Abstract
Locus control regions (LCRs) are complex high-order chromatin structures harbouring several regulatory elements, including enhancers and boundaries. We have analysed the mouse tyrosinase LCR functions, in vitro, in cell lines and, in vivo, in transgenic mice and flies. The LCR-core (2.1 kb), located at –15 kb and carrying a previously described tissue-specific DNase I hypersensitive site, operates as a transcriptional enhancer that efficiently transactivates heterologous promoters in a cell-specific orientation-independent manner. Furthermore, we have investigated the boundary activity of these sequences in transgenic animals and cells. In mice, the LCR fragment (3.7 kb) rescued a weakly expressed reference construct that displays position effects. In Drosophila, the LCR fragment and its core insulated the expression of a white minigene reporter construct from chromosomal position effects. In cells, sequences located 5′ from the LCR-core displayed putative boundary activities. We have obtained genomic sequences surrounding the LCR fragment and found a LINE1 repeated element at 5′. In B16 melanoma and L929 fibroblast mouse cells, this element was found heavily methylated, supporting the existence of putative boundary elements that could prevent the spreading of condensed chromatin from the LINE1 sequences into the LCR fragment, experimentally shown to be in an open chromatin structure.
INTRODUCTION
Eukaryotic genes are generally organised on chromosomes as contiguous but independent units known as expression domains (1,2). Usually, they are represented as chromatin fibres in the form of loops whose ends remain attached to nuclear matrix or scaffold components, thereby preventing the interference between adjacent expression domains (3,4). These expression domains are believed to remain insulated from neighbouring genomic sequences by boundary elements (5), and are thought to include all regulatory elements that are necessary for their proper gene expression (6).
We use the mouse tyrosinase locus as an experimental model to study mammalian expression domains in transgenic animals (7). The mouse tyrosinase gene, located on chromosome 7, encodes the rate-limiting enzyme for melanin biosynthesis and is tightly regulated during development. It is expressed from embryonic day +16.5 in melanocytes, derived from neural crest, and from day +10.5 in RPE cells, derived from the optic cup (8). Minigene tyrosinase constructs were used in mice to rescue the albino phenotype of recipient animals (8,9). These transgenic mice often displayed position effects, manifested as variability in pigmentation levels and variegation (10).
In contrast, the generation of transgenic mice with yeast artificial chromosome (YAC) constructs covering 250 kb of the mouse tyrosinase locus completely rescued the albino phenotype (11), reviewed in (10). These results suggested the existence of important regulatory elements, absent in previous standard constructs, which allow YAC tyrosinase transgenes to overcome position effects (12). One of these elements was identified during the molecular characterisation of the chinchilla-mottled allele (Tyrc–m), and was found ∼15 kb upstream of the tyrosinase gene (13). It contains a DNase I hypersensitive site (HS) that functions as a cell-specific enhancer in vitro and in vivo, when combined with the endogenous tyrosinase promoter (14–16). The relevance of this element was evaluated in vivo by generating deletions within YAC tyrosinase transgenes that allowed the description of a novel locus control region (LCR) in the mouse tyrosinase gene (14,17). The albino phenotype was totally rescued in the presence of the LCR. The absence of the LCR resulted in weaker pigmentation, variegated expression in all cellular types known to express the tyrosinase gene and delayed retinal pigmentation in transgenic mice (18).
LCRs are defined functionally, as they share little, if any, sequence or structural homology. They can overcome position effects in transgenesis, with an expression value per copy comparable with the endogenous alleles (19,20). LCRs are complex high-order chromatin structures containing regulatory sequences that can incorporate several activities, including transcriptional enhancers (20), scaffold/matrix attachment regions (S/MARs) (6) and boundary elements (5). To date, the best-known LCR is that of the human β-globin locus (21). The function of this LCR has been extensively studied in transgenic analyses (19) and at its endogenous genomic site (22,23).
Insulator or boundary elements were first characterised in Drosophila melanogaster and yeast genomes and subsequently found in a number of vertebrates (including chicken, mouse and human genome) and other organisms (5,24–26). Boundaries have been defined functionally by at least one of the following two properties. First, they have a barrier activity, protecting a gene against chromosomal position effects (27), such as the spreading of chromatin condensation and heterochromatinisation into expression domains, as seen in flies (28,29), mammalian cells (30) and yeast (31). Secondly, insulators can also have a blocker activity interfering with the transactivation of a given promoter by a distal enhancer when placed between these two regulatory elements. This has been studied best in the Drosophila boundary elements, such as the scs/scs’ sequences from the hsp70 loci (32) and the Su(Hw) binding region of the gypsy element (33–35).
A few boundary elements have been shown to display both features. This is the case of the best-known insulator in vertebrates, present at the 5′HS4 element of the chicken β-globin locus (36,37), which acts as a blocker, insulating from the activity of enhancers located in the 5′ upstream folate receptor gene (38), and as a barrier, preventing silencing from an upstream region of condensed chromatin (39). These two features appear to be transduced by separate protein activities (40). Analogous insulator elements appear to be present in the homologous human and mouse β-globin loci (41).
Some boundaries have been tested in heterologous systems for gene transfer applications, although with limited success (42–44). Little is known about the molecular mechanisms underlying insulator function for which several models have been proposed (5,45,46). Further in vivo studies in their natural genomic context will be required to help understand the function of boundary elements in ectopic locations.
Here, we have evaluated, in vitro and in vivo, the enhancer and boundary activities of the 3.7 kb EcoRI fragment that contains the mouse tyrosinase LCR. We describe that enhancer functions are operative in front of heterologous promoters in a cell type-dependent and orientation-independent manner. We have discovered a new boundary activity in this region, using transgenic mice and flies. In mice, we show that the LCR fragment can rescue the expression of a luciferase reporter transgene prone to position effects. In Drosophila, we have used the standard white minigene assay, with appropriate positive and negative controls, to show that mouse tyrosinase LCR sequences efficiently protect this reference construct from chromosomal position effects. Further, we have obtained genomic sequences surrounding this LCR and identified a LINE1 (Long Interspersed Elements) repeated element located 5′ upstream from the LCR fragment. We show that this repeated element is methylated in both tyrosinase-expressing and non-expressing cells. The chromatin spanning over the LCR appears more accessible to DNase I digestion in tyrosinase-expressing cells. Finally, results obtained from in vitro enhancer-blocking tests further support the presence of putative boundary elements within this LCR fragment. These elements might contribute to the LCR function by preventing the spreading of condensed chromatin, presumably formed on the upstream LINE1 methylated sequences, thus possibly establishing the putative 5′ boundary of the mouse tyrosinase expression domain.
MATERIALS AND METHODS
Description of plasmids
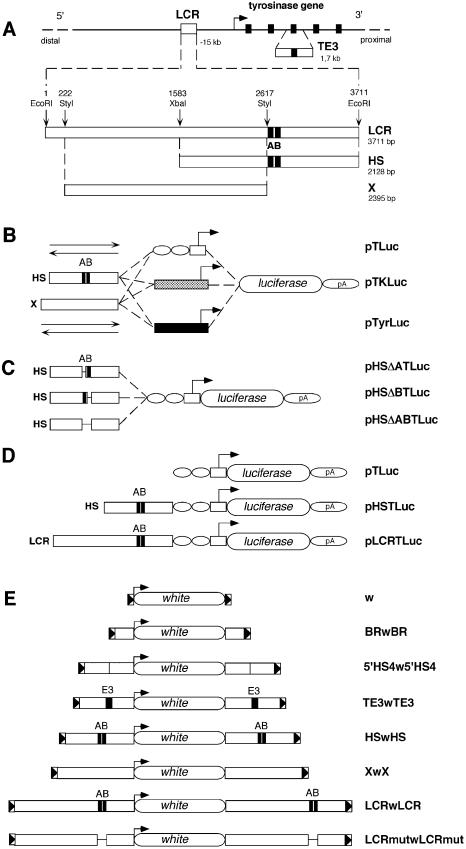
Firefly luciferase gene was obtained from plasmid pDO432 (47) as a 1.8 kb SspI–BamHI fragment and cloned into pW37SP1 vector (4 kb) (48) resulting in plasmid pTLuc (5.75 kb). pW37SP1 contains a TATA box from the promoter of the herpes simplex virus thymidine kinase (HSV-TK) gene (–37/+19) next to two binding sites for the ubiquitous transcription factor SP1, and the splice site and polyadenylation signal from SV40 small-t antigen. pTKLuc (5.8 kb) contains the promoter of the HSV-TK gene (–105/+51) from pBLCAT2 (49). pTyrLuc (5.9 kb) contains the mouse tyrosinase promoter (–270/+9) from pCAT (14). The HS (2.13 kb) and X (2.4 kb) regions are derived from plasmid pTyr14:E6 (14) and were cloned, in either direction, in pTLuc, pTKLuc and pTyrLuc vectors, resulting in pHSTLuc/pSHTLuc (7.87 kb), pXTLuc/pXinvTLuc (8.14 kb), pHSTKLuc/pSHTKLuc (7.93 kb), pXTKLuc/pXinvTKLuc (8.2 kb), pHSTyrLuc/pSHTyrLuc (8.03 kb) and pXTyrLuc/pXinvTyrLuc (8.3 kb) constructs. pHSΔATluc (7.85 kb), pHSΔBTluc (7.85 kb) and pHSΔABTLuc (7.81 kb) deletion constructs were derived from pHSTLuc with the following deletions: (nucleotide positions are given with respect to X76647): ΔA, nucleotides 2622–2654; ΔB, nucleotides 2657–2682; and ΔAB, nucleotides 2622–2682 (50) (see Supplementary Material). In addition, the LCR fragment (3.71 kb), entirely contained in plasmid pTyr14:E6 (14) and the LCR fragment carrying the ΔAB deletion (described above) and/or a deletion of a G-rich box (ΔG; X76647, nucleotides 1–71) were cloned, in either direction, in pTKLuc vector, resulting in pLCRTKLuc/pRCLTKLuc (9.51 kb), pLCRΔGTKLuc/pRCLΔGTKLuc (9.45 kb), pLCRΔABTKLuc/pRCLΔABTKLuc (9.44 kb) and pLCRΔ ABGTKLuc/pRCLΔABGTKLuc (9.38 kb) constructs, respectively.
For the generation of transgenic flies the reference plasmid was pCaSpeR3 (referred in this work as w, 7.8 kb), carrying the white minigene (51). BRwBR plasmid (8.8 kb) was used as reported (52). The rest of the constructs used in flies were derived from pCaSpeRW15* (7.8 kb), a modified version of pCaSpeRW15 (52) with an additional EcoRI site, at the 3′ end of the white minigene. LCR, HS, X, LCRmut, TE3 and 5′HS4 fragments were cloned at either side of pCaSpeRW15* resulting in LCRwLCR (15.2 kb), HSwHS (12 kb), XwX (12.4 kb), LCRmutwLCRmut (15.1 kb), TE3wTE3 (11.2 kb) and 5′HS4w5′HS4 (12.6 kb) constructs, respectively. The 5′HS4 insulator fragment was obtained from pBC1 plasmid (Invitrogen), as a 2.4 kb XbaI fragment containing two copies in tandem of the 1.2 kb 5′HS4 from the chicken β-globin locus (36). The TE3 fragment was obtained from pgTYR-H1.8A as a 1.7 kb HindIII fragment (53).
Plasmid DNAs were purified, using Qiagen Plasmid Maxi Kit (Qiagen) or Concert High Purity Maxiprep Plasmid System (Gibco-BRL, Life Technologies). All plasmids were confirmed by restriction enzyme and sequence analyses. Detailed description of plasmids is available upon request.
Culture and transient transfections of mammalian cells
B16 mouse melanoma, MeWo human melanoma, L929 mouse fibroblasts and D407 human RPE cells (54) were grown as described (14). PC12 rat phaeochromocytoma cells were grown in RPMI medium, supplemented with 10% horse serum (Life Technologies), 5% fetal calf serum and 1 mM sodium pyruvate. All cells included 2 mM glutamine, 103 U/ml penicillin, 1 mg/ml streptomycin and 10 mM HEPES pH 7.4 and were incubated at +37°C, 5% CO2 and 98% humidity.
Transient transfection assays were carried out, in triplicates, using TransFast (Promega), according to the supplier. For each transfection experiment 2.5 µg of experimental Luciferase reporter construct along with 0.5 µg of pCMV-lacZ control plasmid were co-transfected. Luciferase assays were performed using commercial systems (Promega) and measured in a microplate luminometer (Microlumat Plus, EG&G Berthold). β-Galactosidase activity was determined using published procedures (14). Statistical evaluation of transfection data, when indicated, was performed by t-Student paired test (SPSS).
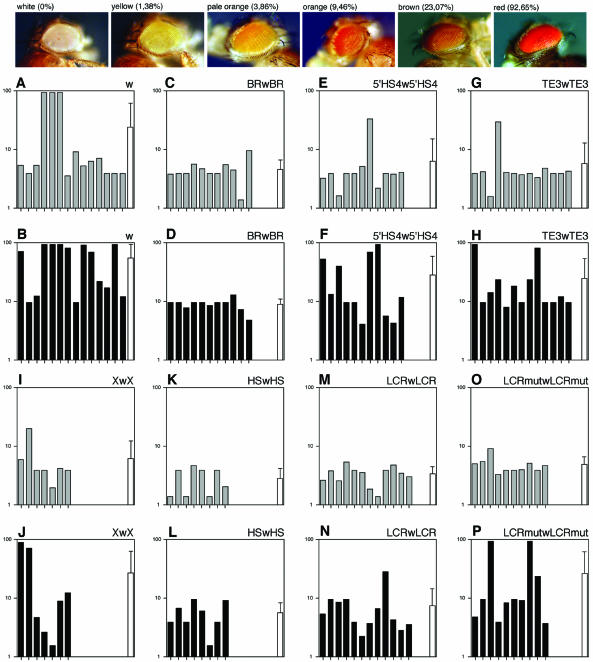
Generation and analysis of transgenic flies
Transgenic flies (D.melanogaster) were made following standard procedures (55), using w1118 or Df ac1 w1118 mutant embryos as hosts (Bloomington Stock Centre, Indiana University, USA). Chromosomal localisation of the insertion was determined by crossing transgenic males with w1118; If/CyO; TM2/MKRS females (56). Copy number was confirmed by Southern blot analysis, using two probes (white and P-element) derived from plasmid pCaSpeRW15. Eye pigmentation was noted in 30–70 homozygous and heterozygous 72 h female flies from each transgenic line. In total, more than 10 000 transgenic flies were generated and observed. However, only single-copy transgenic lines that could be established in heterozygosis and homozygosis, corresponding to 3059 and 3060 individuals, respectively, were used for graphical and statistical analyses. Eye colour was assigned to one of the following five categories: yellow, pale orange, orange, brown and red (Fig. 4). For graphical purpose, the amount of red pigment in eyes was measured as described (57). Statistical analysis of transgenic fly data was done using SPSS.
Figure 4.
Analysis of boundary function of mouse tyrosinase LCR sequences in transgenic D.melanogaster. Protection from chromosomal position effects of mouse tyrosinase LCR sequences was carried out in transgenic flies using the white minigene assay. Top, fly heads, from reference white mutant stock along with representative transgenic individuals for yellow, pale orange, orange, brown and red eye colours are shown. Pictures are taken in air from anaesthetised animals. Percentage of red-pigment in eyes from each category, established by colorimetric methods (57), is indicated (100% = wild-type flies). The average amount of red-eye pigment within each line was estimated taking into account the percentage of individuals from a given colour observed within each line. Mean values of red-pigment content in eyes for each independent transgenic line are depicted graphically as single bars using a logarithm scale (y axis), because most of the perceived eye colour variability takes place below 10% of wild-type pigment level. Phenotypic evaluation of transgenic flies is shown in heterozygous (grey bars) and homozygous (black bars) individuals. A total of 86 transgenic fly lines are shown, grouped per construct and distributed as follows: 14 lines for ‘w’ (A and B); 11 for BRwBR (C and D); 11 for 5′HS4w5′HS4 (E and F); 13 for TE3wTE3 (G and H); 7 for XwX (I and J); 8 for HSwHS (K and L); 12 for LCRwLCR (M and N); and 10 for LCRmutwLCRmut (O and P). In addition, the overall means and standard deviations (SD) for all lines analysed within each construct are depicted as white bars and are also presented, without logarithm transformations, in Table 1.
Generation and analysis of transgenic mice
Transgenic mice were generated using standard procedures (58) using albino recipient mouse strains NMRI or FVB/N (Harlan), as reported before (17). Transgenic founder mice were identified by PCR and Southern blot analyses. Transgenic lines were established and their copy number evaluated in F1 heterozygous individuals by slot-blot analysis (59). All experiments complied with local and European legislation concerning vivisection and the experimentation and use of genetically modified organisms.
To evaluate luciferase activity in mice, six adult (2–3 months of age) hemizygous individuals were analysed from each transgenic line. Protein extracts were prepared from nine organs (dorsal skin, eyes, skeletal muscle, thin gut, kidney, spleen, liver, lung and brain) in cell culture lysis buffer (Promega), following supplier indications. Protein contents were evaluated using Bio-Rad Protein Assay (Bio-Rad). Luciferase assays were performed as described in cell transfection assays. A series of dilutions were assayed from each extract to ensure that measured values were not quenched.
Isolation of a genomic clone containing LCR and surrounding sequences
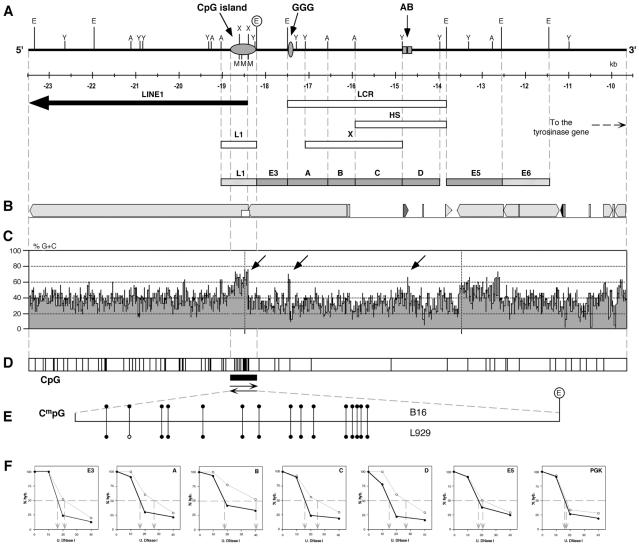
A 129Sv/J mouse genomic library (Stratagene) was screened with a single-copy DNA probe (probe A, 0.9 kb XbaI fragment from pTyr14:E6, shown in Fig. 5) using standard methods (59). Eighteen independent bacteriophage clones were identified, four of which were subjected to three consecutive rounds of screening. One of these clones, λT1, contained a 14 kb insert that included the LCR, was mobilised into a plasmid, pN14n21, and entirely sequenced in both senses of the DNA using specific oligonucleotides (more than 60) and subclones of internal DNA fragments. Details are available upon request.
Figure 5.
Genomic structure and chromatin analysis of DNA sequences surrounding the mouse tyrosinase LCR. (A) Schematic view of the genomic DNA sequence (13 806 bp, GenBank AF364302) containing mouse tyrosinase LCR and neighbouring regions. All restriction enzyme sites for EcoRI (E), StyI (Y), XbaI (A), XhoI (X) and SmaI (M) are shown. Below, the relative distance from the transcription start of tyrosinase gene is indicated in kilobases (nucleotide positions according to Ensembl Project, Mouse Genome Sequencing Consortium; http://www.ensembl.org/Mus_musculus/geneview?gene=ENSMUSG00000004651; derived from BAC clone RP24-459G24 (EMBL accession number AC122517). These nucleotide coordinates have been used to update the position of the 3.7 kb EcoRI fragment, described previously at –12 kb (14,17), according to an original physical restriction site map made from overlapping bacteriophage clones (53). AF364302 is entirely contained within AC122517). A LINE1 element, located at 5′ end of the sequence, is indicated with a thick arrow. Relative position of DNA sub-fragments L1, LCR, HS and X, used for in vivo and in vitro expression analyses, are shown, along with a CG-rich sequence similar to a CpG island (grey horizontal oval), a short G-rich sequence (grey vertical oval) and A-B boxes (grey squares). E3, A, B, C, D and E5 DNA sub-fragments used as single-copy probes in DNase I sensitivity assays (grey boxes) and L1 and E6 repetitive DNA probes (striped boxes) are displayed below as eight adjacent boxes aligned with the sequence map. (B) Repeated elements identified in this sequence by Repeat Masker (A.F.A.Smit and P.Green, unpublished work; http://ftp.genome.washington.edu/cgi-bin/RepeatMasker/) and displayed using PIPMaker symbols (95; http://bio.cse.psu.edu/pipmaker/). Light grey boxes correspond to LINE1-related repeated sequences. Smaller repeats from other families are shown. (C) Percentage of G + C plot along this sequence, using a window size of 30 nucleotides, generated with MacVector (Accelrys). DNA regions rich in G + C content that coincide with the GC-rich stretch of DNA similar to a CpG island, the G-rich and AB the sequences are indicated with arrows. (D) Linear map showing the occurrence of CpG dinucleotides (vertical bars) along this sequence. (E) Bisulphite genomic DNA sequencing of the 5′ CG-rich stretch of DNA similar to a CpG island in mouse melanoma B16 (above) and fibroblasts L929 (below) cells. The position of an EcoRI site, at 3′ end (within a circle), is provided for orientation purposes (AF364302, nucleotide position 5258). The relative positions of in vivo methylated CmpGs and unmethylated CpGs are shown as black and open circles, respectively. (F) Representative DNase I sensitivity analysis with E3, A, B, C, D, E5 (described above) and PGK probes. Nuclear chromatin samples obtained from melanoma B16 (black circles, thick line) and fibroblasts L929 (open circles, thin line) were digested with increasing amounts of DNase I. Thereafter, equivalent amounts of purified digested DNAs were spotted on nylon slot blots and hybridised with the indicated probe. Autoradiograms were quantitated by PhosphorImage analysis and expressed as a percentage of hybridisation signal detected in the undigested DNA sample (assigned to 100%). These percentages are depicted in these seven graphs as a function of DNase I concentration (U.). Fifty per cent hybridisation of samples is indicated with a dashed line. The estimated amount of DNase I required for 50% hybridisation is shown by descending dashed arrows. PGK probe is used as a control and stands for the mouse phosphoglycerate kinase gene promoter, obtained from plasmid pHM2 (GenBank X76683, nucleotides 6557–7064). DNase I general sensitivity assays were repeated three times with comparable results (in general, observed values varied between 5 and 20%, not shown).
DNA methylation analysis
Ten micrograms of HindIII digested genomic DNAs from B16 or L929 cells were denatured and treated with sodium bisulphite. Purification and amplification of transformed DNA was performed essentially as reported (60). Oligo nucleotides BIS1A (5′-ATATaAaaACaTACATATAaCAC CCC-3′; AF364302, nucleotides 5304–5279) and BIS2 (5′-GTGTTttAtTtAttAGAGATtTTAGGG-3′; AF364302, nucleotides 4741–4767) were first used to amplify a 563 bp DNA band [PCR conditions: AmpliTaq (Roche), 10 cycles: 2 min at 94°C, 15 s at 55°C and 4 min at 68°C; followed by 25 cycles: 15 s at 94°C, 15 s at 55°C and 3 min at 68°C, increasing by 20 s the extension step at 68°C in each cycle]. Thereafter, oligonucleotides BIS2 and BIS1B (5′-CCAaaaAATTCCTaAAaCTaATaACC-3′; AF364302, nucleotides 5268–5243) were used for a second round of nested amplification using the same PCR conditions and resulting in a 527 bp DNA band. Transformed nucleotides are written in lowercase. The amplified DNA fragment, from several pooled PCR amplifications, was cloned directly into pCRII (Invitrogen). Four (B16 DNA) and five (L929 DNA) independent clones were sequenced entirely from either direction. A consensus sequence was derived after multiple alignment of individual DNA sequences with the original reference genomic sequence (527 bp; AF364302, nucleotides 4741–5268) using MacVector (Accelrys).
DNAse I sensitivity assays
DNase I sensitivity analysis was performed essentially as described (61). In brief, nuclei were isolated from B16 and L929 cells following described procedures (62), and digested for 10 min at 37°C with increasing amounts of DNase I DPFF (Worthington) (in the range between 0 and 200 U/ml). Following proteinase K digestion, DNA samples were purified by phenol extraction and ethanol precipitation. Fifteen micrograms of each DNA sample, in triplicates, were vacuum-loaded onto a nylon membrane (Hybond-N, Amersham) using a slot-blot apparatus (Minifold II, Schleicher & Schuell). Membranes were hybridised with E3, A, B, C, D, E5 and PGK ramdom prime [α-32P]dCTP-labelled DNA probes (shown in Fig. 5), following standard methods (59). E3, A, B, C, D, E5 and PGK DNA probes were experimentally validated as single-copy probes by standard Southern blot analysis on mouse genomic DNA. Hybridisation signals were quantitated using a PhosphorImager (Molecular Dynamics, Inc.) and analysed using ImageQuant software.
DNA sequences
DNA sequences referred to in this work have been submitted to GenBank and EMBL Databases under accession numbers X76647 and AF364302, respectively. X76647 DNA sequence is included within AF364302 sequence.
RESULTS
Analysis of LCR function: enhancer activity with heterologous promoters in mammalian cells
To functionally dissect the mouse tyrosinase LCR and to understand its fundamental role in the regulation of this locus we sequenced the entire 3.7 kb EcoRI DNA fragment that contained the LCR, extending previously available information. Several internal DNA constructs were prepared and analysed in vitro and in vivo (Fig. 1).
Figure 1.
DNA constructs used for in vitro and in vivo assays. (A) Schematic view of the mouse tyrosinase locus and LCR sequences in mouse chromosome 7. A bent arrow indicates the transcription start site and direction of the mouse tyrosinase gene. Exons (5) are indicated as black rectangles. Not drawn to scale. A 1.7 kb fragment containing mouse tyrosinase exon 3 (TE3) and surrounding intronic sequences is shown below the mouse tyrosinase gene. The 3.7 kb EcoRI fragment containing the mouse tyrosinase LCR, located at –15 kb of the gene is shown below (3711 bp, EMBL Database X76647). Relative position of the 3.7 kb EcoRI DNA fragment with respect to the start of transcription of the tyrosinase gene (–15 kb) has been updated, according to the latest available mouse genome sequence information (see legend for Fig. 5), therefore correcting the position assigned to this LCR fragment in previous reports (14,17). The relative position of restriction enzymes used to subclone internal fragments is indicated along with their size in base pairs (bp). A and B black boxes, referred to previously as HS-1 and HS-2 (14), identify binding sequences for nuclear factors. (B) Three series of backbone plasmids are shown: pTLuc, containing a TATA box from the HSV-TK gene (white box), linked to two sites for the ubiquitous transcription factor Sp1 (white ovals); pTKLuc, containing the promoter of the HSV-TK gene (grey box); and pTyrLuc, carrying the mouse tyrosinase promoter (black box). Upstream of each promoter, the HS and X fragments were cloned in either direction (depicted as two arrows) resulting in the plasmids pHSTLuc, pSHTluc, pXTLuc, pXinvTLuc, pHSTKLuc, pSHTKLuc, pXTKLuc, pXinvTKLuc, pHSTyrLuc, pSHTyrLuc, pXTyrLuc and pXinvTyrLuc, respectively. (C) Plasmids used for functional analysis of deletion mutants within the HS fragment. (D) Constructs used to evaluate boundary activities of mouse tyrosinase LCR sequences in transgenic mice. All constructs shown in (B), (C) and (D) share the firefly luciferase reporter gene and the 3′ splice site and polyadenylation signal from the small t gene of the SV40 genome, indicated as pA. (E) DNA constructs used to evaluate boundary activities of mouse tyrosinase LCR sequences in transgenic flies. ‘w’ represents the reference construct [pCaSpeR3 (51)] containing a white minigene surrounded by 3′P and 5′P sequences from a P element (shown as triangles in a box). BRwBR carries the Su(Hw) BR of gypsy retrotransposon (52), 5′HS4w5′HS4 carries two copies of the 1.2 kb fragment containing the 5′HS4 of the chicken β-globin locus (36), TE3wTE3, HSwHS, XwX, LCRwLCR and LCRmutwLCRmut contain the corresponding mouse tyrosinase LCR DNA fragments.
To evaluate in vitro transcriptional transactivation capacities of mouse tyrosinase LCR sequences in front of heterologous promoters, we made three types of luciferase reporter plasmids. These were driven by a minimal synthetic promoter formed by a TATA box from the HSV-TK gene and two binding sites for the ubiquitous transcription factor Sp1 (48), or the HSV-TK gene promoter (49) (pTLuc and pTKLuc constructs; Fig. 1B). We incorporated the endogenous mouse tyrosinase promoter as a control (pTyrLuc constructs; Fig. 1B), as reported previously (14).
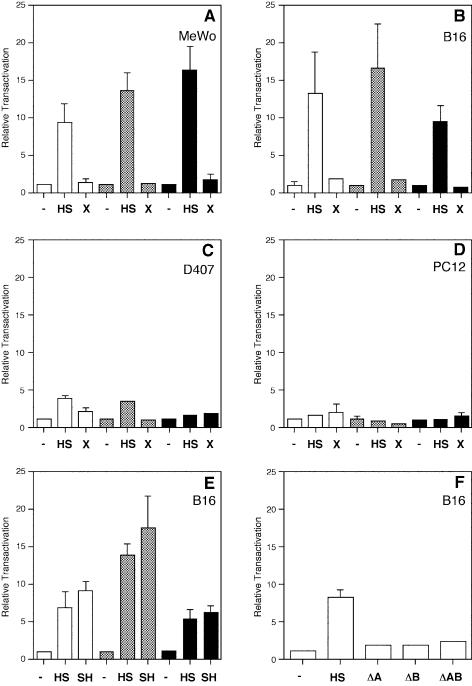
All these constructs were transiently transfected in cell types that potentially express the tyrosinase gene (mouse B16 melanoma, human MeWo melanoma and human D407 RPE cells) and cells that do not express the tyrosinase gene (i.e. rat PC12 phaeochromocytoma cells). Although tyrosinase gene is expressed, in vivo, in RPE cells (8), immortalised RPE cells (such D407) normally lose their capacity to express tyrosinase in most culture conditions and hence, melanin synthesis ceases (54,63).
In the presence of a fragment containing the LCR-core indicated as a putative DNase I tissue-specific HS and two putative binding sites (boxes A and B) for nuclear factors (HS DNA fragment; Fig. 1A) (14), transcriptional enhancement was observed in B16 and MeWo cells, irrespective of the promoter used, and with similar relative transactivation values (Fig. 2A and B). Sequence inspection of A and B boxes had shown the existence of target sequences similar to DNA-binding motifs for the ubiquitous transcription factors CREB (cAMP responsive element binding) and AP-1 (activator protein 1) (14). In D407 cells, transfected constructs with the tyrosinase promoter do not appear to function, in agreement with the endogenous tyrosinase gene being silent (54,63). However, in these cells, heterologous promoters are also capable of being specifically transactivated by the HS DNA fragment, although with lower transactivation values (Fig. 2C), suggesting that cell type-specific trans-acting nuclear factors or tissue-specific modifications of ubiquitous factors that normally bind the HS DNA fragment operate irrespective of the activity of its corresponding tyrosinase promoter.
Figure 2.
Evaluation of the enhancer function of LCR sequences with homologous and heterologous promoters by transient transfection analysis in mammalian cells. Transient transfections of MeWo (A), B16 (B, E and F), D407 (C) and PC12 (D) cells, using HS, SH, X, HSΔA (ΔA), HSΔB (ΔB), HSΔAB (ΔAB) or none (–) LCR-derived fragments in combination with pTLuc (white bars), pTKLuc (grey bars) heterologous promoters and pTyrLuc (black bars) homologous promoter backbone plasmids (Fig. 1). Results are expressed as relative transactivation, arbitrarily assigning to backbone promoter-only plasmids (pTLuc, pTKLuc and pTyrLuc, respectively) the value of x1 and thereafter referring the activity of each construct to its corresponding promoter-only plasmid. In PC12 and D407 cells, tyrosinase promoter constructs produced luciferase reporter expression values close to background. Normalisation of the luciferase reporter values between different transfected constructs is achieved taking into account the activity of a co-transfected lacZ reporter plasmid and the number of pmols of experimental plasmid DNA used in each transfection. See Materials and Methods for plasmid sizes. Relative transactivations are mean values from triplicate experiments (±SD).
In contrast, transcriptional enhancer function of HS was not found in PC12 cells (Fig. 2D). Absence of HS enhancer function was also observed in other cell types tested that do not express the tyrosinase gene such as rat FTO-2B hepatoma, mouse NIH3T3 fibroblasts, mouse L929 fibroblasts, Chinese hamster ovary and human Weri-Rb retinoblastoma cells (not shown), supporting that cell-specific nuclear factors or tissue-specific modifications of ubiquitous factors, presumably binding to box A and box B (14), are required to mediate heterologous transactivation by the HS fragment.
Cell-specific enhancer function associated with the HS fragment was shown to operate irrespective of its orientation with comparable induction values, both in combination with homologous (tyrosinase) and heterologous promoters (Fig. 2E). No significant response was seen with the X fragment (Fig. 1A) with heterologous or homologous promoters (Figs 1B and 2), in either orientation (not shown).
The specific contributions of box A and box B to the enhancer function of the HS fragment was assayed in B16 cells using a heterologous promoter (pTLuc constructs) (Fig. 1C). The presence of both the A and the B boxes is required to activate transcription on heterologous promoters (Fig. 2F), suggesting a pivotal role of both cis-regulatory sequences in the tissue-specific enhancer function of the HS fragment. Similar behaviour was observed previously using homologous promoter sequences (14).
Analysis of LCR function: boundary activity measured as protection from chromosomal position effects in transgenic mice
Previous analyses with standard and artificial-chromosome type constructs in transgenic mice suggested the existence, within the LCR fragment, of a putative boundary activity that could overcome chromosomal position effects. This activity was primarily associated with the tissue-specific enhancer function mapping within the mouse tyrosinase LCR (10,17). The results that we obtained using the HS DNA fragment in combination with heterologous promoters in transiently transfected cells (Fig. 2) prompted us to evaluate these constructs in vivo, in transgenic animals, in an attempt to distinguish between tissue-specific enhancer mediated and boundary effects.
In order to evaluate this boundary function, we analysed whether the HS or the LCR fragments could rescue the weak and variable expression of a reporter transgene driven by a minimal synthetic promoter, prone to position effects (pTLuc, Fig. 1D). In this context, the ‘LCR fragment’ refers to the 3.7 kb EcoRI DNA fragment initially shown to contain a tissue-specific DNase I HS (14), conferring copy number-related expression of tyrosinase minigenes in transgenic mice (14) and later shown to contain a proper LCR activity upon functional analysis of YAC tyrosinase-deletion derivatives in transgenic mice (17). We prepared two additional constructs from pTLuc with the HS (pHSTLuc) or the LCR (pLCRTLuc) fragments cloned 5′ upstream of the minimal promoter (Fig. 1D). We included the putative boundary element (HS or LCR) only at one end (5′) of the constructs because, in standard mouse transgenesis, constructs usually integrate as multicopy tandem arrays. Therefore, most transgene copies would end up shielded by putative boundary elements at either side.
We generated five independent lines of transgenic mice for pTLuc, as well as five for pHSTLuc and eight for pLCRTLuc heterologous constructs. Luciferase activity was measured in a number of organs from adult F1 and F2 individuals of all these transgenic lines. Results from transgenic mouse lines in which luciferase expression could be detected are shown in Figure 3, along with the corresponding transgene copy number for each line. Four out of five pTLuc transgenic mouse lines showed very low but detectable luciferase expression, in at least one tissue (lines 1A to 1D, Fig. 3). In the fifth pTLuc transgenic line (1E, five copies) we could not detect luciferase expression (not shown). These results obtained with pTLuc indicated that this transgene is quite sensitive to chromosomal position effects.
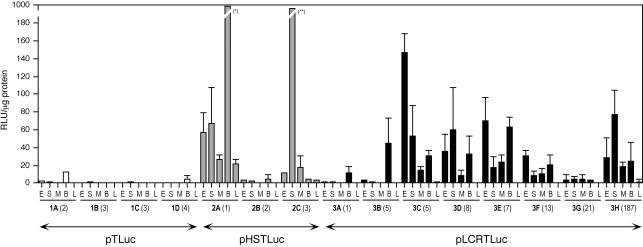
Figure 3.
Analysis of boundary function of mouse tyrosinase LCR sequences in transgenic mice. Analysis of luciferase activity in transgenic mice generated with pTLuc (four lines, 1A to 1D, white bars), pHSTLuc (three lines, 2A to 2C, grey bars) and pLCRTLuc (eight lines, 3A to 3H, black bars), as shown in Figure 1D. Each bar represents the mean value of six adult individuals (±SD) in protein extracts from: eye (E), dorsal skin (S), skeletal muscle (M), brain (B) and liver (L). Transgene copy number is indicated, in parentheses, for each line. Luciferase activity is expressed as relative light units (RLU) per microgram of protein. Values that exceed this scale: (*) 979.52 ± 196.3 and (**) 223.04 ± 154.8 RLU/µg protein.
Three out of five pHSTLuc transgenic mouse lines showed luciferase expression at variable levels. Two pHSTLuc lines (lines 2A and 2C, Fig. 3) showed higher luciferase expression levels than those obtained with pTLuc, whereas a third line (line 2B, Fig. 3) behaved similarly. In the two remaining transgenic pHSTLuc lines (2D and 2E, both single copy) we could not detect luciferase expression (not shown). Indeed, pHSTLuc luciferase expression could be measured in a greater number of tissues, as compared with pTLuc transgenic animals.
Finally, all eight pLCRTLuc transgenic lines showed luciferase expression, at variable levels, thereby rescuing the weak activity of pTLuc reference construct in several tissues (Fig. 3). The highest luciferase levels were measured in brain, eyes, dorsal skin and skeletal muscle protein extracts. The organs in which reporter expression was detected included tyrosinase-expressing tissues (such as eyes and dorsal skin) and tyrosinase-non-expressing tissues (such as brain and skeletal muscle). Other organs analysed showed low reporter expression, such as liver (Fig. 3) or very low reporter expression, such as lung, thin gut, kidney and spleen (not shown). None of these organs (with the exception of eyes and dorsal skin) is known to contain tyrosinase-expressing cells. No correlation was observed between transgene expression levels and number of integrated copies within the host genome with any of the transgenic constructs. Normalisation of transgene expression data to copy number did not result in comparable expression values per copy with any of the transgenic constructs (not shown).
Analysis of LCR function: boundary activity measured as protection from chromosomal position effects in transgenic flies
We decided to extend our analyses of mouse tyrosinase LCR boundary activities, initiated in transgenic mice, using another unrelated species, D.melanogaster, according to previous studies performed with analogous vertebrate sequences (29,36,61). Classically, boundary activities have been analysed in transgenic flies. Therefore, we used the white minigene assay to evaluate whether mouse tyrosinase LCR sequences could insulate the expression of this reporter construct, heavily influenced by chromosomal position effects, in flies (28). The white minigene construct, if efficiently protected from neighbouring chromatin, is expected to generate transgenic flies with a yellow/pale-orange eye colour. However, since P-element-mediated germ-line transformation preferentially targets transgenes to open chromatin regions, at the vicinity of endogenous promoters and/or enhancers (64), unprotected white minigene constructs will usually read these chromosomal position effects and result in transgenic flies with darker eye colour.
Four experimental constructs were prepared carrying the white minigene (51) ‘w’ flanked at both ends by the HS, X, LCR and LCRmut fragments (Fig. 1E). Two additional constructs, BRwBR and 5′HS4w5′HS4, (Fig. 1E) were included as experimental controls, containing previously well-characterised boundary elements from D.melanogaster (BR: Binding Region) of the Su(Hw) protein from the gypsy retrotransposon element (35,52), and vertebrates [5′HS4: 5′ DNase I hypersensitive number 4 from the chicken β-globin locus (36)]. An unrelated DNA fragment from the mouse tyrosinase locus, containing exon 3 and surrounding intronic sequences was included as a negative control (TE3wTE3; Fig. 1E). For this experiment, in contrast to the experimental design devised for transgenic mice, the boundary elements were cloned at either side of the white minigene constructs since we reasoned that standard Drosophila transgenesis, mediated by P-elements, usually proceeds through single-copy integration of constructs (55).
Transgenic flies were obtained for all constructs in a white mutant genetic background. Single-copy lines that could be established and evaluated both in heterozygosity and homozygosity were subsequently chosen for phenotypic analysis (Fig. 4). In general, and as expected, the phenotype of homozygous transgenic flies was stronger than their corresponding heterozygous individuals from which they originate (36). Therefore, we made use of homozygous individuals in order to reveal presence/absence of boundary activities that could not be easily detected in the corresponding heterozygous animals.
Transgenic flies carrying the white minigene ‘w’ reference construct clearly exhibited chromosomal position effects in heterozygosity and homozygosity, resulting in heavily pigmented transgenic lines (bars with high values) and high variability of pigmentation between independent lines (Fig. 4A and B). No position effects were seen in flies carrying the positive control construct BRwBR, resulting in lines with reduced eye pigment (bars with low values) and low variability of pigmentation between independent lines (Fig. 4C and D). The vertebrate positive control, construct 5′HS4w5′HS4, behaved differently. In heterozygous flies nearly all but one line showed weak eye colours, suggesting an effective protection from chromosomal position effects, as reported (36) (Fig. 4E). However, homozygous individuals carrying this construct displayed stronger eye colour. Additionally, there was a greater degree of variation in eye colour between independent transgenic lines (Fig. 4F). The pattern observed with transgenic flies carrying the construct 5′HS4w5′HS4 was similar to that obtained with the flies carrying the TE3wTE3 negative control construct (Fig. 4G and H), which did not show protection from position effects, best seen in homozygous individuals (Fig. 4H).
All four experimental constructs containing different fragments of the mouse tyrosinase LCR sequences: XwX, HSwHS, LCRwLCR and LCRmutwLCRmut showed low eye colour values in heterozygousity (Fig. 4I, K, M and O), although the XwX lines showed the highest variability (Fig. 4I). Corresponding homozygous individuals for HSwHS (Fig. 4L) and LCRwLCR (Fig. 4N) maintained low eye colour values and limited variability, suggesting an effective protection from chromosomal position effects, reproducing a pattern seen in BRwBR transgenic lines (Fig. 4C and D). In contrast, XwX and LCRmutwLCRmut homozygous flies increased their eye colour values and variability (Fig. 4J and P), indicating a reduced protection from chromosomal position effects.
In summary, bona fide boundary activity corresponded to low means and reduced standard deviations of red-pigment content in eyes (seen with BRwBR, HSwHS, LCRwLCR constructs in heterozygosity and homozygosity, and with LCRmutwLCRmut construct in heterozygosity). In contrast, prevalence of chromosomal position effects correlated with higher values for both means and standard devia tions (seen with w, 5′HS4w5′HS4, TE3wTE3, XwX constructs in heterozygosity and homozygosity and with LCRmutwLCRmut construct in homozygosity) (Table 1). Statistical evaluation of all transgenic flies analysed, using the hierarchical log-linear model, supported these observations (see Supplementary Material).
Table 1. Summary of transgenic flies analysed.
| Transgenic Drosophila lines analysed | Heterozygous individuals % red-pigment in eyes (mean ± SD) | Homozygous individuals % red-pigment in eyes (mean ± SD) | |
|---|---|---|---|
| w | 14 | 23.92 ± 37.28 | 54.32 ± 37.59 |
| BRwBR | 11 | 4.54 ± 1.96 | 8.89 ± 1.98 |
| 5′HS4w5′HS4 | 11 | 6.17 ± 8.84 | 28.11 ± 30.48 |
| TE3wTE3 | 13 | 5.70 ± 7.09 | 24.51 ± 28.24 |
| HSwHS | 8 | 2.79 ± 1.37 | 5.53 ± 2.75 |
| XwX | 7 | 6.14 ± 6.00 | 26.77 ± 36.18 |
| LCRwLCR | 12 | 3.32 ± 1.13 | 7.26 ± 6.92 |
| LCRmutwLCRmut | 10 | 4.80 ± 1.67 | 25.71 ± 35.72 |
Percentage of red-pigment in eyes (referred to 100% = wild-type).
Genomic and chromatin structure of DNA sequences surrounding the mouse tyrosinase LCR
The presence of a new boundary activity within the mouse tyrosinase LCR, as deduced from experiments in mice and flies, prompted us to investigate the structure of the DNA sequences surrounding these far-upstream 5′ mouse tyrosinase gene regulatory elements since genomic information was not available in public databases. A bacteriophage clone, λT1, carrying a 14 kb DNA insert including the mouse LCR tyrosinase and neighbouring sequences, was isolated from a mouse genomic library and sequenced (Fig. 5A).
Computer analysis of this genomic sequence indicated an unusually large number of restriction sites cutting in G + C rich sequences (i.e. SmaI, XhoI) and a high content of CpG nucleotide residues located about 1 kb upstream of the 3.7 kb EcoRI LCR fragment. This CG-rich stretch of DNA suggested the presence of a CpG island, an element often associated with promoters (65) (Fig. 5A). Comparisons of this sequence to GenBank Database did not reveal homology with any known gene. However, 5 kb from the 5′ end of this 14 kb sequence exhibited an almost perfect homology (98.4%) to a LINE1 element (GenBank M13002) (66), a member of a retrotransposon-like family of highly repeated DNA sequences (67). This LINE1 element is oriented in opposite direction, with respect to the tyrosinase gene. Additional 3′ sequences were absent in the isolated genomic clone, but this LINE1 element is presumably complete in the mouse genome (Fig. 5A). Further LINE1-related and other repeated DNA sequences were detected within this 14 kb genomic fragment (Fig. 5B).
The overall G + C content of the entire genomic sequence is 37.3%. However, there are several regions with a G + C content exceeding 60%, including the CG-rich stretch of DNA similar to a CpG island located 5′ of the LINE1 element, the A and B boxes, and a short G-rich sequence newly identified at the 5′ end of the LCR (Fig. 5C).
Up to 79 occurrences of CpG dinucleotides were found in the entire genomic sequence. The accumulation of CpG residues was particularly high in the CG-rich stretch of DNA similar to a CpG-island that maps at the 5′ end of the LINE1 element and that contains its promoter sequences (Fig. 5D) (66,67).
We reasoned that the presence of this promoter could interfere with the regulation of tyrosinase gene expression. To test this possibility we evaluated the LINE1 promoter activity in vitro, in transient transfection assays using luciferase reporter constructs. Results obtained suggested a poorly or non-functional promoter function in mouse fibroblasts L929 or B16 melanoma cells (see Supplementary Material).
In addition, we hypothesised that the LINE1 promoter sequence could be methylated in vivo, further impairing promoter function. To analyse this possibility we investigated the methylation status of the CG-rich stretch of DNA similar to a CpG island in vivo, in the tyrosinase locus of B16 and L929 cells, applying bisulphite genomic sequencing protocols (60) (Fig. 5E). All 15 CpG dinucleotides extending over the CG-rich stretch of DNA of the LINE1 element were found methylated in B16 cells. Comparably, all but one CpG dinucleotides were also found methylated in L929 cells, suggesting that this CG-rich stretch of DNA similar to a CpG island was heavily methylated in both expressing and non-expressing tyrosinase cells (Fig. 5E).
The methylation status of the LINE1 5′ adjacent LCR sequences suggested a closed chromatin structure in that area. However, previous analysis had showed DNase I hypersensitivity sites at the LCR-core, indicative of an open chromatin structure (14). Thus, we decided to evaluate the chromatin structure of LCR surrounding sequences. In this regard, we performed DNase I sensitivity assays (61) over the LCR region, in nuclei prepared from B16 and L929 cells, using six Southern blot validated single-copy DNA consecutive probes (E3, A, B, C, D, E5 fragments; Fig. 5A and F). Neighbouring L1 and E6 fragments were excluded from this analysis because they were experimentally shown to contain repeated sequences. Additionally, an unrelated DNA probe (a fragment containing the promoter of the ubiquitous mouse phosphoglycerate kinase gene; PGK, Fig. 5F) was included as a control for the DNase I sensitivity assay. The amount of DNase I required for 50% loss of hybridisation for each probe was estimated from experimental data and compared between the two considered cell types (Fig. 5F). Similar amounts of DNase I were required for both cell types in the PGK control probe, suggesting a comparable chromatin structure of this gene, which is transcribed in these two cells. In contrast, for A, B, C and D LCR-internal probes, there was a much higher difference (up to more than two-times, for probe B) in DNase I sensitivity between both cell types, suggesting that chromatin over the LCR-core (probe D), and its surrounding area (probes A, B and C), was more accessible in B16 cells, where tyrosinase gene is transcribed, than it was in L929 cells, where tyrosinase gene is not expressed. Outside the LCR area, differences in DNase I sensitivity became smaller (probes E3 and E5), suggesting more similar chromatin structures in both cell types.
Analysis of LCR function: evaluation of boundary activities by transient transfection analysis in mammalian cells
The results obtained in vivo (in transgenic mice and flies) and in vitro (methylation pattern of 5′ LCR sequences and chromatin structure over this area) pointed to the existence of an additional boundary element within the mouse tyrosinase LCR. Boundaries can be identified in vivo by barrier or enhancer-blocking assays (24,36,52). However, preliminary information can be derived, in vitro, from transient transfection analysis in mammalian cells, as it has been shown before (37,61).
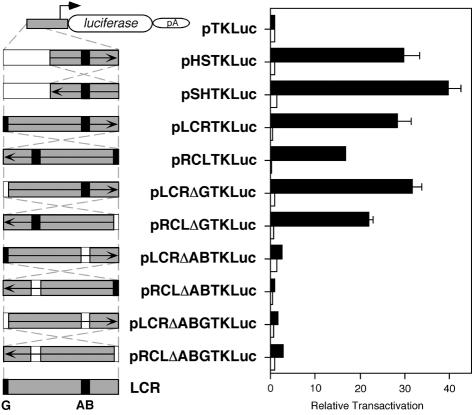
In this regard, we set up to analyse the behaviour of new constructs in transient transfection analysis in B16 and L929 cells. In these constructs, the entire LCR fragment and a series of deletion derivatives were cloned, in either direction, in the reference vector pTKLuc (Fig. 6). In B16 cells, plasmids pHSTKLuc and pLCRTKLuc resulted in comparable transactivation values (P = 0.509). Deletion of the AB boxes completely abolished the enhancer capacity of the entire LCR fragment (shown with pLCRΔABTKLuc and pRCLΔABTKLuc; Fig. 6), as it has been shown before with the HS fragment (Fig. 2F). Inversion of the LCR fragment resulted in a statistical significant ∼41% reduction in transactivation value (shown with pRCLTKLuc as compared to pLCRTKLuc, P = 0.019), further supporting the existence of boundary elements located 5′ from the LCR-core (AB boxes) (Fig. 6). Visual inspection of LCR surrounding sequences had revealed a G-rich stretch at the 5′ end (Fig. 5A and C). We decided to test whether this short sequence could contribute to establish a boundary, according to informatic predictions (TRANSFAC Database) (68), which suggested potential binding of zinc-finger proteins, a family of transcription factors that includes members described to bind to insulators (38,61,69,70). Upon deletion of the G-rich short sequence, construct pLCRΔGTKLuc displayed comparable transactivation value as that of its reference pLCRTKLuc (P = 0.393). In contrast, inversion of the LCR fragment carrying the deletion of the G-rich sequence (construct pRCLΔGTKLuc) showed an intermediate transactivation value, both statistically significant with respect to pLCRTKLuc (P = 0.042) and pRCLTKLuc (P = 0.006). These results suggested that this G-rich sequence could contribute, along with other neighbouring sequences, to the boundary effect observed within the 5′ LCR region. The inversion of the HS fragment resulted in different transactivation values (∼25%), statistically significant between pHSTKLuc and pSHTKLuc (P = 0.010), suggesting the putative existence of additional, although minor, regulatory elements yet to be characterised. Similar trend had been observed before with these two constructs (Fig. 2E), although the higher variability obtained in that experiment prevented statistical significance of those observed differences. Finally, the combined deletion of the G-rich sequence and the AB boxes in pLCRΔABGTKLuc and pRCLΔABGTKLuc constructs behaved similarly as the deletion of AB boxes alone (Fig. 6). All these constructs were also transfected in L929 cells, as a negative control, resulting, as expected, in the absence of transactivation.
Figure 6.
Evaluation of boundary activities associated with tyrosinase LCR sequences by transient transfection analysis in mammalian cells. Transient transfection of backbone promoter-only plasmid pTKLuc, and its derivative constructs: pHSTKLuc, pSHTKLuc, pLCRTKLuc, pRCLTKLuc, pLCRΔGTKLuc, pRCLΔGTKLuc, pLCRΔABTKLuc, pRCLΔABTKLuc, pLCRΔABGTKLuc, pRCLΔABGTKLuc in B16 (black bars) and L929 (white bars) cells. Schemes of these plasmids (left) and the result of transfection in B16 and L929 cells (right) are shown. The AB enhancer-core and G-rich sequences are depicted as black boxes. Grey or white boxes correspond to the DNA fragments present or absent, respectively, in each construct. The arrow depicted inside each DNA fragment represent the 5′ to 3′ direction, with respect to the position of these sequences within the endogenous tyrosinase LCR and gene. Crossed and dashed lines indicate the inversion of the same DNA fragment in their corresponding experimental constructs. Results are expressed as relative transactivation, arbitrarily assigning to the backbone plasmid pTKLuc the value of x1 and thereafter referring the activity of each experimental construct to that value. Normalisation of the luciferase reporter values between different transfected constructs is achieved taking into account the activity of a co-transfected lacZ reporter plasmid and the number of pmols of experimental plasmid DNA used in each transfection. See Materials and Methods for plasmid sizes. Relative transactivations are mean values from triplicate experiments (±SD).
DISCUSSION
The mouse tyrosinase gene is associated with an LCR element, located 15 kb from its promoter (17). To gain insight into the activities present within this LCR, we have dissected and studied enhancer and boundary functions in vitro and in vivo.
An internal fragment of the mouse tyrosinase LCR incorporating the DNase I HS core (HS fragment) is able to transactivate heterologous promoters in a cell-specific and orientation-independent manner. The enhancer function of the HS fragment was observed in all cell types analysed that primarily express tyrosinase (melanocytes and RPE cells; Fig. 2). Additionally, this enhancer function required the presence of both boxes A and B, target sites for nuclear factors that appear to bind members of the ubiquitous transcription factors AP-1 and CREB protein families (14). The fact that a cell-specific enhancer (associated with a cell-specific DNase I HS) could be bound, apparently, by ubiquitous transcription factors can be explained by considering the existence of tissue-specific modifications of those ubiquitous transcription factors, as discussed before (14). Our results extended previous analyses of HS done in combination with the endogenous tyrosinase promoter (14), suggesting the usefulness of tyrosinase enhancer modules with heterologous promoters in gene transfer experiments. Similar results had been obtained with the LCR element from the human β-globin locus (71).
The relevance of A and B boxes in LCR function has been studied in mice (50), using YAC-tyrosinase constructs with analogous deletion-type experiments as those reported here in transiently transfected cells. Transgenic mice obtained with tyrosinase YACs carrying deletions in either box A or B, or both, display more pigmentation than YAC-tyrosinase transgenic animals in which LCR had been deleted (10; P.Giraldo, A.Lavado, M.Nadal and L.Montoliu, unpublished work). These observations, the opposite of what we described in vitro, prompted us to investigate the existence of additional regulatory elements within the LCR regulatory region that could compensate in vivo for the absence of A and B sequences and contribute to establish a proper boundary for this expression domain. In this regard, we evaluated whether LCR tyrosinase sequences could counteract chromosomal position effects on heterologous reporter constructs assayed in transgenic animals, as one of the tests classically described to assay for boundaries.
In transgenic mice, the addition of the entire mouse tyrosinase LCR fragment (pLCRTLuc) to a poorly expressed reporter construct prone to position effects (pTLuc) resulted in elevated luciferase expression, at variable levels, in a variety of organs containing or not tyrosinase-expressing cells, in all lines (eight) analysed (Fig. 3). The smaller HS fragment enhanced but did not guarantee expression of this reference transgenic construct, as two out of five pHSTLuc lines, carrying single-copy transgenes, were silent. In this case, we reasoned that the absence of LCR fragments inserted 3′ of the TLuc reporter construct could be a possible explanation for the observed results. The other three lines showed variable expression levels but in a larger number of organs, as compared with pTLuc animals (Fig. 3). Results with pLCRTLuc mice enhanced and extended the pHSTLuc expression pattern due to the presence of tissue-specific enhancers (AB boxes) and other boundary elements, presumably located outside the HS region, at the 5′ end of the LCR fragment, that could be responsible for the expression rescue of the reporter transgene, although at variable levels, in organs which do not contain tyrosinase-expressing cells (i.e. brain). These analyses extended previous observations in transgenic mice, using the LCR fragment with its endogenous tyrosinase promoter, in which only tissue-specific enhancer copy number-related effects were recorded (14). In addition, new experiments are being undertaken to further evaluate the boundary function of these sequences in vivo in combination with other type of reporter constructs with a clearer tissue-specific expression pattern, in contrast to the pattern observed with pTLuc, which may not be expressed because of the weakness of the promoter and/or the possibility that pTLuc transgenes did not integrate nearby active enhancers.
There was no correlation between the level of the pHSTLuc or pLCRTLuc transgene expression and their copy number. We did not fully reproduce previous observations made when the LCR fragment was fused to 5.5 kb of tyrosinase promoter sequences in tyrosinase minigene constructs analysed in transgenic mice (14), where copy number-related expression was reported, suggesting that the behaviour of the LCR fragment might be different in homologous (tyrosinase minigene constructs) or heterologous (the pTLuc constructs analysed here) environments. In fact, our results would suggest that full LCR activity could only be achieved in a ‘proper’ genomic context, as in the case of YAC-tyrosinase transgenes (11,17), where other regulatory elements are present. Among them, the 5.5 kb proximal tyrosinase promoter sequences [including binding sites for microphthalmia (Mitf), a key transcriptional regulator of the tyrosinase locus (72,73)] that were also present in previous tyrosinase minigene constructs (14).
The same lack of correlation has been reported using boundary elements from other expression domains in combination with heterologous promoters (43,44 but see updated results in 42,74–76). However, the inclusion of boundary elements in gene transfer experiments, such as those reported here, normally increases the probability of expression of transgenes and therefore their use in heterologous constructs is recognised as potentially beneficial (5,7,12,77,78). Indeed, the 5′HS4 boundary element greatly enhances the number of mouse lines expressing the transgenes, irrespectively of the promoter, but it is definitely unable to prevent totally variegation and it does not allow copy number-dependent expression (Louis-Marie Houdebine, personal communication). Similarly, with our tyrosinase LCR elements, efficient protection from chromosomal position effects could not be revealed in transgenic mice, since copy number-dependent and position-independent expression of a poorly expressed reporter construct was not guaranteed. Instead, the elevated general expression observed pointed to the existence of boundary elements within the 5′ upstream regulatory region of the tyrosinase gene that would possibly operate in a tissue-unrelated manner. In addition, the lack of copy number-dependent expression can also be explained, in transgenic lines with high copy numbers as a potential result of heterochromatin-mediated silencing induced by transgene repeats, as initially reported in Drosophila (79). Copy number-dependent expression is a property usually associated with the presence of functional LCR elements (17,19–21) that may not necessarily be associated with functional boundary elements. Indeed, complex higher-order regulatory elements, such as possibly the mouse tyrosinase LCR genomic region, shown to contain a functional LCR (11,14,17) plus additional putative boundary elements (this manuscript), suggest that several activities from multiple composite elements may co-exist and contribute to specify a given eukaryotic expression domain.
To confirm and extend our barrier function analyses of mouse tyrosinase LCR sequences we decided to perform these assays in a more suitable but heterologous species, D.melanogaster, where boundary activities have been classically reported (29,36,61) (Fig. 4).
In transgenic flies, tyrosinase LCR-derived sequences (constructs HSwHS and LCRwLCR) efficiently protected a white minigene construct from chromosomal position effects, both in heterozygous and homozygous animals, to a degree that compared favourably with a well-established D.melanogaster insulator (BR), from the gypsy retrotransposon element (35,52). A similar behaviour was observed with the LCRmutwLCRmut construct, in heterozygous animals. Furthermore, in these experiments, mouse tyrosinase LCR-derived sequences displayed better insulating capacities over chromosomal position effects compared with the reference vertebrate insulator (5′HS4), that behaved differently from what had been reported previously with fewer number of lines analysed and two copies of the 5′HS4 insulator element at either side of white minigene constructs (36). The boundary effect of mouse tyrosinase LCR sequences observed in flies appeared to correlate primarily with its LCR-core, since XwX (in heterozygous and homozygous flies) and LCRmutwLCRmut (in homozygous flies) constructs did not reproduce the pattern observed with HSwHS and LCRwLCR constructs. In flies, the LCR-core (HS fragment) appears to be necessary and sufficient for boundary activity, whereas in mice, the same HS fragment seems to be necessary but not sufficient. This suggests that, in mice, sequences 5′ to the LCR-core also contribute to barrier function. A number of reasons could account for the different behaviour of the LCR fragment in mice and flies, including that proteins associated to 5′ LCR sequences in mice are not functional or even not present in flies. Furthermore, we cannot rule out that there is a fundamental difference in boundary activity in Drosophila as compared with mice, as indicated by differences in the behaviour of 5′HS4w5′HS4, TE3wTE3 and LCRmutwLCRmut constructs in heterozygosity and homozygosity. Other phenomena, such as transvection (80), could also be influencing the results obtained in homozygous individuals, as discussed before for the analysis of the 5′HS4 boundary element of the chicken β-globin locus in transgenic flies (36).
In this study, we used TE3wTE3 as a negative control, instead of using prokaryotic DNA (28,36,57), reasoning that unrelated sequences from the same locus would better control for unspecific barrier activity, as reported previously (29). Indeed, the TE3wTE3 construct behaved similarly as ‘w’ and 5′HS4w5′HS4 constructs in heterozygous and homozygous individuals, supporting the absence of insulating activities in this negative control (Fig. 4, Table 1 and Supplementary Material).
The presence of boundary activities within the mouse tyrosinase LCR had been indirectly addressed during the analysis of standard-type tyrosinase transgenic mice. Indeed, the addition of the entire LCR fragment enhanced the expression of minigene tyrosinase constructs in transgenic mice (14,16). Porter and colleagues demonstrated, in some lines, increased tyrosinase expression by adding the 5′ end of the LCR (15) or the LCR-core alone (16), also indicating the existence of additional regulatory elements in that area. Those results were explained as a product of S/MAR activities, identified previously within the LCR (13). However, we have failed to confirm significant S/MAR activity in the LCR using in vitro comparable experiments and sequence-based S/MAR predictions (not shown). This would not be the first case in which a LCR has been found to be devoid of S/MAR activities (81). Indeed, the implication of enhancers, such as the LCR-core, in the establishment of boundaries, acting as chromatin openers has been proposed as an alternative model to understand insulator function (45). Also, classical insulators have been reported recently to function as transcriptional stimulators (24) and, on the other hand, transcriptional activators have also recently been shown to insulate active loci against chromatin repression (82). In this regard, our data would support the notion that strong enhancers might have been recruited during evolution to contribute to establish boundaries.
Our experiments in flies suggested that most of the boundary activity present within the mouse tyrosinase LCR was associated with its core enhancer (HS fragment). However, we decided to investigate the possible existence of additional boundary elements, outside the enhancer, that could explain why the entire LCR fragment behaved differently than the HS fragment in transgenic mice. In this regard, we sequenced 14 kb of genomic DNA surrounding the mouse tyrosinase LCR, thereby completing 5′ upstream LCR sequences that were absent in public databases. The analysis of this genomic sequence identified an apparently entire LINE1 element, containing a CG-rich stretch of DNA similar to a CpG island, located upstream of the LCR fragment, and oriented in opposite direction with respect to the tyrosinase gene (Fig. 5A). In fact, the presence of LINE1-derived sequences appears to be a characteristic feature of the mouse tyrosinase locus (L.Regales, P.Giraldo, A.Lavado and L.Montoliu, in preparation), and it might have played a role in the generation of some tyrosinase mutant alleles (such as chinchilla-mottled mice, Tyrc–m) (13).
LINE1 sequences are retrotransposon-related repeated DNA elements, 6–7 kb long, carrying two open-reading frames and found in high numbers (∼104–105) in the mouse genome (67,83). Most of these elements (>90%) appear to be truncated in its 5′ end, thereby resulting in inactive copies. LINE1 elements have been associated with the regulation of several mammalian loci and have been proposed to contribute to genomic evolution and mutation (83). In this sense, the transcriptional abilities of LINE1 elements have been correlated with their capacity to induce epigenetic phenomena, due to their potential to interfere with normal transcription of neighbouring genes (84,85).
We investigated whether the LINE1 element located 5′ upstream of the mouse tyrosinase LCR was functional and if it could influence tyrosinase expression. Our findings in vitro indicated that the promoter of this LINE1 element (a CG-rich stretch of DNA similar to a CpG-island) is practically silent in transfection assays (see Supplementary Material). Indeed, we have found that this CG-rich stretch of DNA similar to a CpG island is heavily methylated in vivo, in these two cell types (Fig. 5E). It has been proposed that DNA methylation at promoters of repeated elements could represent a defence mechanism to inactivate their transcription and prevent their mobilisation to ectopic genomic locations (86). Further, DNA methylation has been associated with histone deacetylation (87), and corresponding chromatin condensation (88), that can in turn result in the silencing of transcription (89). Thus, the presence of a heavily methylated LINE1 element next to the tyrosinase LCR could represent a potential danger if the spreading of DNA condensation would reach the LCR-core.
We have shown experimentally, using DNase I sensitivity assays (61), that chromatin surrounding the LCR region is more accessible to DNase I digestion in B16 cells, as compared with L929 cells (Fig. 5F). Differences in DNase I sensitivity extended well beyond the LCR-core in its 5′ region, up to the LINE1 element, suggesting that these sequences upstream of the LCR-core could contribute in establishing a boundary. Therefore, our data would support the existence of a regulatory element between the CG-rich stretch of DNA similar to a CpG-island of the LINE1 element and the LCR-core, with properties of a barrier, capable of preventing potential interferences of upstream genomic sequences with LCR function. Similar barrier functions have been associated with the 5′HS4 insulator from the chicken β-globin locus, coupling high histone acetylation levels and protecting DNA from methylation (90).
To reveal the presence of such additional boundary elements in the 5′ LCR region we decided to evaluate its potential enhancer-blocking capacity, in vitro, using transient transfection into mammalian cells, as reported before (37,61). These analyses first showed that the entire LCR and the HS fragments displayed comparable enhancer activities on a heterologous reporter construct (pTKLuc; Fig. 6). This enhancer activity was mostly dependent on the presence of the AB boxes (Figs 2F and 6). Upon inversion of the LCR fragment, the relative transactivation decreased in a significant manner. This finding suggested the existence of an additional regulatory element, normally located 5′ of the LCR-core, which upon inversion and placement between the enhancer and the promoter, could interfere with the transactivation activity. Sequence inspection of the region between the LINE1 element and the LCR-core highlighted a short G-rich sequence, at the 5′ end of LCR (Fig. 5A and C). Informatic predictions suggested that this sequence could bind transcription factors belonging to the family of zinc-finger nuclear proteins (68). A member of this family, the multifunctional nuclear protein CTCF has been shown to bind to a number of insulator sequences (38,61,69), without an apparent consensus in the target site other than the presence of several G-residues (70). We will analyse the potential association of zinc-finger nuclear proteins with the short G-rich sequence found at the 5′ end of LCR.
To evaluate the putative role of this short G-rich region we prepared several deletion constructs derived from the entire LCR fragment. As expected, the LCR fragment carrying a deletion of the G-rich sequence (LCRΔG), in its natural orientation, retained the enhancer capacity of the entire fragment (Fig. 6). The inversion of this fragment (RCLΔG) resulted again in a decreased enhancer capacity, as compared with LCR and LCRΔG. However, this decrease was different and statistically significant from the inversion of the entire LCR (RCL construct), thus suggesting that both this G-rich region plus other neighbouring sequences within the 5′ region might be required for the observed enhancer-blocking effects. The results obtained with the HS fragment, in its natural and inverted direction (Fig. 6), do not rule out the existence of additional elements within this fragment that will be studied in subsequent experiments. Formally, we cannot exclude an effect of the small differences in distance between the LCR-core and the promoter in these reporter constructs (1 kb in LCR and 2.7 kb in RCL). However, we think that this effect, if present, would not have a major significance since these regulatory elements have been shown to operate in vivo at different distances (from ∼2 to ∼15 kb) with comparable efficiencies (14,16,17), and the relative transactivation of the LCR-core (AB-boxes) to heterologous promoters is comparable with that of the LCR or the HS fragments (not shown). In addition, at present, we cannot exclude that the enhancer function mapping at the AB boxes could not transactivate the heterologous promoter ‘around the clock’, following the circular DNA molecule transiently transfected. Indeed, this might be a potential explanation for the limited enhancer-blocking activity observed upon inverting the entire LCR fragment. Moreover, conventional repressor activity could also be responsible for the observed effects in transiently transfected cells. The comparison of relative transactivaction capacities of pRCLΔABTKLuc and pRCLΔABGTKLuc constructs (Fig. 6) could either be interpreted as a result of boundary or repressor activities mapping at the G-rich sequence. Forthcoming analyses will address the enhancer-blocking capacities of elements found within the mouse tyrosinase LCR in vivo, in transgenic flies and mice, but also in mammalian cells, where different type of constructs will be prepared to discriminate between repressor and enhancer-blocking activities.
Taken together, our results indicate the presence of a putative new boundary activity in the mouse tyrosinase LCR. Several elements can participate in this putative boundary activity including known (i.e. the LCR-core or enhancer) and newly described (i.e. the G-rich region and, possibly, other sequences located 5′ of the LCR-core) sequences. Future experiments will establish the relative contribution of these different elements in the observed barrier and enhancer-blocking effects, as it has been described recently for the chicken β-globin insulator (40).
In this work, we have analysed the putative boundary properties of the 5′ LCR element at either side of reporter transgenic constructs, both in mice and flies. However, this element is only found at the 5′ end of the mouse tyrosinase gene in its native genomic location. Forthcoming analyses will also investigate the presence of analogous sequences, sharing similar boundary properties as those found at the 5′ region of the LCR, in the 3′ region of the mouse tyrosinase gene, which could potentially contribute to specify the mouse tyrosinase expression domain.
The systematic description of new boundary elements in different expression domains, such the tyrosinase locus, is required to extend our still limited knowledge regarding the function and mode-of-action of these sequences and how they could contribute to insulate contiguous transcription units with different expression patterns. However, the unexpected discovery of ubiquitously expressed genes within previously well-established tissue-specific expression domains, reported in the case of the human α-globin (91), the human growth hormone (92,93) and the chicken lysozyme (94) loci, would argue that differentially expressed transcription units might coexist within the same functional chromatin domain. In this sense, it will be instrumental to study and reveal the in vivo role of potential insulators/boundaries before attempting to export their blocker/barrier abilities to ectopic locations in gene transfer experiments. Failure to fully understand these elements may be responsible for the limited success of boundaries usually observed in transgenic experiments.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by funds from Spanish Ministry of Science and Technology (SMST) Bio97-0628, Bio2000-1653, FEDER 2FD1997-2059, and Laboratorios Dr Esteve S.A. to L.M., and funds from SMST PB97-1207, BMC2001-2178 and Comunidad Autónoma de Madrid (CAM) 08.9/0002/98 to A.B. P.G. and L.R. are recipients of PhD fellowships from SMST. A.L. and A.G.D. are recipients from PhD fellowships from CAM. The authors are grateful to P. Geyer for providing pCaSpeR3, pCaSpeRW15 and BRwBR plasmids, R. Hunt for providing D407 cells, S. Montalbán and P. Cozar for assistance with mice, F. Bejarana, A. Parrón and I. González for assistance with flies, M. Cuadrado and F. Antequera for assistance with methylation assays, L. Barrios and C. Manuel for support with statistical analysis, and G. Schütz, J. R. Naranjo, J. J. Sánchez-Serrano, G. Jeffery, K. Harshman, V. G. Corcés, A. West, G. Felsenfeld and L. M. Houdebine for useful comments.
DDBJ/EMBL/GenBank accession nos X76647, AF364302
REFERENCES
- 1.Dillon N. and Grosveld,F. (1994) Chromatin domains as potential units of eukaryotic gene function. Curr. Opin. Genet. Dev., 4, 260–264. [DOI] [PubMed] [Google Scholar]
- 2.Elgin S.C. (1990) Chromatin structure and gene activity. Curr. Opin. Cell. Biol., 2, 437–445. [DOI] [PubMed] [Google Scholar]
- 3.Bell A.C., West,A.G. and Felsenfeld,G. (2001) Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science, 291, 447–450. [DOI] [PubMed] [Google Scholar]
- 4.Laemmli U.K., Kas,E., Poljak,L. and Adachi,Y. (1992) Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev., 2, 275–285. [DOI] [PubMed] [Google Scholar]
- 5.West A.G., Gaszner,M. and Felsenfeld,G. (2002) Insulators: many functions, many mechanisms. Genes Dev., 16, 271–288. [DOI] [PubMed] [Google Scholar]
- 6.Bonifer C. (2000) Developmental regulation of eukaryotic gene loci: which cis-regulatory information is required? Trends Genet., 16, 310–315. [DOI] [PubMed] [Google Scholar]
- 7.Montoliu L. (2002) Gene transfer strategies in animal transgenesis. Clon. Stem Cells, 4, 39–46. [DOI] [PubMed] [Google Scholar]
- 8.Beermann F., Schmid,E. and Schutz,G. (1992) Expression of the mouse tyrosinase gene during embryonic development: recapitulation of the temporal regulation in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 2809–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beermann F., Ruppert,S., Hummler,E., Bosch,F.X., Muller,G., Ruther,U. and Schutz,G. (1990) Rescue of the albino phenotype by introduction of a functional tyrosinase gene into mice. EMBO J., 9, 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraldo P. and Montoliu,L. (2002) Artificial chromosome transgenesis in pigmentary research. Pigment. Cell. Res., 15, 258–264. [DOI] [PubMed] [Google Scholar]
- 11.Schedl A., Montoliu,L., Kelsey,G. and Schutz,G. (1993) A yeast artificial chromosome covering the tyrosinase gene confers copy number-dependent expression in transgenic mice. Nature, 362, 258–261. [DOI] [PubMed] [Google Scholar]
- 12.Giraldo P. and Montoliu,L. (2001) Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res., 10, 83–103. [DOI] [PubMed] [Google Scholar]
- 13.Porter S., Larue,L. and Mintz,B. (1991) Mosaicism of tyrosinase-locus transcription and chromatin structure in dark vs. light melanocyte clones of homozygous chinchilla-mottled mice. Dev. Genet., 12, 393–402. [DOI] [PubMed] [Google Scholar]
- 14.Ganss R., Montoliu,L., Monaghan,A.P. and Schutz,G. (1994) A cell-specific enhancer far upstream of the mouse tyrosinase gene confers high level and copy number-related expression in transgenic mice. EMBO J., 13, 3083–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter S.D., Hu,J. and Gilks,C.B. (1999) Distal upstream tyrosinase S/MAR-containing sequence has regulatory properties specific to subsets of melanocytes. Dev. Genet., 25, 40–48. [DOI] [PubMed] [Google Scholar]
- 16.Porter S.D. and Meyer,C.J. (1994) A distal tyrosinase upstream element stimulates gene expression in neural-crest-derived melanocytes of transgenic mice: position-independent and mosaic expression. Development, 120, 2103–2111. [DOI] [PubMed] [Google Scholar]
- 17.Montoliu L., Umland,T. and Schutz,G. (1996) A locus control region at –12 kb of the tyrosinase gene. EMBO J., 15, 6026–6034. [PMC free article] [PubMed] [Google Scholar]
- 18.Gimenez E., Giraldo,P., Jeffery,G. and Montoliu,L. (2001) Variegated expression and delayed retinal pigmentation during development in transgenic mice with a deletion in the locus control region of the tyrosinase gene. Genesis, 30, 21–25. [DOI] [PubMed] [Google Scholar]
- 19.Grosveld F. (1999) Activation by locus control regions? Curr. Opin. Genet. Dev., 9, 152–157. [DOI] [PubMed] [Google Scholar]
- 20.Li Q., Harju,S. and Peterson,K.R. (1999) Locus control regions: coming of age at a decade plus. Trends Genet., 15, 403–408. [DOI] [PubMed] [Google Scholar]
- 21.Grosveld F., van Assendelft,G.B., Greaves,D.R. and Kollias,G. (1987) Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell, 51, 975–985. [DOI] [PubMed] [Google Scholar]
- 22.Bender M.A., Bulger,M., Close,J. and Groudine,M. (2000) Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell, 5, 387–393. [DOI] [PubMed] [Google Scholar]
- 23.Reik A., Telling,A., Zitnik,G., Cimbora,D., Epner,E. and Groudine,M. (1998) The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol., 18, 5992–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei W. and Brennan,M.D. (2001) The gypsy insulator can act as a promoter-specific transcriptional stimulator. Mol. Cell. Biol., 21, 7714–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrador M. and Corces,V.G. (2002) Setting the boundaries of chromatin domains and nuclear organization. Cell, 111, 151–154. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn E.J. and Geyer,P.K. (2003) Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell. Biol., 15, 259–265. [DOI] [PubMed] [Google Scholar]
- 27.Wilson C., Bellen,H.J. and Gehring,W.J. (1990) Position effects on eukaryotic gene expression. Annu. Rev. Cell. Biol., 6, 679–714. [DOI] [PubMed] [Google Scholar]
- 28.Kellum R. and Schedl,P. (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell, 64, 941–950. [DOI] [PubMed] [Google Scholar]
- 29.Namciu S.J., Blochlinger,K.B. and Fournier,R.E. (1998) Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol., 18, 2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalos M. and Fournier,R.E. (1995) Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol. Cell. Biol., 15, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun F.L. and Elgin,S.C. (1999). Putting boundaries on silence. Cell, 99, 459–462. [DOI] [PubMed] [Google Scholar]
- 32.Kellum R. and Schedl,P. (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol., 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holdridge C. and Dorsett,D. (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol., 11, 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jack J., Dorsett,D., Delotto,Y. and Liu,S. (1991) Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development, 113, 735–747. [DOI] [PubMed] [Google Scholar]
- 35.Geyer P.K. and Corces,V.G. (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev., 6, 1865–1873. [DOI] [PubMed] [Google Scholar]
- 36.Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- 37.Recillas-Targa F., Bell,A.C. and Felsenfeld,G. (1999) Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc. Natl Acad. Sci. USA, 96, 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–396. [DOI] [PubMed] [Google Scholar]
- 39.Prioleau M.N., Nony,P., Simpson,M. and Felsenfeld,G. (1999) An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J., 18, 4035–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recillas-Targa F., Pikaart,M.J., Burgess-Beusse,B., Bell,A.C., Litt,M.D., West,A.G., Gaszner,M. and Felsenfeld,G. (2002) Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl Acad. Sci. USA, 99, 6883–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farrell C.M., West,A.G. and Felsenfeld,G. (2002) Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol., 22, 3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pantano T., Jolivet,G., Prince,S., Menck-Le Bourhis,C., Maeder,C., Viglietta,C., Rival,S. and Houdebine,L.M. (2002) Effect of the rabbit alphas1-casein gene distal enhancer on the expression of a reporter gene in vitro and in vivo. Biochem. Biophys. Res. Commun., 290, 53–61. [DOI] [PubMed] [Google Scholar]
- 43.Potts W., Tucker,D., Wood,H. and Martin,C. (2000) Chicken beta-globin 5′HS4 insulators function to reduce variability in transgenic founder mice. Biochem. Biophys. Res. Commun., 273, 1015–1018. [DOI] [PubMed] [Google Scholar]
- 44.Taboit-Dameron F., Malassagne,B., Viglietta,C., Puissant,C., Leroux-Coyau,M., Chereau,C., Attal,J., Weill,B. and Houdebine,L.M. (1999) Association of the 5′HS4 sequence of the chicken beta-globin locus control region with human EF1 alpha gene promoter induces ubiquitous and high expression of human CD55 and CD59 cDNAs in transgenic rabbits. Transgenic Res., 8, 223–235. [DOI] [PubMed] [Google Scholar]
- 45.Dillon N. and Sabbattini,P. (2000) Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays, 22, 657–665. [DOI] [PubMed] [Google Scholar]
- 46.Gerasimova T.I. and Corces,V.G. (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet., 35, 193–208. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez J.F., Rodriguez,D., Rodriguez,J.R., McGowan,E.B. and Esteban,M. (1988) Expression of the firefly luciferase gene in vaccinia virus: a highly sensitive gene marker to follow virus dissemination in tissues of infected animals. Proc. Natl Acad. Sci. USA, 85, 1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montoliu L., Blendy,J.A., Cole,T.J. and Schutz,G. (1995) Analysis of perinatal gene expression: hormone response elements mediate activation of a lacZ reporter gene in liver of transgenic mice. Proc. Natl Acad. Sci. USA, 92, 4244–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luckow B. and Schutz,G. (1987) CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res., 15, 5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giraldo P., Gimenez,E. and Montoliu,L. (1999) The use of yeast artificial chromosomes in transgenic animals: expression studies of the tyrosinase gene in transgenic mice. Genet. Anal., 15, 175–178. [DOI] [PubMed] [Google Scholar]
- 51.Pirrota V. (1988) In Rodrigues,R.L. and Denhardt,D.T. (eds), Vectors: A Survey of Molecular Cloning Vectors and their Uses. Butterworth, Boston, MA. [Google Scholar]
- 52.Roseman R.R., Pirrotta,V. and Geyer,P.K. (1993) The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J., 12, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruppert S., Muller,G., Kwon,B. and Schutz,G. (1988) Multiple transcripts of the mouse tyrosinase gene are generated by alternative splicing. EMBO J., 7, 2715–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis A.A., Bernstein,P.S., Bok,D., Turner,J., Nachtigal,M. and Hunt,R.C. (1995) A human retinal pigment epithelial cell line that retains epithelial characteristics after prolonged culture. Invest. Ophthalmol. Vis. Sci., 36, 955–964. [PubMed] [Google Scholar]
- 55.Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- 56.Lindsley D.L. and Zimm,G.G. (1992) The Genome of Drosophila melanogaster. Academic Press, Inc., San Diego, CA. [Google Scholar]
- 57.Vazquez J. and Schedl,P. (1994) Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J., 13, 5984–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hogan B., Beddington,R., Constantini,F. and Lacy,E. (1994) Manipulating the Mouse Embryo. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 59.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1999) Current Protocols in Molecular Biology. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 60.Cuadrado M., Sacristan,M. and Antequera,F. (2001) Species-specific organization of CpG island promoters at mammalian homologous genes. EMBO Rep., 2, 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antes T.J., Namciu,S.J., Fournier,R.E. and Levy-Wilson,B. (2001) The 5′ boundary of the human apolipoprotein B chromatin domain in intestinal cells. Biochemistry, 40, 6731–6742. [DOI] [PubMed] [Google Scholar]
- 62.Whitelaw C.B., Harris,S., McClenaghan,M., Simons,J.P. and Clark,A.J. (1992) Position-independent expression of the ovine beta-lactoglobulin gene in transgenic mice. Biochem. J., 286, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abul-Hassan K., Walmsley,R., Tombran-Tink,J. and Boulton,M. (2000) Regulation of tyrosinase expression and activity in cultured human retinal pigment epithelial cells. Pigment Cell Res., 13, 436–441. [DOI] [PubMed] [Google Scholar]
- 64.Bownes M. (1990) Preferential insertion of P elements into genes expressed in the germ-line of Drosophila melanogaster. Mol. Gen. Genet., 222, 457–460. [DOI] [PubMed] [Google Scholar]
- 65.Antequera F. and Bird,A. (1999) CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol., 9, R661–667. [DOI] [PubMed] [Google Scholar]
- 66.Loeb D.D., Padgett,R.W., Hardies,S.C., Shehee,W.R., Comer,M.B., Edgell,M.H. and Hutchison,C.A. (1986) The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol. Cell. Biol., 6, 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hastie N.D. (1996) Highly repeated DNA families in the genome of Mus musculus. In Lyon,M.F., Rastan,S. and Brown,S.D.M. (eds), Genetic Variants and Strains of the Laboratory Mouse, 3rd Edn. Oxford University Press, Oxford, UK, Vol. 2, pp. 1425–1442. [Google Scholar]
- 68.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Pruss,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bell A.C. and Felsenfeld,G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature, 405, 482–485. [DOI] [PubMed] [Google Scholar]
- 70.Ohlsson R., Renkawitz,R. and Lobanenkov,V. (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet., 17, 520–527. [DOI] [PubMed] [Google Scholar]
- 71.Blom van Assendelft G., Hanscombe,O., Grosveld,F. and Greaves,D.R. (1989) The beta-globin dominant control region activates homologous and heterologous promoters in a tissue-specific manner. Cell, 56, 969–977. [DOI] [PubMed] [Google Scholar]
- 72.Ganss R., Schutz,G. and Beermann,F. (1994) The mouse tyrosinase gene. Promoter modulation by positive and negative regulatory elements. J. Biol. Chem., 269, 29808–29816. [PubMed] [Google Scholar]
- 73.Goding C.R. (2000) Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev., 14, 1712–1728. [PubMed] [Google Scholar]
- 74.Guy L.G., Kothary,R., DeRepentigny,Y., Delvoye,N., Ellis,J. and Wall,L. (1996) The beta-globin locus control region enhances transcription of but does not confer position-independent expression onto the lacZ gene in transgenic mice. EMBO J., 15, 3713–3721. [PMC free article] [PubMed] [Google Scholar]
- 75.Ortiz B.D., Cado,D., Chen,V., Diaz,P.W. and Winoto,A. (1997) Adjacent DNA elements dominantly restrict the ubiquitous activity of a novel chromatin-opening region to specific tissues. EMBO J., 16, 5037–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takada T., Lida,K., Akasaka,K., Yasue,H., Torii,R., Tsujimoto,G., Taira,M. and Kimura,H. (2000) Evaluation of heterologous insulator function with regard to chromosomal position effect in the mouse blastocyst and fetus. Mol. Reprod. Dev., 57, 232–237. [DOI] [PubMed] [Google Scholar]
- 77.Geyer P.K. (1997) The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev., 7, 242–248. [DOI] [PubMed] [Google Scholar]
- 78.Neff T., Shotkoski,F. and Stamatoyannopoulos,G. (1997) Stem cell gene therapy, position effects and chromatin insulators. Stem Cells, 15, 265–271. [DOI] [PubMed] [Google Scholar]
- 79.Dorer D.R. and Henikoff,S. (1994) Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell, 77, 993–1002. [DOI] [PubMed] [Google Scholar]
- 80.Kennison J.A. and Southworth,J.W. (2002) Transvection in Drosophila. Adv. Genet., 46, 399–420. [DOI] [PubMed] [Google Scholar]
- 81.Shewchuk B.M., Cooke,N.E. and Liebhaber,S.A. (2001) The human growth hormone locus control region mediates long-distance transcriptional activation independent of nuclear matrix attachment regions. Nucleic Acids Res., 29, 3356–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sutter N.B., Scalzo,D., Fiering,S., Groudine,M. and Martin,D.I.K. (2003) Chromatin insulation by a transcriptional activator. Proc. Natl Acad. Sci. USA, 100, 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smit A.F. (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev., 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 84.Conte C., Dastugue,B. and Vaury,C. (2002) Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol., 22, 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitelaw E. and Martin,D.I. (2001) Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nature Genet., 27, 361–365. [DOI] [PubMed] [Google Scholar]
- 86.Walsh C.P., Chaillet,J.R. and Bestor,T.H. (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet., 20, 116–117. [DOI] [PubMed] [Google Scholar]
- 87.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 88.Antequera F., Macleod,D. and Bird,A. (1989) Specific protection of methylated CpGs in mammalian nuclei. Cell, 58, 509–517. [DOI] [PubMed] [Google Scholar]
- 89.Schubeler D., Lorincz,M.C., Cimbora,D.M., Telling,A., Feng,Y.Q., Bouhassira,E.E. and Groudine,M. (2000) Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure and histone acetylation. Mol. Cell. Biol., 20, 9103–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mutskov V.J., Farrell,C.M., Wade,P.A., Wolffe,A.P. and Felsenfeld,G. (2002) The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev., 16, 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vyas P., Vickers,M.A., Simmons,D.L., Ayyub,H., Craddock,C.F. and Higgs,D.R. (1992) Cis-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell, 69, 781–793. [DOI] [PubMed] [Google Scholar]
- 92.Bennani-Baiti I.M., Jones,B.K., Liebhaber,S.A. and Cooke,N.E. (1995) Physical linkage of the human growth hormone gene cluster and the skeletal muscle sodium channel alpha-subunit gene (SCN4A) on chromosome 17. Genomics, 29, 647–652. [DOI] [PubMed] [Google Scholar]
- 93.Bennani-Baiti I.M., Cooke,N.E. and Liebhaber,S.A. (1998) Physical linkage of the human growth hormone gene cluster and the CD79b (Ig beta/B29) gene. Genomics, 48, 258–264. [DOI] [PubMed] [Google Scholar]
- 94.Chong S., Riggs,A.D. and Bonifer,C. (2002) The chicken lysozyme chromatin domain contains a second, widely expressed gene. Nucleic Acids Res., 30, 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.