Abstract
Molecular tethers play a central role in the organization of the complex membrane architecture of eukaryotic cells. p115 is a ubiquitous, essential tether involved in vesicle transport and the structural organization of the exocytic pathway. We describe two crystal structures of the N-terminal domain of p115 at 2.0 Å resolution. The p115 structures show a novel α-solenoid architecture constructed of 12 armadillo-like, tether-repeat (TR), α-helical tripod motifs. We find that the H1 TR binds the Rab1 GTPase involved in ER to Golgi transport. Mutation of the H1 motif results in the dominant negative inhibition of ER to Golgi trafficking. We propose that the H1 helical tripod contributes to the assembly of Rab-dependent complexes responsible for the tether and SNARE-dependent fusion of membranes.
Keywords: vesicle transport, membrane trafficking, membrane tethering, p115, membrane fusion, SNARE, Rab GTPase, armadillo repeats
Introduction
The membrane architecture of eukaryotic cells comprising the exocytic (endoplasmic reticulum (ER), Golgi, cell surface) and endocytic (endosomes, lysosomes) pathways is organized by the action of cytosolic and membrane-associated proteins comprising the membrome.1 Rab GTPase families regulate the dynamic assembly and disassembly of protein complex hubs that integrate membrome component function with vesicle tethering, docking and fusion.2-4 Tethering is an essential step in membrane trafficking and is now thought to encompass, from a structural point of view, a large group of α-helical rich, coiled-coiled proteins, the Golgins5-14, multi-subunit transport protein particle (TRAPP)15-20 and conserved oligomeric Golgi (COG) complexes21-26, and numerous Rab effectors that contribute to the generation of soluble N-ethylmaleimide sensitive attachment protein receptor (SNARE) fusion complexes.3,4,22,27-32
p115 is a tether protein that has been extensively studied to date.2,12,33 From rotary shadowing34, p115 was suggested to be a myosin-like homodimer with an N-terminal globular head (∼70 kDa) and a smaller (∼30 kDa) C-terminal, extended coil-coiled domain containing four prototypical, heptads repeats (CC1-4) involved in SNARE assembly35 terminated by a short acidic domain that auto-regulates Golgi docking events.36 The yeast homolog of p115, Uso1p, is essential for cell viability and shows genetic and physical interactions with yeast Rab1 (Ypt1)34, thereby stressing a key role for p115 in the normal function of the exocytic pathway required for cell proliferation. In mammalian cells, p115 is involved in transport of cargo from the ER to the Golgi and the structural integrity of the Golgi33,37-47, and for recovery of the Golgi stack following disassembly during mitosis.48-50 p115 function is facilitated by interaction with two other Golgin family tethers, GM130 and giantin.35,39,51-53 Consistent with its central role in membrane organization, p115 is a target for apoptotic caspases 3 and 8 that leads to inactivation and fragmentation of the exocytic pathway and amplification of programmed cell death54,55 Numerous molecular and biochemical studies suggest that cytosolic p115 is anchored to membranes by the Ras-superfamily GTPase Rab1 and is involved in the assembly of SNARE complexes to promote bilayer fusion.35,37,43,47,48,56,57 The structural mechanism(s) by which p115 functions to promote tethering and SNARE assembly is unknown. Herein, we present the crystal structure of the N-terminal globular domain of the p115 dimer and suggest a role the N-terminal H1 tripod in Rab1-mediated ER to Golgi trafficking.
Results
Overall structure
The N-terminal domain of bovine tethering factor p115 (residues 1 to 651) (p115Nt), was expressed using bacterial systems and purified by Ni-nitrilotriacetic acid (NTA) affinity, ion-exchange, and gel filtration chromatography (see Material and Methods). Monomer p115Nt crystallized from a p115 monomer pool based on gel filtration chromatography and diffracted to 2.0 Å. A multiwavelength anomalous diffraction (MAD) data set at 2.7 Å from a seleno-L-methionine derivative was used for the initial phase calculation (Table 1). The p115Nt monomer structure revealed a right-handed, α-solenoid structure58 composed exclusively of helices and loops (Fig. 1A). The overall shape of the p115Nt monomer corresponds to a bowed cylinder 115 Å in length and 55 Å in diameter.
Table 1. Data collection, phasing and refinement statistics.
| Data collection | |||||
|---|---|---|---|---|---|
| Monomer (3grl) | Dimer (3gq2) | ||||
| Native | Se-Met derivative | Native | |||
| Space group | C2221 | P21 | |||
| Unit cell | a = 155.6 Å, | a = 156.4 Å, | a = 78.2 Å, | ||
| b = 174.7 Å, | b = 174.6 Å, | b = 159.8 Å, | |||
| c = 87.5 Å | c = 87.4 Å | c = 81.4 Å, | |||
| β = 115.1° | |||||
| Wavelength (Å) | peak | inflection | remote | ||
| 1.000 | 0.9794 | 0.9796 | 0.9537 | 1.100 | |
| Resolution (Å) | 50.00 – 2.00 | 50.00 – 2.70 | 50.00 – 2.18 | ||
| (2.03 – 2.00)a | (2.75 – 2.70) a | (2.25 – 2.18) a | |||
| Observed reflections | 342,057 | 237,930 | 238,874 | 237,523 | 287,986 |
| Unique reflections | 79,589 | 61,434 | 61,939 | 63,557 | 91,304 |
| Completeness (%) | 98.7 (94.6) a | 99.9 (100.0) a | 100.0 (100.0) a | 100.0 (100.0) a | 96.7 (76.4) a |
| I/σ | 34.3 (1.5) a | 31.6 (5.5) a | 26.2 (3.9) a | 18.6 (2.1) a | 14.3 (1.2) a |
| Rsym (%) | 4.9 (54.6) a | 5.2 (24.9) a | 6.2 (35.0) a | 9.0 (61.7) a | 9.1 (59.3) a |
| Phasing | |||||
| Number of sites | 12 | ||||
| FOM after SOLVE | 0.53 | ||||
| FOM after RESOLVE | 0.70 | ||||
| FOM after DM and phase extension | 0.60 | ||||
| Refinement | |||||
| Resolution (Å) | 40.00 – 2.00 | 20.00 – 2.18 | |||
| Rcryst/Rfree | 17.8/20.4 | 19.9/25.4 | |||
| Number of protein atoms | 4,691 | 9,694 | |||
| Number of solvent atoms | 390 | 607 | |||
| Mean B value of protein (Å2) b | 35.4 | 31.4 | |||
| Mean B value of solvent (Å2) b | 41.0 | 32.3 | |||
| Rmsd bond length (Å) | 0.012 | 0.012 | |||
| Rmsd angle (°) | 1.25 | 1.36 | |||
| Ramachandran plot | 95.8% favored, | 93.4% favored, | |||
| 3.4% allowed | 5.1% allowed | ||||
Rsym = Σ|Ii-<Ii>| / Σ|Ii| where Ii is the scaled intensity of the ith measurement and <Ii> is the mean intensity for that reflection.
Rcryst = Σ| |Fobs|-|Fcalc| | / Σ|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 4.9% of the total reflections chosen at random and omitted from refinement.
Highest resolution shell.
These values represent the residual B that following TLS refinement.
Fig. 1.
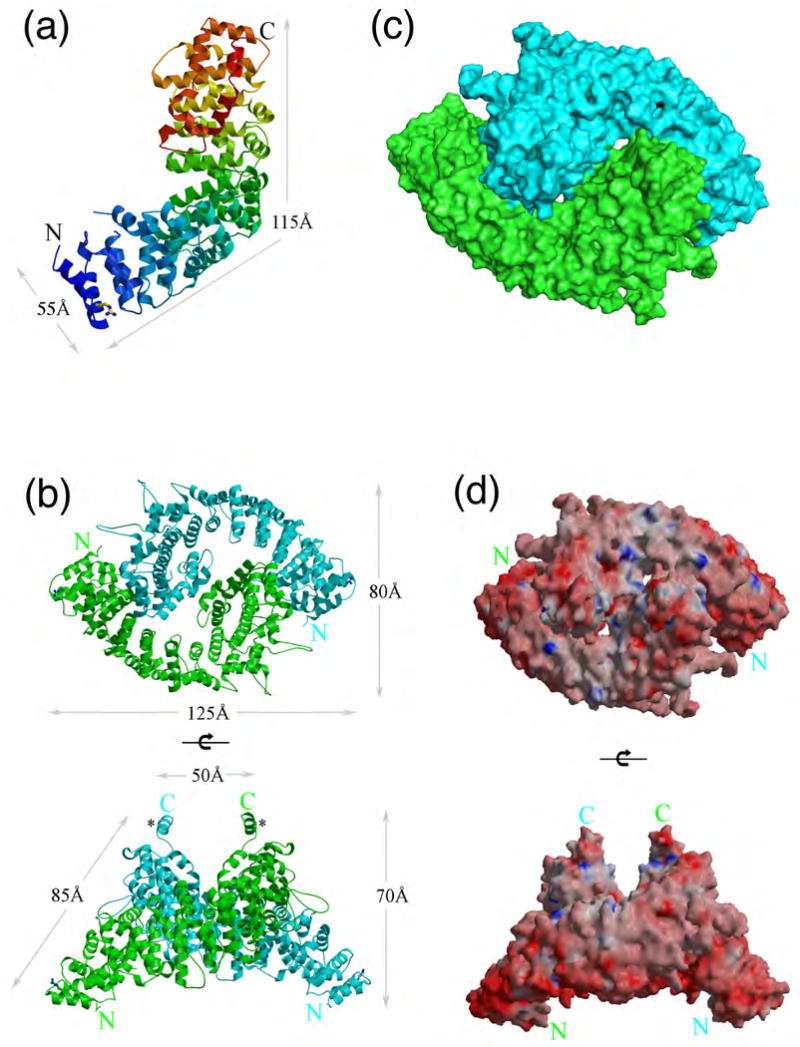
Structure of p115Nt. (a) Overall architecture of p115Nt monomer in a ribbon representation. The color of ribbon changes from blue to red gradually from N- to C-terminus of p115Nt. Arg39 involved in stabilization of the N-terminal region is shown in ball-and-stick representation. (b) p115Nt dimer in a ribbon representation. The two chains of the dimer are shown in green and cyan, respectively. Top view of p115Nt dimer with the C-termini pointing toward the viewer (upper panel); side view (lower panel). The additional C-terminal helix acting as the thumb in the left-hand-shake like dimer structure is indicated by the asterisk (*) in side view. (c) Surface representation of the dimer in the same orientation and colors as in 1b. (d) Electrostatic surface potential of p115Nt dimer. Red surfaces indicate negative potential and blue positive (±15 kit/e); top view (upper panel), side view (lower panel). Figures 1 and 2 were prepared with MOLSCRIPT/BOBSCRIPT115, Raster3D116 and Pymol112.
p115Nt crystals were also obtained from a dimer pool of p115Nt, based on gel filtration chromatography, that diffracted to 2.18 Å resolution. The dimer has two molecules in the asymmetric unit, while the monomer structure has one molecule in the asymmetric unit. The dimer assembly is not observed after applying the symmetry operators in the monomer crystals. Superposition of the monomer onto each chain of the dimer did not reveal any major conformation changes [root-mean-square-deviations (rmsds) range from 1.5 Å (main chain) to 1.8 Å (all atoms)].59 The p115Nt dimer reveals a Yin and Yang assembly consisting of two single α-solenoids that adopt a left hand-shake like structure, with the α-solenoid acting as the palm and an additional helix at the C-terminus, which is disordered in the p115Nt monomer, acting as the thumb in the dimer structure (Figs. 1B, 1C). The buried surface area of the p115 homodimer interface is 810 Å2 per protomer60,61 and contains three distinct groups of residues around Arg195-Asn243, Glu528-Glu533, and Ser585-Gln595 (Fig. 2). A total of 111 interactions are made in the dimer interface, including 4 salt-bridges, 23 hydrogen bonds, and 84 van der Waals contacts (Fig. 2).62-64 The surface electrostatic potential65 of the p115 dimer revealed both negatively and positively charged regions (Fig. 1D), possibly reflecting distinct, ligand-binding surfaces. We did not observed density for residues 1-18 for either monomer or dimer crystals, suggesting that they may be disordered.
Fig. 2.
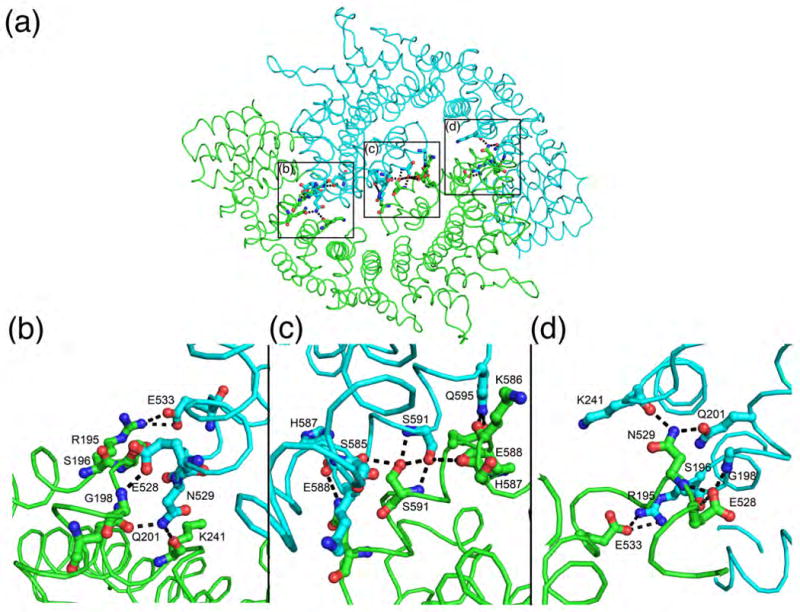
Salt-bridges and hydrogen bond interactions involved in p115Nt dimerization. Hydrogen bonds and salt bridges are shown as dashed lines, with interacting residues in ball-and stick representation.
Structurally similar proteins
A search for structurally homologous proteins using the Dali server66 revealed that importin α, a key transport protein in selective nuclear import67-69, had the highest similarity to p115 (Z score of 23.0 for subunits of the dimer) (Table S1). Importin α contains a tandem array of ten helical, tripod-like, armadillo-consensus sequence repeats, organized in a right-handed α/α/α-solenoid. High similarity was also observed for the α-solenoid armadillo repeat of β-catenin70, the armadillo-like repeat of the chaperone HspBP171, and, more weakly, for importin β that contains helical-hairpin (α1-loop-α2) HEAT repeats that form an α/α solenoid (Table S1).72-74
Structural organization of p115 repeat motifs
Each subunit of the dimer contains 12 tandem, triple-helical (α1-α2-α3) repeats that forms the right-handed α/α/α-solenoid, referred to herein as the tether-repeat (TR) motif (Fig. 3A, Table S2). The p115 TR is similar to that of armadillo repeats despite the fact that each repeat sequence is different with only the distinct H1 and H2 TR motifs being evolutionarily conserved, suggesting that H1 and H2 TRs have functionally different roles in docking and/or fusion (Table S2; see below). The first 11 TRs form a continuous right-handed superhelix, and an incomplete C-terminal tether repeat (lacking helix 3) forms a cap region (Fig. 3A). The p115Nt subunit is composed of TRs that are arranged roughly parallel, with neighboring repeats separated by an average translation of 11 Å. A rotation of approximately 30° (for the first 11 TRs) between adjacent repeats results in a right-handed, superhelical twist along the entire length of the molecule (Fig. 3A). The tight and contiguous packing of the helices leads to an extended hydrophobic core along the length of the solenoid. Packing of TRs is further stabilized by hydrogen bonds and salt bridges from hydrophilic residues that are accessible to solvent.
Fig. 3.
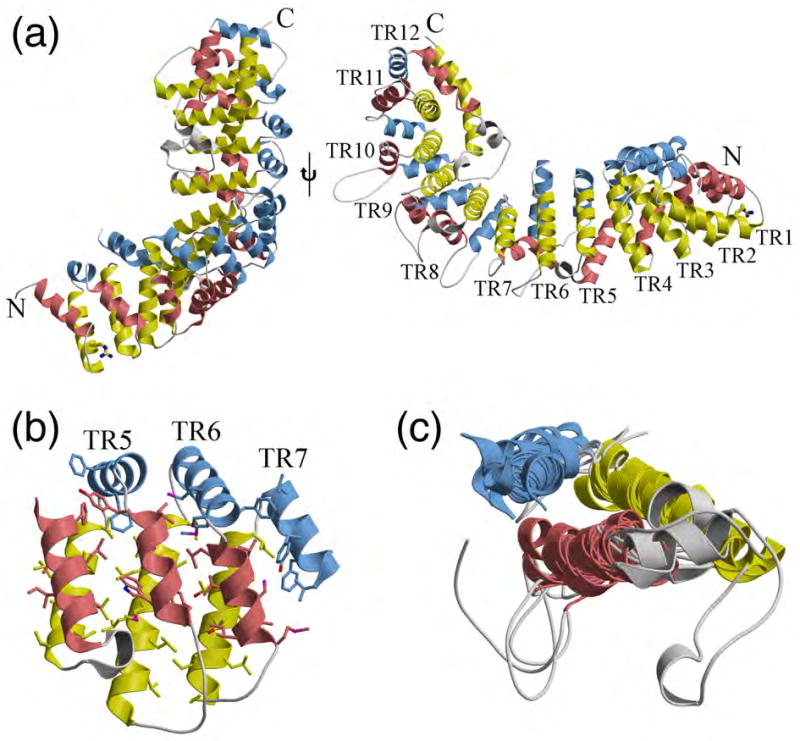
Tether repeat (TR) motif. (a-b) Structural features of the 12 TRs in the p115Nt monomer. Except for the C-terminal TR, each TR includes three α helices, which are shown in salmon, yellow and light blue, respectively. The insertions between any adjacent helices are colored gray. Arg39 is shown in ball-and-stick representation; side view (left panel), top view (right panel). (b) The conformation of three adjacent tether repeats (5th, 6th and 7th) highlights the tight packing of the conserved hydrophobic core throughout the solenoid, where the hydrophobic residues involved in stabilizing the solenoid are shown in ball-and-stick. (c) Superimposition of the 12 TR motifs in p115Nt revealing the conserved structural organization.
Despite the high variability in the amino-acid sequences within each TR, they share very similar overall structures (Fig. 3B). The concave (inner) surface of the p115 N-terminal superhelix is formed from the longest helix 2 (yellow) of the 11 TRs. The convex (outer) surface contains more diverse secondary structure elements, including a variety of different length loops and short helices, which is formed primarily from helices 1 (salmon) and 3 (light blue) of the TRs and their connecting regions. The loop insertions between any two adjacent helices vary from 0 to 23 residues, except for the 5th, 8th and 12th TRs that have additional helical insertions between helices 1 and 2. The rmsds between pairs of TRs range from 1.1 to 3.2 Å (main chain). Superimposition of TRs 1-11 reveals a remarkably conserved structural fold comprising a helical tripod (rmsd- 1.87 Å (main chain) (Fig. 3C). The incomplete TR found at the C-terminus also follows the right-handed, superhelical twist formed by previous 11 TRs, but with a rotation of approximately 75° instead of 30° from the 11th TR (Fig. 4). This incomplete TR is stabilized through extensive interactions to the concave (inner) surface (helix 2) of tether repeats 8, 9, 10 and 11, which results in this motif acting as a C-terminal cap region of the superhelix formed by the previous 11 TRs, and provides a means to link to the extended coiled-coiled, C-terminal region of p115 (residues 652 to 961), likely through the linking residues 666-700 that exhibit a low coiled-coiled, predicted score (PAIRCOIL2).75,76
Fig. 4.
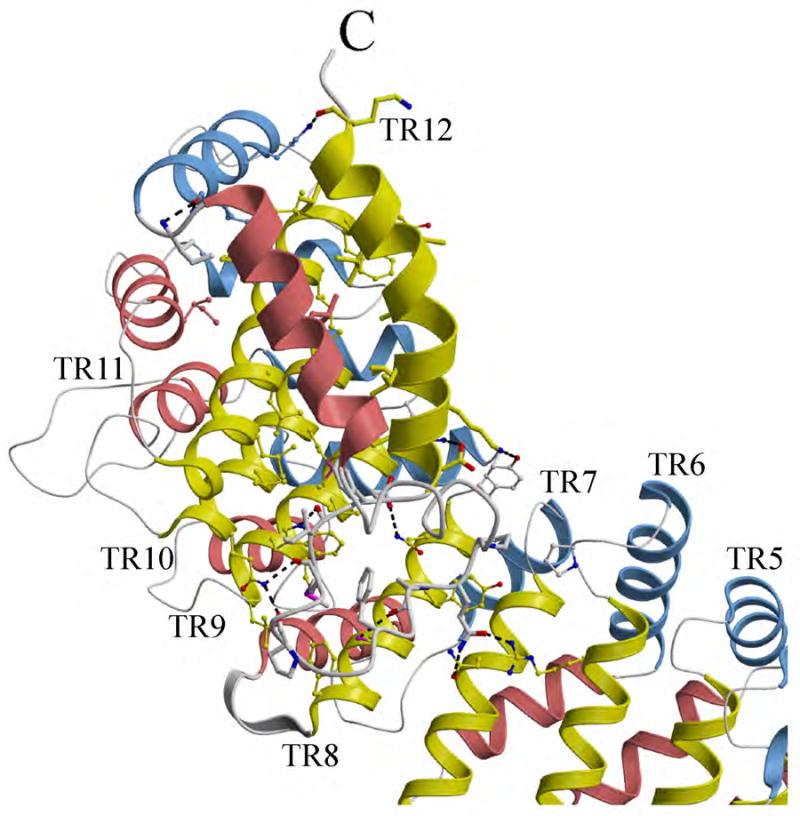
Conformation of the incomplete tether motif at the C-terminus of p115Nt. The 12th TR motif is shown in thicker ribbon representation than other tether repeats where it functions as a C-terminal cap for the α-solenoid. Residues involved in stabilization of the helical domains and the connecting loops of TR12 are represented by ball-and-stick figures. Hydrogen bonds are indicated by dashed lines.
Diversity in the TR sequence structure
Each TR consists of 32-44 residues that form three helices, with helix 1 consisting of 9-14 residues, helix 2 consisting of 14-21 residues and helix 3 consisting of 7-15 residues (Table S2). Variable leucine-rich motifs present in each TR roughly corresponds to xnLLx1-2LLx1-2LLxn for helices 1 and 2, and xnLLxn for helix 3, where xn can be up to 23 residues for connections between helices and L represents obligate hydrophobic residues whose side chains point into the solenoid (Fig. 3, Table S2). These conserved hydrophobic residues can include Leu, Ile, Cys, Ala, Val, Gly, Met, Phe and Pro. The organization for each of the p115 TRs can be weakly identified by using the conserved residues of the armadillo-consensus repeat (Table S2), but are sufficiently divergent to be defined to as ‘armadillo-like’ TRs that contribute to the formation of right-handed α/α/α-solenoids. Despite the lack of sequence conservation, the variable, leucine-rich motifs that define the p115 TRs can be detected in a number of tethering protein family members (Table S2 and Discussion).
The H1 TR motif directs p115-Rab1-GTP complex formation
p115 contains two sequence-conserved regions that, from a structural perspective, can be referred to as the H1-TR (residues 21-54) and H2-TR (residues 200-252), with yeast Uso1p being the most divergent member (Fig. 5). Glu201 and Lys241 in the H2 region contribute to the stability of the dimer interface (Fig. 2). Previous biochemical studies utilizing monomeric forms of the N-terminal domain of p115 (residues 1-651) did not detect Rab1 binding to the globular head domain.35,36 In contrast, we found that Rab1-GTP, but not Rab1-GDP, bound strongly to the full-length p115 dimer and to the dimeric form of p115Nt globular domain, but not to monomeric p115Nt, based on gel-filtration, with the latter result being consistent with previous reports35,36 (Fig. 6A).
Fig. 5.
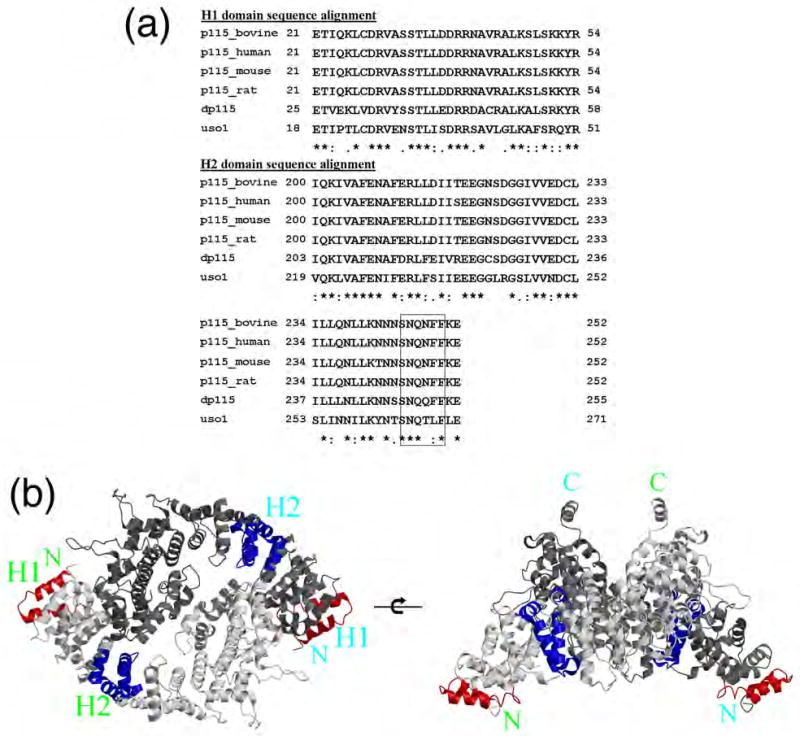
Sequence alignment and structural orientation of p115 conserved subdomains H1 and H2. (a) Sequence alignment of H1 and H2 regions. While yeast Uso1p is more divergent than observed within higher eukaryotes, its level of conservation suggests significant overlap in functional properties related to the function of domains H1 and H2 in vesicle trafficking. (b) Location of H1 and H2 domains in p115 dimer structure, with H1 and H2 shown in red and dark blue, respectively; top view (left panel), side view (right panel).
Fig. 6.
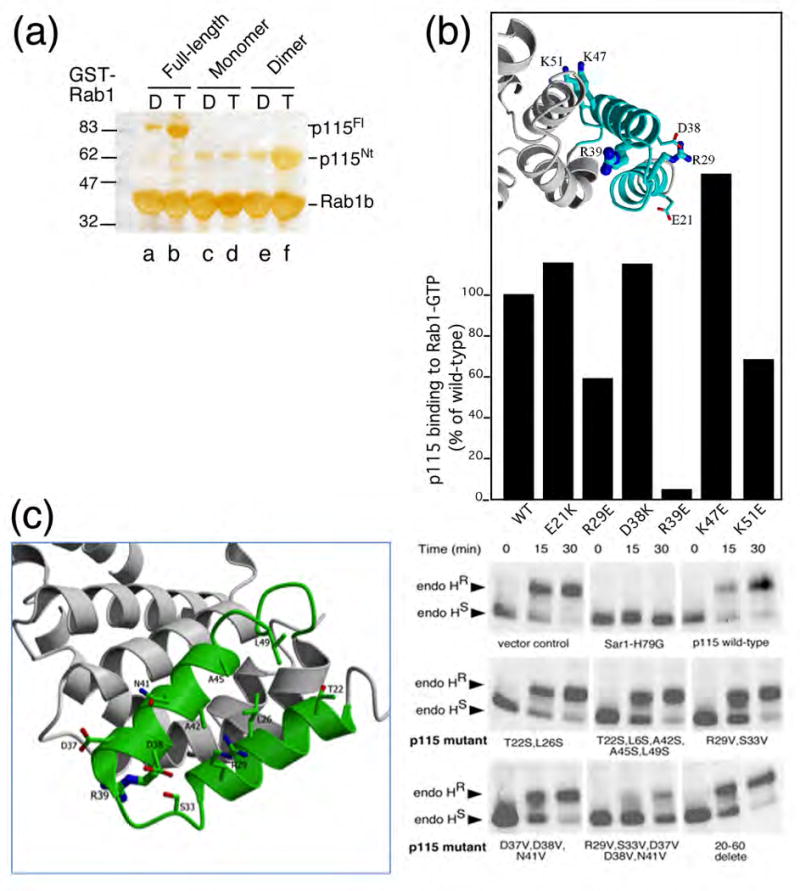
Role of H1 TR in Rab1 function in ER to Golgi trafficking. (a) Binding of Rab1-GTP, but not GDP, to the full-length p115 dimer and to the dimeric form of p115Nt globular domain, but not to monomeric p115Nt. GST-Rab1-GDP- (control) (lanes, a, c, e) and GST-Rab1-GTP bound to GST-beads (lanes b,d,f) were incubated with recombinant full-length wild-type (p115Fl) (lanes a,b) and mutant monomer (c,d) or dimer (e,f) p115Nt as described in the Materials and Methods. Shown is a silver stained gel of Rab1 (lanes a-f), p115Fl (lanes a, b) or p115Nt (lanes c-f) retained on beads. (b) Effect of mutation of H1 residues on Rab1 binding to p115Fl. p115 mutants were generated and p115Fl interaction with Rab1 was performed as described in Materials and Methods. p115 binding reported as % of total wild-type p115Fl bound relative to total GST-Rab1 coupled to beads. (Inset) Structural orientation of residues mutated for biochemical studies are shown as ball-and-stick figures. (c). Effect of mutations in p115 (left panel) on trafficking of VSV-G (right panel). Transport of VSV-G from the ER to the Golgi was followed by pulse-chase with [35]S-Met for the indicated time and treatment of cell lysates with endoglycosidase (Endo) H followed by SDS-PAGE as described in Materials and Methods. The reduction of processing to the endo H resistant glycoform reflects decreased delivery to Golgi compartments. Sar1-H79G, or the indicated wild-type or mutant full-length p115 construct was over-expressed ∼10-fold relative to endogenous p115 using a vaccinia transient expression system as described in Materials and Methods.
To further assess the potential role of residues in the evolutionarily conserved H1 TR motif involved in Rab1-GTP binding, we mutated selected charged residues in the H1 TR and expressed recombinant p115Nt protein for analysis of binding in vitro (Fig. 3; Fig. 6B). Mutation of Arg39 in helix 2 to Glu (R39E) completely disrupted Rab1-GTP binding in vitro to full-length p115, a result consistent with its orientation and key role in structural stabilization of the H1 helical tripod (Fig. 6B). Interaction with Rab1-GTP was partially disrupted by mutation of Arg29 to Glu (R29E) in helix 1 facing the solvent on the face opposite the interface between H1 and H2 (Fig. 6B). Thus, the N-terminal H1-TR can confer Rab1 interaction in vitro. Interestingly, mutation of Glu21 to lysine, which was recently shown to disrupt the interaction between p115 and the β-COPI subunit of the COPI coat complex77, had no effect on Rab1 interaction in vitro, suggesting that H1 has an additional binding site for other proteins, or that β-COPI has a sequence or structural motif similar to Rab1 (Fig. 6B).
The H1 TR is required for ER to Golgi traffic
To test whether the H1 TR is involved in Rab1-dependent trafficking of cargo between the ER and the Golgi, we examined whether p115 H1 TR mutants, which would likely function as dimers, given the extensive C-terminal interactions, could function as dominant negative inhibitors of trafficking. For this purpose, mutants were overexpressed in cells expressing vesicular stomatitis virus glycoprotein (VSV-G), a type 1 transmembrane protein that is efficiently transported between the ER and the Golgi in a p115-dependent fashion.36,37,39,46,56,78 Overexpression (∼10-fold), using a vaccinia transient expression system in the presence of the Sar1-H79G dominant negative mutant that inhibits COPII vesicle formation79,80, prevents export VSV-G from the ER and acquisition of endoglycosidase H (endo H) resistance, a hallmark of processing by cis Golgi α-mannosidases and glycosidases81-83 (Fig. 6C). Overexpression of the N-terminal (1-650) or C-terminal p115 domains (651 to 960) (data not shown) or full-length p115 protein (Fig. 6C) had little effect on VSV-G trafficking from the ER to the Golgi as indicated by acquisition of endo H resistance, suggesting that the intact protein is required for function and that excess p115 does not inhibit the activity of interacting components likely functional at the bilayer. Moreover, overexpression of single mutants that were tested for interaction with Rab1 in vitro (Fig. 6B) (data not shown), nor deletion of the H1 TR motif (residues 20-60) from the full-length protein, generated a dominant negative phenotype (Fig. 6C). The inability of the R39E mutation to bind Rab1 (Fig. 6A) is consistent with the inability of this mutant to function as a dominant negative inhibitor, particularly if recognition of Rab1 through the H1 motif is the first step in p115 recruitment to membranes. These results suggest that a dominant negative interaction cannot arise by complete ablation of the Rab1 binding site. In contrast, mutants that may partially perturb the interaction of the H1 domain with Rab1 without disrupting the structural organization of the H1 domain (see Discussion) may show a dominant negative phenotype. Indeed, mutants harboring the R29V-S33V-D37V-D38V-N41V combination showed a strong dominant negative effect on VSV-G processing to the endo H resistant form (Fig. 6C). Analysis of binding of this mutant to Rab1 in vitro (Fig. 6B) was not possible given its poor expression in E. coli. Because mutants expressing the R29V-S33V or D37V-D38V-N41V combinations were not dominant negative in vivo (Fig. 6C), we conclude that residues 29 through 41 provide a platform involving both α1 and α2 helical domains for interaction with Rab1 and possibly other factors. The H1 tripod may facilitate the assembly and/or disassembly of tethering-fusion complexes in ER to Golgi trafficking and Golgi integrity.
Discussion
Structural basis for p115 tether function
We have demonstrated that the evolutionarily conserved tether p115 has a C-terminal head domain assembled from α-helical-tripod TRs. The p115 TR α-solenoid is structurally related to the well-studied, superhelical α-solenoid importin α, and β-catenin proteins that are constructed of tri-helical armadillo repeats.70,73,84 Importin α belongs to a large family of related proteins that serve as adaptors for targeting many proteins to the nucleus.85 Importin α is a monomeric helicoidal protein, and binds protein ligands through conserved basic nuclear localization signals (NLSs) to its highly flexible internal and external α-helical faces.86,87 Similarly, monomeric β-catenin utilizes its α-helical faces to organize the assembly of complexes involved in cell signaling.84,88 In contrast, p115 forms a dimeric helicoidal protein where the internal faces are largely restricted to maintaining an energetically stable dimer interface in the cytosol. Thus, p115 has utilized the interaction surface of the helicoidal head domain to largely limit interactions to its external faces, of which only two TRs, H1 and H2, are highly conserved. Intriguingly, the H2-TR has recently been proposed to bind the conserved oligomeric Golgi (COG) complex.23 COGs are multimeric complexes that are believed to play a critical role in Golgi structure and retrograde trafficking of a number Golgi glycan processing enzymes.25 Disruption of COG function leads to a number of inherited glycosylation disorders.21 While we found that overexpression of a variety of mutants in the H2 domain had no dominant effect on VSV-G transport to the Golgi (data not shown), the complete deletion of the H2 domain results in Golgi fragmentation and a reduction in ER to Golgi transport23, consistent with an important role for both of the conserved H1 (shown herein) and H223 domains in p115 function.
While H2 binds COG23, we found that the N-terminal H1 TR can bind Rab1. Previous observations demonstrated that Rab1 binds to a coiled-coil region (CC1) (residues 650-961) that lack predicted TRs and is distal to the N-terminal globular domain reported herein.35 This CC1 domain (residues 650-780) is thought to form a cryptic binding site masked by the acidic tail domain (residues 875-920) that, in the absence of the Golgin tether GM130 that displaces the acidic domain, would prevent Rab1 binding.36,78 An interaction of Rab1 with a coiled-coil domain is not unlike the recent structural complex reported for the yeast Sec4 Rab GTPase bound to a coiled-coil motif in the Sec4 GEF89,90 or Rab6-interaction with GCC185.7 Interestingly, the CC1 region was inactive in Rab1 binding when isolated as a monomeric CC1 fragment.35 One possibility is that the CC1 region may only function as a dimer, as observed for Rab1 binding to the N-terminal globular domain herein. It remains possible that both the Rab1 and CC1 interaction domains in the monomeric state are more unfolded and/or flexible in solution when compared to the dimer and, therefore, unable to productively bind Rab1. The identification of the H1 TR domain as a binding partner for Rab1 raises the possibility that two different Rab1 binding regions may have differential functions in the sequential events that direct tethering and SNARE assembly which facilitate vesicle docking and fusion.35,36,78,91,92. We suggest that the H1 domain may possibly initiate these events given the complexities associated with GM130 exposing the CC1 motif 36,78. Although the E21 residue in the H1 TR has recently been reported to interact with the β-subunit of COPI77, we were unable to detect an effect of this mutant on the interaction with Rab1. This is consistent with the fact Rab1-H1 interacting domain likely involves residues 29-41.
While TRs define the α-solenoid structural organization of the N-terminal globular head domain, they lack sequence conservation except for notable leucine-rich motifs that can be best described as armadillo-like features (Table S2). Although an extensive database analysis using a variety of current alignment algorithms failed to reveal a consensus motif that could be used to define a TR family/superfamily (Rotkiewicz and Godzik, unpublished), it is of note that upon visual inspection, a leucine-rich pattern separated by sequences of variable length, as found for p115Nt, can be detected in a broad spectrum of tether proteins that are involved in exocytic and endocytic trafficking pathways (Table S2). While it is difficult to assign statistical significance to such manual assignments, it is interesting to note that the leucine-rich pattern separated by sequences of variable length are nearly exclusively found in predicted coiled-coiled regions75 of Golgins and other Rab effectors where binding to Rab, Arf, and Arl GTPases has been demonstrated.28 One possibility is that these regions, despite our current inability to recognize them, comprise a broad family of evolutionarily related, α-helical rich subdomains that adopt structural features to facilitate GTPase-regulated membrane trafficking. Consistent with this hypothesis is the presence of both di-helical and tri-helical interaction domains found in a number of Rab-effector complexes28 (Table S2; Fig. 7B). Here, two out of the three α-helices appear to be involved in generating a GTPase-binding platform, whereas the third helix contributes to structural orientation of the platform. These differences suggest that GTPase interaction with tethering components can involve a multiplicity of structural motifs involving α-helical domains that may have different functional consequences.93
Fig. 7.
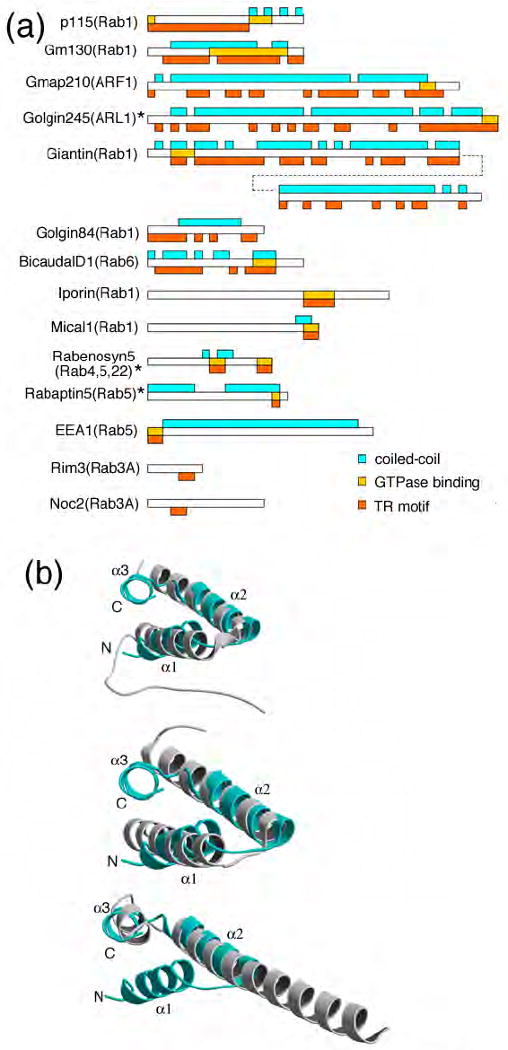
Conservation of TR structure. (a) Predicted coiled-coil regions are shown in cyan boxes above each protein; putative TR containing regions (see Table S2 and text of Discussion for further details) that overlap with predicted coiled-coil regions, are shown in orange boxes below each protein. The region binding either ARL1, ARF1 or Rab GTPases is shown by the gold boxes within the protein. Asterisk (*) indicates TR motifs with known structure. (b) Superimposition of TR motifs of p115Nt and structurally characterized Rab GTPase effector complexes: (upper panel) helices 1 and 2 in the H1 TR of p115Nt (cyan) with rabenosyn-5 (441-501, PDB-1ZOK) (gray); (middle panel) helices 1 and 2 in the H1 TR of p115Nt (cyan) with Rabenosyn-5 (734-784, PDB-1ZOJ) (gray); and (lower panel) helices 2 and 3 in H1 of p115Nt (cyan) and Rabaptin5 (804-849, PDB-1TU3) (gray).
Homology model of the Rab1-p115 complex
The structure of the small GTPase ARL1 bound to the C-terminal, α-helical-rich fragment corresponding to the GRIP domain in the tether Golgin 245 has been determined.94,95 Like Rab1, ARL1 belongs to the Ras-superfamily of GTPases that retains a highly conserved core fold that is augmented with unique switch and interswitch regions that direct specific function.96-98 We performed a structural alignment of the p115 H1 TR with the Golgin 245 GRIP domain bound to ARL194,95 (PDP codes 1R4D, 1UPT). Superimposition of consensus hydrophobic residues of the α1 and α2 helices of the GRIP domain with the α1 and α2 helices of p115Nt H1-TR yielded an rmsd of 0.8 Å (main chain), that indicated similar structural configurations (Fig. 8A). Because ARL1 and Rabs have nearly identical core GTPase folds that bind guanine nucleotide, we were able to align the Rab5 nucleotide binding domain (PBD code 1TU3) with that of ARL1 (PDB code 1R4D) (Fig. 8B). By mutation of the Rab5 switch domain residues to the corresponding Rab1 switch region residues, we could generate a model for the p115Nt-Rab1 complex (Fig. 8C) that illustrated the role of R39 in stabilizing the structure of the H1 TR, and R29, and potentially D38 for interaction with Rab1 (Fig. 8D). This alignment yields an orientation consistent with the interpretation that the switch and interswitch regions of Rab1 are involved in binding the H1 TR in p115. The orientation of the truncated C-terminus of Rab1 (Fig. 8C), suggests that binding of either one or two Rab1 molecules to the lipid bilayer can be readily accommodated by insertion of the prenyl lipids covalently attached to conserved cysteine-residues at the terminus of the C-terminus (Fig. 8E).
Fig. 8.
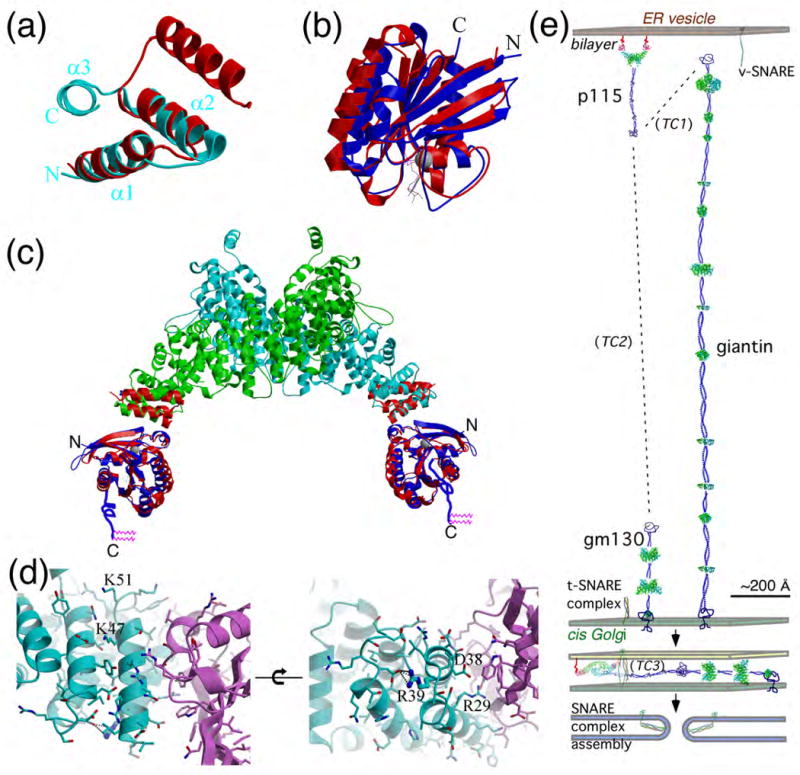
A model for the Rab1-GTP:p115 complex in membrane tethering. (a) Superimposition of helices 1 and 2 of the TR like-motif in GRIP domain (red) (from ARL1-GTP-GRIP complex, PDB-1R4A) with H1 of p115Nt (cyan) based on superimposition of consensus hydrophobic residues. (b) Superimposition of ARL1-GTP and Rab5-GTP (PDB-1TU3), with ARL1-GTP (red) and Rab5-GTP (green). The switch and interswitch regions that interact with the GRIP or H1 TR motifs are facing the viewer. (c) Superimposition of the ARL1(GTP)-GRIP (PDB-1R4A) (red) with the H1 domain of p115Nt and Rab5-GTP (PDB-1TU3) (blue) illustrating a possible molecular model for the Rab1-GTP-p115 complex. The unstructured 23-residue hypervariable domain of Rab1 containing two C-terminal geranylgeranyl prenyl lipids (pink) is illustrated with blue lines. (d) Rab1-GTP-p115 complex interface. Rab1-GTP model (purple) was generated by mutation of corresponding residues of Rab5-GTP (cyan) based on sequence alignment. Indicated residues in p115 were mutated for analysis of their effects on Rab1 binding, as described in the text. Mutation of Asp38 to Lys had no effect on Rab1 binding (Fig. 6b) consistent with its orientation in the homology model. In contrast, while Arg39 does not have direct interactions with Rab1, it is likely required for stabilization of the H1 TR domain and thereby strikingly disrupts interactions with Rab1 when mutated (Fig. 6b). Arg29 likely interacts with the switch regions to alter Rab1 binding (Fig. 6b). Lys47 and Lys51 may affect stabilization of the TR1 domain and have differential effects on Rab1 binding (Fig. 6b). (e) Structural model of p115 ER-Golgi tether-fusion pathway. Binding of p115 by Rab1-GTP37,45 to the bilayer through the C-terminal prenyl lipids of Rab1 links p115 to ER to Golgi transport vesicles.37,38 The first step in recognizing the Golgi at a distance is by interaction with the Golgi-associated, extended tether giantin to form tethering complex 1 (TC1) through interaction with the acidic-C-terminal domain of p115.36,51,100,117 In the subsequent tethering complex (TC2), p115 is bound to the more compact, Golgi-associated GM130 tether, again through the p115 acidic C-terminal domain.35,39,100 Close juxtaposition of the vesicle bilayer to the Golgi bilayer by TC2 promotes SNARE assembly (TC3) and fusion.35,37,43,57 The molecular and biochemical events directing transitions between TC1-3 are unknown. p115, giantin and GM130 were structurally modeled based on the distribution of potential TRs (green icons based on Table S2) and predicted coiled-coiled motifs. The v- and t-SNARE partial and complexed structures are illustrated based on the intact SNARE complex (PDB-1N7S). All protein components illustrated are approximately calibrated to their predicted physical dimensions.
Our knowledge of the structure of the globular head region, combined with the extensive biochemical analyses of p115 function in trafficking and SNARE assembly37-41,43,44, now provides an opportunity to model the membrane tethering and docking events in ER to Golgi trafficking that involve p115 (Fig. 8E). p115 is recruited to activated Rab1-GTP on the donor vesicle through the H1 TR domain and likely is retained on the bilayer by the flexible 23-residue, hypervariable, prenylated C-terminus of Rab1.99 Because p115 interacts with the highly elongated tether giantin that is attached to the acceptor cis Golgi compartment51,100,101, we raise the possibility that tethering can initially occur at a distance of up to 200 nm from the bilayer. Following this step, p115 may be handed off to the more structurally compact GM130 tether35,37,39,57 (Fig. 8E). This would serve to not only physically draw the membrane surfaces together, but, based on current models35,78, is expected to accelerate the release of the autoinhibitory acidic domain found at the C-terminus p115 to allow interaction with Rab1 on the acceptor membrane with the CC1 region of p115, thereby stabilizing association of opposing membranes.35-38,78 This physical model of tethering explains the observation that, while giantin and GM130 are regulatory, they are not essential for either Golgi assembly or ER to Golgi transport.42,56,100 Rather, tethering at a distance may improve the overall efficiency of trafficking in higher eukaryotes by reducing a 3-dimensional walk in the cytosol to a more restricted 2-dimensional walk along the surface of the Golgi.102 Finally, p115-dependent SNARE assembly at the CC1 region would direct bilayer fusion (Fig. 8E). We conclude that the dimeric solenoid structure of the N-terminal region of p115 described herein provides a Rab1-dependent binding platform on the nascent vesicle to initiate a cascade of tethering-fusion events culminating in the transfer of cargo between the ER and the Golgi.
Materials and Methods
Preparation of monomer and dimer p115 N-terminal domain
The N-terminal domain of bovine p115 from residue 1 to 651 was cloned into pET-28a vector. The protein was expressed with E. coli BL21 (DE3) cell and purified using Ni-NTA (Qiagen), ion-exchange (Mono Q, Pharmacia), gel filtration chromatography (Superdex 200, Pharmacia) and thrombin digestion. The purified protein was concentrated to 10 mg/ml in 50 mM Tris-HCl at pH 8.5. Se-Met protein was expressed with B834 (DE3) cell in Se-Met containing minimal media, and purified under reducing conditions containing 10 mM β-mercaptoethanol. The protein used to prepare the p115 dimer was generated, as described for the monomer, with the exception that the protein was engineered to contain a tandem N-terminal HA/hexaHis tag in a pET-11d vector. Protein samples were designated monomer or dimer based on their gel filtration profiles.
Crystallization
Monomer crystals were obtained from 1.2 M NaH2PO4/K2HPO4, pH 5.5. Se-Met protein crystals were grown from similar conditions with 10 mM β-mercaptoethanol. Dimer crystals were obtained from 10% MPD, 18% PEG 4000 and 0.1 M Tris-HCl, pH 8.4. The native monomer data set was collected at SSRL 9-2 to 2.0 Å resolution, and a three-wavelength MAD data set with Se-Met derivative was collected at ALS 8.2.1 to 2.7 Å. The dimer data set was collected at ALS 5.0.2 to 2.18 Å. All data sets were processed with the program HKL2000103. Phases to 2.7 Å were obtained from the MAD data set with the program SOLVE, and density modification was performed by RESOLVE104. Phases were extended with the native data set (2.0 Å) with the program DM.105 The automatic model building program ARP/wARP106 and manual building were used to build both main chain and side chains from the phase-extended map. The resulting model was initially refined with CNS107 and completed with REFMAC5 using TLS refinement.108 Manual model fitting was carried out using the programs O109 and COOT.110 The dimer phases were solved from native data set with the molecular replacement program Molrep105 using the monomer structure as the search model. The stereochemical quality of the models were verified using AutoDepInputTool111 MolProbity112, and WHATIF 5.0113. Data collection and refinement statistics are summarized in Table 1. The dimer crystals have one dimer in the asymmetric unit, while the monomer crystals have one monomer in the asymmetric unit. The dimer configuration is not observed from any of the symmetry operators in the monomer crystals.
Binding of p115 to Rab1
25 μg recombinant GST-Rab1 protein was immobilized to 5 μl GST beads (GE Healthcare, Piscataway, NJ) in reaction buffer (25 mM Hepes-KOH (pH 7.4), 100 mM NaCl, 1mM MgCl2, 1mM DTT) followed by nucleotide exchange with GTPγS or GDP respectively.37 50 μg of purified full length p115 was incubated in the reaction buffer plus 1 mM nucleotide at 4 °C overnight followed by 3× wash with reaction buffer. Protein bound to the beads was eluted with denaturing buffer (10 mM Tris-HCl (pH 7.0), 1% SDS, 1% β-ME) at 37 °C for 10 min, and analyzed by SDS-PAGE with silver staining to determine purity.
Generation of p115 N-terminal domain mutants
Point mutations in the p115 H1 domain (aa 21-54) were engineered using using pFastBAC1-hexaHis-p115 wild-type as the template and complimentary mutagenic oligomers with the Quikchange Kit (Stratagene, #200518). Following sequence confirmation, mutagenized p115 fragments (amino acids 1-456) were excised by Nde I-Bsu36 I digest and ligated back into the similarly digested FastBAC-hexaHis-p115 template. DH10BAC. E. coli was transposed with the pFastBAC1-hexaHis-mutant p115 plasmids to generate mutant 6His-p115 bacmids using the BAC-to-BAC Baculovirus Expression System (GibcoBRL/Invitrogen Corp.). Sf9 insect cells were transfected with the bacmids to produce recombinant mutant hexaHis-p115 baculoviruses which were then used to infect TN5 insect cells for protein expression.
Transport in vivo
[35S]Met pulse-chase labeling and vaccinia transient expression and quantification was performed as described.114
Accession codes
Coordinates and structure factors have been deposited in the PDB with accession codes 3gq2 (dimer) and 3grl (monomer).
Supplementary Material
Acknowledgments
This work was supported by grants NIH GM42336 and GM33301 (WEB), and CA58896 (IAW). We acknowledge the helpful support of staff members at SSRL-92, ALS8-2-1 and ALS5-0-2. This is TSRI manuscript #67854.
Abbreviations used
- TR
tether repeat
- COG
conserved oligomeric Golgi
- TRAPP
transport protein particle
- SNARE
soluble N-ethylmaleimide factor attachment protein receptors
- VSV-G
vesicular stomatitis virus glycoprotein
- TC
tethering complex
Footnotes
Supplementary Data: Supplementary data associated with this article can be found, in the on-line version, at doi:xx.yyy/j.jmb.2009.zz.nnn
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gurkan C, Lapp H, Alory C, Su AI, Hogenesch JB, Balch WE. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell. 2005;16:3847–3864. doi: 10.1091/mbc.E05-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am J Physiol Cell Physiol. 2006;290:11–26. doi: 10.1152/ajpcell.00293.2005. [DOI] [PubMed] [Google Scholar]
- 3.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- 5.Drin G, Morello V, Casella JF, Gounon P, Antonny B. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 2008;320:670–673. doi: 10.1126/science.1155821. [DOI] [PubMed] [Google Scholar]
- 6.Diao A, Frost L, Morohashi Y, Lowe M. Coordination of golgin tethering and SNARE assembly: GM130 binds syntaxin 5 in a p115-regulated manner. J Biol Chem. 2008;283:6957–6967. doi: 10.1074/jbc.M708401200. [DOI] [PubMed] [Google Scholar]
- 7.Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–298. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosing M, Ossendorf E, Rak A, Barnekow A. Giantin interacts with both the small GTPase Rab6 and Rab1. Exp Cell Res. 2007;313:2318–2325. doi: 10.1016/j.yexcr.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 9.Lieu ZZ, Derby MC, Teasdale RD, Hart C, Gunn P, Gleeson PA. The golgin GCC88 is required for efficient retrograde transport of cargo from the early endosomes to the trans-Golgi network. Mol Biol Cell. 2007;18:4979–4991. doi: 10.1091/mbc.E07-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- 11.Stefano G, Renna L, Hanton SL, Chatre L, Haas TA, Brandizzi F. ARL1 plays a role in the binding of the GRIP domain of a peripheral matrix protein to the Golgi apparatus in plant cells. Plant Mol Biol. 2006;61:431–449. doi: 10.1007/s11103-006-0022-y. [DOI] [PubMed] [Google Scholar]
- 12.Short B, Haas A, Barr FA. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim Biophys Acta. 2005;1744:383–395. doi: 10.1016/j.bbamcr.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Lieu ZZ, Lock JG, Hammond LA, La Gruta NL, Stow JL, Gleeson PA. A trans-Golgi network golgin is required for the regulated secretion of TNF in activated macrophages in vivo. Proc Natl Acad Sci U S A. 2008;105:3351–3356. doi: 10.1073/pnas.0800137105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luke MR, Houghton F, Perugini MA, Gleeson PA. The trans-Golgi network GRIP-domain proteins form alpha-helical homodimers. Biochem J. 2005;388:835–841. doi: 10.1042/BJ20041810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacher M, Kim YG, Lavie A, Oh BH, Segev N. The TRAPP Complex: Insights into its Architecture and Function. Traffic. 2008;9:2032–2042. doi: 10.1111/j.1600-0854.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kummel D, Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252. [DOI] [PubMed] [Google Scholar]
- 17.Cai Y, Chin HF, Lazarova D, Menon S, Fu C, Cai H, Sclafani A, Rodgers DW, De La Cruz EM, Ferro-Novick S, Reinisch KM. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummel D, Muller JJ, Roske Y, Henke N, Heinemann U. Structure of the Bet3-Tpc6B core of TRAPP: two Tpc6 paralogs form trimeric complexes with Bet3 and Mum2. J Mol Biol. 2006;361:22–32. doi: 10.1016/j.jmb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, Walz T, Oh BH, Sacher M. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 2006;127:817–830. doi: 10.1016/j.cell.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Kummel D, Muller JJ, Roske Y, Misselwitz R, Bussow K, Heinemann U. The structure of the TRAPP subunit TPC6 suggests a model for a TRAPP subcomplex. EMBO Rep. 2005;6:787–793. doi: 10.1038/sj.embor.7400463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeevaert R, Foulquier F, Jaeken J, Matthijs G. Deficiencies in subunits of the Conserved Oligomeric Golgi (COG) complex define a novel group of Congenital Disorders of Glycosylation. Mol Genet Metab. 2008;93:15–21. doi: 10.1016/j.ymgme.2007.08.118. [DOI] [PubMed] [Google Scholar]
- 22.Chavas LM, Ihara K, Kawasaki M, Torii S, Uejima T, Kato R, Izumi T, Wakatsuki S. Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a. Structure. 2008;16:1468–1477. doi: 10.1016/j.str.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 23.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8:270–284. doi: 10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 24.Cavanaugh LF, Chen X, Richardson BC, Ungar D, Pelczer I, Rizo J, Hughson FM. Structural analysis of conserved oligomeric Golgi complex subunit 2. J Biol Chem. 2007;282:23418–23426. doi: 10.1074/jbc.M703716200. [DOI] [PubMed] [Google Scholar]
- 25.Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Ungar D, Oka T, Vasile E, Krieger M, Hughson FM. Subunit architecture of the conserved oligomeric Golgi complex. J Biol Chem. 2005;280:32729–32735. doi: 10.1074/jbc.M504590200. [DOI] [PubMed] [Google Scholar]
- 27.Lupashin V, Sztul E. Golgi tethering factors. Biochim Biophys Acta. 2005;1744:325–339. doi: 10.1016/j.bbamcr.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki M, Nakayama K, Wakatsuki S. Membrane recruitment of effector proteins by Arf and Rab GTPases. Curr Opin Struct Biol. 2005;15:681–689. doi: 10.1016/j.sbi.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 30.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong G, Medkova M, Novick P, Reinisch KM. A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol Cell. 2007;25:455–462. doi: 10.1016/j.molcel.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiba T, Koga H, Shin HW, Kawasaki M, Kato R, Nakayama K, Wakatsuki S. Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc Natl Acad Sci U S A. 2006;103:15416–15421. doi: 10.1073/pnas.0605357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waters MG, Clary DO, Rothman JE. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sapperstein SK, Walter DM, Grosvenor AR, Heuser JE, Waters MG. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc Natl Acad Sci U S A. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shorter J, Beard MB, Seemann J, Dirac-Svejstrup AB, Warren G. Sequential tethering of Golgins and catalysis of SNAREpin assembly by the vesicle-tethering protein p115. J Cell Biol. 2002;157:45–62. doi: 10.1083/jcb.200112127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beard M, Satoh A, Shorter J, Warren G. A cryptic Rab1-binding site in the p115 tethering protein. J Biol Chem. 2005;280:25840–25848. doi: 10.1074/jbc.M503925200. [DOI] [PubMed] [Google Scholar]
- 37.Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: Requirement for p115 at a pre-Golgi VTC stage. J Cell Biol. 1999;147:1205–1222. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moyer BD, Allan BB, Balch WE. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis--Golgi tethering. Traffic. 2001;2:268–276. doi: 10.1034/j.1600-0854.2001.1o007.x. [DOI] [PubMed] [Google Scholar]
- 40.Nelson DS, Alvarez C, Gao YS, Garcia-Mata R, Fialkowski E, Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao X, Ballew N, Barlowe C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998;17:2156–2165. doi: 10.1093/emboj/17.8.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puthenveedu MA, Linstedt AD. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J Cell Biol. 2001;155:227–238. doi: 10.1083/jcb.200105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barroso M, Nelson DS, Sztul E. Transcytosis-associated protein (TAP)/p115 is a general fusion factor required for binding of vesicles to acceptor membranes. Proc Natl Acad Sci U S A. 1995;92:527–531. doi: 10.1073/pnas.92.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morsomme P, Riezman H. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev Cell. 2002;2:307–317. doi: 10.1016/s1534-5807(02)00133-8. [DOI] [PubMed] [Google Scholar]
- 46.Sohda M, Misumi Y, Yoshimura S, Nakamura N, Fusano T, Sakisaka S, Ogata S, Fujimoto J, Kiyokawa N, Ikehara Y. Depletion of vesicle-tethering factor p115 causes mini-stacked Golgi fragments with delayed protein transport. Biochem Biophys Res Commun. 2005;338:1268–1274. doi: 10.1016/j.bbrc.2005.10.084. [DOI] [PubMed] [Google Scholar]
- 47.Bentley M, Liang Y, Mullen K, Xu D, Sztul E, Hay JC. SNARE status regulates tether recruitment and function in homotypic COPII vesicle fusion. J Biol Chem. 2006;281:38825–38833. doi: 10.1074/jbc.M606044200. [DOI] [PubMed] [Google Scholar]
- 48.Shorter J, Warren G. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine TP, Rabouille C, Kieckbusch RH, Warren G. Binding of the vesicle docking protein p115 to Golgi membranes is inhibited under mitotic conditions. J Biol Chem. 1996;271:17304–17311. doi: 10.1074/jbc.271.29.17304. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura N, Lowe M, Levine TP, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 51.Lesa GM, Seemann J, Shorter J, Vandekerckhove J, Warren G. The amino-terminal domain of the golgi protein giantin interacts directly with the vesicle-tethering protein p115. J Biol Chem. 2000;275:2831–2836. doi: 10.1074/jbc.275.4.2831. [DOI] [PubMed] [Google Scholar]
- 52.Seemann J, Jokitalo EJ, Warren G. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol Biol Cell. 2000;11:635–645. doi: 10.1091/mbc.11.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malsam J, Satoh A, Pelletier L, Warren G. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- 54.Chiu R, Novikov L, Mukherjee S, Shields D. A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J Cell Biol. 2002;159:637–648. doi: 10.1083/jcb.200208013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee S, Chiu R, Leung SM, Shields D. Fragmentation of the Golgi apparatus: an early apoptotic event independent of the cytoskeleton. Traffic. 2007;8:369–378. doi: 10.1111/j.1600-0854.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 56.Puthenveedu MA, Linstedt AD. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis. Proc Natl Acad Sci U S A. 2004;101:1253–1256. doi: 10.1073/pnas.0306373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gmachl MJ, Wimmer C. Sequential involvement of p115, SNAREs, and Rab proteins in intra-Golgi protein transport. J Biol Chem. 2001;276:18178–18184. doi: 10.1074/jbc.M101513200. [DOI] [PubMed] [Google Scholar]
- 58.Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 59.Martin ACR. Program ProFit [Google Scholar]
- 60.Connolly ML. The molecular surface package. J Mol Graph. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 61.Gelin BR, Karplus M. Side-chain torsional potentials: effect of dipeptide, protein, and solvent environment. Biochemistry (Mosc) 1979;18:1256–1268. doi: 10.1021/bi00574a022. [DOI] [PubMed] [Google Scholar]
- 62.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 63.Sheriff S, Hendrickson WA, Smith JL. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- 64.Sheriff S, Silverton EW, Padlan EA, Cohen GH, Smith-Gill SJ, Finzel BC, Davies DR. Three-dimensional structure of an antibody-antigen complex. Proc Natl Acad Sci U S A. 1987;84:8075–8079. doi: 10.1073/pnas.84.22.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 66.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 67.Conti E. Structures of importins. Results Probl Cell Differ. 2002;35:93–113. doi: 10.1007/978-3-540-44603-3_5. [DOI] [PubMed] [Google Scholar]
- 68.Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 69.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 70.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 71.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 72.Chook YM, Blobel G. Structure of the nuclear transport complex karyopherin-beta2-Ran × GppNHp. Nature. 1999;399:230–237. doi: 10.1038/20375. [DOI] [PubMed] [Google Scholar]
- 73.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 74.Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 75.Gruber M, Soding J, Lupas AN. Comparative analysis of coiled-coil prediction methods. J Struct Biol. 2006;155:140–145. doi: 10.1016/j.jsb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 76.McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- 77.Guo Y, Punj V, Sengupta D, Linstedt AD. Coat-tether interaction in Golgi organization. Mol Biol Cell. 2008;19:2830–2843. doi: 10.1091/mbc.E07-12-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satoh A, Warren G. In situ cleavage of the acidic domain from the p115 tether inhibits exocytic transport. Traffic. 2008;9:1522–1529. doi: 10.1111/j.1600-0854.2008.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aridor M, Fish KN, Bannykh S, Weissman J, Roberts TH, Lippincott-Schwartz J, Balch WE. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol. 2001;152:213–229. doi: 10.1083/jcb.152.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rowe T, Aridor M, McCaffery JM, Plutner H, Nuoffer C, Balch WE. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J Cell Biol. 1996;135:895–911. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown WJ, Plutner H, Drecktrah D, Judson BL, Balch WE. The lysophospholipid acyltransferase antagonist CI-976 inhibits a late step in COPII vesicle budding. Traffic. 2008;9:786–797. doi: 10.1111/j.1600-0854.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bannykh SI, Plutner H, Matteson J, Balch WE. The role of ARF1 and rab GTPases in polarization of the Golgi stack. Traffic. 2005;6:803–819. doi: 10.1111/j.1600-0854.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- 83.Aridor M, Bannykh SI, Rowe T, Balch WE. Cargo can modulate COPII vesicle formation from the endoplasmic reticulum. J Biol Chem. 1999;274:4389–4399. doi: 10.1074/jbc.274.7.4389. [DOI] [PubMed] [Google Scholar]
- 84.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 85.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 86.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 87.Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 88.Choi HJ, Huber AH, Weis WI. Thermodynamics of beta-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 89.Sato Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the Sec4p.Sec2p complex in the nucleotide exchanging intermediate state. Proc Natl Acad Sci U S A. 2007;104:8305–8310. doi: 10.1073/pnas.0701550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sato Y, Shirakawa R, Horiuchi H, Dohmae N, Fukai S, Nureki O. Asymmetric coiled-coil structure with Guanine nucleotide exchange activity. Structure. 2007;15:245–252. doi: 10.1016/j.str.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Satoh A, Malsam J, Warren G. Tethering assays for COPI vesicles mediated by golgins. Methods Enzymol. 2005;404:125–134. doi: 10.1016/S0076-6879(05)04013-9. [DOI] [PubMed] [Google Scholar]
- 93.Hayes GL, Brown FC, Haas AK, Nottingham RM, Barr FA, Pfeffer SR. Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol Biol Cell. 2009;20:209–217. doi: 10.1091/mbc.E08-07-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu M, Lu L, Hong W, Song H. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat Struct Mol Biol. 2004;11:86–94. doi: 10.1038/nsmb714. [DOI] [PubMed] [Google Scholar]
- 95.Panic B, Perisic O, Veprintsev DB, Williams RL, Munro S. Structural basis for Arl1-dependent targeting of homodimeric GRIP domains to the Golgi apparatus. Mol Cell. 2003;12:863–874. doi: 10.1016/s1097-2765(03)00356-3. [DOI] [PubMed] [Google Scholar]
- 96.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 97.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pfeffer SR. Structural clues to Rab GTPase functional diversity. J Biol Chem. 2005;280:15485–15488. doi: 10.1074/jbc.R500003200. [DOI] [PubMed] [Google Scholar]
- 99.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Linstedt AD, Jesch SA, Mehta A, Lee TH, Garcia-Mata R, Nelson DS, Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J Biol Chem. 2000;275:10196–10201. doi: 10.1074/jbc.275.14.10196. [DOI] [PubMed] [Google Scholar]
- 101.Sonnichsen B, Lowe M, Levine T, Jamsa E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013–1021. doi: 10.1083/jcb.140.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Orci L, Perrelet A, Rothman JE. Vesicles on strings: morphological evidence for processive transport within the Golgi stack. Proc Natl Acad Sci U S A. 1998;95:2279–2283. doi: 10.1073/pnas.95.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 104.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 105.Collaborative. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 106.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 107.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 108.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 109.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 110.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 111.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 112.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 114.Yoo JS, Moyer BD, Bannykh S, Yoo HM, Riordan JR, Balch WE. Non-conventional trafficking of the cystic fibrosis transmembrane conductance regulator through the early secretory pathway. J Biol Chem. 2002;277:11401–11409. doi: 10.1074/jbc.M110263200. [DOI] [PubMed] [Google Scholar]
- 115.Kraulis PJ. MOLSCRIPT: A Program to Produce Both Detailed and Schematic Plots of Protein Structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 116.Merritt EA, Bacon DJ. Raster3D: Photorealistic Molecular Graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 117.Alvarez C, Garcia-Mata R, Hauri HP, Sztul E. The p115-interactive proteins GM130 and giantin participate in endoplasmic reticulum-Golgi traffic. J Biol Chem. 2001;276:2693–2700. doi: 10.1074/jbc.M007957200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


