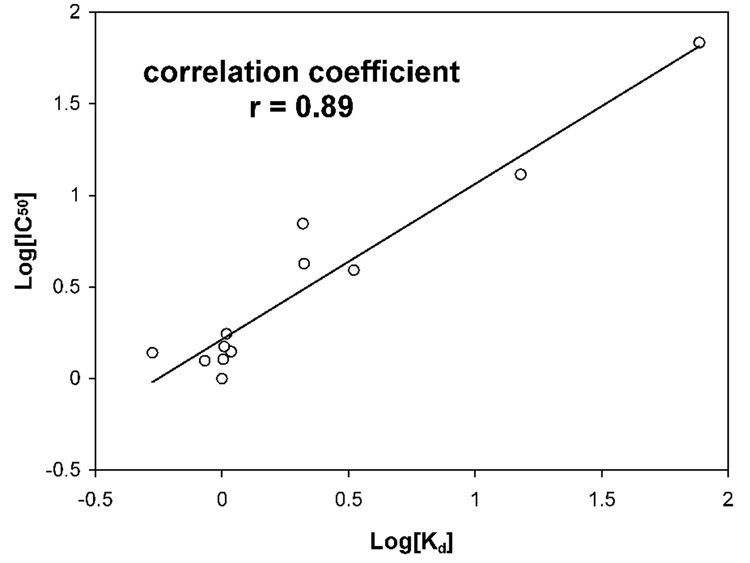
Surface plasmon resonance (SPR) was used to measure the binding affinity of the pro peptides for HNP1. Shown in Figure 2 are four representative association and dissociation kinetics (sensorgrams) of the wild type pro peptide, Δ19, E15A, and DE/NQ on immobilized HNP1. To alleviate subjectivity often associated with SPR data analysis, we also quantified solution interactions between HNP1 and the pro peptide analogs using a previously described, SPR-based competition assay21. Representative competition binding curves for the wild type pro peptide, Δ19, E15A, and DE/NQ are shown in Figure 3. The dissociation equilibrium constants (Kd) derived from direct binding assays (on a low-density chip) and IC50 values obtained from competition binding assays are summarized in Table 1. Kd and IC50 values for VVVL/4A are missing due to lack of HNP1 binding. In addition, the Kd value for W30A could not be reliably determined because the kinetic data did not fit to a 1:1 kinetic binding model. Otherwise, the Kd values are in general 2–10 fold lower than the corresponding IC50 values. With W30A and VVVL/4A excluded, relative binding affinities for HNP1 of the remaining 11 pro peptide analogs versus the wild type pro peptide, determined by the two different techniques, correlated reasonably well as shown in Figure 4.
Figure 2.
Representative direct binding of wild type, E15A, Δ19, and DE/NQ pro peptides to the wild type HNP1, monitored by surface plasmon resonance (Biacore). Measurements were conducted on a CM4 chip with HNP1 immobilized. Concentrations ranging from 2 to 0.125 µM were used for wild type and E15A pro peptides; higher concentrations ranging from 20 to 1.25 µM were used for analogs Δ19 and DE/NQ.
Figure 3.
Representative competitive binding of HNP1 to the wild type pro peptide in the presence of wild type (circle), E15A (empty square), Δ19 (diamond), and DE/NQ (filled square) pro peptides, monitored by surface plasmon resonance (Biacore). Measurements were conducted on a CM4 chip with the wild type pro peptide immobilized. HNP1 of 100 nM was incubated with varying concentrations of pro peptide, followed by an injection of the incubation mixture. Free HNP1 in the solution was deducted, based on the initial rate (slope) of association, from a standardized calibration curve established by kinetic measurements of HNP1 injected alone at different concentrations.
Table 1.
Binding parameters for HNP1 interacting with its pro peptide and various pro-peptide analogs, determined by SPR-based direct binding assays (Kd) and competition binding assays (IC50).
| Kd (µM) | Kd, mut/Kd, wt | IC50 (µM) | IC50,mut/IC50,wt | |
|---|---|---|---|---|
| Wild type Mutants | 0.200 | 0.574 ± 0.010 | ||
| E1A | 0.106 | 0.53 | 0.794 ± 0.070 | 1.38 |
| D8A | 0.217 | 1.08 | 0.808 ± 0.196 | 1.40 |
| E9A | 0.202 | 1.01 | 0.732 ± 0.026 | 1.27 |
| E15A | 0.422 | 2.11 | 2.44 ± 0.54 | 4.25 |
| D20A | 0.417 | 2.08 | 4.03 ± 0.12 | 7.02 |
| E23A | 0.835 | 4.17 | 2.25 ± 0.24 | 3.91 |
| D31A | 0.204 | 1.02 | 0.861 ± 0.101 | 1.50 |
| E32A | 0.208 | 1.04 | 1.01 ± 0.07 | 1.76 |
| W30A | n.r.a | n.r. | 2.47 ± 0.33 | 4.30 |
| Δ9 | 0.171 | 0.855 | 0.72 ± 0.16 | 1.26 |
| Δ19 | 15.4 | 77.0 | 39.3 ± 3.3 | 68.4 |
| DE/NQ | 3.03 | 15.2 | 7.48 ± 0.89 | 13.0 |
| VVVL/4A | n.b.b | n.b. |
n.r., not reported due to erratic curve fitting
n.b., no binding detected.
Figure 4.
Correlation between the binding affinities of HNP1 for selected pro peptides (W30A and VVVL/4A excluded), measured by SPR-based direct binding assays and competitive binding assays.
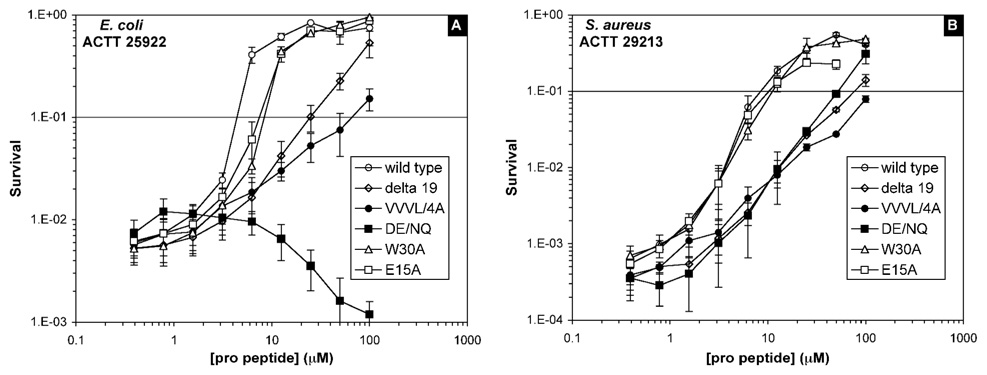
In addition to the SPR-based binding measurements, antibacterial assays were performed to evaluate the ability of various pro peptide analogs (0.4–100 µM) to inhibit the bactericidal activity of 10 µM mature HNP1 against E. coli and S. aureus. Shown in Figure 5 is a representative dose-dependent defensin inhibition by the wild type pro peptide, Δ19, E15A, DE/NQ, W30A, and VVVL/4A. HNP1 alone at 10 µM killed about 99% of input E. coli cells (a 2-log reduction in survival) and approximately 99.9% input S. aureus cells (a 3-log reduction in survival). The pro peptides, except for DE/NQ, alone, showed no bactericidal activity against either E. coli or S. aureus (data not shown). However, DE/NQ was bactericidal only against E. coli presumably due to its gained cationicity. When added to 10 µM HNP1, DE/NQ caused a dose-dependent enhancement in E. coli killing, as evidenced by a downward survival curve shown in Figure 5A. By contrast, addition of the wild type pro peptide or any other pro peptide analog to 10 µM HNP1 resulted in a dose-dependent increase in bacterial survival due to defensin inhibition. To facilitate quantitative comparison, we defined EC90 as the effective concentration of pro peptide at which 90% of input cells were killed by 10 µM HNP1 - equivalent to a one-log reduction in bacterial survival. Tabulated in Table 2 are EC90 values generated from the vCC assays for the wild type pro peptide and all 13 pro peptide analogs. The higher the EC90 value of a pro peptide is, the weaker its inhibition of HNP1. Not surprisingly, the wild type pro peptide, with the lowest EC90 values for both E. coli and S. aureus, is among the strongest inhibitors of HNP1.
Figure 5.
Inhibition of the antibacterial activity of HNP1 by pro peptides at 37 °C. The data are from two independent vCC assays for E. coli (panel A) and S. aureus (panel B) with 10 µM HNP1, titrated by the wild type pro peptide (empty circle), Δ19 (diamond), VVVL/4A (filled circle), DE/NQ (filled square), E15A (empty square), or W30A (triangle).
Table 2.
Effective concentrations (EC90, µM) of pro peptide at which 90% of input cells were killed by 10 µM HNP1, determined by vCC assays. The values in the top half of the table were the means of two separate experiments; the entries in the bottom half of the table were averages of three independent experiments performed at a later date.
| S. aureus | E. coli | |||
|---|---|---|---|---|
| EC90 | ratio | EC90 | ratio | |
| WT | 7.8 ± 1.1 | 1.0 | 3.6 ± 0.1 | 1.0 |
| E15A | 9.6 ± 0.5 | 1.2 | 6.8 ± 0.5 | 1.9 |
| W30A | 10.9 ± 1.8 | 1.4 | 7.0 ± 0.0 | 1.9 |
| Δ19 | 74.5 ± 7.8 | 9.6 | 25.6 ± 4.2 | 7.1 |
| DE/NQ | 51.8 ± 1.6 | 6.6 | n.a | |
| VVVL/4A | 137 ± 20 | 18 | 66.4 ± 20.2 | 18 |
| WT | 4.1 ± 0.3 | 1.0 | 2.3 ± 0.3 | 1.0 |
| E1A | 5.8 ± 0.8 | 1.4 | 3.3 ± 0.3 | 1.4 |
| D8A | 5.0 ± 0.6 | 1.2 | 2.8 ± 0.2 | 1.2 |
| E9A | 4.8 ± 0.5 | 1.2 | 3.0 ± 0.7 | 1.3 |
| E15A | 5.7 ± 0.7 | 1.4 | 2.9 ± 0.5 | 1.3 |
| D20A | 8.9 ± 0.5 | 2.2 | 3.3 ± 0.3 | 1.4 |
| E23A | 5.4 ± 1.0 | 1.3 | 3.6 ± 0.3 | 1.6 |
| D31A | 4.7 ± 0.7 | 1.1 | 4.3 ± 1.4 | 1.9 |
| E32A | 4.4 ± 0.5 | 1.1 | 3.2 ± 0.6 | 1.4 |
| Δ9 | 4.9 ± 0.6 | 1.2 | 3.2 ± 0.2 | 1.4 |
| Δ19 | 55.6 ± 22.0 | 14 | 12.6 ± 1.0 | 5.5 |
Effects of truncation
Deletion of the first nine amino acid residues (EPLQARADE) of the pro peptide had little impact on its ability to bind and inhibit HNP1. The Kd and IC50 values of Δ9 for HNP1, 0.17 µM and 0.72 µM, were similar to those of the wild type pro peptide, 0.20 µM and 0.57 µM, respectively. The Δ9 peptide and the wild type pro peptide also showed indistinguishable potency with respect to their inhibition of the bactericidal activity of HNP1, as reflected by similar EC90 values for either E. coli or S. aureus (Table 2). These results suggest that the first nine residues of the pro peptide, including one Asp and two Glu, are unlikely to be involved in inter-molecular interactions with the defensin.
Further truncation by ten amino acid residues (VAAAPEQIAA), however, had markedly different outcomes. The affinity of Δ19 for HNP1 decreased by almost two orders of magnitude compared with the full-length pro peptide (Table 1). The ability of Δ19 to inhibit HNP1 in bactericidal activity assays also significantly weakened, as evidenced by an approximately one-order-of-magnitude increase in EC90 for S. aureus and E. coli (Table 2). Since the ten amino acid residues deleted are largely of hydrophobic nature, these findings suggest that hydrophobic interactions between the pro peptide and HNP1 are important. Additional support to this conclusion comes from charge neutralization studies detailed below.
Effects of charge
Based on the ratios of Kd and IC50 values of the pro peptide analogs to those of the wild type, acidic residues Asp and Glu can be divided into three groups: (1) residues with little impact on HNP1 binding (0.5 < Kd and IC50 ratios < 2); (2) residues with modest impact on HNP1 binding (2 < Kd and IC50 ratios < 10); and (3) residues that significantly impact HNP1 binding (Kd and IC50 ratios > 10). Five acidic residues out of a total of eight, Glu1, Asp8, Glu9, Asp31 and Glu32, belong to the first group (Table 1), and likely make no direct contact with the defensin. Consistent with this finding is the result on the Δ9 pro peptide, where deletion of the first nine amino acid residues, including Glu1, Asp8 and Glu9, caused virtually no change in the ability of Δ9 to bind or inhibit HNP1.
Mutation to Ala of the remaining three acidic residues in the pro peptide, Glu15, Asp20 and Glu23, showed modest effects on HNP1 binding. Ala-substitutions at those positions caused, on average, a 3-fold (by direct binding) or a 5-fold (by competition binding) decrease in binding affinity for HNP1, suggesting that Glu15, Asp20 and Glu23 may be involved in specific electrostatic interactions with cationic residues in HNP1. Although HNP1 contains four cationic Arg residues, since one Arg forms a structurally important salt bridge within the molecule with a Glu residue20, only three are available for potential charge-charge interactions with Glu15, Asp20 and Glu23 of the pro peptide. It is worth noting that the moderately deleterious effect of an E15A mutation was insufficient to account for the huge difference in Kd and IC50 between Δ19 and the wild type pro peptide, underscoring the importance of the hydrophobic residues in the vicinity of Glu15 for HNP1 binding.
Neutralizing all eight negative charges (DE/NQ) decreased the binding affinity of the pro peptide for HNP1 roughly by 15-fold. The weakened binding could be fully accounted for by individual mutations of the three moderately important anionic residues Glu15, Asp20 and Glu23 combined, further supporting that the other five acidic residues, Glu1, Asp8, Glu9, Asp31 and Glu32, are functionally dispensable. This finding also indicates that although important, electrostatic interactions are not a dominant force dictating the binding of the pro peptide to HNP1.
The results from SPR-based binding assays are largely consistent with those from vCC-based functional inhibition assays. On the basis of their respective EC90 values (Table 2), the relative potencies of the pro peptides inhibiting HNP1 are: (wild type, Δ9, E1A, D8A, E9A, D31, E32A) ~ (E15A, D20A, E23A) ~ W30A > DE/NQ > Δ19 > VVVL/4A. However, the mutational effects seen from SPR-based binding studies were significantly attenuated in antibacterial activity assays. The ratios of EC90 values of all eight singly charge-mutated pro peptides to that of wild type ranged from 1.1 to 2.2 (Table 2), making it hard to differentiate E15A, D20A and E23A from the remaining five acidic residues. Nevertheless, the DE/NQ pro peptide displayed an EC90 value for S. aureus consistent with the conclusions drawn from the binding studies.
Effects of hydrophobicity
The central region (residues 10–30) of the pro peptide is mostly hydrophobic, containing 16 aliphatic residues, one Gln, one Ser, and the three anionic residues Glu15, Asp20 and Glu23. Results from Δ9 and Δ19 suggested that the hydrophobic residues surrounding Glu15 of the pro peptide are important for HNP1 binding. Perhaps more strikingly, replacement of the conserved hydrophobic motif Val24Val25Val26Leu28 by four Ala residues largely abolished HNP1 binding. The loss of binding of VVVL/4A to HNP1 in SPR-based direct and competition binding assays is consistent with a significantly reduced inhibition of the bactericidal activity of the defensin by the pro peptide analog. As shown in Table 2, for both E. coli and S. aureus the EC90 values of VVVL/4A increased approximately 18-fold compared with the wild type pro peptide. In fact, VVVL/4A was the weakest pro peptide inhibitor of HNP1 in the panel tested. These results further demonstrate the importance of hydrophobic interactions between HNP1 and its pro peptide.
Direct binding assays to obtain a reliable Kd value for W30A were not possible because the data, for unknown reasons, did not fit to a 1:1 kinetic binding model. The result from competition binding assays indicated that the only Trp residue in the pro peptide region is modestly important for HNP1 binding, as evidenced by a 4-fold increase in IC50 as a result of the Trp-to-Ala mutation. Antimicrobial activity assays showed that while the W30A mutation increased the EC90 value of the pro peptide by a factor of 2 for E. coli, the increase in EC90 for S. aureus was marginal, confirming that the importance of Trp30 for HNP1 binding is likely on par with one of the three moderately important anionic residues (Glu15, Asp20 and Glu23).
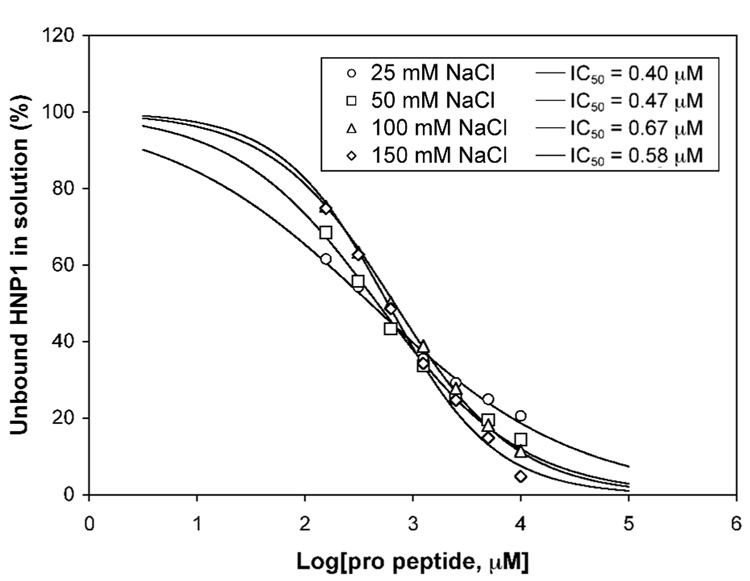
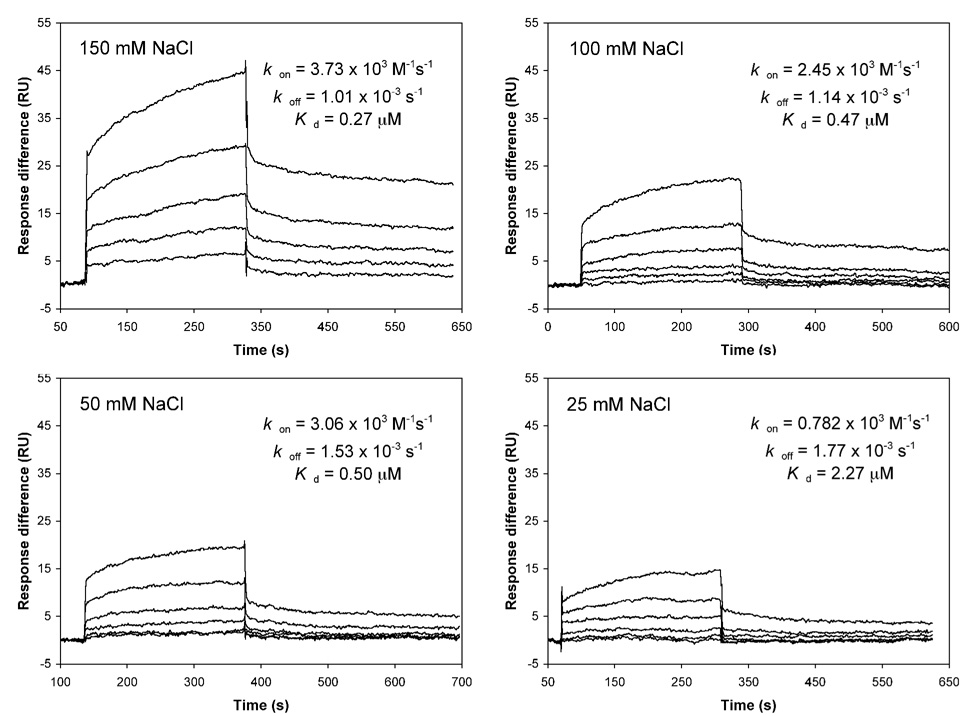
The standard buffer used in SPR experiments contained 150 mM NaCl, which is generally required to disrupt potential non-specific interactions on the chip surface. A high salt concentration may unnecessarily weaken electrostatic interactions between HNP1 and its pro peptide due to increased charge-shielding (or masking) of cationic and anionic side chains by counter ions in solution. To evaluate the effects of salt on the pro peptide binding to HNP1, we also performed SPR measurements at different NaCl concentrations (25, 50, 100, 150 mM) for the wild type pro peptide using both direct binding (on a high-density chip) and competition binding assays. The results are shown in Figure 6 and Figure 7. The IC50 values obtained from competition binding assays, averaging at 0.53 ± 0.10 µM, are largely independent of salt concentration (Figure 6). However, the slope became steeper with increased salt concentration, indicative of a salt-enhanced stronger binding. Salt-enhanced interactions between the pro peptide and HNP1 were more evident in direct binding assays (Figure 7). The Kd value increased by a factor of 2 (from 0.27 µM to 0.5 µM) as the NaCl concentration decreased from 150 mM to 50 mM. A further decrease in NaCl to 25 mM weakened the interaction by an additional 4-fold, attributable mainly to a slower association rate kon. High salt generally weakens electrostatic interactions and strengthens hydrophobic interactions22; 23; 24. This is because desalvation, required for two attracting protein surfaces, becomes more favorable at higher salt concentration for non-polar surface but less favorable for polar surfac22. The fact that an increase in ionic strength enhances binding provides strong evidence supporting hydrophobic rather than electrostatic interactions as the dominant force mediating defensin recognition by its pro peptide. Opposite results would otherwise have ensued22; 23; 24. It should be pointed out, though, that antibacterial activity assays were performed at low salt concentration as high salt is known to inhibit the killing of bacteria by defensins – a process dictated largely by electrostatic interactions between peptide and microbe1; 25; 26; 27.
Figure 6.
Competitive binding of 100 nM HNP1, at different salt concentrations, to immobilized wild type pro peptide in the presence of varying concentrations of the same pro peptide, monitored by surface plasmon resonance (Biacore). Free HNP1 in the solution was deducted from the slopes of sensor grams as described in the legend of Figure 3.
Figure 7.
Direct binding of varying concentrations of wild type pro peptide to immobilized HNP1 in the presence of different concentrations of salt, monitored by surface plasmon resonance (Biacore).
It is generally accepted that hydrophobic interactions are the dominant force in protein folding and stability28; 29. Due to a large difference in heat capacity between the folded and unfolded states of a protein, the enthalpy and entropy are strong functions of temperature, and the free energy of folding is temperature dependent30. Hydrophobic interactions increase in strength with increasing temperature – a process largely driven by entropy at ambient temperature30; 31. We also measured the binding affinity of the pro peptide for HNP1 at 4 °C and 37 °C using the direct SPR binding assay, and found that the Kd value did decrease with temperature, albeit a small difference (0.24 µM at 4 °C and 0.18 µM at 37 °C). It is plausible that the enhancement in binding affinity of the pro peptide for HNP1 was largely cancelled out by an entropy penalty imposed by the disordering of the pro peptide at higher temperatures.
Effects of truncation, charge and hydrophobicity in the context of proHNP1
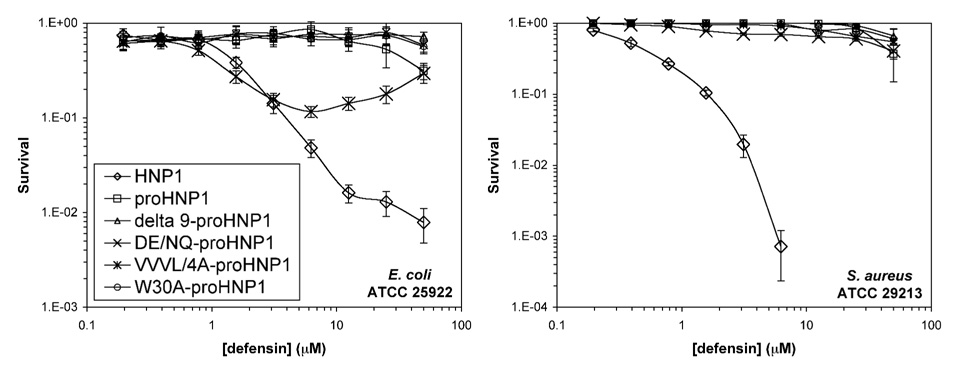
To evaluate the effects of truncation, charge and hydrophobicity of the pro peptide in the context of proHNP1, we compared the ability of wild type proHNP1, Δ9-proHNP1, DE/NQ-proHNP1, W30A-proHNP1 and VVVL/4A-proHNP1 to kill E. coli and S. aureus. Wild type HNP1 was used as a positive control, and the data are plotted in Figure 8. As was previously reported15; 16; 19, the wild type proHNP1 was inactive against both strains. Interestingly, none of the four proHNP1 analogs showed any bactericidal activity against S. aureus, and none, except for DE/NQ-proHNP1, was effective against E. coli within the concentration range used. The marginal bactericidal activity of DE/NQ-proHNP1 was likely a result of the E. coli-killing activity of the pro peptide itself due to gained cationicity. However, the activity of DE/NQ-proHNP1 was significantly less pronounced than that of either HNP1 or the DE/NQ pro peptide, indicative of a reciprocal intra-molecular functional inhibition.
Figure 8.
Survival curves of E. coli and S. aureus exposed to full-length wild type proHNP1 and its analogs. Each curve is the mean of two independent experiments.
Discussion
Defensins kill bacteria through membrane disruption – a process dictated by the interplay of cationicity and hydrophobicity26; 32; 33. The pro regions of α-defensins inhibit the antimicrobial activity of defensins both intra- and inter-molecularly15; 16; 18. This report provides us with a rare glimpse at the molecular basis for the recognition of HNP1 by its pro peptide. Several conclusions can be drawn from functional characterizations of a series of pro peptide analogs interacting with HNP1. First, the majority of negatively charged residues in the pro peptide region are not directly involved in interactions with HNP1. The three acidic residues in the pro region, Glu15, Asp20 and Glu23, are likely candidate partners of three free Arg residues from HNP1. However, electrostatic interactions, while moderately important energetically, are not a dominant force. Second, the first 9 amino acid residues at the N-terminus are functionally dispensable, whereas the succeeding 10 residues of hydrophobic nature are critical for HNP1 binding. Nevertheless, the more important recognition determinants appear to be the conserved hydrophobic motif Val24Val25Val26Leu28 and, to a significantly lesser extent, the adjacent aromatic residue Trp30. Third, disruption of intra-molecular interactions between the pro peptide and the C-terminal defensin domain has limited impact on the bactericidal activity, or lack thereof, of proHNP1. Neutralization of all negative charges in the pro peptide region only partially activates defensin activity – a finding in partial accord with the results reported by Ouellette and colleagues on proCryptdin-417. Taken together, our findings demonstrate that hydrophobic rather than electrostatic interactions are the dominant force mediating the interactions between HNP1 and its pro peptide – a premise perhaps relevant to the observations made by Ganz and colleagues that the hydrophobic residues in the pro region are important for correct proHNP1 trafficking and sorting in neutrophils14.
The conclusions from our study are counter-intuitive, contrasting the commonly held view that electrostatic interactions between the oppositely charged N-terminal pro peptide and C-terminal mature defensin domain result in charge neutralization and inhibition of defensin function. Ouellette and colleagues recently demonstrated with mouse proCryptdin-4 that replacement of all acidic residues by Gly in the pro region converted the functionally inactive pro α-defensin into a fully active molecule with respect to bacterial killing and membrane permeabilization, suggesting that electrostatic force-mediated charge neutralization within proCryptdin-4 is responsible for its lack of activity17. Similarly, the interactions between HNP1 and its pro peptide are considered to be of electrostatic nature15, although definitive biochemical and/or structural evidence is still lacking.
The interacting mode for pro peptide and defensin appears different between proCryptdin-4 and proHNP1. The proCryptdin-4 pro peptide is extremely polar and highly anionic, comprising 4 Gly, 26 polar residues including 11 anionic residues and 2 cationic residues, and only 9 hydrophobic residues sparsely distributed across the entire sequence. By contrast, the proHNP1 pro peptide contains 25 hydrophobic residues, 1 Gly, and 19 polar residues including 8 anionic residues and 4 cationic residues. The difference in amino acid composition between cryptdin-4 and HNP1 is equally striking. While the highly cationic cryptdin-4 consists of 8 Arg, 2 Lys and 1 Glu, HNP1 is composed of 1 Glu and only 4 Arg. On the basis of the amino acid compositions of proCryptdin-4 and proHNP1, it is conceivable that electrostatic forces play a major role in mediating both inter- and intra-molecular interactions between cryptdin-4 and its pro peptide. Replacement of Asp and Glu residues in the pro peptide region should disrupt such interactions, exposing the previously sequestered cationic residues in the C-terminal cryptdin-4 domain for microbial killing17; 34.
We recently demonstrated that the proHNP1 pro peptide catalyzes defensin oxidative folding in vitro, either intra- or inter-molecularly, through two independent mechanisms: solubilization of and interaction with HNP119. Surprisingly, however, a charge-reversing mutant of the pro peptide, where Arg/Lys residues were changed to Asp, and Asp/Glu residues to Lys, showed similar catalytic efficiency. Further, the negatively charged wild type pro peptide and its cationic, charge-reversing analog bound to HNP1 with similar affinities. This report provides an important clue and explanation for the previous observations – that is, hydrophobic interactions between HNP1 and its pro peptide are more important than charge-charge interactions. Our findings reported here also shed light on the propensity of misfolding/aggregation of Δ19-proHNP1 – likely resulting from a combination of weakened intramolecular hydrophobic interactions and poor solubility of the polypeptide.
But, how can we reconcile with the Ouellette’s report on proCryptdin-4 our finding that Δ9-proHNP1, W30A-proHNP1 and VVVL/4A-proHNP1 were totally ineffective in the killing of E. coli and S. aureus, whereas DE/NQ-proHNP1 was only partially active against E. coli? First, disruption of intra-molecular interactions is always strongly disfavored entropically. Therefore, it is plausible that none of the mutations constructed in the context of proHNP1, including the most promising candidate VVVL/4A-proHNP1, was sufficiently disruptive to intra-molecular pro peptide-defensin interactions. Second, the lack of bactericidal activity of proHNP1 is more complex than previously thought. We have shown that charge neutralization is not a prerequisite for defensin inactivation19. For example, covalent attachment of a 10-residue poly Ser sequence to the N-terminus of HNP1 was sufficient to fully inactivate its bactericidal activity against both E. coli and S. aureus19. We suspect that in some pro α-defensins specific charge-charge or hydrohobic interactions are not critical for keeping the C-terminal defensin domain inactive because a covalently attached pro segment itself, depending on its chemical nature, can exert greater influence on defensin oligomerization and defensin-membrane interactions and, ultimately, defensin activity. Third, cryptdin-4 – a much more potent bactericidal peptide than HNP1 due to its significantly higher number of cationic residues – may be more resilient than HNP1 to functionally deleterious modifications.
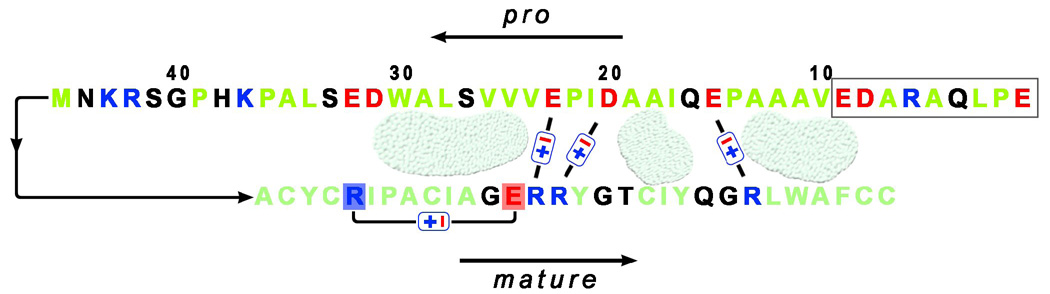
Finally, the hydrophobic mode of interaction for HNP1 and its pro peptide is consistent with known structural features of human α-defensins. Due to small size, α-defensins are structurally stabilized primarily by three disulfides with little or no hydrophobic packing in the core. As a result, most non-polar residues including some disulfide bonds are either fully exposed or partially solvent-accessible. This should provide sufficiently large hydrophobic surfaces for interactions with the non-polar residues from the pro peptide. Perhaps not coincidently, HNP1 carries, on one side of the molecule, three net cationic charges positioned at 14, 15 and 24 (HNP1 numbering) along the second and third β-strands. (The only other cationic residue, Arg5, forms a conserved salt bridge with Glu13 important for defensin stability20.) Although highly speculative, Arg14, Arg15 and Arg24 on HNP1 can conceivably interact with the three functionally important anionic residues from the pro peptide, i.e., Glu15, Asp20 and Glu23, providing additional specificity for the recognition of HNP1 by its pro peptide (Figure 9). Structural studies are needed to unveil at the molecular level residues involved in the defensin-pro peptide interactions.
Figure 9.
Schematic representation of stabilizing interactions between the pro peptide (the top sequence) and mature HNP1 (the bottom sequence). The basic, acidic, and hydrophobic residues are colored in blue, red, and light-green, respectively. The first nine residues of the pro peptide, marked with a box, were shown in this study as dispensable for HNP1 binding and inhibition. Three acidic residues, Glu15, Asp20, and Glu23, that stabilize an interaction with HNP1, may participate in the electrostatic interactions (i.e. salt bridges) with three accessible arginines of mature defensin. Hypothetical hydrophobic contacts between the pro peptide and HNP1 are indicated by grainy light-green shapes. For clarity, every tenth residue of the pro peptide is numbered, and two charged residues of HNP1 (Arg5 and Glu13) forming an intra-molecular salt bridge, thus inaccessible for intermolecular interactions, are also indicated.
Experimental procedures
Synthesis of HNP1 and pro peptide analogs
Peptide synthesis was carried out on an Applied Biosystems 433A synthesizer using an in-house Boc chemistry35; 36, tailored from the HBTU activation and DIEA in situ neutralization protocol previously developed by Kent and coworkers37. Crude peptides, after chain assembly and hydrogen fluoride cleavage/deprotection, were purified by C18 reversed phase high performance liquid chromatography (RP-HPLC). Reduced HNP1 was folded using the oxidative folding protocol described previously35, followed by RP-HPLC purification. Purified peptides were quantified spectroscopically at 280 nm using molar extinction coefficients calculated according to a published algorithm38, and their molecular masses were verified by electrospray ionization mass spectrometry (ESI-MS).
Synthesis of proHNP1 analogs using native chemical ligation
Synthesis of proHNP1 and various proHNP1 analogs using native chemical ligation39; 40 was essentially as described previously16. Briefly, the N-terminal thioester peptides were synthesized on Trityl-SCH2CH2COO-Leu-OCH2-phenylacetamidomethyl resin, to which the C-terminal defensin domain starting with an N-terminal Cys residue was ligated, resulting in a full-length wild type proHNP1 and four proHNP1 analogs. Oxidative folding of proHNP1 and its analogs was carried out as previously described16; 20. To verify correct pro defensin folding, mature HNP1 was cleaved off the folded products by CNBr using a previously detailed protocol16, and analyzed by analytical RP-HPLC and ESI-MS.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) – based kinetic measurements were performed on a Biacore 3000 (Biacore AB, Uppsala, Sweden). The experiments were conducted at 25 °C in HBS-EP buffer (10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20, pH 7.4). HNP1 equivalent to 180 response units (RUs), prepared in 10 mM acetate buffer, pH 5.5, was immobilized to a CM4 sensor chip using the coupling procedures recommended by the manufacture. A reference surface was prepared by a similar procedure without injection of HNP1. The pro peptide and its analogs were injected at 30 µl min−1 for 4 min, followed by a 5-min dissociation. Salt-dependent binding affinity of the wild type pro peptide was determined at different NaCl concentrations of 25, 50, 100 and 150 mM on a similarly prepared high-density (CM4) chip with HNP1 immobilized at 1036 RUs.
A parallel competition assay was also carried out for the synthetic pro peptides. Briefly, 800 RUs of the wild type pro peptide were immobilized (in 10mM acetate buffer, pH 4.0) to a CM4 sensor chip using the amine coupling chemistry. Kinetic analysis of the binding to the wild type pro peptide by 100 nM HNP1, either alone or in the presence of a varying concentration of each pro peptide analog, was carried out at 25 °C in HBS-EP buffer. Defensin HNP1 of 100 nM was incubated at room temperature for 15 min with different concentrations of pro peptide analogs, and the incubation mixture injected at a flow rate of 20 µl/min for 2 min, followed by a 3-min dissociation. The concentration of free HNP1 in solution (not complexed with pro peptide analogs) was deduced, based on the initial rate (slope) of association, from a standardized calibration curve established by kinetic measurements of HNP1 injected alone at different concentrations. Non-linear regression analysis was performed using GraphPad Prism 4 to give rise to IC50 values, concentrations of pro peptide at which 50% of HNP1 was sequestered in pro peptide – HNP1 complexes. Salt-dependent binding affinity of the wild type pro peptide for HNP1 was also determined at varying NaCl concentrations of 25, 50, 100, and 150 mM.
Antibacterial activity assays
To evaluate functional inhibition of HNP1 by its pro peptide and pro peptide analogs, antimicrobial assays against E. coli ATCC 25922 and S. aureus ATCC 29213 were conducted using a previously detailed 96-well turbidimetric method dubbed virtual colony counting41. A twofold dilution series of pro peptide, ranging from 0.4–100 µM, was mixed with 10 µM HNP1 for 30 min in 10 mM sodium phosphate, pH 7.4, and subsequently incubated at 37 °C for 2h with E. coli or S. aureus (1 × 106 CFU/ml), followed by addition of twice-concentrated Mueller-Hinton broth (2 × MHB) and 12-h kinetic measurements of bacterial growth at 650 nm. In addition, direct antimicrobial activities of wild type proHNP1 and four proHNP1 analogs were determined using the virtual colony counting protocol41.
Acknowledgements
We thank Drs. Jing Li and Marzena Pazgier of IHV for useful discussion. This research was supported by the National Institutes of Health Grants AI072732 and AI061482 (to W.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 2.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 3.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 4.Cunliffe RN. Alpha-defensins in the gastrointestinal tract. Mol Immunol. 2003;40:463–467. doi: 10.1016/s0161-5890(03)00157-3. [DOI] [PubMed] [Google Scholar]
- 5.Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- 6.Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI. Primary structures of three human neutrophil defensins. J Clin Invest. 1985;76:1436–1439. doi: 10.1172/JCI112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganz T, Selsted ME, Szklarek D, Harwig SS, Daher K, Bainton DF, Lehrer RI. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76:1427–1435. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones DE, Bevins CL. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. Febs Letters. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 9.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. The Journal of Biological Chemistry. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 10.Valore EV, Ganz T. Posttranslational processing of defensins in immature human myeloid cells. Blood. 1992;79:1538–1544. [PubMed] [Google Scholar]
- 11.Cunliffe RN, Rose FR, Keyte J, Abberley L, Chan WC, Mahida YR. Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut. 2001;48:176–185. doi: 10.1136/gut.48.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, Yadav SP, Crabb JW, Ganz T, Bevins CL. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 13.Porter EM, Poles MA, Lee JS, Naitoh J, Bevins CL, Ganz T. Isolation of human intestinal defensins from ileal neobladder urine. FEBS Lett. 1998;434:272–276. doi: 10.1016/s0014-5793(98)00994-6. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Ganz T. The pro region of human neutrophil defensin contains a motif that is essential for normal subcellular sorting. Blood. 1995;85:1095–1103. [PubMed] [Google Scholar]
- 15.Valore EV, Martin E, Harwig SS, Ganz T. Intramolecular inhibition of human defensin HNP-1 by its propiece. J Clin Invest. 1996;97:1624–1629. doi: 10.1172/JCI118588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z, Prahl A, Powell R, Ericksen B, Lubkowski J, Lu W. From pro defensins to defensins: synthesis and characterization of human neutrophil pro alpha-defensin-1 and its mature domain. J Pept Res. 2003;62:53–62. doi: 10.1034/j.1399-3011.2003.00068.x. [DOI] [PubMed] [Google Scholar]
- 17.Weeks CS, Tanabe H, Cummings JE, Crampton SP, Sheynis T, Jelinek R, Vanderlick TK, Cocco MJ, Ouellette AJ. Matrix metalloproteinase-7 activation of mouse paneth cell pro-alpha-defensins: SER43 down arrow ILE44 proteolysis enables membrane-disruptive activity. J Biol Chem. 2006;281:28932–28942. doi: 10.1074/jbc.M602041200. [DOI] [PubMed] [Google Scholar]
- 18.Satchell DP, Sheynis T, Shirafuji Y, Kolusheva S, Ouellette AJ, Jelinek R. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes. Prosegment inhibition of peptide association with biomimetic membranes. J Biol Chem. 2003;278:13838–13846. doi: 10.1074/jbc.M212115200. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Li X, Ericksen B, de Leeuw E, Zou G, Zeng P, Xie C, Li C, Lubkowski J, Lu WY, Lu W. Impact of pro segments on the folding and function of human neutrophil alpha-defensins. J Mol Biol. 2007;368:537–549. doi: 10.1016/j.jmb.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Li X, de Leeuw E, Ericksen B, Lu W. Why is the Arg5-Glu13 salt bridge conserved in mammalian alpha-defensins? J Biol Chem. 2005;280:43039–43047. doi: 10.1074/jbc.M510562200. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Zou G, Yuan W, Lu W. Defining the native disulfide topology in the somatomedin B domain of human vitronectin. J Biol Chem. 2007;282:5318–5326. doi: 10.1074/jbc.M611396200. [DOI] [PubMed] [Google Scholar]
- 22.Curtis RA, Steinbrecher C, Heinemann M, Blanch HW, Prausnitz JM. Hydrophobic forces between protein molecules in aqueous solutions of concentrated electrolyte. Biophys Chem. 2002;98:249–265. doi: 10.1016/s0301-4622(02)00071-6. [DOI] [PubMed] [Google Scholar]
- 23.Mathew S, Arandjelovic S, Beyer WF, Gonias SL, Pizzo SV. Characterization of the interaction between alpha2-macroglobulin and fibroblast growth factor-2: the role of hydrophobic interactions. Biochem J. 2003;374:123–129. doi: 10.1042/BJ20021655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wintrode PL, Privalov PL. Energetics of target peptide recognition by calmodulin: a calorimetric study. J Mol Biol. 1997;266:1050–1062. doi: 10.1006/jmbi.1996.0785. [DOI] [PubMed] [Google Scholar]
- 25.Zou G, de Leeuw E, Li C, Pazgier M, Zeng P, Lu WY, Lubkowski J, Lu W. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J Biol Chem. 2007;282:19653–19665. doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]
- 26.Fujii G, Selsted ME, Eisenberg D. Defensins promote fusion and lysis of negatively charged membranes. Protein Sci. 1993;2:1301–1312. doi: 10.1002/pro.5560020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehrer RI, Ganz T, Szklarek D, Selsted ME. Modulation of the in vitro candidacidal activity of human neutrophil defensins by target cell metabolism and divalent cations. J Clin Invest. 1988;81:1829–1835. doi: 10.1172/JCI113527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 29.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 30.Privalov PL, Gill SJ. Stability of protein structure and hydrophobic interaction. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 31.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 32.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Straus SK, Hancock RE. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL, Hagen SJ, Ouellette AJ. Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem. 2002;277:5219–5228. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z, Powell R, Lu W. Productive folding of human neutrophil α-defensins in vitro without the pro-peptide. J Am Chem Soc. 2003;125:2402–2403. doi: 10.1021/ja0294257. [DOI] [PubMed] [Google Scholar]
- 36.Wu Z, Ericksen B, Tucker K, Lubkowski J, Lu W. Synthesis and characterization of human alpha-defensins 4–6. J Pept Res. 2004;64:118–125. doi: 10.1111/j.1399-3011.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 37.Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB. In situ neutralization in Boc-chemistry solid phase peptide synthesis. Rapid, high yield assembly of difficult sequences. Int J Pept Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 38.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 40.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 41.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]