Abstract
DNA methylation is an epigenetic mechanism involved in many biological functions in prokaryotes and eukaryotes. Contrary to mammalian DNA, which is thought to contain only 5-methylcytosine (m5C), bacterial DNA contains two additional methylated bases, namely N6-methyladenine (m6A), and a more recently discovered minor base N4-methylcytosine (m4C). These modified bases are involved in the protection of bacterial DNA from the action of specific endonucleases via the host-specific restriction-modification system which is regarded as a defense mechanism against bacteriophage infection. However, if the main function of m5C and m4C in bacteria is the protection against restriction enzymes, the roles of m6A are multiple and include for example the regulation of virulence and the control of many bacterial DNA functions such as the replication, repair, expression and transposition of DNA. Hence, in regard to the multiple roles of m6A in bacteria, and to the well known tendency for m5C to deaminate in thymine, the selection of the mutagenic m5C instead of m6A in mammals as the only methylated base may seem surprising. However, even if adenine methylation is usually considered as a bacterial DNA feature, the presence of m6A is not restricted to prokaryotic DNA since this methylated base has been found in protist and plant DNAs. Furthermore, indirect evidence suggests the presence of m6A in mammal DNA, raising the possibility that this base has remained undetected due to the low sensitivity of the analytical methods used. This points to the importance to consider m6A as the sixth element of DNA.
Keywords: Adenine, analogs & derivatives, classification, metabolism, Animals, DNA, chemistry, DNA Methylation, Eukaryotic Cells, Phylogeny
Keywords: DNA methylation, epigenetics, N6-methyladenine, 5-methylcytosine
I. Introduction
Although four bases are required for DNA synthesis, DNA contains several additional methylated bases, namely 5-methylcytosine, N6-methyladenine, N4-methylcytosine, which result from the post-replicative modification of DNA by DNA methylases (1–3). However, if the most popular modified base is 5-methylcytosine (m5C), recent data point to the biological importance of N6-methyladenine (m6A) as the other methylated base. Thus, adenine methylation is essential for the viability of several bacteria (4–9). Moreover, accumulating evidence suggests that the presence of m6A is not limited to eubacterial DNA but also occurs in at least some archaeabacteria and eukaryotic cells where its role remains largely unknown (Fig. 1) (10–14). This review focuses on m6A as the sixth base of DNA and aims to be a source of information and inspiration for the development of new ideas and hypotheses on the possible functions of m6A in eukaryotic cells.
Fig. 1.
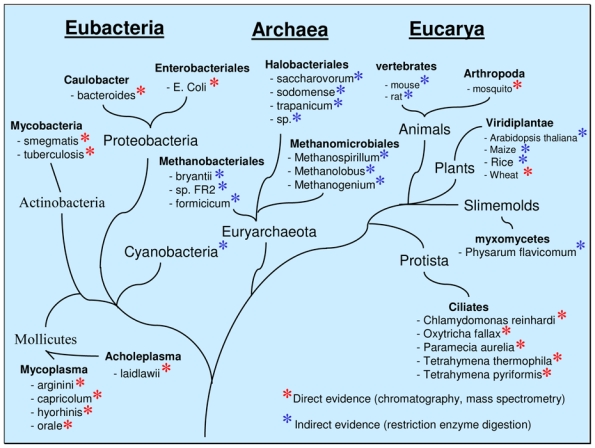
Schematic representation of the phylogenetic distribution of m6A in DNA.
II. DNA adenine methylation in prokaryotes
In bacterial DNA the function of adenine methylation has historically been associated with the protection of DNA from the action of specific endonucleases via the host-specific restriction/modification system. In this system, which is regarded as a defense mechanism against bacteriophage infection (15,16), cytosine and adenine methylation of bacterial DNA protects it from the action of the corresponding restriction endonuclease, whereas unmethylated sites of foreign DNA such as bacteriophage DNA are cleaved (Fig. 2A). Interestingly, DNA adenine methyltransferases can also be encoded by viral DNA as has been shown for bacteriophages T4, Mx8 (17, 18) and for the archeal viruses φ.Ch1 and SNDV (19, 20). In addition to these DNA adenine methyltransferases related to the restriction/modification system, there are at least two DNA adenine methyltransferases which lack a cognate restriction enzyme. These DNA methylases named Dam and CcrM, methylate GATC and GANTC sequences respectively. They differ in their distribution since Dam methylation is found primarily in the members of the gamma division of Protobacteria and in some Archeabacteria (21), whereas CcrM is relatively widespread in the eubacteria alpha division (22, 23). In E. coli, Dam is involved in the replication, mismatch repair and transposition of DNA (Fig. 2B) (24, 25), and in the control of gene expression (26). Hence, Dam mutant are characterized by a pleiotropic phenotype including for example enhanced sensitivity to DNA-damaging agents, higher mutability and increased recombination frequency. However, E. coli lacking Dam activity are viable, but dam is an essential gene in Vibrio cholerae and Yersinia pseudotubercolosis (5). The other known DNA adenine methyltransferase lacking a corresponding restriction endonuclease is CcrM. CcrM is a DNA methyltransferase originally described in Caulobacter cresecentus as a “cell cycle-regulated methyltransferase” (26). It is essential for viability in Caulobacter crescentus, Rhizobium meliloti, Agrobacterium tumefaciens, Brucella abortus (4, 7, 8). Like Dam, CcrM regulates gene expression (6), and control the initiation of DNA replication (23, 28). In addition to their multiple functions which have led Dam and CcrM to be considered as “cell cycle regulators” (23), these DNA adenine methyltransferases are also involved in bacterial pathogenicity as they control virulence gene expression and secretion of virulence determinants (Fig. 2C) (5, 29–32). These points are of particular interest since they suggest that DNA adenine methylation could be a new target for antibiotics (33, 34).
Fig. 2. Some established and putative functions of m6A in DNA.
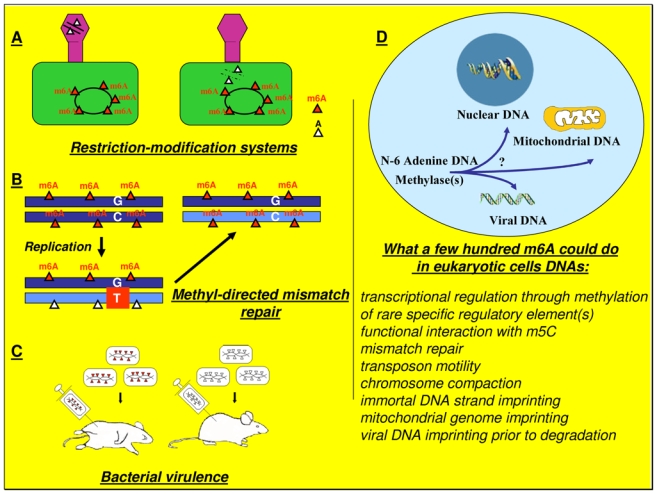
A) Restriction-modifications systems defend bacteria from invasions by viral DNA. These systems are composed of a bacterial restriction enzyme which cuts a specific sequence in phage DNA, and a cognate adenine or cytosine DNA methyltransferase which recognizes the same target site that the restriction enzyme and protects bacterial DNA from its own restriction enzyme(s). B) In E. coli, adenine methylation is also used to discriminate the transiently unmethylated newly synthesized complementary strand during DNA replication. This discrimination is the basis of the methyl-directed mismatch repair which makes correction on the newly synthesized strand only. C) Alteration in the levels of DNA adenine methylation attenuates the virulence of a number of pathogens. D) Speculative representation of the biological functions of putative eucaryotic N-6 adenine-specific DNA methylase(s).
III. DNA adenine methylation in eukaryotes
M6A in protistDNA
It has long been known that m6A is present in DNA from several unicellular eukaryotes, including members of the genera Chlamydomonas, Chlorella, Oxytricha, Paramecium, Tetrahymena (Fig. 2) (14, 35–41). Moreover, genes encoding both restriction endonucleases and their cognate adenine DNA methyltransferases have been found in Chlorella viruses, suggesting a role for adenine methylation during viral infection (42, 43). A main feature of ciliate protozoans such as Paramecia and Tetrhaymena, is the presence in the same cell of two nuclei, a diploid germ line micronucleus and a polyploid somatic macronucleus whose transcription supports cell growth, differentiation and proliferation (41). Interestingly, m6A is detected in macronucleus DNA only. Unfortunately, the exact functions of this adenine methylation are far less understood in protist than in bacteria. Studies performed on Tetrahymena have demonstrated that de novo adenine methylation of DNA is not random, but occurs according to a specific pattern and preferentially in linker DNA (38, 40, 44). Morever, it has been reported that in Physarum flavicomumrecent, cyst DNA but not growing cell DNA contains m6A (45), whilst recent data suggest that in Paramecium, adenine methylation could be involved in the excision of internal eliminated sequences (IESs) (46).
M6A in plant and animal DNA
The general assumption that m6A is not found in the DNA of higher eukaryotes originates from experiments performed more than thirty years ago which had a detection limit around 0.1% - 0.01% (47–49) and which detected m5C as the only methylated base. This selection of m5C instead of m6A to control crucial regulatory biological processes such as genomic imprinting, X-chromosome inactivation, gene expression and embryonic development may seem surprising if we consider the multiple roles of m6A in bacteria, and the well known tendency for m5C to deaminate in thymine (50). A possible explanation lies in the fact that most of the m5C in mammalian DNA is found in transposons (51), a finding that has led to the suggestion that the presence of m5C in mammalian DNA could provide a host defense mechanism against parasitic DNA through the repressive effects of m5C on gene expression and through the accumulation of mutations resulting from the spontaneous deamination of m5C in thymine (52). Hence, the large amount of transposons (>45%) found in human DNA could account for the relative abundance of m5C which could have masked, by its overrepresentation, the presence of small amounts of m6A. In this respect it is noteworthy that experiments designed to determine the base composition of mammalian DNA were, in addition to their low sensitivity, performed on a limited number of tissue or cell samples. Consequently the occurrence of m6A during development, programmed cell death, aging or in pathologic conditions such as proliferative, degenerative or infectious diseases has not been extensively investigated. Furthermore, and in spite of common opinion, several data have reported the presence of m6A in the DNA of higher eukaryotes. Thus, m6A has been detected in plastid, mitochondrial and nuclear plant DNA (10, 13, 53–55), and in mosquito DNA (56). Regarding the presence of m6A in mammalian DNA, indirect evidence obtained using restriction enzymes sensitive to adenine methylation suggests the presence of m6A in the mouse Myo-D1 gene and in the rat type 2 steroid 5α-reductase gene (11, 12). In the case of the rat type 2 steroid 5α-reductase gene, the restriction pattern is correlated to its expression (11). Unfortunately, direct evidence for the presence of m6A in mammals, based on its physical detection by mass spectrometry, is still lacking. Indeed, it seems that the relative high abundance of 5mC in mammalian DNA has focused attention on the role of m5C to the detriment of the m6A quest. In this regard, it should be pointed out that at an overall content of <0.001% m6A can be biologically significant if it occurs, for example, in a regulated fashion on specific mammalian gene regulatory elements (Fig. 2D). Hence, according to the size of the human genome (3.3 × 109 bp) and to the number of genes (~30 × 103), the presence of a few hundred m6A can be sufficient to play a crucial role in the control of biological processes such as cell differentiation or morphogenesis. Interestingly, evidence suggests that m6A does affect the regulation of gene expression in mammalian cells. Thus, in mammalian cells, the artificial presence of m6A can affect the binding of a nuclear factor to its responsive element (57), decrease the activity of adenoviral E1A promoter (58), or generate a steroid hormone response element (59). Incidentally, this point may be of special concern since all the plasmids currently used in transient gene expression experiments are subjected to adenine methylation as a consequence of the bacterial Dam and CcrM activities of E. coli (60, 61). In addition, several reports have also demonstrated the influence of m6A on the activity of plant gene promoters (62–64), while addition of m6A to mammalian cell cultures induces cell differentiation in several cell lines (65, 66).
III. Perspective and concluding remarks
The fact that the essential roles played by DNA adenine methyltransferases in bacterial viability and virulence have been recognized only recently underscores the importance to investigate the presence and biological functions of adenine methylation in eukaryotic DNA. Thus, a precise knowledge of the adenine methylation status in human DNA is of crucial concern for the development of new antibiotics targeting bacterial DNA adenine methyltransferases. Moreover, the possibility that DNA adenine methylation could occur during the life-cycle of some parasites and virus should also be reconsidered, since, for example, the reported lack of m6A in the DNA of Plasmodium falciparum or adenovirus has been based on the use of low sensitive methods (67, 68). In this regard, if we speculate that one of the functions of adenine methylation in eukaryotic cells is a defense mechanism which marks foreign DNA in order to ensure its degradation, then, preparations of viral DNA used to quantify the presence of m6A are inevitably made of molecules which have escaped this methylation/degradation process and which are therefore described as unmethylated. Another possible function of adenine methylation could be to mark the immortal DNA strand suggested to be present in adult stem cells that divide by asymmetric mitosis (69–71). Thus, if we consider that the object of the immortal DNA strand is to protect the genome of stem cells from mutations, then m6A is a conceivable alternative to the use of the mutagenic m5C (50). Hence, the presence of m6A in a very rare population of cells could provide another explanation of the difficulty in detecting m6A in mammals. A prerequisite to the “natural” methylation of adenine in mammalian DNA is the existence of at least one adenine-N6-DNA methyltransferase gene in mammalian genomes. In contrast, to cytosine DNA methyltransferases, which belong to a family of conserved enzymes, bacterial adenine-N6 methyltransferase are much more heterogeneous. Besides a weakly conserved F_G_G amino acid motif shared by all Mtases, DNA adenine-methyltransferases only contain one moderately conserved (D/N)PP(Y/F) motif (72). On the basis of computer analysis several putative DNA adenine methyltransferases have been identified in human and murine genomes (66, 73). Whether these genes encode true DNA adenine methylases or are the fossils of a restriction-modification system present either in the putative archea-like ancestor of eukaryotic cells or ancestral bacterial endosymbiont at the origin of mitochondria should warrant further investigations.
Table 1.
| Organism | % MeAde | Reference |
|---|---|---|
| Eubacteria | ||
| Caulobacter bacteroides | 1.0 | 89 |
| Enterobacteriales E. coli | 1.4–2.0 | 2 |
| Mycobacteria smegmatis | 3.0 | 90 |
| Mycobacteria tuberculosis | 0.45–0.5 | 90 |
| Mlycoplasma arginini | 2.0 | 91 |
| Mycoplasma capricolum | 0.2 | 91 |
| Mycoplasma hyorhinis | 2.0 | 91 |
| Mycoplasma orale | 0.9 | 91 |
| Acholeplasama laidlawii | 0.5 | 91 |
| Eucarya | ||
| Oxytricha fallax | 0.6–0.7 | 43 |
| Paramecia Aurelia | 2.5 | 14 |
| Chlamydomonas reinhardi | 0.50 | 41 |
| Tetrahymena pyriformis | 0.65 – 0.80 | 41,48 |
| Tetrahymena thermophila | 0.80 | 44 |
| Viridiplantae, Wheat | 0.09 – 10 (mt) | 61 |
| 0.46 – 0.55 (n) | 61 | |
| Arthropoda, Aedes albopictus | 0.1 | 63 |
mt: mitochondrial, n: nuclear
Acknowledgments
We thank Pr. AL. Benabid for his support and Dr. K. Herbert for critical reading of the manuscript. DW is supported by the Fondation Jérôme Lejeune
References
- 1.Doskocil J, Sormo’Va Z. Biochim Biophys Acta. 1965 Mar 15;95:513–5. [PubMed] [Google Scholar]
- 2.Dunn DB, Smith JD. Biochem J. 1958 Apr;68:627–36. doi: 10.1042/bj0680627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich M, et al. Nucleic Acids Res. 1985 Feb 25;13:1399–412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahng LS, Shapiro L. J Bacteriol. 2001 May;183:3065–75. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Julio SM, et al. Infect Immun. 2001 Dec;69:7610–5. doi: 10.1128/IAI.69.12.7610-7615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reisenauer A, Shapiro L. Embo J. 2002 Sep 16;21:4969–77. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson GT, et al. J Bacteriol. 2000 Jun;182:3482–9. doi: 10.1128/jb.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright R, Stephens C, Shapiro L. J Bacteriol. 1997 Sep;179:5869–77. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens C, Reisenauer A, Wright R, Shapiro L. Proc Natl Acad Sci U S A. 1996 Feb 6;93:1210–4. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashapkin VV, Kutueva LI, Vanyushin BF. FEBS Lett. 2002 Dec 18;532:367–72. doi: 10.1016/s0014-5793(02)03711-0. [DOI] [PubMed] [Google Scholar]
- 11.Reyes EM, Camacho-Arroyo I, Nava G, Cerbon MA. J Androl. 1997 Jul–Aug;18:372–7. [PubMed] [Google Scholar]
- 12.Kay PH, et al. Gene. 1994 Dec 30;151:89–95. doi: 10.1016/0378-1119(94)90636-x. [DOI] [PubMed] [Google Scholar]
- 13.Pintor-Toro JA. Biochem Biophys Res Commun. 1987 Sep 30;147:1082–7. doi: 10.1016/s0006-291x(87)80181-x. [DOI] [PubMed] [Google Scholar]
- 14.Cummings DJ, Tait A, Goddard JM. Biochim Biophys Acta. 1974 Nov 20;374:1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- 15.Luria SE, Human ML. J Bacteriol. 1952 Oct;64:557–69. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertani G, Weigle JJ. J Bacteriol. 1953 Feb;65:113–21. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlagman SL, Hattman S. Gene. 1983 May–Jun;22:139–56. doi: 10.1016/0378-1119(83)90098-7. [DOI] [PubMed] [Google Scholar]
- 18.Magrini V, et al. J Bacteriol. 1997 Jul;179:4254–63. doi: 10.1128/jb.179.13.4254-4263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranyi U, Klein R, Lubitz W, Kruger DH, Witte A. Mol Microbiol. 2000 Mar;35:1168–79. doi: 10.1046/j.1365-2958.2000.01786.x. [DOI] [PubMed] [Google Scholar]
- 20.Arnold HP, Ziese U, Zillig W. Virology. 2000 Jul 5;272:409–16. doi: 10.1006/viro.2000.0375. [DOI] [PubMed] [Google Scholar]
- 21.Lodwick D, Ross HN, Harris JE, Almond JW, Grant WD. J Gen Microbiol. 1986 Nov;132:3055–9. doi: 10.1099/00221287-132-11-3055. [DOI] [PubMed] [Google Scholar]
- 22.Barbeyron T, Kean K, Forterre P. J Bacteriol. 1984 Nov;160:586–90. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reisenauer A, Kahng LS, McCollum S, Shapiro L. J Bacteriol. 1999 Sep;181:5135–9. doi: 10.1128/jb.181.17.5135-5139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barras F, Marinus MG. Trends Genet. 1989 May;5:139–43. doi: 10.1016/0168-9525(89)90054-1. [DOI] [PubMed] [Google Scholar]
- 25.Lobner-Olesen A, Skovgaard O, Marinus MG. Curr Opin Microbiol. 2005 Apr;8:154–60. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Oshima T, et al. Mol Microbiol. 2002 Aug;45:673–95. doi: 10.1046/j.1365-2958.2002.03037.x. [DOI] [PubMed] [Google Scholar]
- 27.Zweiger G, Marczynski G, Shapiro L. J Mol Biol. 1994 Jan 14;235:472–85. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 28.Boye E, Lobner-Olesen A. Cell. 1990 Sep 7;62:981–9. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 29.Braaten BA, Nou X, Kaltenbach LS, Low DA. Cell. 1994 Feb 11;76:577–88. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 30.Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. Science. 1999 May 7;284:967–70. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Del Portillo F, Pucciarelli MG, Casadesus J. Proc Natl Acad Sci U S A. 1999 Sep 28;96:11578–83. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low DA, Weyand NJ, Mahan MJ. Infect Immun. 2001 Dec;69:7197–204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashhoon N, et al. J Biol Chem. 2004 Dec 10;279:52075–81. doi: 10.1074/jbc.M408182200. [DOI] [PubMed] [Google Scholar]
- 34.Wahnon DC, Shier VK, Benkovic SJ. J Am Chem Soc. 2001 Feb 7;123:976–7. doi: 10.1021/ja003285o. [DOI] [PubMed] [Google Scholar]
- 35.Hattman S, Kenny C, Berger L, Pratt K. J Bacteriol. 1978 Sep;135:1156–7. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorovsky MA, Hattman S, Pleger GL. J Cell Biol. 1973 Mar;56:697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rae PM, Spear BB. Proc Natl Acad Sci U S A. 1978 Oct;75:4992–6. doi: 10.1073/pnas.75.10.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pratt K, Hattman S. Mol Cell Biol. 1981 Jul;1:600–8. doi: 10.1128/mcb.1.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capowski EE, Wells JM, Harrison GS, Karrer KM. Mol Cell Biol. 1989 Jun;9:2598–605. doi: 10.1128/mcb.9.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison GS, Findly RC, Karrer KM. Mol Cell Biol. 1986 Jul;6:2364–70. doi: 10.1128/mcb.6.7.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez JC, Callejas S, Borniquel S, Martin-Gonzalez A. Int Microbiol. 2000 Sep;3:139–46. [PubMed] [Google Scholar]
- 42.Zhang Y, Nelson M, Nietfeldt JW, Burbank DE, Van Etten JL. Nucleic Acids Res. 1992 Oct 25;20:5351–6. doi: 10.1093/nar/20.20.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Que Q, et al. Gene. 1997 May 6;190:237–44. doi: 10.1016/s0378-1119(96)00862-1. [DOI] [PubMed] [Google Scholar]
- 44.Karrer KM, VanNuland TA. Nucleic Acids Res. 2002 Mar 15;30:1364–70. doi: 10.1093/nar/30.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu CM, Henney HR., Jr Biochem Cell Biol. 1990 Jun;68:944–8. doi: 10.1139/o90-139. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama S, Endoh H. Genes Genet Syst. 2003 Dec;78:391–8. doi: 10.1266/ggs.78.391. [DOI] [PubMed] [Google Scholar]
- 47.Vanyushin BF, Tkacheva SG, Belozersky AN. Nature. 1970 Mar 7;225:948–9. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- 48.Lawley PD, Crathorn AR, Shah SA, Smith BA. Biochem J. 1972 Jun;128:133–8. doi: 10.1042/bj1280133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunthert U, Schweiger M, Stupp M, Doerfler W. Proc Natl Acad Sci U S A. 1976 Nov;73:3923–7. doi: 10.1073/pnas.73.11.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole A, Penny D, Sjoberg BM. Nat Rev Mol Cell Biol. 2001 Feb;2:147–51. doi: 10.1038/35052091. [DOI] [PubMed] [Google Scholar]
- 51.Bestor TH. Hum Mol Genet. 2000 Oct;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 52.Yoder JA, Walsh CP, Bestor TH. Trends Genet. 1997 Aug;13:335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 53.Vanyushin BF, Alexandrushkina NI, Kirnos MD. FEES Lett. 1988;233:397–9. [Google Scholar]
- 54.Dhar MS, Pethe VV, Gupta VS, Ranjekar PK. Theor Appl Genet. 1990;80:402–8. doi: 10.1007/BF00210080. [DOI] [PubMed] [Google Scholar]
- 55.Ngernprasirtsiri J, Akazawa T. Eur J Biochem. 1990 Dec 12;194:513–20. doi: 10.1111/j.1432-1033.1990.tb15646.x. [DOI] [PubMed] [Google Scholar]
- 56.Adams RL, McKay EL, Craig LM, Burdon RH. Biochim Biophys Acta. 1979 Jun 20;563:72–81. doi: 10.1016/0005-2787(79)90008-x. [DOI] [PubMed] [Google Scholar]
- 57.Tronche F, Rollier A, Bach I, Weiss MC, Yaniv M. Mol Cell Biol. 1989 Nov;9:4759–66. doi: 10.1128/mcb.9.11.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knebel D, Doerfler W. J Mol Biol. 1986 May 20;189:371–5. doi: 10.1016/0022-2836(86)90518-8. [DOI] [PubMed] [Google Scholar]
- 59.Truss M, Bartsch J, Chalepakis G, Beato M. Nucleic Acids Res. 1992 Apr 11;20:1483–6. doi: 10.1093/nar/20.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berger F, Canova C, Benabid AL, Wion D. Nat Biotechnol. 1999 Jun;17:517. doi: 10.1038/9778. [DOI] [PubMed] [Google Scholar]
- 61.Allamane S, et al. Biochem Biophys Res Commun. 2000 Oct 5;276:1261–4. doi: 10.1006/bbrc.2000.3603. [DOI] [PubMed] [Google Scholar]
- 62.Sugimoto K, Takeda S, Hirochika H. Plant J. 2003 Nov;36:550–64. doi: 10.1046/j.1365-313x.2003.01899.x. [DOI] [PubMed] [Google Scholar]
- 63.Rogers JC, Rogers SW. Plant J. 1995 Feb;7:221–33. doi: 10.1046/j.1365-313x.1995.7020221.x. [DOI] [PubMed] [Google Scholar]
- 64.Graham MW, Larkin PJ. Transgenic Res. 1995 Sep;4:324–31. doi: 10.1007/BF01972529. [DOI] [PubMed] [Google Scholar]
- 65.Ratel D, et al. Biochem Biophys Res Commun. 2001 Jul 20;285:800–5. doi: 10.1006/bbrc.2001.5240. [DOI] [PubMed] [Google Scholar]
- 66.Charles MP, et al. Biochem Biophys Res Commun. 2004 Feb 6;314:476–82. doi: 10.1016/j.bbrc.2003.12.132. [DOI] [PubMed] [Google Scholar]
- 67.Pollack Y, Kogan N, Golenser J. Exp Parasitol. 1991 May;72:339–44. doi: 10.1016/0014-4894(91)90079-c. [DOI] [PubMed] [Google Scholar]
- 68.Wienhues U, Doerfler W. J Virol. 1985 Oct;56:320–4. doi: 10.1128/jvi.56.1.320-324.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cairns J. Nature. 1975 May 15;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 70.Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cancer Res. 2002 Dec 1;62:6791–5. [PubMed] [Google Scholar]
- 71.Potten CS, Owen G, Booth D. J Cell Sci. 2002 Jun 1;115:2381–8. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 72.Jeltsch A, Christ F, Fatemi M, Roth M. J Biol Chem. 1999 Jul 9;274:19538–44. doi: 10.1074/jbc.274.28.19538. [DOI] [PubMed] [Google Scholar]
- 73.Shorning BY, Vanyushin BF. Biochemistry (Mosc) 2001 Jul;66:753–62. doi: 10.1023/a:1010260612109. [DOI] [PubMed] [Google Scholar]


