Abstract
The modification of chromatin structure is an important regulatory mechanism for developmental gene expression. Differential expression of the mammalian ISWI genes, SNF2H and SNF2L, has suggested that they possess distinct developmental roles. Here we describe the purification and characterization of the first human SNF2L-containing complex. The subunit composition suggests that it represents the human ortholog of the Drosophila nucleosome-remodeling factor (NURF) complex. Human NURF (hNURF) is enriched in brain, and we demonstrate that it regulates human Engrailed, a homeodomain protein that regulates neuronal development in the mid–hindbrain. Furthermore, we show that hNURF potentiates neurite outgrowth in cell culture. Taken together, our data suggess a role for an ISWI complex in neuronal growth.
Keywords: BPTF/Engrailed/ISWI/NURF/SNF2L
Introduction
Modification of histone tails or altering the placement of nucleosomes within the context of a promoter can lead to profound effects on gene expression and, ultimately, on cell function and/or differentiation. As such, mutations in genes encoding chromatin-modifying proteins cause a wide range of human developmental disorders. Such a wide range of defects probably reflects the specificity of the individual complexes that these proteins form (Hendrich and Bickmore, 2001). Indeed, chromatin-remodeling complexes have been identified in a number of species from yeast to humans, and many of these complexes show a high level of conservation in their composition and function. In this regard, Drosophila melanogaster has been shown to be a model organism to define these complexes. For example, three chromatin-remodeling complexes containing the ISWI protein as the ATPase catalytic subunit were identified initially in Drosophila and include NURF (nucleosome-remodeling factor), ACF (ATP-utilizing chromatin assembly and remodeling factor) and CHRAC (chromatin assembly complex), and it is likely that each complex performs unique functions (Tsukiyama et al., 1995; Ito et al., 1997; Varga-Weisz et al., 1997).
Mammalian genomes encode two genes with close homology to Drosophila ISWI, SNF2H and SNF2L (Okabe et al., 1992; Aihara et al., 1998; Lazzaro and Picketts, 2001). In vivo, SNF2H is ubiquitously expressed, with highest expression in populations of actively dividing cells during development and in adults (Lazzaro and Picketts, 2001). Biochemically, SNF2H is the ATPase catalytic core subunit of a variety of complexes including WCRF (WSTF-related chromatin-remodeling factor)/hACF (Bochar et al., 2000; LeRoy et al., 2000), RSF (remodeling and spacing factor) (LeRoy et al., 1998), WICH (WSTF-ISWI chromatin-remodeling complex) (Bozhenok et al., 2002), NoRC (nucleosome-remodeling complex) (Strohner et al., 2001), huCHRAC (Poot et al., 2000) and SNF2H–cohesin (Hakimi et al., 2002). The SNF2H complexes all possess nucleosome spacing activity that results in the formation of regular ordered arrays. Moreover, several of these complexes associate with heterochromatin and facilitate its replication. Taken together, it is suggestive of a role for SNF2H complexes in the establishment and maintenance of heterochromatin and, by extension, gene repression.
Despite a growing number of SNF2H-containing complexes, relatively little is known about the closely related SNF2L protein. Specific antibodies to SNF2L suggest that a small amount of HuCHRAC or WCRF/hACF comprises SNF2L rather than SNF2H, although an SNF2L-containing complex has not been purified (Bozhenok et al., 2002). The absence of a mammalian NURF complex has led to speculation that this may be an SNF2L-specific complex involved in transcriptional activation, yet this remains to be determined. Furthermore, the brain-enriched expression pattern of SNF2L suggests a neurocentric role for this putative complex (Lazzaro and Picketts, 2001).
To address the physiological function of SNF2L, here we report the purification of an SNF2L complex to homogeneity, which corresponds to the human ortholog of Drosophila NURF (dNURF). We demonstrate that human NURF (hNURF) has nucleosome-, and to a lesser extent, DNA-stimulated ATPase activity, and that it can remodel a chromatin template in vitro. Moreover, the brain-enriched hNURF complex interacts at the promoter of two developmentally important genes to activate expression. Importantly, we also demonstrate that this complex may induce neurite outgrowth, thereby suggesting a developmental consequence of its activity.
Results
SNF2L exists in a multiprotein complex
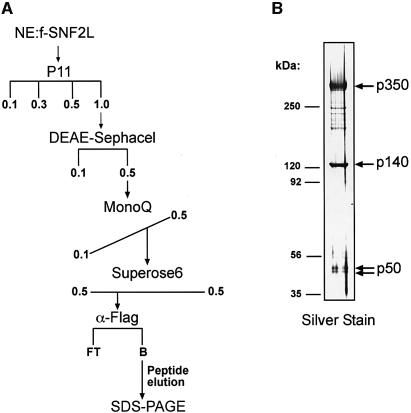
To facilitate the purification of SNF2L complexes, we generated HEK293 cell lines that expressed Flag epitope-tagged human SNF2L (f-SNF2L). This cell line was used due to its detectable but low endogenous levels of SNF2L and its propensity to express neuronal-specific genes (data not shown) (Shaw et al., 2002). We enriched for the SNF2L-containing complex(es) by chromatographically fractionating nuclear extract derived from the f-SNF2L cell line according to the scheme shown (Figure 1A). The enriched fractions were subjected to affinity purification using anti-Flag antibodies. The bound fraction was eluted with a peptide corresponding to the Flag epitope and analyzed by SDS–PAGE followed by silver stain. This analysis revealed three prominent polypeptides of ∼350, 140 and a 50 kDa doublet (Figure 1B) as well as a few substoichiometric bands that proved to be breakdown products of the p350 protein (data not shown).
Fig. 1. Biochemical isolation of the hNURF SNF2L-associated complex. (A) Purification scheme. fSNF2L-HEK293 nuclear extract was fractionated by chromatography as described in Materials and methods. The horizontal and diagonal lines indicate stepwise and gradient elution, respectively. (B) Fractions immunoprecipitated from Superose 6 elutions using M2 anti-Flag beads were resolved on an SDS-polyacrylamide (4–12%) gel, and proteins were visualized by silver staining. Molecular masses of marker proteins (left) and polypeptide masses of associated subunits (right) are indicated.
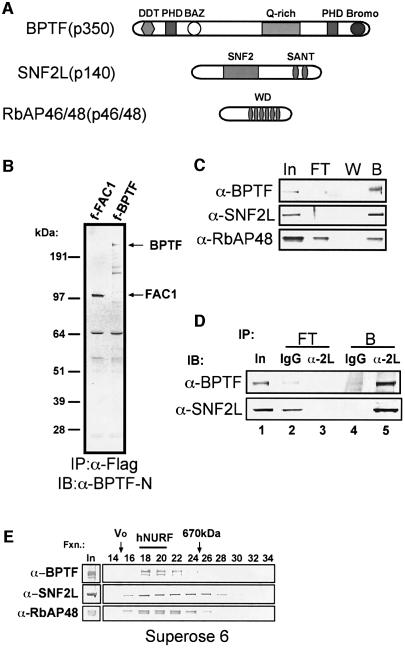
Each subunit of the SNF2L complex was excised from the gel, digested with trypsin, and subjected to sequencing by ion trap mass spectrometry. Forty-nine peptides confirmed our predictions that the 140 kDa band consisted entirely of the SNF2L transgene. Forty-six peptides identified the 350 kDa band as the bromodomain PHD finger transcription factor (BPTF) gene product, while 10 peptides identified the 50 kDa doublet as the retinoblastoma-associated protein 48 and 46 (RbAP48/46) gene products (Figure 2A).
Fig. 2. Composition of the novel SNF2L-associated complex. (A) Schematic of the SNF2L complex subunits. (B) Specificity of BPTF-N antibody for cDNAs expressing Fac1 (BPTF 1–2622) and full-length BPTF. (C) Immunoblot analysis of fractions from M2 anti-Flag beads resolved on an SDS–polyacrylamide (4–12%) gel (I, input; FT, flow through; W, wash; B, bound). Antibodies used for immunoblot are indicated (left). (D) Endogenous hNURF immunoprecipitated from HEK293 partially purified nuclear extract. Immunoblot analysis of immunoprecipitation was resolved on an SDS–polyacrylamide (4–12%) gel. Antibodies used for immunoprecipitation (IP) are indicated across the top, while antibodies used for immunoblot (IB) are on the left. Input represents 5% of IP. Flow through (FT) and bound (B) fractions are as indicated. (E) hNURF molecular mass is ∼1 MDa. Immunoblot of elutions from the Superose 6 gel filtration column from partially purified HEK293 nuclear extract. Fractions (fxn) as well as void (Vo) and thyroglobulin (670 kDa) molecular weight markers are indicated (top).
The SNF2L-associated complex is the human ortholog of dNURF
BPTF and RbAP48/46 are the mammalian orthologs of Drosophila NURF301 and NURF55 polypeptides, respectively. Along with ISWI, these proteins represent three of the four components of the dNURF complex (Martinez-Balbas et al., 1998; Xiao et al., 2001). Therefore, we designated our SNF2L complex as hNURF. The fourth component of dNURF, the NURF38 pyrophosphatase (Gdula et al., 1998), was not identified in our Flag-purified complex despite multiple attempts at purification (data not shown).
BPTF is a known protein with two characterized alternatively spliced gene products. The full-length gene encodes a predicted 311 kDa protein containing a bromodomain, two PHD fingers, three LXXLL putative nuclear receptor-binding motifs, a DDT DNA association domain, a BAZ domain present in many ISWI binding partners, and a glutamine-rich region (Jones et al., 2000) (Figure 2A). Interestingly, the BPTF gene has been localized to chromosome 17q23, which is a hotspot for chromosomal changes in neuroblastomas and is a prognostic factor for rapid disease progression (Lastowska et al., 2001). A truncation of BPTF, called fetal Alzheimer’s clone 1 (FAC1), contains the first 2643 nucleotides of BPTF and was isolated in a screen for proteins in Alzheimer’s disease senile plaques (Bowser et al., 1995). Subsequently, FAC1 was shown to be upregulated in motor neurons during development and in the neurodegenerative disease amyotrophic lateral sclerosis (Mu et al., 1997). FAC1 was also shown to exhibit sequence-specific DNA binding activity and may function in transcriptional regulation (Jordan-Sciutto et al., 1999).
To characterize hNURF further, we cloned the 8295 bp cDNA encoding BPTF. Antibodies were raised against an N-terminal peptide unique to BPTF, and were characterized using ectopically expressed Flag-BPTF as well as a truncated form of BPTF corresponding to the FAC1 alternative splice form (Figure 2B). We confirmed the presence of BPTF and RbAP48 in the original SNF2L immunoprecipitation experiment using the BPTF-N antibodies and commercially available RbAP48 antibodies (Figure 2C). The absence of BPTF in the unbound fraction of the immunoprecipitation suggested that the majority of BPTF in the cell associated exclusively with SNF2L. A fraction of RbAP48 remaining in the unbound fraction represented the RbAP48 in other known complexes such as NuRD (Zhang et al., 1999), E(Z) (Czermin et al., 2002), Sin3 (Zhang et al., 1998) and others.
We isolated hNURF using an ectopic, highly expressed Flag-SNF2L. To confirm the presence of the endogenous hNURF complex, we subjected partially fractionated naïve HEK293 nuclear extract to immunoprecipitation of endogenous SNF2L followed by western analysis for the presence of BPTF. As predicted, SNF2L antibodies immunoprecipitated a significant amount of endogenous SNF2L and BPTF as compared with control antibodies (Figure 2D, compare lanes 4 and 5). This experiment allowed us to ask whether endogenous BPTF is primarily associated with SNF2L, or if it is incorporated into other non-SNF2L complexes. We assayed the unbound fractions of the immunoprecipitations and observed that SNF2L immunodepleted BPTF from this fraction compared with IgG (Figure 2D, compare lanes 2 and 3). This suggests that the majority of cellular BPTF is associated with SNF2L.
The predicted molecular weight of the hNURF complex assuming one copy of each subunit is ∼500 kDa. To confirm this, we measured the molecular weight of the hNURF complex using gel filtration chromatography. We showed that hNURF subunits co-eluted at ∼1 MDa (Figure 2E), which is twice the predicted mass. This observation is in agreement with predicted versus measured molecular weights for other ISWI-containing complexes including WCRF/hACF (Bochar et al., 2000), huChRAC (Poot et al., 2000) and WICH (Bozhenok et al., 2002).
One-step purification of hNURF
The purification scheme employed for the isolation of hNURF (Figure 1A) permitted the characterization of the subunit composition of the complex. However, we found that enzymatic assays on the purified complex were difficult, mainly due to an inherent degradation of the BPTF subunit, presumably from endogenous proteolytic activity. The lability of BPTF was also seen in its Drosophila ortholog, NURF301 (Xiao et al., 2001), resulting in a smaller size (215 kDa) and difficulty in sequencing the peptide. This led us to develop a novel one-step protocol for the isolation of hNURF.
We generated an HEK293 stable cell line expressing a Flag epitope-tagged BPTF cDNA. Nuclear extract from the Flag-BPTF cell line was prepared, subjected to immunopreciptation with Flag beads, washed stringently, and eluted with Flag peptide. SDS–PAGE fractionation of immunoprecipitated proteins followed by silver staining demonstrates the presence of an electrophoretically pure hNURF complex (Figure 3) containing a BPTF subunit with minimal proteolysis. We confirmed the identity of the subunits by western analysis using BPTF, SNF2L, RbAP48 and RbAP46 antibodies. As in our previous purification, both silver (Figure 3) and colloidal staining (data not shown) failed to demonstrate the presence of the human ortholog of one of the dNURF subunits, the p38 pyrophosphatase.
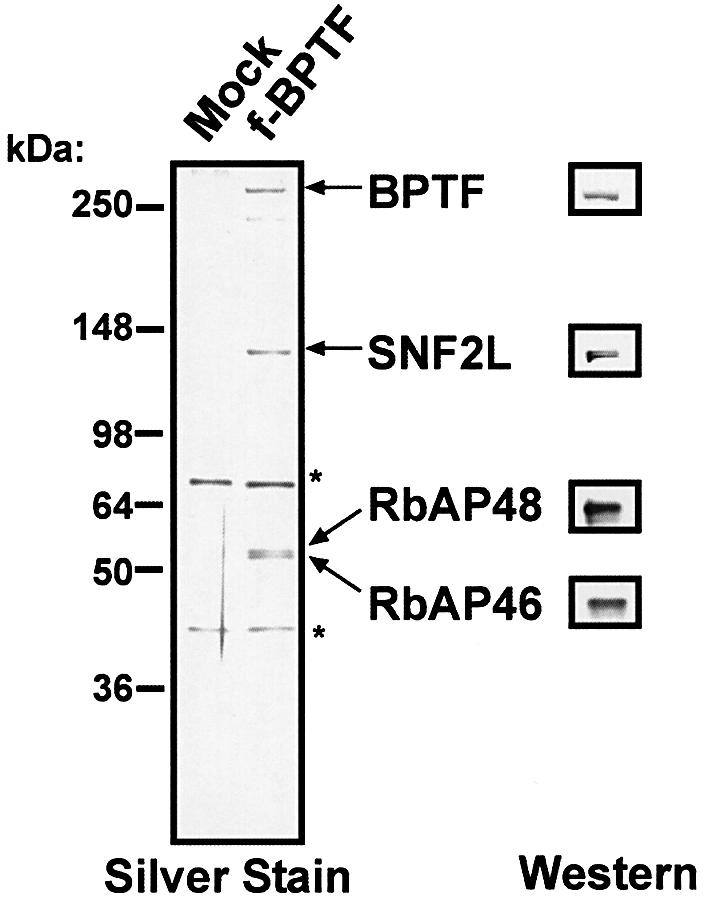
Fig. 3. hNURF purified with a one-step purification protocol. Mock and Flag-BPTF eluted fractions were fractionated by 8–16% SDS–PAGE followed by silver staining and western analysis. BPTF, SNF2L, RbAP48 and RbAP46 poplypeptides are indicated on the right. Western blot analysis performed in parallel confirms the identification of the polypeptides. Polypeptides marked with asterisks are contaminants also present in the mock elution. Molecular weight markers are indicated on the left.
hNURF remodels chromatin
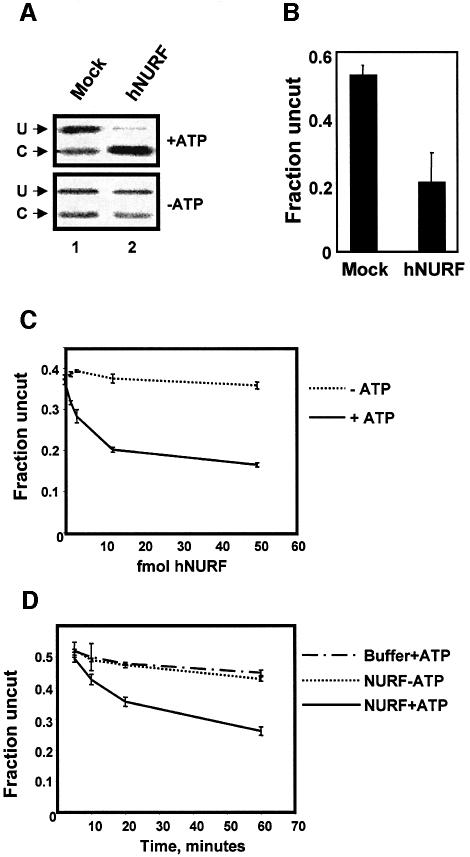
The hNURF complex, by virtue of its SNF2L subunit, is predicted to function as a chromatin-remodeling complex. To confirm this, we performed chromatin-remodeling assays comparing purified hNURF with mock immunoprecipitations from HEK293 nuclear extract. The chromatin-remodeling activity of hNURF was assessed using the restriction enzyme accessibility assay, which relies on the ability of DNA incorporated into chromatin to be refractory to restriction endonuclease digestion. Upon addition of a chromatin-remodeling protein (or complex), mobilization of chromatin at the restriction endonuclease site is facilitated, resulting in an ATP-dependent decrease in undigested array.
In the presence of an elution from a mock immunoprecipitation, we find that ∼50% of the DNA remained uncut (U) and thus refractory to restriction enzyme cleavage (Figure 4A, lane 1). The cut (C) band represented the basal level of accessible DNA in the assay. Upon addition of purified hNURF, we found a potent decrease in the level of uncut (U) DNA along with the concomitant increase in cut (C) DNA (Figure 4B, lane 2). Consistent with the fact that chromatin remodeling is coupled to release of energy from ATP hydrolysis, hNURF decreased the levels of the uncut (U) DNA in an ATP-dependent manner (Figure 4B, lane 2, compare top and bottom panels). Densitometric analysis of restriction enzyme accessibility assays performed in triplicate demonstrated a significant (∼2.5-fold) decrease in the fraction of uncut DNA (Figure 4B).
Fig. 4. hNURF is a chromatin remodeler. (A) hNURF remodels chromatin. Autoradiograph of the restriction endonuclease-coupled remodeling assay demonstrating chromatin-remodeling activity. Mock IP failed to demonstrate any decrease in the uncut (U) array in an ATP-dependent manner (compare top and bottom panel of lane 1). Addition of purified hNURF results in a qualitative decrease in the uncut (U) array compared with mock (compare lanes 1 and 2, top panel). This activity is ATP dependent (compare lane 2 top and bottom panels). (B) hNURF addition results in an ∼2.5-fold decrease in the fraction of the uncut array. Densitometric analysis of restriction endonuclease-coupled remodeling. The graph represents the averages of the quantified fraction of the uncut array (U/U + C) from three experiments. Standard errors for the experiments are as indicated in the graph. (C) hNURF exhibits dose-dependent chromatin-remodeling activity. Graphical representation of densitometric analysis of assays over 0, 1.25, 2.5, 5 and 50 fmol of hNURF with or without ATP. Points represent the average of three reactions, with standard error as indicated. (D) Time course of hNURF-mediated chromatin remodeling. Restriction enzyme accessibility for buffer + ATP, 50 fmol hNURF-ATP or 50 fmol hNURF + ATP. Points represent the average of three reactions, with standard error as indicated.
By titrating in increasing amounts of hNURF and maintaining saturating amounts of ATP and nucleosomal array, we found that as few as 1.25 fmol of hNURF remodeled the nucleosomal array in an ATP-dependent manner (Figure 4C). We then measured the rate of hNURF-mediated remodeling. At all time points, hNURF demonstrated ATP-dependent remodeling activity indicated by the decrease in the fraction of uncut array compared with hNURF without ATP or with restriction enzyme alone (Figure 4D). We calculated the specific activity of hNURF in the remodeling assay at 15 U/mg of SNF2L (1 U = 1 pmol of array digested/min). This translates to ∼1 pmol of hNURF remodeling 2 fmol of array per min. This activity is of the same order as the published measured activity of another chromatin- remodeling complex, ySWI/SNF, using the identical assay (Logie and Peterson, 1997).
hNURF possesses intrinsic ATPase activity
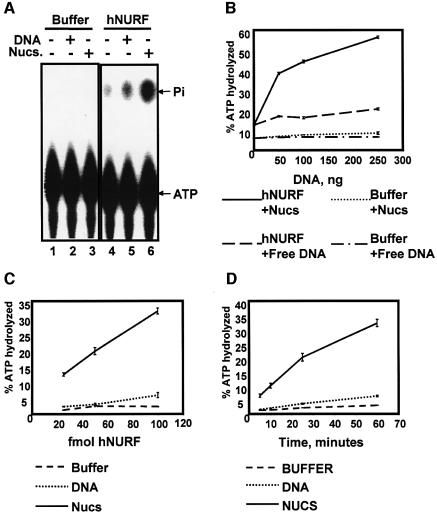
Recombinant monomeric ISWI proteins and ISWI- containing complexes such as WCRF/hACF and ChRAC exhibit an intrinsic ATPase activity that is activated by addition of naked DNA and further potentiated by the addition of nucleosomes (Varga-Weisz, 1997; Bochar et al., 2000; Aalfs et al., 2001). Among ISWI complexes, dNURF is unique in that its ATPase activity is exclusively nucleosome dependent (Xiao et al., 2001). We predicted that hNURF would behave similarly to its Drosophila counterpart. In fact, we found that hNURF exhibited a slight but measurable DNA-dependent ATPase activity (Figure 5A, compare lanes 5 and 4). However, the hNURF ATPase activity was far more pronounced upon addition of nucleosomes (Figure 5A, compare lanes 6 and 5). Indeed, upon titration of the two hNURF substrates, we found that nucleosomes consistently displayed a stronger potentiation of ATPase activity (Figure 5B). These data, along with previously published ATPase assays using dNURF (Xiao et al., 2001), suggest that the physiological substrate of the NURF complex may indeed be nucleosomes rather than free DNA.
Fig. 5. hNURF exhibits intrinsic nucleosome-dependent ATPase activity. (A) hNURF exhibits DNA- and nucleosome-stimulated ATPase activity. ATPase activity assays performed on [γ-32P]ATP followed by separation of free phosphate (Pi) from ATP by PEI-cellulose TLC and autoradiography of TLC plates. hNURF activity is stimulated by supplementing the reaction with DNA (50 ng) and is potentiated further by nucleosomes (50 ng); compare lanes 4, 5 and 6. (B) hNURF activity responds to increasing amounts of nucleosomes. A 25 fmol concentration of hNURF or buffer was incubated with increasing amounts (0, 50, 100 or 250 ng) of either free DNA or free nucleosomes. Points represent the average of three reactions, with standard error as indicated. (C) hNURF exhibits a dose-dependent ATPase activity in the presence of nucleosomes. ATPase activity of three different hNURF concentrations (20, 50 and 100 fmol) with buffer, free DNA or nucleosomal DNA. Points represent the average of three reactions, with standard error as indicated. (D) Time course of hNURF ATPase activity. hNURF (100 fmol) was incubated with buffer, free DNA or nucleosomal DNA, and reactions were assayed for ATPase activity at the indicted times. Points represent the average of three reactions, with standard error as indicated.
Further characterization of hNURF revealed that its ATPase activity was dose dependent, with as little as 20 fmol of hNURF displaying potent nucleosome- dependent ATPase activity (Figure 5C). The specific activity of the hNURF nucleosome-stimulated ATPase activity determined by measuring the rate of ATP hydrolysis (Figure 5D) was calculated at 200 U/mg of SNF2L (1 U = 1 pmol of ATP hydrolyzed per min). Consistent with the minimal responsiveness to DNA, the specific activity of the DNA-stimulated ATPase activity was found to be ∼100-fold lower than with nucleosomes.
SNF2L and BPTF co-localize within adult mouse brain to hippocampus and cerebellum
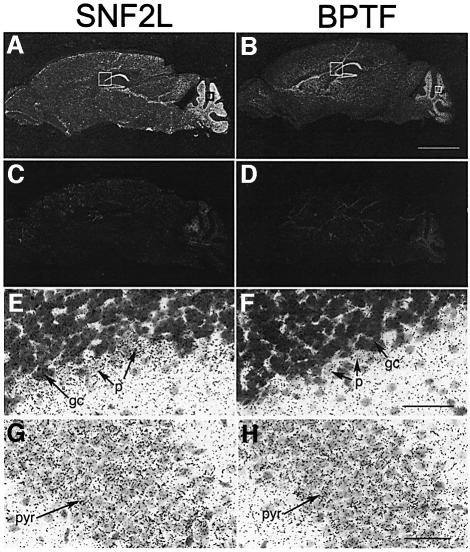
Mouse SNF2L has been shown to exhibit a restricted expression pattern, which included higher expression levels in the central nervous system (Lazzaro and Picketts, 2001). Since the majority of BPTF appeared to be associated exclusively with SNF2L in HEK293 cells, we postulated that the BPTF gene should have a similar expression profile to SNF2L in the brain. As such, adult mouse brain was analyzed for expression of SNF2L and BPTF by RNA in situ hybridization. Our results show that the BPTF transcript is highly expressed throughout the hippocampus and in the cerebellum, which is nearly identical to the SNF2L expression profile (Figure 6A and B) (Lazzaro and Picketts, 2001). The specificity of the hybridization is demonstrated by the lack of signal with sense SNF2L and BPTF probes (Figure 6C and D, respectively). Higher magnification of cresyl violet-counterstained sections showed that the SNF2L and BPTF transcripts are both localized within the granule and Purkinje cell layer of the cerebellum (Figure 6E and F) and in the pyramidal neurons throughout the hippocampus (Figure 6G and H). The BPTF and SNF2L expression profiles suggest that the hNURF complex is involved in the regulation of cerebellar and hippocampal function. In addition, analysis of BPTF and SNF2L reveals a similar pattern of expression during embryonic development (Supplementary figure 1 available at The EMBO Journal Online).
Fig. 6. SNF2L and BPTF show overlapping expression in adult mouse brain. Adjacent sections from an adult mouse brain were hybridized with SNF2L (A, E and G) and BPTF (B, F and H) antisense RNA probes. Both SNF2L and BPTF were highly expressed throughout all regions of the hippocampus (CA1–4) and in the granule cell layer of the cerebellum (A and B dark field). The specificity of SNF2L and BPTF antisense probes is demonstrated by the lack of hybridization of corresponding sense probes (C and D). Higher magnifications of sections shown in (A) and (B) counterstained with cresyl violet demonstrate the expression of SNF2L and BPTF in the granule and Purkinje cell layer of the cerebellum (E and F) and in the pyramidal neurons of the hippocampus (G and H). Scale bars, 2 mm in (B); 50 µm in (F) and (H); gc, granule cells; p, Purkinje neurons; CA1 pyr, CA1 pyramidal neurons.
In vivo localization of hNURF to human engrailed promoters
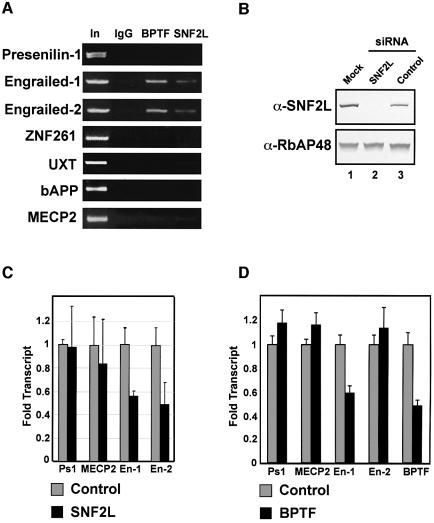
The mammalian homologs of the Drosophila segment polarity genes engrailed-1 (en-1), engrailed-2 (en-2), wnt-7b and Pax-2 have been shown to have a crucial role in cerebellar development (Millen et al., 1995). Mice lacking either en-1 or en-2 demonstrate mid–hindbrain malformations and cerebellar dysgenesis, respectively (Wurst et al., 1994; Millen et al., 1995). Interestingly, expression of Drosophila engrailed was severely compromised in flies mutated for ISWI or NURF301 expression (Deuring et al., 2000; Badenhorst et al., 2002). These results and the overlapping expression of SNF2L and BPTF that we observed in the cerebellum prompted us to examine the human en-1 and en-2 genes as possible targets of the hNURF complex. To this end, we employed chromatin immunoprecipitation (ChIP) assays using antibodies against two components of the hNURF complex, SNF2L and BPTF. To demonstrate specificity, we assayed for their presence at other promoters important in various biological processes including neuronal development (Johnston et al., 2001; Tanzi and Bertram, 2001). While our antibodies failed to show an interaction between hNURF and presenilin-1, znf261, uxt, βAPP or Mecp2 promoters, we successfully demonstrated a specific interaction with the promoters of both En-1 and En-2 (Figure 7A). The regions demonstrating the strongest hNURF binding correspond to ∼1 kb upstream on the en-1 and en-2 transcription start sites. Interestingly, while the en-1 promoter is still relatively uncharacterized, the en-2 promoter was found to have an upstream activation element proximal to the hNURF-binding sites (McGrew et al., 1999).
Fig. 7. hNURF localizes to and regulates human Engrailed. (A) ChIP assays localize hNURF specifically to engrailed-1 (en-1) and engrailed-2 (en-2) promoters. Chromatin from HEK293 cells was subjected to formaldehyde cross-linking followed by sonication and immunoprecipitation with non- specific, BPTF and SNF2L antibodies. After extensive washes, bound DNA was eluted and subjected to PCR with the indicated promoter primers. A specific and reproducible signal at the engrailed promoters comparable with input (In) was detected in the BPTF and SNF2L IPs but not in the non-specific IgG. Input fractions represent 1% of the total IP. Multiple primer pairs from each promoter were employed. Non-specific promoters include presenilin-1, znf261, UXT, APP and MECP2 as indicated. A representative set is displayed. (B) siRNA-mediated depletion of SNF2L. HEK293 cells transfected with either mock, SNF2L-specific siRNAs or non-specific siRNAs. Extract from cells 72 h after transfection were prepared and separated by SDS–PAGE followed by western analysis for SNF2L levels. The SNF2L-specific siRNA depletes SNF2L to undetectable levels compared with mock and non-specific siRNAs (compare lane 2 with lanes 1 and 3, top panel). Lower panel: loading control for protein levels determined by RbAP48 levels. (C) Depletion of SNF2L leads to a significant decrease in en-1 and en-2 transcripts. Total RNA was extracted from SNF2L-depleted and control HEK293 cells and subjected to reverse transcription followed by real-time PCR. Levels of transcripts were quantified and normalized to β-actin levels using the ABI7000 sequence detection system. Genes lacking hNURF at their promoters as determined by ChIP were used as controls. The bar graph depicts the average of three independent experiments. Standard errors are indicated. (D) Depletion of BPTF leads to a significant decrease in en-1 transcript. Experiments were performed as in (C). The bar graph depicts the averages of three independent experiments. Standard errors are indicated.
hNURF regulates expression of endogenous human engrailed
The localization of the hNURF complex at the en-1 and en-2 promoters prompted us to assess the role of hNURF in regulating their expression. Small interfering (si)RNAs that are specific for SNF2L were generated and transfected into HEK293 cells, and expression of SNF2L was checked by western blot. The SNF2L siRNAs reduced expression of SNF2L to below detectable levels, while control siRNAs had no effect (Figure 7B).
Using the siRNAs for SNF2L, we then assayed for transcript levels of endogenous en-1 and en-2 in the presence or absence of hNURF. Total RNA was extracted from SNF2L siRNA- and control siRNA-transfected cells, reverse transcribed to generate cDNAs, and subjected to real-time PCR. Transcript levels were quantified and normalized to β-actin levels using the ABI7000 sequence detection software. Transcript levels of PS1 and MECP2, which lack hNURF at their promoters, are unaffected by depletion of SNF2L. Importantly, en-1 and en-2 transcript levels show a significant reduction (2-fold) in response to depletion of SNF2L (Figure 7C). We attempted similar experiments using siRNAs against BPTF, and demonstrated a significant decrease in en-1 transcript level (Figure 7D). However, we were unable to affect en-2 transcript levels. This may be due to the fact that the siRNAs against BPTF reduced its concentration by only 2-fold (Figure 7D), which may not be sufficient to alter en-2 expression. Supporting this notion are reports in the literature demonstrating substantial differences in en-1 and en-2 regulation (Hanks et al., 1995).
Taken together, our data reveal that hNURF associates with the promoters of En-1 and En-2 and can modulate expression of en-1 and probably en-2. Furthermore, since loss of En-1 and En-2 causes defects in the formation of the cerebellum (Joyner and Martin, 1987; Wurst et al., 1994; Millen et al., 1995), these data suggest a role for hNURF in the regulation of genes required for normal brain development.
SNF2L promotes neurite outgrowth
The functional maturation of the developing nervous system relies, in part, on the formation of axonal and dendritic processes that extend from neuronal cell bodies, and on the ability of these processes to form proper synaptic connections. Several factors, including the Engrailed homeodomain protein (Cosgaya et al., 1998), are known to promote outgrowth of neuritic processes. Since the two human orthologs of Engrailed are regulated by hNURF, we asked whether SNF2L would promote neurite outgrowth.
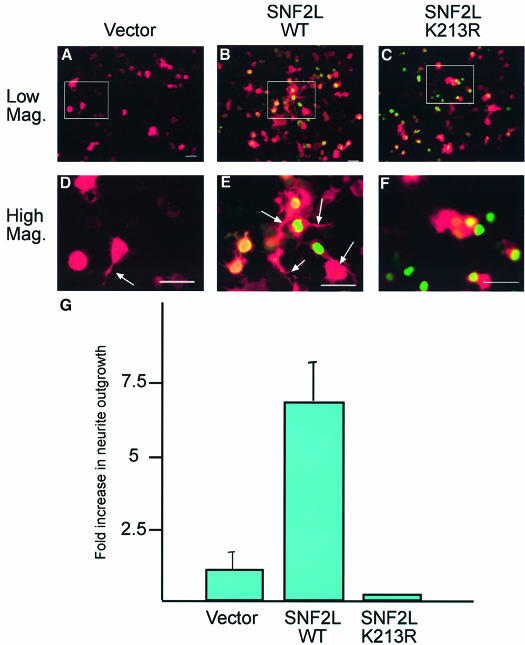
To investigate this, we utilized the N1E-115 mouse neuroblastoma cell line, which can be induced to differentiate morphologically to a neuronal phenotype and extend neurites when cultured in conditions of reduced serum (Hirose et al., 1998). We asked whether uninduced N1E-115 cells undergo differentiation following transfection with SNF2L. Cells were transfected with empty vector, SNF2L-WT, or SNF2L-K213R, which contains a point mutation in the nucleotide-binding P-loop motif, abrogating all ATPase activity. Co-transfection of a β-galactosidase vector permitted visualization of neurites by immunofluorescence microscopy. At 30 h post-transfection, a comparison of the ability of each transfected plasmid to promote morphological differentiation and neurite outgrowth was assessed by counting cells with neurite extensions measuring more than twice the length of the cell body.
Cells transfected with empty vector exhibited a very low, but measurable, rate of uninduced neurite outgrowth (Figure 8A and D). However, transfection of wild-type SNF2L resulted in a significant potentiation of neurite outgrowth (Figure 8, compare B and E with A and D). This ability to promote neurite outgrowth was dependent on the enzymatic activity of SNF2L since the SNF2L-K213R ATPase-dead mutant virtually abolished all neuronal outgrowth (Figure 8, compare C and F with B and E). Similarly, the specificity for SNF2L was demonstrated further by a failure of SNF2H to induce neurite extension (data not shown). Quantification of these results demonstrates that, indeed, transfection of SNF2L-WT promotes neurite outgrowth while SNF2L-K213R renders outgrowth undetectable (Figure 8F). The promotion of neurite extension that we observed following transfection of SNF2L is similar to that observed for other factors that modulate neurite outgrowth (Kobayashi et al., 2002; Li et al., 2002; Abe et al., 2003). We believe that the ectopically expressed SNF2L acts via incorporation into the NURF complex; however, we cannot rule out the possibility that the neurite outgrowth phenotype could have resulted from unincorporated SNF2L. While these data were derived from a tissue culture-based neurite outgrowth assay, they lend further support to the hypothesis that SNF2L may play a role in the differentiation and maturation of neurons during development.
Fig. 8. SNF2L promotes neurite outgrowth. N1E-115 cells were co-transfected transiently with vector (A and D), f-SNF2L-WT (B and E) or f-SNF2L-K213R (C and F) along with a β-galactosidase expression plasmids in a 10:1 ratio. After transfection, cells were cultured for 30 h in normal (non-inducing) growth medium and then fixed for immunofluorescence staining (Flag = green, β-galactosidase = red). Differentiated cells were identified as cells in which the length of the neurite extensions was at least twice the diameter of the cell body. The fold increase in neurite outgrowth in f-SNF2L-transfected cells compared with vector-transfected cells was calculated from cell counts (600 cells) obtained from three different experiments (G) and is statistically significant at P < 0.005. Scale bars, 50 µm.
Discussion
The Drosophila ISWI protein exists in three multiprotein complexes, namely, ACF, CHRAC and NURF (Tsukiyama et al., 1995; Varga-Weisz et al., 1997; Ito et al., 1999). Mammalian complexes corresponding to ACF and CHRAC have been purified and contain the SNF2H protein (Bochar et al., 2000; Poot et al., 2000). Additional unique mammalian ISWI complexes have also been purified, including RSF, WICH, NoRC and SNF2H–cohesin (LeRoy et al., 2000; Strohner et al., 2001; Bozhenok et al., 2002; Hakimi et al., 2002), and these all comprise the SNF2H protein. Despite the growing list of mammalian ISWI complexes, a NURF equivalent or complexes containing the related protein SNF2L are notably absent. Here, we describe the purification of the first human SNF2L complex. The subunit composition suggests that it represents the human ortholog of the dNURF complex. In this regard, the hNURF complex also contains BPTF and RbAP46/48. Surprisingly, hNURF does not contain the inorganic pyrophosphatase protein NURF38. Nonetheless, the biochemical activity of hNURF is similar as it displayed predominantly nucleosome-stimulated ATPase activity, as well as potent chromatin-remodeling activity on oligonucleosomal arrays.
The brain-enriched expression profile of SNF2L prompted us to examine a role for hNURF in neuronal physiology. We showed that SNF2L chromatin-remodeling activity could induce neurite outgrowth in a tissue culture-based assay, and that this was specific to SNF2L-containing ISWI complexes since SNF2H expression did not result in a similar induction (data not shown). The conversion of a neuroblast to a differentiated neuron will require the modification of chromatin structure at numerous genes, for both activation and repression, and it is not likely to be restricted to the NURF complex. Nonetheless, our studies suggest that hNURF has a role in this process and, thus, identification of target genes will help elucidate the molecular pathways. In this regard, we clearly demonstrate that hNURF can regulate the mammalian engrailed genes, through a direct interaction at the promoters of these two homeotic loci. The murine engrailed genes are critical regulators of mid–hindbrain development, as ablation leads to animals that are missing most of the colliculi and cerebellum (Wurst et al., 1994). Although engrailed was identified previously as a NURF target gene through the characterization of flies harboring mutant ISWI or NURF301 genes (Deuring et al., 2000; Badenhorst et al., 2002), a neural defect was not appreciated due to the early lethality of these animals. As such, this may represent a novel function for the NURF complex.
The effect of chromatin-remodeling complexes on development is a well-established phenomenon. Linkages between chromatin remodeling and developmental disorders include ATRX and mental retardation (Picketts et al., 1996), SMARCAL1 and Schimke immuno-osseous dysplasia (Boerkoel et al., 2002), CSB and Cockayne syndrome (Citterio et al., 2000), and SNF2H and William’s syndrome (Bochar et al., 2000). Based on our results, we hypothesize that the hNURF complex may represent another connection of a chromatin-remodeling protein to disorders of development, and we are confident that the hNURF complex regulates other developmentally important genes. In this regard, the analysis of flies ablated for the NURF complex also suggests a role for the hNURF complex in hematopoietic development and the regulation of chromosome structure (Badenhorst et al., 2002). However, such studies in mammals must await further dissection using in vivo model systems.
Materials and methods
Purification of the hNURF complex
hNURF was purified from 350 mg of fSNF2L-HEK293 nuclear extract. Nuclear extract was loaded on a 100 ml column of phosphocellulose (P11, Whatman, USA) and fractionated stepwise by the indicated KCl concentrations in buffer A [20 mM Tris–HCl pH 7.9, 0.2 mM EDTA,10 mM β-mercaptoethanol, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride (PMSF)]. The 1.0 M KCl fraction (25 mg) of P11 was loaded on a 5 ml DEAE–Sephacel column (Amersham Biosciences, Sweden) and eluted with 0.5 M KCl. The 0.5 M KCl elution (12.5 mg) was dialyzed to 0.1 M KCl in buffer A containing 1 µg/ml aprotinin, leupeptin and pepstatin, and loaded on a Mono Q HR 5/5 column (Amersham Biosciences, Sweden). The column was resolved by using a linear 10 column volume gradient of 100–500 mM KCl in buffer A containing 1 µg/ml aprotinin, leupeptin and pepstatin. SNF2L-containing fractions were fractionated on a Superose 6 HR 10/30 column (Amersham Biosciences, Sweden) equilibrated in 0.5 M KCl in buffer A containing 0.1% NP-40, 1 µg/ml aprotinin, leupeptin and pepstatin. Fractions 18–20 were subjected to immunoaffinity purification using M2 anti-Flag antibody-conjugated agarose beads (Sigma, USA). Beads were washed with 0.5 M KCl in IP buffer [20 mM Tris–HCl pH 7.9, 0.2 mM EDTA, 1 mM dithiothreitol (DTT), 10% glycerol, 0.2 mM PMSF] containing 1 µg/ml aprotinin, leupeptin and pepstatin, and bound fractions were eluted with 400 µg/ml of Flag peptide (Sigma, USA) in 0.1 M KCl in IP buffer. Purified hNURF was subjected to SDS–PAGE followed by silver staining and immunoblot. Alternatively, nuclear extract was prepared from Flag-BPTF-expressing cells and subjected to immunoprecipitation as indicated. Beads were washed with 0.5 M KCl and 0.5% NP-40 for 10 min, followed by 0.75 M KCl and 0.1% NP-40 in IP buffer for 10 min, followed by 1.0 M KCl with 0.1% NP-40 in IP buffer for 10 min. Beads were subjected to elution as described for the previous purification.
Mass spectrometric peptide sequencing
Excised bands were subjected to N-terminal sequence analysis as described (Bochar et al., 2000).
In situ hybridization
For in situ hybridization experiments, an adult mouse brain cDNA was used to amplify by PCR a 475 bp fragment corresponding to a segment of the 3′-coding region and untranslated region of human BPTF. The mouse BPTF fragment was cloned into pBluescript KS as a template for RNA probes. Brains dissected from adult mice perfused with 4% paraformaldehyde in 100 mM NaPO4 pH 7.4 were processed for cyosectioning. Antisense and sense Snf2l and Bptf RNA probes were labeled with [33P]UTP (2000 Ci/mmol) (Amersham Biosciences, Sweden) by in vitro transcription from plasmid templates using T3 and T7 RNA polymerases (Promega, USA). Cryosections were processed for hybridization, hybridized with probes and exposed to autoradiographic emulsion as described previously.
Restriction endonuclease-coupled remodeling assay
Reconstitution of nucleosomal arrays and chromatin-remodeling assays were performed as described (Logie and Peterson, 1997). More details are provided in the Supplementary data.
Purification of nucleosomes
Nucleosomes were purified from HeLa cell nuclear pellet as described (Utley et al., 1996). More details can be found in the Supplementary data.
ATPase assays
Measurement of ATPase activity was performed in 10 µl reactions under the following conditions: 20 mM Tris–HCl, 60 mM KCl, 4% glycerol, 4 mM MgCl2, 1 mM cold ATP, 1 µCi of [γ-32P]ATP and 2 nmol of hNURF or an equivalent volume of elution buffer. Where indicated, the reaction was supplemented with 50 ng of naked DNA or nucleosomes. Reactions were performed at 30°C for 1 h. Free phosphate and ATP were separated by TLC on PEI-cellulose plates (J.T.Baker, USA). A 1 µl aliquot of the reaction was spotted onto a plate and TLC was carried out in 1 M formic acid, 0.5 M LiCl. Plates were then allowed to dry and were visualized by exposure to a phosporimager cassette (Molecular Dynamics, Sweden) for densitometric analysis, or film (Kodak, USA). For rate assays, reaction mix was pre-warmed to 30°C prior to addition of hNURF.
Chromatin immunoprecipitation (ChIP)
ChIP experiments and buffers were performed as described in the Upstate ChIP protocol. More details are available in the Supplementary data.
Quantitative PCR
Transcript levels were determined by quantitative PCR using an ABI7000 (Applied Biosystems, USA). PCR was performed using the Sybr Green 2x Master Mix (Applied Biosystems, USA) as per the manufacturer’s protocol. Unknowns were amplified in triplicate. Positive control reactions were performed to establish a standard curve for each primer pair for transcript level normalization. β-actin served as an internal control.
Neurite outgrowth assay
N1E-115 cells were seeded at 50% confluency 24 h prior to transfection. Cells were transfected with pCDNA3 vector, pCDNA3-f-SNF2L-WT or pCDNA3-f-SNF2L-K213R and β-galactosidase expression plasmids in a 10:1 ratio. Cells were maintained in normal growth medium for 30 h post-transfection. Cells were then fixed for immunofluorescent staining. Transfected cells and neurites were visualized by Flag and β-galactosidase antibodies. Differentiated cells were identified as cells in which the length of neurite extensions was at least twice the diameter of the cell body. For quantification experiments, 600 cells were counted.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank D.Bochar for comments and experimental advice, J.Neveu, J.Asara and R.Robinson for their expertise in HPLC and mass spectrometry, Dr Robert Bowser for Flag-FAC1 cDNA, the National Cell Culture Center (Minneapolis, MN) for propagation of fSNF2L-293 cells, and the Wistar Institute protein microchemistry/mass spectrometry facility. R.S. was supported by grants from the NIH (GM61204) and the American Cancer Society. O.G.B. was supported by the NIH cancer grant (CA09171-28). M.A.L. was supported by the Ontario Mental Health Foundation. D.J.P. is a CIHR New Investigator with work supported by a CIHR grant (MOP53224).
References
- Aalfs J.D., Narlikar,G.J. and Kingston,R.E. (2001) Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem., 276, 34270–34278. [DOI] [PubMed] [Google Scholar]
- Abe T., Kato,M., Miki,H., Takenawa,T. and Endo,T. (2003) Small GTPase Tc10 and its homologue RhoT induce N-WASP-mediated long process formation and neurite outgrowth. J. Cell Sci., 116, 155–168. [DOI] [PubMed] [Google Scholar]
- Aihara T., Miyoshi,Y., Koyama,K., Suzuki,M., Takahashi,E., Monden,M. and Nakamura,Y. (1998) Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet. Cell Genet., 81, 191–193. [DOI] [PubMed] [Google Scholar]
- Badenhorst P., Voas,M., Rebay,I. and Wu,C. (2002) Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev., 16, 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar D.A., Savard,J., Wang,W., Lafleur,D.W., Moore,P., Cote,J. and Shiekhattar,R. (2000) A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl Acad. Sci. USA, 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel C.F. et al. (2002) Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat. Genet., 30, 215–220. [DOI] [PubMed] [Google Scholar]
- Bowser R., Giambrone,A. and Davies,P. (1995) FAC1, a novel gene identified with the monoclonal antibody Alz50, is developmentally regulated in human brain. Dev. Neurosci., 17, 20–37. [DOI] [PubMed] [Google Scholar]
- Bozhenok L., Wade,P.A. and Varga-Weisz,P. (2002) WSTF–ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J., 21, 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E., Van Den Boom,V., Schnitzler,G., Kanaar,R., Bonte,E., Kingston,R.E., Hoeijmakers,J.H. and Vermeulen,W. (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair–transcription-coupling factor. Mol. Cell. Biol., 20, 7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgaya J.M., Aranda,A., Cruces,J. and Martin-Blanco,E. (1998) Neuronal differentiation of PC12 cells induced by engrailed homeodomain is DNA-binding specific and independent of MAP kinases. J. Cell Sci., 111, 2377–2384. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi,R., McCabe,D., Seitz,V., Imhof,A. and Pirrotta,V. (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell, 111, 185–196. [DOI] [PubMed] [Google Scholar]
- Deuring R. et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell, 5, 355–365. [DOI] [PubMed] [Google Scholar]
- Gdula D.A., Sandaltzopoulos,R., Tsukiyama,T., Ossipow,V. and Wu,C. (1998) Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev., 12, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M.A., Bochar,D.A., Schmiesing,J.A., Dong,Y., Barak,O.G., Speicher,D.W., Yokomori,K. and Shiekhattar,R. (2002) A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature, 418, 994–998. [DOI] [PubMed] [Google Scholar]
- Hanks M., Wurst,W., Anson-Cartwright,L., Auerbach,A.B. and Joyner,A.L. (1995) Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science, 269, 679–682. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hendrich B. and Bickmore,W. (2001) Human diseases with underlying defects in chromatin structure and modification. Hum. Mol. Genet., 10, 2233–2242. [DOI] [PubMed] [Google Scholar]
- Hirose M. et al. (1998) Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J. Cell Biol., 141, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Bulger,M., Pazin,M.J., Kobayashi,R. and Kadonaga,J.T. (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell, 90, 145–155. [DOI] [PubMed] [Google Scholar]
- Ito T., Levenstein,M.E., Fyodorov,D.V., Kutach,A.K., Kobayashi,R. and Kadonaga,J.T. (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev., 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M.V., Jeon,O.H., Pevsner,J., Blue,M.E. and Naidu,S. (2001) Neurobiology of Rett syndrome: a genetic disorder of synapse development. Brain Dev., 23, S206–S213. [DOI] [PubMed] [Google Scholar]
- Jones M.H., Hamana,N. and Shimane,M. (2000) Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics, 63, 35–39. [DOI] [PubMed] [Google Scholar]
- Jordan-Sciutto K.L., Dragich,J.M. and Bowser,R. (1999) DNA binding activity of the fetal Alz-50 clone 1 (FAC1) protein is enhanced by phosphorylation. Biochem. Biophys. Res. Commun., 260, 785–789. [DOI] [PubMed] [Google Scholar]
- Joyner A.L. and Martin,G.R. (1987) En-1 and En-2, two mouse genes with sequence homology to the Drosophila engrailed gene: expression during embryogenesis. Genes Dev., 1, 29–38. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Taniura,H. and Yoshikawa,K. (2002) Ectopic expression of necdin induces differentiation of mouse neuroblastoma cells. J. Biol. Chem., 277, 42128–42135. [DOI] [PubMed] [Google Scholar]
- Lastowska M., Van Roy,N., Bown,N., Speleman,F., Roberts,P., Lunec,J., Strachan,T., Pearson,A.D. and Jackson,M.S. (2001) Molecular cytogenetic definition of 17q translocation breakpoints in neuroblastoma. Med. Pediatr. Oncol., 36, 20–23. [DOI] [PubMed] [Google Scholar]
- Lazzaro M.A. and Picketts,D.J. (2001) Cloning and characterization of the murine imitation switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem., 77, 1145–1156. [DOI] [PubMed] [Google Scholar]
- LeRoy G., Orphanides,G., Lane,W.S. and Reinberg,D. (1998) Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science, 282, 1900–1904. [DOI] [PubMed] [Google Scholar]
- LeRoy G., Loyola,A., Lane,W.S. and Reinberg,D. (2000) Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem., 275, 14787–14790. [DOI] [PubMed] [Google Scholar]
- Li X., Meriane,M., Triki,I., Shekarabi,M., Kennedy,T.E., Larose,L. and Lamarche-Vane,N. (2002) The adaptor protein Nck-1 couples the netrin-1 receptor DCC (deleted in colorectal cancer) to the activation of the small GTPase Rac1 through an atypical mechanism. J. Biol. Chem., 277, 37788–37797. [DOI] [PubMed] [Google Scholar]
- Logie C. and Peterson,C.L. (1997) Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J., 16, 6772–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Tsukiyama,T., Gdula,D. and Wu,C. (1998) Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl Acad. Sci. USA, 95, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew L.L., Takemaru,K., Bates,R. and Moon,R.T. (1999) Direct regulation of the Xenopus engrailed-2 promoter by the Wnt signaling pathway and a molecular screen for Wnt-responsive genes, confirm a role for Wnt signaling during neural patterning in Xenopus. Mech. Dev., 87, 21–32. [DOI] [PubMed] [Google Scholar]
- Millen K.J., Hui,C.C. and Joyner,A.L. (1995) A role for En-2 and other murine homologues of Drosophila segment polarity genes in regulating positional information in the developing cerebellum. Development, 121, 3935–3945. [DOI] [PubMed] [Google Scholar]
- Mu X., Springer,J.E. and Bowser,R. (1997) FAC1 expression and localization in motor neurons of developing, adult and amyotrophic lateral sclerosis spinal cord. Exp. Neurol., 146, 17–24. [DOI] [PubMed] [Google Scholar]
- Okabe I., Bailey,L.C., Attree,O., Srinivasan,S., Perkel,J.M., Laurent,B.C., Carlson,M., Nelson,D.L. and Nussbaum,R.L. (1992) Cloning of human and bovine homologs of SNF2/SWI2: a global activator of transcription in yeast S.cerevisiae. Nucleic Acids Res., 20, 4649–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts D.J., Higgs,D.R., Bachoo,S., Blake,D.J., Quarrell,O.W. and Gibbons,R.J. (1996) ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum. Mol. Genet., 5, 1899–1907. [DOI] [PubMed] [Google Scholar]
- Poot R.A., Dellaire,G., Hulsmann,B.B., Grimaldi,M.A., Corona,D.F., Becker,P.B., Bickmore,W.A. and Varga-Weisz,P.D. (2000) HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J., 19, 3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G., Morse,S., Ararat,M. and Graham,F.L. (2002) Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J., 16, 869–871. [DOI] [PubMed] [Google Scholar]
- Strohner R., Nemeth,A., Jansa,P., Hofmann-Rohrer,U., Santoro,R., Langst,G. and Grummt,I. (2001) NoRC—a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J., 20, 4892–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi R.E. and Bertram,L. (2001) New frontiers in Alzheimer’s disease genetics. Neuron, 32, 181–184. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Daniel,C., Tamkun,J. and Wu,C. (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell, 83, 1021–1026. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Owen-Hughes,T.A., Juan,L.J., Cote,J., Adams,C.C. and Workman,J.L. (1996) In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol., 274, 276–291. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P.D., Wilm,M., Bonte,E., Dumas,K., Mann,M. and Becker,P.B. (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature, 388, 598–602. [DOI] [PubMed] [Google Scholar]
- Wurst W., Auerbach,A.B. and Joyner,A.L. (1994) Multiple developmental defects in Engrailed-1 mutant mice: an early mid–hindbrain deletion and patterning defects in forelimbs and sternum. Development, 120, 2065–2075. [DOI] [PubMed] [Google Scholar]
- Xiao H., Sandaltzopoulos,R., Wang,H.M., Hamiche,A., Ranallo,R., Lee,K.M., Fu,D. and Wu,C. (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell, 8, 531–543. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng,H.H., Erdjument-Bromage,H., Tempst,P., Bird,A. and Reinberg,D. (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev., 13, 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]