Abstract
The genomic revolution has provided the first glimpses of the architecture of regulatory networks. Combined with evolutionary information, the “network view” of life processes leads to remarkable insights into how biological systems have been shaped by various forces. This understanding is critical because biological systems, including regulatory networks, are not products of engineering but of historical contingencies. In this light, we attempt a synthetic overview of the natural history of regulatory networks operating in the development and differentiation of multicellular organisms. We first introduce regulatory networks and their organizational principles as can be deduced using ideas from the graph theory. We then discuss findings from comparative genomics to illustrate the effects of lineage-specific expansions, gene-loss, and non-protein-coding DNA on the architecture of networks. We consider the interaction between expansions of transcription factors, and cis regulatory and more general chromatin state stabilizing elements in the emergence of morphological complexity. Finally, we consider a case study of the Notch sub-network, which is present throughout Metazoa, to examine how such a regulatory system has been pieced together in evolution from new innovations and pre-existing components that were originally functionally distinct.
INTRODUCTION
The history of biology has been marked by considerable “provincialism”, despite the availability of a unifying framework in the form of the evolutionary theory for at least the past 150 years (Darwin, 1859; Mayr, 1982) (as Dobzhanksy remarked: “Nothing in Biology Makes Sense Except in the Light of Evolution”ref for this? 1973? Yes – I’ve put the reference in the back). For a good portion of this period, most major disciplines within biology emerged and operated in relative isolation before being integrated into the overarching framework of the science. During this phase, evolutionary studies, taxonomy, and ecology formed relatively isolated pursuits of the naturalists in the Darwinian tradition, whereas genetics, developmental biology, and biochemistry followed their own largely independent traditions (Mayr, 1982). However, by the second half of the previous century there were several partial unifications centered on genetics – the neo-Darwinian synthesis that successfully combined genetics and the evolutionary theory and the rise of developmental genetics that provided the first glimpses of how genes cooperated to specify the forms of multicellular organisms (Gould, 2002; Huxley, 1942; Raff, 1996). The first hints of a more fundamental unification were seen with the beginnings of molecular biology – it provided a means of understanding genes and their products at a molecular level, thereby bridging the gap between the phenotype and its underlying biochemical basis (Morange, 1998). One of the consequences of this was the emergence of the so called “evo-devo” field, which sought to incorporate evolutionary principles to explain aspects of animal development and explain the emergence of the diversity of animal form(Arthur, 2002; Raff, 1996). A primary result from these studies was the identification of numerous evolutionarily conserved pathways that determined tissue differentiation and pattern formation throughout Metazoa, despite their apparent morphological disparity. The flip-side of this discipline was the relatively narrow focus on few conserved genetic pathways, rather than objectively addressing the mechanisms behind the biological diversity as specified in the total gene complement of organisms (Arthur, 2002; Kirschner and Gerhart, 2005; Raff, 1996).
Starting in 1995, there was a veritable revolution in biology, with the complete sequencing of the first genome of an organism (Fleischmann et al., 1995). In the coming years not only was the monumental task of sequencing the genomes of most model organisms (1998; 2000; Adams et al., 2000; Aparicio et al., 2002; Blattner et al., 1997; Goffeau et al., 1996; Kunst et al., 1997; Sodergren et al., 2006), including humans (2004; Venter et al., 2001), achieved, but sequencing of the genomes of several experimentally less-tractable organisms was also completed (Abad et al., 2008; Ivens et al., 2005; Loftus et al., 2005). These developments opened up unprecedented new avenues: (1) they allowed researchers to break free from the constraints of conventional forward-genetics studies. Organisms could now be studied on the basis of their complete gene sets rather than on the basis of limited prior hints from other model organisms. (2) The use of powerful computational methods to analyze protein and nucleic acid sequences and structures helped develop high confidence predictions regarding biological function directly from genome sequence. In many cases such predictions based on evolutionary principles and the statistical power of sequence analysis went far beyond what could be inferred through naive experimental genetic or biochemical explorations of the same protein or nucleic acid molecule (Altschul et al., 1997; Durbin, 1998). (3) The birth of genomics allowed the first robust reconstructions of evolutionary relationships between organisms. It also enabled the identification of the genomic correlates of major morphological transitions in evolution, such as emergence of eukaryotes and the origins of multicellularity (Aravind and Subramanian, 1999; Aravind et al., 2000; Doolittle, 1999; Lespinet et al., 2002). (4) Genomics also provided the foundation for a whole class of high-throughput studies on cellular and developmental processes that tried to address the function of every gene in a given organism. These studies took several forms – generation of large-scale gene-knockout repositories (Giaever et al., 2002; Moerman and Barstead, 2008), condition/tissue-specific gene expression maps (Hughes et al., 2000; Jongeneel et al., 2005; Murray et al., 2004), determination of complete protein-protein interaction maps of several organisms (Gavin et al., 2006; Giot et al., 2003; Krogan et al., 2006; Li et al., 2004; Rual et al., 2005) (Ewing et al., 2007; LaCount et al., 2005), identification of transcription factor-target gene interactions (Gama-Castro et al., 2008; Harbison et al., 2004; Lee et al., 2002; Luscombe et al., 2004; Sierro et al., 2008; Teichmann and Babu, 2004), determination of parts of the proteome subject to various post-translational modifications (Peng et al., 2003; Ptacek et al., 2005), and interactions between genes and regulatory RNAs (Amaral et al., 2008). While these studies are far from complete, they have already produced data on an unprecedented scale and are promising to change the way all aspects of biology are addressed.
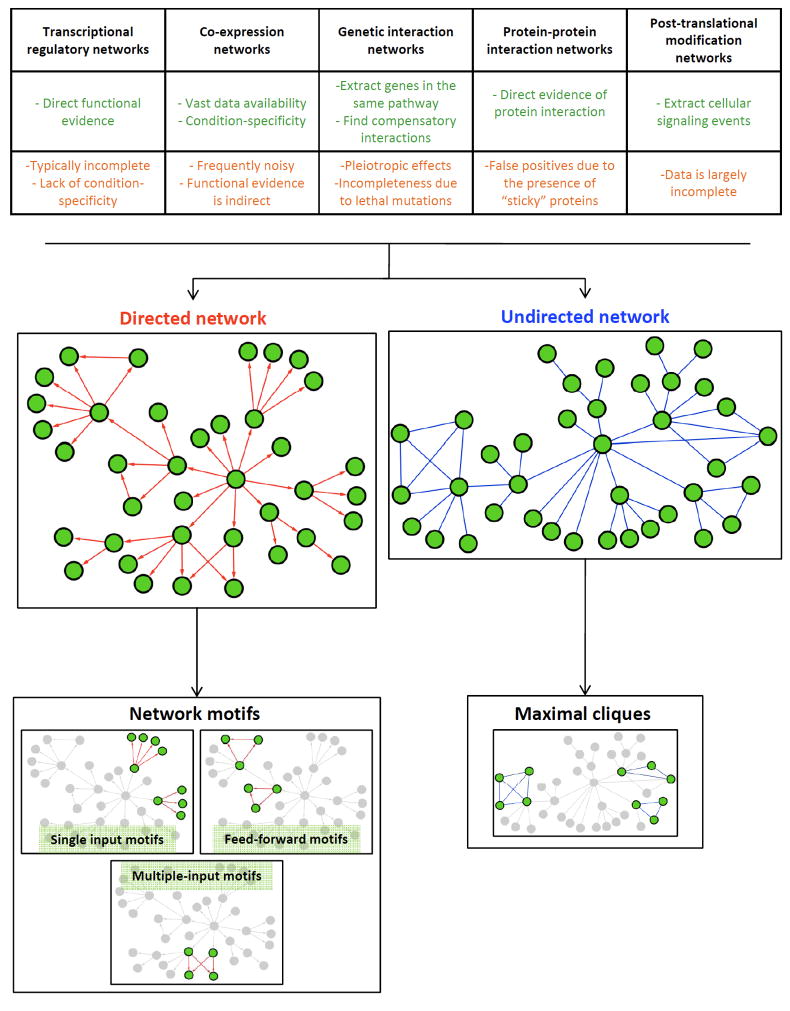
One hope is that the combination of these studies might allow a unification of the seemingly independent disciplines within biology, beyond what has been previously achieved (Kirschner and Gerhart, 2005). In particular, it is hoped that evolution, biochemistry, and development could be brought together successfully to explain the diversity of multicellular forms. A notable aspect of moving towards such a unified view has been the development of the network representation of biological data (Balaji et al., 2006a; Barabasi and Oltvai, 2004; Gianchandani et al., 2006; Russell and Aloy, 2008; Shen-Orr et al., 2002). Networks or graphs represent various entities such as genes, proteins, or other metabolites as nodes, which are then connected by edges, which represent an abstraction of a particular form of association or interaction (Fig. 1). Such interactions between nodes may take many forms, such as regulatory interactions between a gene and a transcription factor or a regulatory RNA, protein-protein interactions, genetic interactions between two genes, an abstraction representing the post-translational modification of one protein by another, a reaction linking two successive compounds in a biochemical pathway or the linkage of individual domains in a polypeptide (Fig. 1). The immediate advantage of such representations is that they can be explored for patterns and features by the human eye, while at the same time being amenable to computational operations. This latter set of operations has been inspired by methods from the graph theory, and is of enormous value in extracting previously concealed information regarding the system as a whole (Balaji et al., 2006a; Barabasi and Bonabeau, 2003; Barabasi and Oltvai, 2004; Gianchandani et al., 2006; Russell and Aloy, 2008; Shen-Orr et al., 2002). Thus, one can for the first time explore how the surrounding context of pathways affect the behavior of an individual pathway, which might have been put together from painstaking genetic or molecular studies. Another less-appreciated, but vital aspect, of network representations has been the ability to interface them with conventional evolutionary studies. Such investigations previously concentrated on the evolution of the nodes of the networks, i.e., proteins or nucleic acids. But they can now be integrated with the evolutionary changes relating to their biological roles, i.e., the edges which represent their interactions.
Figure 1.
The upper panel shows the main technologies employed for generating high-throughput data that are further used to compute the regulatory networks. Some important advantages and disadvantages of each technique are emphasized. Depending on the nature of the data, directed or undirected networks can be inferred. The modular structure of the network can also be explored through the detection of motifs (in directed networks) and cliques (in undirected networks).
The success of the above approach, often termed the “systems” approach, in the past decade has resulted in an abundance of these network representations, especially for the unicellular models such as Saccharomyces cerevisiae and Escherichia coli, and to a certain extent the multicellular animals including humans and parasites, such as Plasmodium falciparum (Balaji et al., 2006a; Barabasi and Oltvai, 2004; Gianchandani et al., 2006; LaCount et al., 2005; Russell and Aloy, 2008; Shen-Orr et al., 2002). However, in multicellular forms developmental process and spatial differentiation have presented technical difficulties for the complete application of the systems approach. Despite obvious differences between multicellular forms and the unicellular models, there are underlying commonalities of great significance. Firstly, in the context of development, though multicellular forms exhibit spatial and temporal differentiated states, these have cognates in the temporal differentiation exhibited by the unicellular models – i.e., the same cell of a unicellular organism assumes very different metabolic and physiological states over time (rather than space) in the course of encountering different environmental inputs. Secondly, evolutionary studies show that various multicellular lineages observed in animals, amoebozoans, plants and fungi have closely related to unicellular sister-groups, which appear to approximate the ancestral condition from which multicellularity emerged (James et al., 2006; Pawlowski and Burki, 2009; Ruiz-Trillo et al., 2008). Hence, the principles of biological network structure and dynamics gleaned from unicellular models, when combined with the more sparse data from multicellular forms, could illuminate several aspects pertaining to the provenance and expressions of multicellularity.
In this review we attempt to combine concepts related to the organization of various regulatory networks with evolutionary inferences derived from comparative genomics to present a synthetic view of some aspects of the origin and diversification of multicellular forms. Our intention is not to comprehensively list the conclusions of all studies in this direction since the coming to fore of the systems approach. Instead, we seek to highlight key points, including some that have been relatively neglected, and then present their potential in understanding aspects of the biology of multicellular forms. To achieve this we layout the review in three broad and apparently distinct sections: (1) we first introduce types of regulatory networks and the principles that can be deduced from them. (2) We then consider the major conclusions emerging from comparative genomics to provide the evolutionary context for the nodes and edges in networks. (3) Finally, we consider a case study to illustrate how an actual regulatory sub-network pertinent to tissue differentiation in animals has been pieced together in evolution.
REGULATORY NETWORKS
Regulatory works and their types
As mentioned above, a wide range of biological networks have been reconstructed, chiefly differing in the abstraction specified by their edges (Fig. 1). Among these there are generic networks, which encompass all genes or their protein products, such as the genetic interaction networks (GInet) (Collins et al., 2007; Li et al., 2004) or the protein-protein interaction networks (PPInet) (Gavin et al., 2006; Krogan et al., 2006; Rual et al., 2005), and more restricted networks connecting transcription factors to their target genes (Tnet) (Balaji et al., 2006a; Harbison et al., 2004; Luscombe et al., 2004; Vermeirssen et al., 2007) or regulatory RNA-target gene networks (Ke et al., 2003). In the current article we primarily consider regulatory networks. While there is some fuzziness in defining these networks, there is no difficulty in recognizing such a network. A regulatory network can be defined as a network where the nodes are either genes or their products, and the edges signify transcriptional, post-transcriptional, or post-translational control of one node by another. It might also more abstractly signify two genes interacting in a regulatory cascade, commonly termed signaling pathways, due to genetic epistasis or physical interaction involving their products. We usually do not consider certain “structural interactions”, for example, interactions of proteins and RNAs in constituting the mature ribosome, in a regulatory network. The archetypal examples of regulatory networks are Tnets that capture the regulation of genes at the transcriptional level (Balaji et al., 2006a; Harbison et al., 2004; Luscombe et al., 2004; Vermeirssen et al., 2007). Tnets are directed networks (Barabasi and Bonabeau, 2003) – the edges in this network always go from a transcription factor (TF) to a target gene (TG) (Fig. 1). Comparable to the Tnet is a regulatory network with edges connecting regulatory RNAs to their target genes (Ke et al., 2003). Another similar type of regulatory network is that between kinases, phosphatases and their target proteins that are subject to phosphorylation or dephosphorylation (Fiedler et al., 2009; Ptacek et al., 2005). In a sense these phoshorylation networks are sub-networks of the conventional protein-protein interaction networks.
A more complex form of a regulatory network is the ubiquitin network (Venancio et al., 2009), which depicts interactions between components of the Ub-system, i.e., ubiquitin/ubiquitin-like proteins (e.g., SUMO), the conjugation/de-conjugation enzymes, the proteasome, and various other accessory components. This regulatory network too overlaps with the more generic PPInet and GInet (Venancio et al., 2009). Edges in networks such as these are typically depicted as undirected, because there might not be a sense of polarity in all of these interactions (Fig. 1) (Barabasi and Bonabeau, 2003). In principle, various individual regulatory networks can also be combined to produce composite networks. Other more abstract regulatory networks are derivatives from primary networks that connect different regulatory proteins with edges by virtue of shared targets. The best known of these is the co-regulatory network derived from the Tnet by connecting transcription factors, which share common target genes, and is very useful in understanding cooperation between regulatory proteins (Balaji et al., 2006a; Balaji et al., 2006b).
Currently, regulatory network reconstructions with the best coverage and quality in terms of both nodes and edges are only available for unicellular forms, such as S. cerevisiae and E. coli. PPInets for metazoans, such as Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens, with reasonable coverage(Giot et al., 2003; Li et al., 2004; Rual et al., 2005), derived mainly from high-throughput yeast two-hybrid studies, have become available, but the situation is less satisfactory for Tnets (Vermeirssen et al., 2007). Networks reconstructed from large-scale data are good in terms of coverage, but suffer from false positives to varying extents due to recovery of spurious interactions (Fig. 1) (Yu et al., 2008). Technical issues, in addition to the inherent complexities of developmental and differentiation processes, which affect the reconstruction of such networks in multicellular systems, are: (1) difficulties due to the complex gene structure, including large introns, alternative splicing, and presence of composite transcription regulatory elements that are often at great distances from the genes they regulate (Maniatis and Reed, 2002). (2) The complexities of chromatin organization, which influence more conventional regulatory interactions between TFs and TGs (Iyer et al., 2008). (3) The still incompletely understood processes, such as DNA modifications, chromatin protein modifications, and signaling pathways (Iyer et al., 2008). On a more positive note, we do possess detailed studies on specific developmental regulatory networks in animals, e.g., the Notch network or the TGF network (Kitisin et al., 2007; Kopan and Ilagan, 2009), or in plants, e.g., leaf and floral development (Lewis et al., 2006), with information on interactions between transcription factors and their intricately tangled target elements. Currently, networks reconstructed from unicellular models are best for inferring large-scale or bulk properties of regulatory networks, whereas those from multicellular models are best for detailed case studies.
General structural properties of regulatory networks
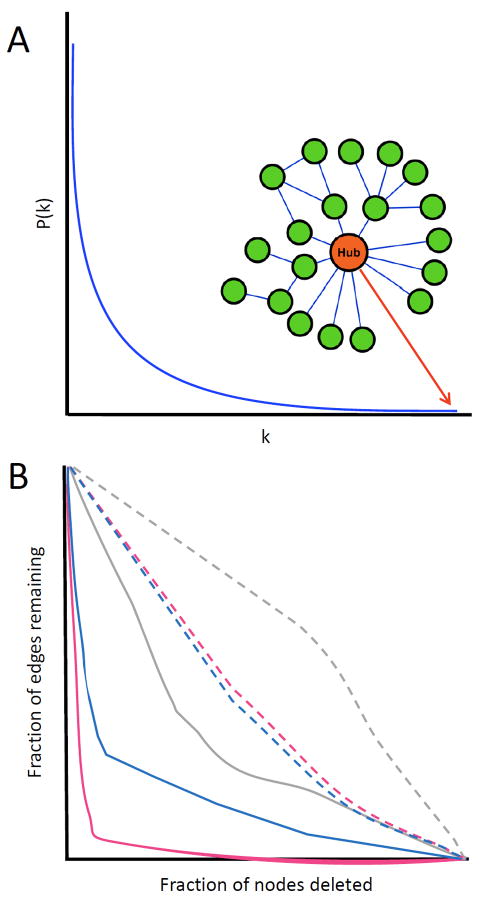
Right from the earliest studies in this regard a fundamental unity in the organization of disparate biological networks has been repeatedly noted (Balaji et al., 2006a; Barabasi and Bonabeau, 2003; Barabasi and Oltvai, 2004; Gianchandani et al., 2006; Russell and Aloy, 2008; Shen-Orr et al., 2002). In global terms they have a nested or self-similar structure that appears to hold over several levels of organization – a structure that can be approximately described as fractal (Fig. 1). The number of edges that connect to a node is termed its degree. When the number of nodes in a network possessing a particular degree is plotted, one gets a distribution (the degree distribution) that is best fitted by the power-law equation of the form n(x)=axk; where n is the number of nodes with a particular degree and x is the degree (Fig. 2). The ‘a’ and ‘k’ in the equations are constants unique to each power-law distribution (Balaji et al., 2006a; Barabasi and Bonabeau, 2003; Barabasi and Oltvai, 2004; Gianchandani et al., 2006; Russell and Aloy, 2008; Shen-Orr et al., 2002). This distribution implies that regulatory networks are similar in properties at all levels in which they exist and are hence scale-free. In reality they are only approximations of the genuinely scale-free structure seen in theoretical networks because, unlike them, biological networks have a well-defined stop – the nodes in the network above or beyond which there are no further levels (Fig. 2). A consequence of the power-law distribution of degrees is that there are few nodes with numerous connections (termed hubs), but most nodes have very few connections – thus hubs dominate the network in terms of connectivity(Barabasi and Albert, 1999; Barabasi and Oltvai, 2004) (Fig. 2).
Figure 2.
A. The degree distribution of the regulatory networks is typically well-approximated by a power-law equation. In the graph, degree (k) is the number of regulatory connections between the nodes, while P(k) indicates the probability of a gene with a given number of such connections. An example of hub in a small network is also shown, along with its position in the degree distribution.
B. Network susceptibility to attack (loss of hubs, solid lines) and failure (random loss of genes, dashed lines). Three networks are represented for comparison purposes: transcriptional (gray), ubiquitin (pink) and protein-protein interactions (blue).
Beyond their distinctive global structure, regulatory networks are also characterized by peculiar structures at their lower levels. In directed regulatory networks, like the Tnet, they are termed motifs (Shen-Orr et al., 2002). Three basic types of network motifs have been identified (Fig. 1): (1) single input motifs, where a given target gene receives inputs only from a single transcription factor; (2) multiple input motifs, where a given target gene receives inputs from two or more transcription factors; (3) feed-forward motifs, where target genes receive inputs from at least two transcription factors, with the additional condition of one of the TFs in the motif also regulating the other TF (Fig. 1). Single input motifs specialize in coordinating expression of various genes required in a particular response, enforce an order in gene expression, and are also the basis for immediate transcriptional responses (Shen-Orr et al., 2002). Multiple input motifs are the key players in integrating responses to different signaling pathways with respect to gene expression. Finally, feed-forward motifs are critical for responding to persistent signals and filtering noise (Shen-Orr et al., 2002). Thus, relative proportions of such motifs in a Tnet are of considerable significance in terms of the regulatory flux passing through the network (Shen-Orr et al., 2002). In undirected networks, such as the PPInet, GInet, and their derivative regulatory networks, such as the ubiquitin-network, a different kind of low-level structure is observed – the dense sub-graph (Yu et al., 2006). These are subsets of nodes in the network that are highly connected relative to the rest of the network. Such regions in networks are determined by identification of structures called cliques (Fig. 2). Formally, a clique is the maximum number of nodes having all possible edges between themselves (i.e., the largest group of nodes, which forms a polygon with edges corresponding to all its sides and diagonals being present) (Yu et al., 2006). The clustering of genes into a clique is suggestive of functional coherence between them or some type of functional interaction between their products. Thus, identification of cliques in regulatory networks is a useful tool for the prediction of functions of poorly-characterized genes linked in a clique with functionally characterized genes by way of the “guilt by association principle”(Balaji et al., 2006a; Balaji et al., 2006b; Yu et al., 2006).
The concept of centrality developed in the graph theory helps in assessing the “importance”, particular genes or proteins in the structure of regulatory networks. Two common measures of centrality of node in a network are degree and betweenness (Barabasi and Albert, 1999; Barabasi and Oltvai, 2004; Brandes, 2001). As described above, the degree is a simple description of how connected a node is, and the most central elements by this measure are the hubs (Barabasi and Albert, 1999; Barabasi and Oltvai, 2004). In directed graphs like Tnets there two types of degrees, namely the in- and out-degrees, that respectively denote the number of genes a TF regulates and the number of regulatory inputs a particular target gene receives from different TFs (Balaji et al., 2006a; Balaji et al., 2006b). In Tnets, TFs which are hubs, typically termed global regulators, influence a vast number of genes and thereby set numerous transcriptional programs in motion (Balaji et al., 2008). TFs with lower connectivity, in contrast, appear to be required for the fine tuning of a transcriptional program by regulating smaller sets of genes (Balaji et al., 2006a; Balaji et al., 2006b). Betweenness is a different kind of centrality measure that represents the number of shortest paths in the network that include a node (Brandes, 2001). Hence, the shortest paths between all pairs of nodes should be calculated in order to compute the betweenness score of a node. Although degree and betweenness of a node in a network are typically correlated, they can illuminate different aspects of the networks. Certain nodes with high betweenness may not be hubs, but play a significant role in connecting various disparate parts of the regulatory networks (Yoon et al., 2006).
Biological significance of the structure of regulatory networks
Scale-free structures similar to biological regulatory networks are also encountered in various unrelated systems – the world-wide-web, the physical structure of the Internet, and the network of human sexual relationships (Amaral et al., 2000; Barabasi and Bonabeau, 2003). This has led researchers to propose generalized evolutionary explanations for the origin of such structures across these disparate systems. The simplest of these merely assumes that (1) a network grows by addition of new nodes and edges, and (2) the edges show preferential attachment to nodes with higher pre-existing degree. Thus, in such an evolutionary scenario, the “rich get richer” and there is a tendency for formation of few hubs and numerous poorly connected nodes (Amaral et al., 2000; Barabasi and Bonabeau, 2003; Gama-Castro et al., 2008; Harbison et al., 2004; Lee et al., 2002; Luscombe et al., 2004; Sierro et al., 2008; Teichmann and Babu, 2004; Yook et al., 2002). This and related simulations can reproduce the structure of biological networks; however, there is no clear test yet which establishes such a mechanism to indeed be the cause for the emergence of such a structure in regulatory networks. Irrespective of the mechanistic model for their origin, structural properties of regulatory networks have profound implication for biological systems. An aspect of network structure, which is important with respect to evolution, is the modularity that is observed at the lower levels of organization in the form of motifs or cliques (Fig. 2). It indicates that particular motifs or cliques can be linked to a set of functionally distinct nodes within a given regulatory network as a natural consequence of its scale-free structure (Balaji et al., 2006a; Balaji et al., 2006b; Gama-Castro et al., 2008; Harbison et al., 2004; Lee et al., 2002; Luscombe et al., 2004; Sierro et al., 2008; Teichmann and Babu, 2004). Thus, a particular regulatory mechanism specified by a clique or a motif can be easily recruited in various functional contexts. This provides the basis for understanding the prevalent observation that similar regulatory sub-networks can be used in both unicellular and multicellular forms or reused within different tissues in multicellular forms. Another, more direct consequence of the scale-free structure is the remarkable resilience of such regulatory networks to random loss of nodes or failure (Albert et al., 2000) (Fig. 1). This is because most nodes in scale-free network have few links; hence, disrupting one of them at random is unlikely to break down the network. In contrast, disruption of hubs, termed attack, can break apart a regulatory network more easily, as hubs account for a large number of the connections in a network (Albert et al., 2000) (Fig. 1).
The biological correlate of this network property is the ability of regulatory systems to withstand random failures from disruption of genes due to mutation or chemical action. This in part explains why across the evolutionary tree the number of genes whose disruption results in lethality is less than 15%–20% of the total number of genes in an organism(Giaever et al., 2002). While resilience to failure is common to different types of regulatory networks, they still show marked quantitative differences in this particular property (Fig. 2). Tnets in general are more resilient to failure compared to other regulatory networks that entirely or predominantly depend on protein-protein or genetic interactions (Balaji et al., 2006a). Some regulatory networks, like the ubiquitin network, are also far more susceptible to attacks than others (Venancio et al., 2009). Genetic studies have also suggested that eukaryotes in particular can be quite resilient to mutation of transcription factors, including those lacking close paralogs (Balaji et al., 2008; Hu et al., 2007). This suggests that the Tnet is a particularly robust regulatory network, beyond what would be expected on the basis of gene redundancy. The construction of co-regulatory networks based on Tnets shows that there is underlying architecture indicative of indirect backup, where multiple unrelated TFs can potentially cover for each other in this regulatory system (Balaji et al., 2006a; Balaji et al., 2006b). Existence of such an “over-engineered” backup system in the form of the architecture of the co-regulatory network is likely to be a major determinant of the “evolvability” of the gene expression in organisms (see below). Differences in relative tolerance of networks to failure and attack also have a bearing on both the evolution of such networks – versions more tolerant to failure and/or attack appear to evolve more rapidly between organisms and might contribute to regulatory diversity between organisms (Balaji et al., 2006a; Balaji et al., 2006b).
To be able to use the information from regulatory networks in understanding problems pertaining to multicellularity we need to place them in the context of the evolution of such organisms. For this we turn to comparative genomics in the next section and try to understand how components of regulatory systems have originated and evolved.
EVOLUTION OF REGULATORY SYSTEMS
Multiple origins of multicellularity
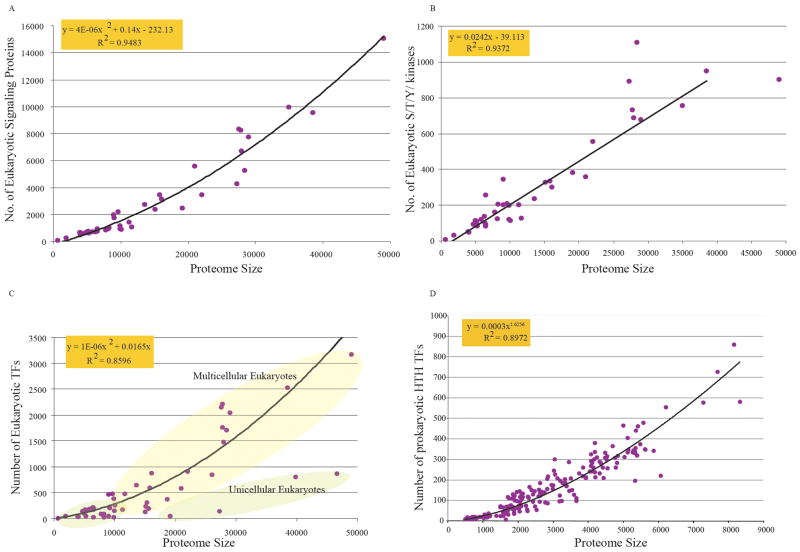
The presence of multicellular forms across the tree of life has suggested that this morphological principle emerged on multiple occasions in course of evolution (Kirschner and Gerhart, 2005; Raff, 1996; Rokas, 2008). The major independent emergences of the multicellularity among eukaryotes include: (1) Animals; (2) Fungi; (3) Amoebozoan slime molds; (4) Plants; (5) Chromist algae (e.g. brown algae) (6) chromist oomycetes (mildews or water molds); and (7) Heterolobosean or amoeboflagellate (acrasid slime molds). Among bacteria too multiple emergence of multicellular forms have been noted, for example, among myxobacteria (delta-proteobacteria), actinobacteria, cyanobacteria, and acidobacteria, and amongst archaea at least one multicellular form lineage has been observed in the form of Methanosarcina (Mayerhofer et al., 1992; Shapiro and Dworkin, 1997). At face value it would appear that these multiple origins have little in common, but comparative genomics has revealed certain shared features at the molecular level. It has been observed that the number of specific transcription factors scales non-linearly with increase in proteome size (Anantharaman et al., 2007a; van Nimwegen, 2003) (Fig. 3). In the case of bacteria, multicellular forms typically have both large proteomes and a more than linear increase in the fraction of the proteome that comprises of specific transcription factors (Aravind et al., 2005). In the case of eukaryotes, larger proteome size does not necessarily imply a multicellular morphology – the largest eukaryotic proteomes are currently seen in unicellular forms such as ciliates (Eisen et al., 2006) and Trichomonas (Carlton et al., 2007) (Fig. 3). However, just as in the case of multicellular bacteria, the multicellular eukaryotes have a much higher fraction of their proteome devoted to specific transcription factors than unicellular forms of comparable size (Iyer et al., 2008). In particular this trend is exemplified to the greatest degree in animals, which apparently have the most complex multicellular morphologies. Thus, across the tree of life, a greater than proportional increase in the number of specific transcription factors appears to be a correlate of multicellularity (Fig. 3).
Figure 3.
(A). Nonlinear scaling of total number of signaling proteins in eukaryotes with proteome size along with the best-fit curve. One hundred seventy signaling domains were studied in 43 completely sequenced eukaryotic genomes. (B). Scaling of serine/threonine/tyrosine (S/T/Y) kinases in eukaryotes with proteome size with the best-fit curve. (C). Scaling of eukaryotic transcription factors with proteome size with the best-fit curve. Note that multicellular forms have higher numbers of specific transcription factors. (D). Scaling of prokaryotic specific transcription factors with the HTH domain from complete genomes with the best-fit curve.
In the case of multicellular bacteria, a significantly higher number of serine/threonine/tyrosine kinases (S/T/Y kinases) and phospho-peptide-binding FHA domains has been observed than their morphologically less organized counterparts (Perez et al., 2008). Thus, a case could be made that in bacteria the emergence of complex phosphorylation networks was a notable correlate of the origin of multicellularity. However, all eukaryotes have expanded protein kinase repertoires. Hence, no comparable trend to the bacteria is observed with respect to S/T/Y kinases, or for that matter with most families of signaling proteins, which display largely linear or mild power-law scaling with respect to overall proteome size of eukaryotic organisms (Fig. 3) (Anantharaman et al., 2007a). Interestingly, bacteria that display multicellular organization or temporal morphological development consistently encode an interesting array of proteins in the genomes which, in addition to the S/T/Y kinases and FHA domains, includes STAND superfamily ATPases, caspase-like proteases, and TIR (Toll-interleukin receptor) domain proteins (Anantharaman et al., 2007a). Among eukaryotes too, such regulatory proteins with the above set of domains are particularly prevalent in the proteomes of multicellular forms. In both multicellular animals (e.g., nematode Ced4 and human APAF1, caspase-1) and plants (e.g., disease resistance gene N and metacaspases) proteins with the same domains have been implicated in specific signaling pathways pertaining to cell death, pathogen response, and tissue remodeling (Aravind et al., 2001; Chamaillard et al., 2003; Ting et al., 2008). This suggests that apoptosis-related signaling pathways based on these protein domains might be a notable common denominator among several multicellular lineages across the three superkingdoms of life. This can be interpreted within the evolutionary framework under the kin selection hypothesis (Hochberg et al., 2008). In a multicellular organism, the cells being clonal are effectively an ensemble of kin. Thus, some cells “sacrificing” themselves via apoptosis for the highly increased fitness of sister cells in the ensemble might provide the dying cells with greater inclusive fitness than if they remained in the unicellular state. In terms of regulatory networks, animal model systems indicate that these proteins functionally interact to form distinct modules in regulatory networks primarily devoted to mediation of apoptosis (Aravind et al., 2001; Chamaillard et al., 2003; Ting et al., 2008). In bacteria, in addition to co-occurring in the genomes of organisms displaying multicellularity or developmental complexity, genes encoding such proteins also tend to cluster together in predicted operons, suggesting functional interactions in similar networks (Aravind et al., 2005). Thus, proteins, which mediate apoptosis or related cell-death processes, are likely to comprise related regulatory network modules that are common to phylogenetically distant organisms sharing multicellularity. Based on their phyletic patterns, it appears that lateral transfer of these interacting genes, encoding proteins with functions related to apoptosis, enabled development of multicellularity in different organisms. Likewise, several extracellular protein domains involved in adhesion also appear to have been disseminated by lateral transfer across bacteria and multicellular eukaryotes, and might have provided a common functional mechanism for cellular assembly (Anantharaman et al., 2007a).
Other subtle and less-recognized molecular features with considerable bearing on the structure of regulatory networks also distinguish eukaryotic multicellularity. Domain architectures, i.e., the way individual protein domains are linked in a polypeptide, can be converted into a network representation. In this representation, domains are conceived as nodes of the domain architecture network and the adjacent occurrence of two domains in a polypeptide is indicated as an edge joining the two domains (Anantharaman et al., 2007a; Iyer et al., 2008). The total domain architecture network is the ensemble of all such connections between domains across a set of proteins under consideration. The domain architecture network can tell us how simple or complex the architectures of a particular set of proteins are in a given organism. When these networks were computed for signaling proteins across eukaryotes, it was observed that multicellular forms often tend to have greater complexity in these networks (more nodes and edges between them) than their unicellular relatives (Anantharaman et al., 2007; Iyer et al., 2008). This trend was even more marked for proteins involved in chromatin structure and dynamics, such as histone-modifying and chromatin remodeling proteins (termed chromatin proteins to distinguish them from specific transcription factors) (Anantharaman et al., 2007a; Iyer et al., 2008). The increased complexity of the domain architecture network of signaling and chromatin proteins suggests that there are more likely to be a concomitant increase in number of interactions between proteins, because combining multiple domains in a polypeptide allows for more combinatorial interactions. In particular, in the case of chromatin proteins, increased architectural complexity is suggestive of increased ability to add epigenetic marks via histone and DNA modifications, and subsequently “read” those marks via specific interactions (Anantharaman et al., 2007a; Iyer et al., 2008). This could have major role in maintaining multiple differentiated cellular states via epigenetic control.
Hence, at least in case of the better-studied eukaryotic multicellular forms, we notice that two sets of regulatory networks are likely to have concomitantly grown larger and more complex. Firstly, the disproportionate increase in the number of specific transcription factors indicates that the Tnet greatly increased in complexity (Fig. 3). Secondly, the emergence of more complex domain architectures among signaling and chromatin proteins suggests that the emergence of multicellularity was accompanied by expansion of signaling and protein-modification networks relative to their unicellular counterparts (Anantharaman et al., 2007a; Iyer et al., 2008). Nevertheless, given the overall approximately scale-free architecture of regulatory networks, the general principles of network organization (as elucidated above) would remain consistent across unicellular and multicellular forms, despite the expansion in the latter. Importantly, the general results pertaining to the modularity of regulatory networks (see above) suggest how modules, such as those pertaining to apoptotic functions, are likely to have been ported across distantly related lineages via lateral transfer. Finally, it should be noted that despite the multiple origins of eukaryotic multicellularity, four of these are concentrated in a particular monophyletic clade of eukaryotes termed the crown group, which includes animals, fungi, slime molds, and plants (Iyer et al., 2008; Pawlowski and Burki, 2009; Rokas, 2008; Ruiz-Trillo et al., 2008). The chromists, which also display multicellular forms, have emerged through a secondary photosynthetic endosymbiosis with the plant lineage (Bhattacharya et al., 2004). Interestingly, these chromists encode several regulatory proteins, including transcription factors, which appear to have been acquired from the plant endosymbiont (Iyer et al., 2008). These lineages also generally possess a higher normalized count of chromatin proteins and transcription factors than other eukaryotes (Iyer et al., 2008). Taken together these observations raise the possibility that the genome of the ancestral crown group eukaryote already possessed certain features that enabled some form of facultative multicellularity, perhaps comparable to what is observed today in amoebozoan slime molds. The base-level multicellularity was probably reinforced in some lineages with further expansions of transcription factors, chromatin proteins, and adhesion proteins, while it was attenuated in others via extensive gene loss (for example, related saprophytic life style in fungi).
Lineage-specific gene expansions
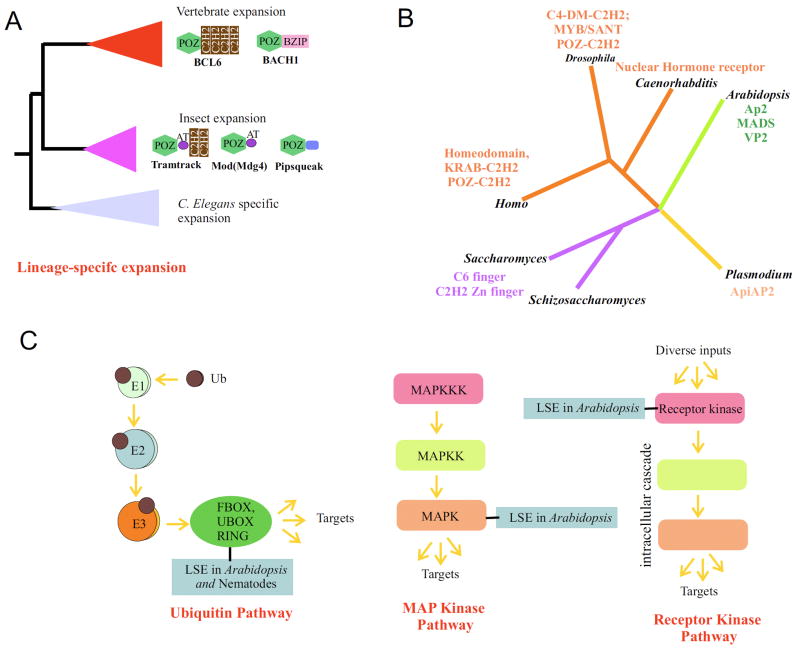
One of the most striking revelations from the comparative genomics of eukaryotes has been the discovery of the phenomenon of lineage-specific expansions of protein families (LSEs) (Lander et al., 2001; Lespinet et al., 2002). A LSE is defined as the expansion of a family of proteins in a particular lineage after its divergence from a reference sister lineage (Lespinet et al., 2002) (Fig. 4). One of the classical examples of lineage-specific expansions is that of the family of transcription factors with the POZ (also called BTB) domain (Aravind and Koonin, 1999a; Lespinet et al., 2002) These transcription factors have an N-terminal POZ domain combined with a C-terminal DNA-binding domain that is usually a C2H2 Zn-finger (Spokony and Restifo, 2007). Both vertebrates and insects have large numbers of these transcription factors (over 50 paralogs per genome). However, phylogenetic analysis reveals that these expansions happened independently in the insect and vertebrate lineages, after they separated from their common ancestor (Aravind and Koonin, 1999a) – the vertebrate POZ domain transcription factors group with each other to the exclusion of the insect versions, and likewise the insect versions group with themselves to the exclusion of the vertebrate forms (Fig. 4). Development of an algorithm to systematically detect LSEs and their case-by-case analysis across the eukaryotes revealed that they are one of the most important forces that shape the contours of proteomes (Lespinet et al., 2002). Anywhere between 20% (e.g., in yeasts) to 80% (e.g., plants and vertebrates) of the eukaryotic proteomes are comprised of families of lineage-specifically expanded families. Further these LSEs account for nearly one half of all the paralogous clusters of proteins encoded in a eukaryotic proteome (Lespinet et al., 2002). Categorization of the LSEs suggests that they are particularly prevalent in certain cellular functions. These include proteins involved in responses to stress, pathogen/parasite and xenobiotic, proteins placed at the termini of signaling cascades (e.g., E3s in the ubiquitin-based pathways and MAP kinases of the phosphorylation cascades), transcription factors, and chemoreceptors (Lespinet et al., 2002). This pattern of function-wise enrichment of the LSEs is consistently retained across eukaryotic phylogeny and has thus become a powerful tool for predicting functions among uncharacterized proteins when combined with sequence analysis.
Figure 4.
(A). A simplified phylogenetic tree of the POZ domain transcription factors illustrating the concept of the lineage-specific expansion. Note that the transcription factors from a given lineage are closer to its paralogs than to those from other lineages. The domain architectures of selected proteins are shown to the right. C2H2- Zinc-finger domain; AT- AT-hook DNA binding domain; Bzip- basic zipper domain; the C-terminal domain in pipsqueak is a helix-turn-helix domain. (B). Examples of lineage-specific gene expansions in various transcription factor families in different eukaryotic lineages which are labeled by genus name. All LSEs from one lineage are colored in the same away. The Myb/SANT domains expanded in Drosophila represent a family of helix-turn-helix DNA transcription factors typified by the Zeste protein. (C). The ubiquitin and kinase pathways are shown to illustrate LSEs occurring in component proteins of the pathway that are at termini.
One of the most important aspects of LSEs with respect to evolution of regulatory networks is the preponderance of this phenomenon among transcription factor families (Coulson and Ouzounis, 2003; Iyer et al., 2008; Lander et al., 2001; Lespinet et al., 2002) (Fig. 4). The most prevalent family of transcription factors in a given proteome is widely different across different eukaryotic lineages, including within different animal lineages (Fig. 4): Nuclear hormone receptor-type zinc finger transcription factors are the most prevalent transcription factors among nematodes, the KRAB-type C2H2 zinc fingers in vertebrates, and AP2, VP1, and MYB domain transcription factors in angiosperm plants (Coulson and Ouzounis, 2003; Iyer et al., 2008; Lander et al., 2001; Lespinet et al., 2002). Developmental genetics in model systems have shown that many key developmental processes are regulated by transcription factors belonging to these LSEs, rather than those inherited relatively unchanged from the last common ancestor of all animal or plant lineages. A striking case of this is the POZ domain transcription factors in Drosophila, which as noted above belong to an insect-specific LSE. Gene products of this expansion regulate a diverse range of developmental decisions at the transcription level in contexts such as axonal path-finding (Lola) (Giniger et al., 1994), morphological diversity of neuronal dendrites (abrupt) (Kim et al., 2006; Ryner et al., 1996), specification of neurons that determine sexual orientation (Fruitless) (Ito et al., 1996), specification of cell-fates in the eye (Tramtrack69) (Lai and Li, 1999), the development of distinctive external genitalia (ken and barbie) (Lukacsovich et al., 2003), epithelial morphogenesis (ribbon)(Shim et al., 2001), and early oogenesis (Pipsqueak) (Horowitz and Berg, 1996), to name just a few representatives. This functional “colonization” of a large number of disparate functions after the emergence of a LSE suggests they might have a particularly important role in the diversification of morphology in multicellular forms. The occurrence of such LSE also implies that Tnets undergo massive reorganization and rewiring with the emergence of new lineages (Babu et al., 2006). This is also consistent with studies on Tnets, which indicate that hubs are routinely displaced by new transcription factors or that hubs are lost and new hubs emerge in their place. This plasticity of Tnets is potentially attributable to its innate robustness due to the presence of internal backup, which allows the replacement of old transcription factors by new ones emerging from an LSE (Balaji et al., 2006b). Further, representatives of the LSE of POZ domain transcription factors in Drosophila are typically positioned downstream of master regulators of antero-posterior patterning (e.g., the Hox proteins), or function with the chromatin proteins (e.g., polycomb and trithorax group proteins) involved in maintaining the boundaries of the antero-posterior gene expression (Ghosh et al., 2001; Pagans et al., 2002; Zhang et al., 2006; Zhu et al., 2006). Hence, transcription factors which are generated by LSEs appear to be fitted in the regulatory network hierarchy usually in terminal locations, thereby supporting their role in generation of morphological diversity within the framework of an otherwise conserved generic antero-posterior body plan.
Similar patterns are observed in the case of certain other proteins undergoing LSEs in the Ub/Ubl conjugation network (U-net) (Venancio et al., 2009) and the protein phosphorylation networks (Fiedler et al., 2009; Ptacek et al., 2005). In the U-net the most prominent LSEs are concentrated among components of E3 enzymes of the pathway, such as RING, Ub-box, and F-box domains (Fig. 4) (Lespinet et al., 2002). The E3s are the terminal enzymes in the conjugation cascade which finally transfer the Ub to specific substrates (Hochstrasser, 2009). In contrast, E1 and E2 enzymes tend to show no LSEs and are largely vertically conserved across large sections of the eukaryotic phylogenetic tree (Anantharaman et al., 2007a). This indicates that the E3 LSEs in the U-net aid in directing a conserved stem pathway of E1s and E2s towards a diversity of substrates that differ from lineage to lineage. In the case of the F-boxes, LSEs might have a specific role in targeting various pathogen proteins for Ub-mediated development (Thomas, 2006). However, the LSEs of RING domains participating in developmental regulation are supported by studies in both animals and plants (Serrano et al., 2006). Similarly, in the case of phosphorylation networks, LSEs are noted among kinases of the Calcium-dependent kinase and MAP kinase families in plants, casein-kinase and soluble tyrosine families in nematodes, and various receptor kinases in various plant and animal lineages (Lespinet et al., 2002) (Fig. 4). Protein phosphatases also show LSEs in angiosperm plants and to a certain extant in vertebrates (Lespinet et al., 2002; Popescu et al., 2009). Many of these expanded families of kinases again belong to the termini of signaling cascades – MAP kinases phosphorylate specific substrates, whereas the receptor kinases are usually extreme upstream responders to primary extracellular signals (Lespinet et al., 2002; Popescu et al., 2009). Thus, these kinase LSEs are also likely to have played a major role in harnessing a conserved core pathway to various extrinsic signals or targeting them to different sets of substrates in different lineages (Fig. 4).
Prevalence of LSEs in specific transcription factors and termini of signaling cascades, especially in multicellular forms, has been a major factor in the rewiring of parts of regulatory networks. In contrast to the emphasis on lineage-specific adaptations, conventional “evo-devo” studies have repeatedly shown conserved regulatory cores in networks. These might be shared across Metazoa, and are required for antero-posterior and dorso-ventral axis patterning, specification of tissues developing from different germ layers, asymmetric cell-division, and signaling between different germ layers (Arthur, 2002; Davidson and Erwin, 2006; De Robertis, 2008; Kopan and Ilagan, 2009; Raff, 1996). Likewise, in plants certain conserved regulatory networks have been implicated in developmental pathways for morphological elements, such as flowers and leaves, and differentiation of tissues (Endress and Doyle, 2007; Krizek and Fletcher, 2005; Reinhardt, 2005; Tsukaya, 2006). Hence, by combining insights from these evo-devo studies and those offered by LSEs discerned from comparative genomics, we may conclude that: (1) core modules of regulatory networks specifying general morphological landmarks of a major lineage of multicellular organisms (e.g., plants or animals) are indeed more widely conserved; (2) however, beyond these generic modules, the regulatory networks are extensively refashioned due to generation of new nodes by LSEs, thereby allowing adaptive radiations via lineage-specific alterations of patterning and biochemistry of specific tissues.
Gene loss and horizontal transfer
Another force which comparative genomics has revealed to play a major role in the re-organization of regulatory networks is gene loss. Studies on patterns of gene loss suggest that beyond a general background of sporadic gene losses there are discernable patterns of concerted loss in which functionally connected genes tend to be lost as a unit (Aravind et al., 2000; Edvardsen et al., 2005; Miller et al., 2005). This latter form of gene loss is a reflection of the modular network architecture. Loss of a key gene can render an entire module of a regulatory network dysfunctional. By virtue of the scale-free network architecture, other genes in the module typically might not have many extraneous functional connections, and are effectively superfluous as the loss of the key gene has already attenuated the role of the module. Hence, there is good chance that the other genes in the module are also lost subsequently. Massive losses of this type are observed in fungi, particularly in forms like yeasts (Liti and Louis, 2005; Wapinski et al., 2007). Given that multicellularity was already present in the ancestral fungus (James et al., 2006), such gene loss is likely to have been a major player in the regression of yeasts to a more unicellular condition. Similar losses are also seen across Metazoa (Edvardsen et al., 2005; Miller et al., 2005) – in extreme cases, such losses appear to similarly result in regression of the multicellular animal form to a more unicellular condition, as seen in Myxozoa (Kent et al., 2001). In other cases the loss might be correlated with different degrees of morphological simplification. Availability of genomic sequences of basal animals, such as cnidarians, Trichoplax, and sponges, shows that nematodes have lost several modules of regulatory networks, such as the hedgehog, NFkB, and certain apoptotic signaling modules (Burglin, 2008; Miller et al., 2007; Srivastava et al., 2008; Zmasek et al., 2007). These losses might have a role in the absence of prominent lateral appendages and developed photoreceptors in nematodes, such as C. elegans.
A regulatory system, which is often subject to gene loss, is the RNA-interference (RNAi) network that performs the key role of negatively regulating genes at the post-transcriptional level. This system is highly developed in plants, and certain fungi and animals, but has been repeatedly lost in many of the intervening sister lineages (Anantharaman et al., 2002; Aravind et al., 2000; Cerutti and Casas-Mollano, 2006). For example, it has been entirely lost in the yeast, S. cerevisiae, but is present in a largely intact form in other fungi such as Schizosaccharomyces pombe, Neurospora crassa, and mushrooms. In animals this system shows an interesting pattern of multiple partial losses. The nematodes have a complete RNAi network with both the small RNA-processing system (e.g., Dicer and Drosha), the mRNA targeting nucleases of the Argonaute family, and the propagators of siRNAs via RNA-dependent replication (the RNA-dependent RNA polymerase/RdRP) (Anantharaman et al., 2002; Aravind et al., 2000; Cerutti and Casas-Mollano, 2006). Some insects and vertebrates lack the siRNA replicating part of this network encompassing the RdRP system (Anantharaman et al., 2002; Cerutti and Casas-Mollano, 2006). Genomics studies have shown that the RdRP is however present in the genome of the basal chordate, amphioxus (Branchiostoma), suggesting that there have been at least two independent losses of this subnetwork, one in the line leading to vertebrates and one in insects. A point emphasized by losses in the RNAi network is that regulatory networks might have an intrinsic robustness due to indirect back-up from other functionally comparable systems. In the case of the RNAi system, the chromatin-level gene silencing and post-translational protein degradation systems perform biochemically unrelated actions that are nevertheless effectively functionally related, i.e., they modulate the levels of gene products (Lorentzen and Conti, 2006; Zofall and Grewal, 2006). The availability of such a backup enables the loss of certain systems depending on the evolutionary forces acting on an organism. These evolutionary forces usually derive from organismal lifestyles that have been termed K-selected or r-selected in the evolutionary theory (Stearns, 1976). Typically, organisms that adopt a lifestyle characterized by rapid growth rates and reproductive cycles tend to evolve more streamlined regulatory systems through gene loss (r-selection). On the other hand, organisms that are strong competitors in heavily populated niches and that invest heavily in fewer offspring tend to have more complex regulatory systems with lesser gene loss. In the latter case, the retention of more regulatory systems potentially allows them to compete better by fine-tuning the regulation of their genes.
Right from its earliest days comparative genomics suggested that lateral gene-transfer might play an important role in shaping the composition of genes in an organism (Boucher et al., 2003; Gogarten et al., 2002). We have already discussed how the lateral transfer of protein domains involved in apoptotic networks and adhesion might have played a role in evolution of multicellularity in different lineages. Throughout eukaryotic evolution, lateral transfers, especially from bacteria, have played a role in the emergence of novel regulatory mechanisms. Such transfers have played important roles in the evolution of cell-cell communication and transcription regulation in multicellular organisms (Anantharaman et al., 2007a). For example, key receptors in animals, such as the acetylcholine receptor-type ligand-gated ion channels and the nitric oxide receptor, have their ultimate origins in receptors laterally transferred from bacteria to eukaryotes at different points prior to the radiation of metazoans (Tasneem et al., 2005). Similarly, NMDA and metabotropic glutamate receptors have ligand-binding domains evolutionarily derived from small-molecule sensors in bacterial signaling systems (Tasneem et al., 2005). In plants, DNA-binding domains of two notable transcription factor families, which include several developmental regulators, namely the Vp1 and AP2 families, appear to have been derived from DNA-binding domains found in bacteria (Babu et al., 2006; Iyer et al., 2008). Most bacterial versions are encoded by bacterial transposons or mobile restriction-modification systems. Thus, it is quite likely that the DNA-binding domains of these transcription factors entered the plant lineage, early evolution, via transfers from bacteria, probably via the medium of mobile elements (Babu et al., 2006; Iyer et al., 2008). On a related note, across eukaryotes, mobile elements such as transposons have contributed greatly to the evolution of transcription factors (Babu et al., 2006). Transposases of most transposons contain sequence-specific DNA-binding domains that can provide the precursor for the innovation of a novel DNA-binding domain for a transcription factor (Babu et al., 2006; Iyer et al., 2008; Lin et al., 2007). Thus, catalytically inactive transposons remnants, which retain their DNA-binding domains, have been the progenitors of several specific transcription factors, including certain major developmental regulators (Babu et al., 2006; Iyer et al., 2008; Lin et al., 2007). Such lateral transfers or remnants of transposases have the ability of delivering entirely functional “pre-made” regulatory molecules. During the emergence and subsequent elaboration of multicellularity, such transfers appear to have provided important raw material for the origin of new regulatory proteins which were incorporated at various points in pre-existing regulatory networks.
The conundrum of the dissociation between morphology and proteomic complexity: non-protein coding segments of genomes
Within multicellular lineages, organizational complexity has developed along very different lines. In some cases there have been regressions to more unicellular-like states (see above), whereas in other cases there has been enormous increase in complexity in terms of morphology and tissue types. Such trends are visible in the course of the evolution of plant, animal, and fungal lineages, but have been best studied in the metazoans. Genome sequences of early-branching metazoan lineages, particularly the cnidarians, have brought forth a conundrum. Cnidarians are accepted to be organizationally less-complex than vertebrates and arthropods (probably even nematodes), from the view point of tissue differentiation and morphology (Schierwater et al., 2009). However, analysis of cnidarian proteomes suggests that they possess all the major regulatory networks seen in vertebrates (Burglin, 2008; Putnam et al., 2007; Zmasek et al., 2007). In fact, certain subsets of these have been lost in insects (aspects of interferon-signaling-type pathways such as the IRF family transcription factors) and nematodes (e.g., certain parts of the apoptosis network) (Burglin, 2008; Zmasek et al., 2007). Thus, a largely complete complement of typically “animal-like” regulatory networks was already present at the base of the metazoan tree, but this did result in a corresponding organizational complexity (Burglin, 2008; Zmasek et al., 2007). Rather, organizational complexity continued to emerge in the course of metazoan evolution, despite loss of some of these pathways in lineages such as arthropods and nematodes (Schierwater et al., 2009). Comparative genomics has pointed to a number of other factors such as new LSEs and increased complexity of the antero-proterior patterning regulators (the Hox cluster), that emerged only in the organizationally more complex metazoans (Arthur, 2002; Hueber and Lohmann, 2008; Lespinet et al., 2002; Schierwater et al., 2009; Zmasek et al., 2007). Although these factors could play important roles, it should be noted that cnidarians possess their own LSEs and a comparable or larger proteome size than the more complex metazoans (Putnam et al., 2007). This does raise the question as to whether there are other factors behind the rise of organizational complexity in metazoa.
Recently, attention has turned towards non-protein coding segments of the genomes in search for solutions to the above conundrum. The amount of non-coding segments greatly varies across different eukaryotes, but most multicellular lineages are highly enriched in these segments (Taft et al., 2007). In vertebrates and certain plant lineages the amount of non-protein coding DNA greatly outstrips the amount of protein-coding DNA (Taft et al., 2007). There are many ways in which non-coding DNA could make a notable difference to the behavior of regulatory systems. The most obvious of these, which merely extends the existing diversity of proteins, is the increase in the number of introns. Increased introns in animal lineages has been accompanied by the prevalence of alternative splicing in which a great variety of proteins can be generated from a single gene (Maniatis and Tasic, 2002). Striking examples of these include the arthropod DSCAM (Down Syndrome Cell Adhesion Molecule) and vertebrate neurexins, which show an enormous diversity of proteins generated from a single gene (Craig and Kang, 2007; Wojtowicz et al., 2007; Yu et al., 2009). In the former example, over 18,000 distinct DSCAM isoforms differing in their extracellular Ig domains are produced via alternative splicing from a single gene in Drosophila (Wojtowicz et al., 2007; Yu et al., 2009). In the case of both neurexins and DSCAM, the diversity of isoforms are predominantly expressed in neurons and play an important role in neuronal wiring and specific synapse formation. Similarly, the earlier mentioned POZ domain transcription factor, lola, involved in axon path specification, undergoes extensive alternative splicing in Drosophila to spawn at least 19 different transcription factors of which 17 differ in their DNA-binding domains (Goeke et al., 2003). Thus, the increased alternative splicing might have a role in the emergence of increased complexity, at least in neural development which is typified by an enormous array of specific axon trajectories and synaspses (Holland and Short, 2008).
Other non-protein coding segments of the genome have instead been found to specify a panoply of non-coding regulatory RNAs. These range in size from large spliced or non spliced RNAs, such as Xist in vertebrates and Rox1/2 in arthropods, to medium-sized RNAs such as NRON, and a whole group of much smaller processed transcripts, such as piwi RNAs (piRNAs) and even smaller microRNAs (miRNAs) (Amaral et al., 2008; Carninci et al., 2005; Nakaya et al., 2007; Ponting et al., 2009). While the functions of this entire spectrum of non-coding RNAs still remain under investigation, there have been considerable advances in understanding the role of the piRNAs and miRNAs (Amaral et al., 2008; Grimson et al., 2008). Both of these classes of RNAs primarily function via RNAses of the argonaute family in the degradation of targeted transcripts (Cerutti and Casas-Mollano, 2006; Grimson et al., 2008). Whereas the piRNAs function mainly in defense against mobile elements such as transposons, the latter group appears to have a major regulatory role. The origin of protein components of the miRNA generating and utilizing network predate the separation of plants and animals (Anantharaman et al., 2002; Cerutti and Casas-Mollano, 2006). Conventional animal-type miRNAs have been detected in sponges and cnidarian, suggesting that they were already functional in the earliest metazoans (Grimson et al., 2008). However, it has been noted that there has been a progressive expansion of miRNAs in the course of animal evolution, with more organizationally complex metazoans having much larger complements of miRNAs (Heimberg et al., 2008; Ruby et al., 2007). Computational analysis of miRNA-binding sites has revealed that in vertebrates in particular, concomitant with the expansion of their miRNA complement, a major fraction of the protein-coding genes has come under the control of these RNAs, (Chen and Rajewsky, 2006; Lewis et al., 2005). Some workers have gone as far as to suggest a correlation between the number of miRNAs encoded in a metazoan lineage and the tissue complexity of that lineage (Technau, 2008). At face value it appears that the expansion of the regulatory RNA network, especially the sub-system based on miRNAs, might have a major role in providing a new layer of regulation over and beyond that offered by the conventional protein-dependent regulatory processes (Chen and Rajewsky, 2006; Heimberg et al., 2008). While this might be a relevant contributing factor to the emergence of organizational complexity in metazoans, it needs to be kept in mind that this system has been lost or considerably attenuated in certain animal lineages, such as the sea-squirt, Oikopleura, and Trichoplax (Fu et al., 2008; Grimson et al., 2008; Heimberg et al., 2008). Hence, it remains to be seen if the miRNA-based regulatory network is in large part a reinforcement or a back-up for the more conserved protein-based regulatory networks involved in silencing, rather than being a novel ensemble of control steps.
Finally, there is genuine non-coding DNA which exerts its regulatory influence by providing binding sites for specific transcription factors as well as chromatin proteins. The former set of binding sites includes conventional promoter elements and more distant enhancer and silencer elements (Bonn and Furlong, 2008; Busser et al., 2008; Weinstock, 2007; Zinzen and Furlong, 2008). The latter set includes the elements such as insulators, boundary elements, and sequences bound by trithorax and polycomb group proteins, all of which play a major role in higher order dynamics of chromatin structure, and consequently gene expression (Breiling et al., 2007; Valenzuela and Kamakaka, 2006). A simple comparison of a fungal genome like yeast with that of an animal like Drosophila reveals that regulatory elements are close to the gene and typically simple in the former and can be enormously complex in the latter (Bonn and Furlong, 2008; Harbison et al., 2004). Evidence for this increased complexity of transcription regulatory elements comes from many direct studies on such elements in developmental genes in various animals, as well as indirect studies from genetic polymorphism-phenotype association studies in humans and other animals (Bonn and Furlong, 2008; Busser et al., 2008; Verlaan et al., 2009; Weinstock, 2007; Zinzen and Furlong, 2008). In humans such association studies show that a large number of single nucleotide polymorphisms with extensive phenotypic consequences do not affect the protein-coding parts of genes. Rather, they affect non-coding regions, often at great distances from the coding segment, indicating the presence of a rich landscape of regulatory sites that have profound consequences on gene expression (Verlaan et al., 2009). Indeed, the DNA-binding domains of various lineage-specifically expanded TFs in multicellular forms are often very similar and unlikely to have very distinct binding specificities. However, they do widely differ in expression patterns suggesting that they have notable differences in their regulatory elements and this differential expression is a major aspect of their functional differentiation (Babu et al., 2006; Hoey and Levine, 1988).
The importance of cis regulatory elements as major players in organizational complexity has been highlighted by classical case studies such as those on the even-skipped gene (eve) in Drosophila and the Endo16 gene in the sea urchin, Strongylocentrotus purpuratus (Howard and Davidson, 2004; Istrail and Davidson, 2005). The former gene is expressed early in embryonic development in seven circumferential stripes that play a role in defining the territories of the future segments in the Drosophila body plan (Stanojevic et al., 1989; Stanojevic et al., 1991; Veitia, 2008). This expression pattern in the form of seven stripes is exquisitely orchestrated via the action of five cis regulatory elements. Of these, three elements control the emergence of one eve stripe each, where as the remaining two control two stripes each. One lesson from these eve elements is that the precise spatial emergence of a pattern is controlled via both the positive and negative regulatory actions of specific transcription factors binding their target sites on these elements. For example, in the case of stripe #2 the transcriptional activators Bicoid and Hunchback activate the expression of eve (Hoey and Levine, 1988; Howard and Davidson, 2004; Stanojevic et al., 1989; Stanojevic et al., 1991). However, these transcription factors themselves are broadly distributed and the precise spatial restriction of eve expression is brought about by two negative regulators, giant and kruppel, that respectively limit the anterior and posterior expression boundaries of eve (Stanojevic et al., 1991). Based on these observations, specific transcription factor binding sites in the regulatory elements controlling the expression of the eve gene have been compared to AND and NOT logical operators at the DNA level (Howard and Davidson, 2004). It is also clear that different non-coding regulatory elements could exert influence at many different levels in the network hierarchy (Bonn and Furlong, 2008; Busser et al., 2008; Weinstock, 2007; Zinzen and Furlong, 2008). The core promoter elements allow the driving of the basic expression of a gene. The cis regulatory elements coordinate with the core promoter to establish a spatial or temporal expression pattern by acting as logic gates that parse the ambient transcription factor environment. However, this initial expression pattern could be lost over subsequent cell cycles. The maintenance of these patterns over multiple cell cycles is then mediated by regulatory elements that recruit binding of chromatin-level modifiers (Breiling et al., 2007; Valenzuela and Kamakaka, 2006). These protein complexes “fix” a certain expression state by either maintaining an open chromatin configuration usually (trithorax group proteins) or a condensed chromatin state (polycomb group proteins). The proteins binding the insulator elements prevent the propagation of these chromatin states from one region to another (Bushey et al., 2008). Without changes to the actual protein-coding sequence of a gene, alterations to regulatory regions can modify the position of a gene in the regulatory network by changing its linkages to upstream transcription factors (Busser et al., 2008; Veitia, 2008). Thus, the spatial and temporal location in which a protein performs its function can be drastically altered, potentially resulting in changes to the morphological complexity by using the same underlying protein complement. Both the extensive LSEs of transcription factors and the increased domain architectural complexity of chromatin proteins seen in multicellular forms could be seen as a “pre-adaptation” with which the diversifying non-coding regulatory elements interacted to produce organizational diversity in certain lineages (Copley, 2008).
THE MAKING OF SUB-NETWORKS – A CASE STUDY
The Notch system
In the final part of this review we take up a well-known metazoan regulatory sub-network as a case study. We discuss the provenance of its various components and how different evolutionary forces and network organization principles have acted together in assembling this sub-network (Fig. 5). The Notch subnetwork is conserved throughout metazoa and appears to be a shared derived character that sets metazoans apart from their closest sister groups such as the choanoflagellate, Monosiga (King et al., 2008). Components of the Notch sub-network have been worked out in considerable detail across different metazoan models, such as human/mouse, Drosophila, and Caenorhabditis (Kopan and Ilagan, 2009), and it interfaces with other signaling sub-networks, such as epidermal growth factor receptor (EGFR) and Wnt, within the larger regulatory network involved in tissue differentiation (Sahlgren and Lendahl, 2006). These studies have shown that the Notch sub-network by itself is essentially a selector system that helps in choosing the execution of a particular sub-network from among different preexisting sub-networks in a cell. This action of the Notch system thus helps in choosing between activation and suppression of cell proliferation, cell death, and survival, and between alternative differentiated states (Kopan and Ilagan, 2009). This last function is especially important in asymmetric cell-divisions, wherein the Notch system helps the daughter cells adopt distinct differentiated states; for example, in ectodermal differentiation Notch helps in the asymmetric divisions accompanying the separation of epithelial and neural fates (Guo et al., 1996).
Figure 5.
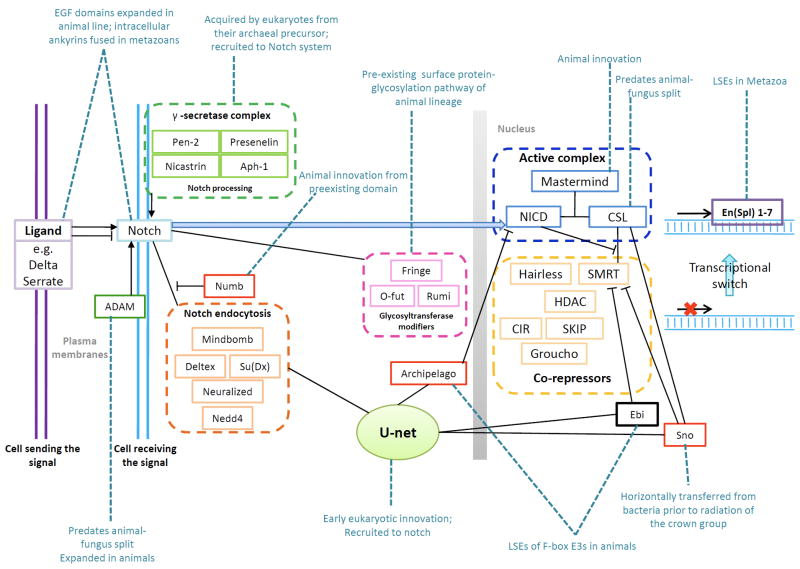
A Graphical representation of the Notch System. The notch ligands may be membrane-bound or soluble proteins. Upon ligand binding, an intracellular part of the protein (NICD) is released by proteolytic processing which is shown separately. Other regulatory processes impinging on the Notch sub-network are shown in boxes to indicate their co-functional linkage. The network is festooned with labels indicating the evolutionary history of different components.
The Notch system is centered on the receptor-ligand pair of Notch and one of its many related ligands prototyped by Drosophila, serrate or delta (D’Souza et al., 2008; Kopan and Ilagan, 2009). These ligands are also typically membrane-bound proteins, thus making Notch signaling dependent on the mechanical force acting on the Notch protein due to direct cell-cell interactions (Kopan and Ilagan, 2009). There are certain other soluble Notch ligands that either act as negative regulators or cooperate with a membrane-bound version to increase the strength of the ligand-Notch interaction. When associated with its ligand, the Notch protein is processed by multiple membrane-associated cleavage steps mediated by the ADAM family of metalloproteases (Bray, 2006) and presenilins (γ-secretase complex) (Selkoe and Wolfe, 2007) that then release an intracellular fragment of the protein which translocates to the nucleus (Kopan and Ilagan, 2009). In the nucleus it interacts with a transcription factor of the CSL (CBF1/RBPjκ/Su(H)/Lag-1) family. By default the CSL transcription factor associates with a co-repressor complex to negatively regulate gene expression and also recruits the histone chaperone ASF1 to promote condensed chromatin that further modulates gene expression (Kovall, 2008). Upon interaction with the Notch intracellular fragment, the CSL transcription factor switches its association from the co-repressor to recruit the co-activator MAM, which together recruit chromatin modifying and remodeling factors to promote transcriptional activation of target genes (Kopan and Ilagan, 2009). One of the targets of the CSL transcription factor conserved throughout Metazoa are the Enhancer of split (en(Spl)) transcription factors with bHLH DNA-binding domains that initiate a further transcriptional cascade by binding their target sites (Davis and Turner, 2001; Neves and Priess, 2005). This core pathway is dependent on a number of other regulatory inputs (Kopan and Ilagan, 2009; Lai et al., 2000). One of these is O-linked glycosylation of the EGF domains found in the Notch extracellular region by the fucosyltransferases, OFUT1 and Fringe, and the glucosyltransferase, Rumi (Haines and Irvine, 2003; Kopan and Ilagan, 2009; Stanley, 2007). These modifications alter the strength of the ligand-Notch interaction and have an effect on the downstream signal flux through the Notch pathway. The Notch system also intersects with the ubiquitin network in many ways, which results in altered stability or function of different components (Nichols et al., 2007). Ligands of Notch undergo endocytosis due to mono-ubiquitination mediated by the E3 enzymes, Neuralized and Mindbomb, and this process through an as yet unclear mechanism produces a more active form of the ligand (Kopan and Ilagan, 2009). The Notch protein is also subject to ubiquitination by E3s, such as Deltex, Itch, Cbl, and Nedd4, which might target it for lysosomal degradation or recycling (Kopan and Ilagan, 2009). This aspect of regulation is also central to the crosstalk between Notch and other signaling sub-networks such as EGFR. The protein, Phyllopod, activated by EGFR signaling, is an adaptor for the E3 Ebi, which helps in directing Notch to the early endocytotic vesicles and thereby favoring its lysosomal degradation (Nagaraj and Banerjee, 2009).
Origin and diversification of the Notch system
Despite the depth of our understanding of components of the Notch system and their operation in different metazoans, their origins and evolutionary diversification have to be determined to understand how such a system has come together to play such key roles in differentiation. The principal innovation in the emergence of this system occurred in the base of the metazoan tree in the form of the Notch receptor and its ligands (Kasbauer et al., 2007). The extracellular domains of both Notch and its ligands are comprised primarily of EGF domains. EGF domain proteins had already extensively proliferated even before the emergence of metazoans in the common ancestor shared with their sister group, the choanoflagellates (King et al., 2008). In choanoflagellates we already encounter proteins related to Notch, which have a gigantic extracellular region with numerous EGF repeats; however, they differed from Notch in lacking the distinctive intracellular ankyrin repeat modules (e.g., MONBRDRAFT_27644, gi:167527456). Ankyrin repeats, like those present in Notch, are also found fused to DNA-binding domains in transcription factors, such as the NFkB family and the SPT23 family, which share a common evolutionary origin with the CSL family of transcription factors (Hoppe et al., 2000; Iyer et al., 2008). These transcription factor families are unified by the presence of a specialized immunoglobulin fold domain, the TIG domain which interacts with the ankyrin repeats (Aravind and Koonin, 1999b; Hoppe et al., 2000; Kovall, 2007). This suggests an ancient functional association between the TIG domain transcription factors and ankyrin repeats. Comparative genomics reveals that CSL transcription factors were already present in the common ancestor of animals and fungi, long before the emergence of Notch and its ligands (Aravind and Subramanian, 1999). The fusion of ankyrin repeats to the cytoplasmic tail of a large EGF repeat protein that had emerged in the animal lineage prior to the radiation of metazoans (e.g., the above version found in choanoflagellates) appears to have given rise to a new signaling receptor that could now communicate with preexisting nuclear TIG domain transcription factors such as CSL. This observation indicates that the Notch-sub-network was built stepwise in evolution by superimposition of a newly derived membrane receptor on to an already existent transcription sub-network regulated by a CSL transcription factor.
Linkage to several other sub-systems with very distinct origins appears to have played an additional role in transforming this core into the recognizable Notch system. The ubiquitin-network proteins involved in endocytotic processes controlling the Notch system, in part, represent the adaptation of an older eukaryotic protein trafficking and degradation system to regulate this signaling system (Venancio et al., 2009). Presenilins and the associated γ-secretase complex is an ancient membrane-protein processing complex that was inherited by the eukaryotes from their archaeal ancestors (VA and LA unpublished). In contrast, the ADAM metalloproteases underwent a major expansion in the animal lineage in relation to the extracellular matrix that emerged along with multicellularity (Andreini et al., 2005). Thus, proteolytic systems with very distinct origins appear to have come together in the ancestral animal to mediate the processing of the Notch receptor (Fig. 5). Similarly, emergence of multicellularity in animals appears to have been accompanied by the expansion of different O-linked glycosylation enzymes that modified serines and threonines in adhesion-related domains found in cell-surface proteins (Anantharaman et al., 2007b). This expansion might have primarily been an adaptation for regulating cell-adhesion through glycosylation, but the presence of this system also allowed it to be adapted as a regulatory influence on the incipient Notch system. Taken together, these observations suggest that major parts of the Notch system were: (1) already available modules minimally adapted in biochemical terms for a new role or (2) emerged as part of a more general series of molecular adaptations that accompanied the origin of multicellularity. However, there were components of the Notch system that appear to represent innovations specific to Metazoa: the MAM domain of the MAM co-activators and the specialized SPRY domain found only in the Neuralized family E3s that are specific to this system are the two main instances. Even in these cases, the innovations were not too drastic because the MAM domain is merely a long bent α-helix and domains of the Neuralized have been derived from preexisting SPRY domains through rapid divergence (Kovall, 2007; Kuang et al., 2009).
The two prominent genome-shaping forces of lineage-specific expansion and gene loss also play an important role in the history of the Notch pathway in metazoan evolution (Fig. 5). Gene loss and degradation are observed in the nematode lineage. The co-activator of the CSL transcription factor, MAM, is apparently lost in Caenorhabditis. Likewise, the Notch-ligand endocytosis regulator, Mindbomb, appears to have lost the N-terminal Herc2 and ZZ domains found in other metazoan lineages (Kasbauer et al., 2007) (LA, unpublished). LSEs have impacted the evolution of many nodes in the Notch sub-network at various points in it organization. A notable LSE of the Notch ligands is seen in Caenorhabditis with at least 15 distinct ligands (Kopan and Ilagan, 2009). A smaller LSE of Notch ligands and of Notch itself is observed in vertebrates (Kopan and Ilagan, 2009). This LSE of ligands has enabled the transmission of signals encompassing a whole range of relative strengths via the same receptors. In Drosophila there is a lineage-specific expansion of a remarkable Zn-finger domain of the treble-clef fold, the C4DM domain (also called ZAD), with four conserved cysteines (Lander et al., 2001). Members of this family of domains appear to be adaptors that link a variety of targets to ubiquitination by E3s (Jauch et al., 2003). Phyllopod, which connects the EGFR and Notch networks (Nagaraj and Banerjee, 2009), is a member of this LSE. Thus, phyllopod offers an example of how the emergence of a neomorphic protein through an LSE has resulted in the strengthening of the linkage between two pathways. One of the conserved targets of the notch pathway, the En(Spl) transcription factors also show striking independent LSEs in various lineages. In Drosophila melanogaster, the LSE in the En(Spl) has resulted in 7 distinct paralogs (Knust et al., 1992). A similar LSE is seen in Caenorhabditis elegans, wherein the En(Spl) cognate has undergone an LSE to spawn 6 distinct paralogs, many of which contain two bHLH DNA-binding domains (Neves and Priess, 2005). These paralogs are not found in Brugia or even other Caenorhabditis species, suggesting that is a relatively recent LSE followed by rapid sequence divergence (L. Aravind, unpublished). This LSE of the En(Spl) genes has probably played a role in diversifying the targets that are under the control of the Notch signaling system. In part this diversification of the function of the En(Spl) transcription factors has occurred not via differences in their DNA-binding properties but their cis regulatory elements (Maeder et al., 2009).
In conclusion, the evolutionary dissection of the Notch pathway illustrates some of the principles derived from regulatory network structure in action. Firstly, it emphasizes the modularity of the networks and how the emergence of a key linking element with a high betweenness (in this case Notch) can bring together disparate modules into a single network. The biochemical basis for this property of Notch is the fact that it evolved through fusion of two distinct sets of domains, which respectively mediate intracellular and extracellular interactions. Furthermore, this analysis shows how genes or part thereof, which are considered to be critical in one system can be lost in another – when viewed in light of the resistance of regulatory networks to failure (i.e., random node loss) such losses are not unexpected. The example also illustrates how LSEs act at various levels to allow a core module of a network to receive inputs and deliver outputs that greatly differ in their consequences or be combined with signals from other sub-networks (Nagaraj and Banerjee, 2009). Finally, these LSEs combined with the diversification of cis regulatory regions actually illustrate how a conserved sub-network like Notch has been used in generating unique morphologies in different animal lineages.
CONCLUDING REMARKS
The triumphs of the genomic revolution and the consequent impact on the way biology is done have allowed us for the first time to apprehend the architecture and functions of regulatory networks. When combined with evolutionary information, we obtain a remarkable view of how biological systems have actually been shaped by various forces. This information has considerable implications for how processes such as development and differentiation in multicellular forms are addressed. Primarily, it allows one to see these processes not in isolation, but in the context of both their histories and place in the overall network of molecular interactions. This is important because all biological systems, including regulatory networks, are not products of engineering but of the contingencies of historical processes (De Robertis, 2008; Gould, 2002; Kirschner and Gerhart, 2005; Raff, 1996). It has been typical to take an engineering approach to the analysis of regulatory networks in the past, due to lack of evolutionary information. In the “post-genomic era” this need not be the case (Balaji et al., 2006a; Barabasi and Oltvai, 2004; Busser et al., 2008). Hence, we can transcend the view of regulatory networks restricted to particular model organisms and instead view them as evolving entities across the evolutionary tree. In the first place, this approach helps in objectively tackling features of the model organisms themselves. In the example of the Notch system it was observed that Phyllopod is a part of an LSE that is unique to insects; hence, there would be no point to search for it in other metazoan models. Thus, understanding a regulatory network in evolutionary terms helps in discriminating between universal and non-universal components of a system and delineating its functional core. Moreover, such an approach also helps us in trying to identify features of regulatory networks that might be of interest in new models or experimentally less-tractable organisms for which genome sequences are available (Grimson et al., 2008; Veitia, 2008). In such cases using concepts, such as LSEs and gene loss, identification of cis regulatory elements, and clues from domain architectures, one could focus on aspects of the network that are likely to be critical in organism-specific functions (Lespinet et al., 2002; Weinstock, 2007). This could potentially lead to uncovering of the mechanisms by which sub-networks have been involved in generating a lineage-specific phenotypic novelty.
However, considerable work still remains to be done. None of the large-scale studies in multicellular eukaryotes or prokaryotes has reached the level of detail achieved by the individual efforts such as those that explain the formation of even-skipped stripes in Drosophila or such other spatio-temporal patterns (Howard and Davidson, 2004; Istrail and Davidson, 2005). Achieving that level of detail in regulatory network reconstruction is certainly impeded by technical hurdles, as mentioned in the beginning of this review. However, the rapid advances in high-throughput methods, including sequencing, optics, and micro-fabrication technology indicate that these roadblocks may be overcome sooner than later (Imelfort et al., 2009; Joos and Bachmann, 2009; Nygaard and Hovig, 2009; Todt and Blohm, 2009). Hence, it does appear that direct comparisons of entire regulatory networks of different multicellular forms are a clear possibility in the near future. While the general principles that have been discussed in this article will remain guiding principles, it is very likely that several unexpected findings come up from these comparisons.
Acknowledgments
The authors of this review are supported by the Intramural Research Program of the NIH, National Library of Medicine. Given the vast number of articles in the topic under consideration, we have been unable to cite all of them due to obvious space constraints. We do acknowledge the enormous labor of workers in this field and apologize for not being able to cite them all.
References
- Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26(8):909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406(6794):378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral LA, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci U S A. 2000;97(21):11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319(5871):1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol. 2007a;61:453–475. doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Balaji S, Aravind L. Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: insights from comparative genomics. Int Rev Cytol. 2007b;262:1–74. doi: 10.1016/S0074-7696(07)62001-4. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30(7):1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C, Banci L, Bertini I, Elmi S, Rosato A. Comparative analysis of the ADAM and ADAMTS families. J Proteome Res. 2005;4(3):881–888. doi: 10.1021/pr0500096. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297(5585):1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29(2):231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Aravind L, Dixit VM, Koonin EV. Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science. 2001;291(5507):1279–1284. doi: 10.1126/science.291.5507.1279. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J Mol Biol. 1999a;285(4):1353–1361. doi: 10.1006/jmbi.1998.2394. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999b;287(5):1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Aravind L, Subramanian G. Origin of multicellular eukaryotes - insights from proteome comparisons. Curr Opin Genet Dev. 1999;9(6):688–694. doi: 10.1016/s0959-437x(99)00028-3. [DOI] [PubMed] [Google Scholar]
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A. 2000;97(21):11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur W. The emerging conceptual framework of evolutionary developmental biology. Nature. 2002;415(6873):757–764. doi: 10.1038/415757a. [DOI] [PubMed] [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34(22):6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J Mol Biol. 2006a;360(1):213–227. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Balaji S, Iyer LM, Aravind L, Babu MM. Uncovering a hidden distributed architecture behind scale-free transcriptional regulatory networks. J Mol Biol. 2006b;360(1):204–212. doi: 10.1016/j.jmb.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Balaji S, Iyer LM, Babu MM, Aravind L. Comparison of transcription regulatory interactions inferred from high-throughput methods: what do they reveal? Trends Genet. 2008;24(7):319–323. doi: 10.1016/j.tig.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Bonabeau E. Scale-free networks. Sci Am. 2003;288(5):60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bonn S, Furlong EE. cis-Regulatory networks during development: a view of Drosophila. Curr Opin Genet Dev. 2008;18(6):513–520. doi: 10.1016/j.gde.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbo CL, Case RJ, Doolittle WF. Lateral gene transfer and the origins of prokaryotic groups. Annu Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- Brandes U. A faster algorithm for betweenness centrality. Journal of Mathematical Sociology. 2001;25(2):163–177. [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Breiling A, Sessa L, Orlando V. Biology of polycomb and trithorax group proteins. Int Rev Cytol. 2007;258:83–136. doi: 10.1016/S0074-7696(07)58002-2. [DOI] [PubMed] [Google Scholar]
- Burglin TR. Evolution of hedgehog and hedgehog-related genes, their origin from Hog proteins in ancestral eukaryotes and discovery of a novel Hint motif. BMC Genomics. 2008;9:127. doi: 10.1186/1471-2164-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32(1):1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser BW, Bulyk ML, Michelson AM. Toward a systems-level understanding of developmental regulatory networks. Curr Opin Genet Dev. 2008;18(6):521–529. doi: 10.1016/j.gde.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315(5809):207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50(2):81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamaillard M, Girardin SE, Viala J, Philpott DJ. Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 2003;5(9):581–592. doi: 10.1046/j.1462-5822.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446(7137):806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Copley RR. The animal in the genome: comparative genomics and evolution. Philos Trans R Soc Lond B Biol Sci. 2008;363(1496):1453–1461. doi: 10.1098/rstb.2007.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson RM, Ouzounis CA. The phylogenetic diversity of eukaryotic transcription. Nucleic Acids Res. 2003;31(2):653–660. doi: 10.1093/nar/gkg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17(1):43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27(38):5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection: or, The preservation of favoured races in the struggle for life. 1. ix. London: John Murray, Albemarle Street; 1859. pp. 502–532. [501] folded leaf of plate p. [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311(5762):796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20(58):8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Evo-devo: variations on ancestral themes. Cell. 2008;132(2):185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, Theodosius Nothing in Biology Makes Sense Except in the Light of Evolution. American Biology Teacher. 1973;35:125–129. [Google Scholar]
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284(5423):2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Durbin R. Biological sequence analysis: probabilistic models of proteins and nucleic acids. xi. Cambridge, UK New York: Cambridge University Press; 1998. p. 356. [Google Scholar]
- Edvardsen RB, Seo HC, Jensen MF, Mialon A, Mikhaleva J, Bjordal M, Cartry J, Reinhardt R, Weissenbach J, Wincker P, et al. Remodelling of the homeobox gene complement in the tunicate Oikopleura dioica. Curr Biol. 2005;15(1):R12–13. doi: 10.1016/j.cub.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4(9):e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK, Doyle JA. Floral phyllotaxis in basal angiosperms: development and evolution. Curr Opin Plant Biol. 2007;10(1):52–57. doi: 10.1016/j.pbi.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136(5):952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269(5223):496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Fu X, Adamski M, Thompson EM. Altered miRNA repertoire in the simplified chordate, Oikopleura dioica. Mol Biol Evol. 2008;25(6):1067–1080. doi: 10.1093/molbev/msn060. [DOI] [PubMed] [Google Scholar]
- Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36(Database issue):D120–124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. Embo J. 2001;20(10):2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gianchandani EP, Brautigan DL, Papin JA. Systems analyses characterize integrated functions of biochemical networks. Trends Biochem Sci. 2006;31(5):284–291. doi: 10.1016/j.tibs.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Giniger E, Tietje K, Jan LY, Jan YN. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development. 1994;120(6):1385–1398. doi: 10.1242/dev.120.6.1385. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goeke S, Greene EA, Grant PK, Gates MA, Crowner D, Aigaki T, Giniger E. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat Neurosci. 2003;6(9):917–924. doi: 10.1038/nn1105. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science. 1996;274(5287):546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19(12):2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Gould SJ. The structure of evolutionary theory. xxii. Cambridge, Mass: Belknap Press of Harvard University Press; 2002. p. 1433. [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17(1):27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4(10):786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431(7004):99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105(8):2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg ME, Rankin DJ, Taborsky M. The coevolution of cooperation and dispersal in social groups and its implications for the emergence of multicellularity. BMC Evol Biol. 2008;8:238. doi: 10.1186/1471-2148-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458(7237):422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988;332(6167):858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Short S. Gene duplication, co-option and recruitment during the origin of the vertebrate brain from the invertebrate chordate brain. Brain Behav Evol. 2008;72(2):91–105. doi: 10.1159/000151470. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102(5):577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Horowitz H, Berg CA. The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development. 1996;122(6):1859–1871. doi: 10.1242/dev.122.6.1859. [DOI] [PubMed] [Google Scholar]
- Howard ML, Davidson EH. cis-Regulatory control circuits in development. Dev Biol. 2004;271(1):109–118. doi: 10.1016/j.ydbio.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39(5):683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30(10):965–979. doi: 10.1002/bies.20823. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102(1):109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Huxley J. Evolution, the modern synthesis. Vol. 645. New York: Harper; 1942. p. 641. [Google Scholar]
- Imelfort M, Batley J, Grimmond S, Edwards D. Genome sequencing approaches and successes. Methods Mol Biol. 2009;513:345–358. doi: 10.1007/978-1-59745-427-8_18. [DOI] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci U S A. 2005;102(14):4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci U S A. 1996;93(18):9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int J Parasitol. 2008;38(1):1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443(7113):818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jauch R, Bourenkov GP, Chung HR, Urlaub H, Reidt U, Jackle H, Wahl MC. The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein-protein interaction module. Structure. 2003;11(11):1393–1402. doi: 10.1016/j.str.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Jongeneel CV, Delorenzi M, Iseli C, Zhou D, Haudenschild CD, Khrebtukova I, Kuznetsov D, Stevenson BJ, Strausberg RL, Simpson AJ, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15(7):1007–1014. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos T, Bachmann J. Protein microarrays: potentials and limitations. Front Biosci. 2009;14:4376–4385. doi: 10.2741/3534. [DOI] [PubMed] [Google Scholar]
- Kasbauer T, Towb P, Alexandrova O, David CN, Dall’armi E, Staudigl A, Stiening B, Bottger A. The Notch signaling pathway in the cnidarian Hydra. Dev Biol. 2007;303(1):376–390. doi: 10.1016/j.ydbio.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7(4):516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- Kent ML, Andree KB, Bartholomew JL, El-Matbouli M, Desser SS, Devlin RH, Feist SW, Hedrick RP, Hoffmann RW, Khattra J, et al. Recent advances in our knowledge of the Myxozoa. J Eukaryot Microbiol. 2001;48(4):395–413. doi: 10.1111/j.1550-7408.2001.tb00173.x. [DOI] [PubMed] [Google Scholar]
- Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20(20):2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451(7180):783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J. The plausibility of life: resolving Darwin’s dilemma. xiii. New Haven: Yale University Press; 2005. p. 314. [Google Scholar]
- Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. Tgf-Beta signaling in development. Sci STKE. 2007;2007(399):cm1. doi: 10.1126/stke.3992007cm1. [DOI] [PubMed] [Google Scholar]
- Knust E, Schrons H, Grawe F, Campos-Ortega JA. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992;132(2):505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr Opin Struct Biol. 2007;17(1):117–127. doi: 10.1016/j.sbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Kovall RA. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene. 2008;27(38):5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet. 2005;6(9):688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuang Z, Yao S, Xu Y, Lewis RS, Low A, Masters SL, Willson TA, Kolesnik TB, Nicholson SE, Garrett TJ, et al. SPRY domain-containing SOCS box protein 2: crystal structure and residues critical for protein binding. J Mol Biol. 2009;386(3):662–674. doi: 10.1016/j.jmb.2008.12.078. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390(6657):249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, Schoenfeld LW, Ota I, Sahasrabudhe S, Kurschner C, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438(7064):103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Posakony JW. The enhancer of split complex of Drosophila includes four Notch-regulated members of the bearded gene family. Development. 2000;127(16):3441–3455. doi: 10.1242/dev.127.16.3441. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Li Y. Tramtrack69 is positively and autonomously required for Drosophila photoreceptor development. Genetics. 1999;152(1):299–305. doi: 10.1093/genetics/152.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298(5594):799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12(7):1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis MW, Leslie ME, Liljegren SJ. Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol. 2006;9(1):59–65. doi: 10.1016/j.pbi.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318(5854):1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Louis EJ. Yeast evolution and comparative genomics. Annu Rev Microbiol. 2005;59:135–153. doi: 10.1146/annurev.micro.59.030804.121400. [DOI] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433(7028):865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E. The exosome and the proteasome: nano-compartments for degradation. Cell. 2006;125(4):651–654. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lukacsovich T, Yuge K, Awano W, Asztalos Z, Kondo S, Juni N, Yamamoto D. The ken and barbie gene encoding a putative transcription factor with a BTB domain and three zinc finger motifs functions in terminalia development of Drosophila. Arch Insect Biochem Physiol. 2003;54(2):77–94. doi: 10.1002/arch.10105. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431(7006):308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Megley C, Eastman DA. Differential expression of the Enhancer of split genes in the developing Drosophila midgut. Hereditas. 2009;146(1):11–18. doi: 10.1111/j.1601-5223.2008.02094.x. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416(6880):499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418(6894):236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Mayerhofer LE, Macario AJ, de Macario EC. Lamina, a novel multicellular form of Methanosarcina mazei S-6. J Bacteriol. 1992;174(1):309–314. doi: 10.1128/jb.174.1.309-314.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. The growth of biological thought: diversity, evolution, and inheritance. ix. Cambridge, Mass: Belknap Press; 1982. p. 974. [Google Scholar]
- Miller DJ, Ball EE, Technau U. Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 2005;21(10):536–539. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, Agata K, Bosch TC. The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss. Genome Biol. 2007;8(4):R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Barstead RJ. Towards a mutation in every gene in Caenorhabditis elegans. Brief Funct Genomic Proteomic. 2008;7(3):195–204. doi: 10.1093/bfgp/eln016. [DOI] [PubMed] [Google Scholar]
- Morange M. A history of molecular biology. Cambridge, Mass: Harvard University Press; 1998. p. 336. [Google Scholar]
- Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell. 2004;15(5):2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj R, Banerjee U. Regulation of Notch and Wingless signalling by phyllopod, a transcriptional target of the EGFR pathway. Embo J. 2009;28(4):337–346. doi: 10.1038/emboj.2008.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8(3):R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8(6):867–879. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Weinmaster G. Notch signaling--constantly on the move. Traffic. 2007;8(8):959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- Nygaard V, Hovig E. Methods for quantitation of gene expression. Front Biosci. 2009;14:552–569. doi: 10.2741/3262. [DOI] [PubMed] [Google Scholar]
- Pagans S, Ortiz-Lombardia M, Espinas ML, Bernues J, Azorin F. The Drosophila transcription factor tramtrack (TTK) interacts with Trithorax-like (GAGA) and represses GAGA-mediated activation. Nucleic Acids Res. 2002;30(20):4406–4413. doi: 10.1093/nar/gkf570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J, Burki F. Untangling the phylogeny of amoeboid protists. J Eukaryot Microbiol. 2009;56(1):16–25. doi: 10.1111/j.1550-7408.2008.00379.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Perez J, Castaneda-Garcia A, Jenke-Kodama H, Muller R, Munoz-Dorado J. Eukaryotic-like protein kinases in the prokaryotes and the myxobacterial kinome. Proc Natl Acad Sci U S A. 2008;105(41):15950–15955. doi: 10.1073/pnas.0806851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23(1):80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438(7068):679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Raff RA. The shape of life: genes, development, and the evolution of animal form. xxiii. Chicago: University of Chicago Press; 1996. p. 520. [Google Scholar]
- Reinhardt D. Regulation of phyllotaxis. Int J Dev Biol. 2005;49(5–6):539–546. doi: 10.1387/ijdb.041922dr. [DOI] [PubMed] [Google Scholar]
- Rokas A. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17(12):1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25(4):664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Russell RB, Aloy P. Targeting and tinkering with interaction networks. Nat Chem Biol. 2008;4(11):666–673. doi: 10.1038/nchembio.119. [DOI] [PubMed] [Google Scholar]
- Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, Hall JC, Taylor BJ, Wasserman SA. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87(6):1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- Sahlgren C, Lendahl U. Notch signaling and its integration with other signaling mechanisms. Regen Med. 2006;1(2):195–205. doi: 10.2217/17460751.1.2.195. [DOI] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 2009;7(1):e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131(2):215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Serrano M, Parra S, Alcaraz LD, Guzman P. The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol. 2006;62(4):434–445. doi: 10.1007/s00239-005-0038-y. [DOI] [PubMed] [Google Scholar]
- Shapiro JA, Dworkin M. Bacteria as multicellular organisms. xiii. New York: Oxford University Press; 1997. p. 466. [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31(1):64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Shim K, Blake KJ, Jack J, Krasnow MA. The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development. 2001;128(23):4923–4933. doi: 10.1242/dev.128.23.4923. [DOI] [PubMed] [Google Scholar]
- Sierro N, Makita Y, de Hoon M, Nakai K. DBTBS: a database of transcriptional regulation in Bacillus subtilis containing upstream intergenic conservation information. Nucleic Acids Res. 2008;36(Database issue):D93–96. doi: 10.1093/nar/gkm910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314(5801):941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokony RF, Restifo LL. Anciently duplicated Broad Complex exons have distinct temporal functions during tissue morphogenesis. Dev Genes Evol. 2007;217(7):499–513. doi: 10.1007/s00427-007-0159-y. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454(7207):955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stanley P. Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol. 2007;17(5):530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Kruppel in Drosophila. Nature. 1989;341(6240):331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254(5036):1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- Stearns SC. Life-history tactics: a review of the ideas. Q Rev Biol. 1976;51(1):3–47. doi: 10.1086/409052. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29(3):288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6(1):R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U. Evolutionary biology: Small regulatory RNAs pitch in. Nature. 2008;455(7217):1184–1185. doi: 10.1038/4551184a. [DOI] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36(5):492–496. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- Thomas JH. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16(8):1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8(5):372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- Todt S, Blohm DH. Immobilization chemistries. Methods Mol Biol. 2009;529:81–100. doi: 10.1007/978-1-59745-538-1_6. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. Mechanism of leaf-shape determination. Annu Rev Plant Biol. 2006;57:477–496. doi: 10.1146/annurev.arplant.57.032905.105320. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19(9):479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- Veitia RA. One thousand and one ways of making functionally similar transcriptional enhancers. Bioessays. 2008;30(11–12):1052–1057. doi: 10.1002/bies.20849. [DOI] [PubMed] [Google Scholar]
- Venancio TM, Balaji S, Iyer LM, Aravind L. Reconstructing the ubiquitin network - cross-talk with other systems and identification of novel functions. Genome Biol. 2009;10(3):R33. doi: 10.1186/gb-2009-10-3-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Verlaan DJ, Ge B, Grundberg E, Hoberman R, Lam KC, Koka V, Dias J, Gurd S, Martin NW, Mallmin H, et al. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 2009;19(1):118–127. doi: 10.1101/gr.084798.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo CA, Babon JA, Sequerra R, Doucette-Stamm L, Barabasi AL, Walhout AJ. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 2007;17(7):1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449(7158):54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Weinstock GM. ENCODE: more genomic empowerment. Genome Res. 2007;17(6):667–668. doi: 10.1101/gr.6534207. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130(6):1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook SH, Jeong H, Barabasi AL. Modeling the Internet’s large-scale topology. Proc Natl Acad Sci U S A. 2002;99(21):13382–13386. doi: 10.1073/pnas.172501399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, Blumer A, Lee K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics. 2006;22(24):3106–3108. doi: 10.1093/bioinformatics/btl533. [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Paccanaro A, Trifonov V, Gerstein M. Predicting interactions in protein networks by completing defective cliques. Bioinformatics. 2006;22(7):823–829. doi: 10.1093/bioinformatics/btl014. [DOI] [PubMed] [Google Scholar]
- Yu HH, Yang JS, Wang J, Huang Y, Lee T. Endodomain diversity in the Drosophila Dscam and its roles in neuronal morphogenesis. J Neurosci. 2009;29(6):1904–1914. doi: 10.1523/JNEUROSCI.5743-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10(6):719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127(2):409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Furlong EE. Divergence in cis-regulatory networks: taking the ‘species’ out of cross-species analysis. Genome Biol. 2008;9(11):240. doi: 10.1186/gb-2008-9-11-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8(10):R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Grewal SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–496. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]