Abstract
Proper segregation of chromosomes during mitosis requires that the products of chromosome replication are paired together—termed sister chromatid cohesion. In budding yeast, Ctf7p/Eco1p is an essential protein that establishes cohesion between sister chromatids during S phase. In fission yeast, Eso1p also functions in cohesion establishment, but is comprised of a Ctf7p/Eco1p domain fused to a Rad30p domain (a DNA polymerase) both of which are independently expressed in budding yeast. In this report, we identify and characterize the first candidate human ortholog of Ctf7p/Eco1p, which we term hEFO1p (human Establishment Factor Ortholog). As in fission yeast Eso1p, the hEFO1p open reading frame extends well upstream of the C-terminal Ctf7p/Eco1p domain. However, this N-terminal extension in hEFO1p is unlike Rad30p, but instead exhibits significant homology to linker histone proteins. Thus, hEFO1p is a unique fusion of linker histone and cohesion establishment domains. hEFO1p is widely expressed among the tissues tested. Consistent with a role in chromosome segregation, hEFO1p localizes exclusively to the nucleus when expressed in HeLa tissue culture cells. Moreover, biochemical analyses reveal that hEFO1p exhibits acetyltransferase activity. These findings document the first characterization of a novel human acetyltransferase, hEFO1p, that is comprised of both linker histone and Ctf7p/Eco1p domains.
INTRODUCTION
Faithful transmission of chromosomes requires that chromosomes are replicated and that the resulting sister chromatids are paired together. This pairing, or sister chromatid cohesion, occurs concomitantly with chromosome replication during S phase (1). During mitosis, cohesion ensures that one chromatid associates with microtubules from the spindle pole opposite that of its sister chromatid. At anaphase onset, cohesion is inactivated, allowing each sister to move along the spindle apparatus into the newly forming daughter cells (2,3).
In budding yeast, several structural cohesion proteins (cohesins) have been identified: Smc1p, Smc3p, Mcd1p/Scc1p, Irr1p/Scc3p and Pds5p (4–10). One current model is that these cohesins form rings that encompass or tether together sister chromatids (11,12). Deposition cohesion factors Scc2p (Mis4p in fission yeast) and Scc4p are required for the proper association of structural cohesins with chromatin, with deposition occurring from G1/S through most of mitosis (7,13,14). Ctf7p/Eco1p is an acetyltransferase that represents a third class of cohesion proteins, the establishment factors (7,15,16). Although loss of Ctf7p/Eco1p (herein termed Ctf7p) function leads to precocious sister chromatid separation and cell death, Ctf7p is not required to maintain cohesion nor deposit cohesins onto DNA. Instead, budding yeast Ctf7p appears to establish cohesion by coupling the cohesion machinery to DNA replication.
Several lines of evidence reveal that cohesion establishment is coupled to DNA replication. First, Ctf7p is required exclusively during S phase when DNA replication occurs (7,16). Second, cells defective for Ctf7p function are rescued by elevated levels of PCNA (proliferating cell nuclear antigen), a sliding clamp that promotes processive DNA replication (16,17). Third, Ctf7p physically associates with each of three independent replication factor C (RFC) complexes (18). RFC complexes load PCNA-like sliding clamps onto double-stranded DNA, at least a subset of which are known to function in sister chromatid cohesion (18–22). The characterization of Trf4p (Pol σ) provided the first evidence that a DNA polymerase could also function in sister chromatid cohesion (23,24). More recently, the large subunit of budding yeast DNA polymerase ε (Pol2) was found to associate with Pol σ and participate in cohesion (25).
Despite the essential roles of budding yeast Ctf7p in chromosome segregation and cell viability, nothing is known concerning the Ctf7p cohesion establishment pathway in higher eukaryotes. Recent findings from yeast reveal that Ctf7p function is conserved through evolution. For instance, in budding yeast, CTF7 is located on chromosome VI and encodes 281 amino acids (7,16). RAD30, encoding a DNA repair polymerase (Pol η), is located on chromosome IV and encodes 632 amino acids (26–28). In fission yeast, CTF7 and RAD30 sequences are genetically fused to produce ESO1. Eso1p is comprised of 872 amino acids and functions in both cohesion establishment and DNA polymerase functions, indicating that both Ctf7p and Rad30p activities are preserved (29,30). In this report, we identify the first full-length human ortholog of Ctf7p and characterize its enzymatic activity, localization and novel genetic composition.
MATERIALS AND METHODS
Cell growth and database methods
Growth and sporulation media for yeast were described previously (31). Yeast and bacterial strains and transformations were performed as described, with minor modifications (32–36). HeLa cell cultures were maintained in MEM supplemented with 10% fetal bovine serum, Earle’s BSS, l-glutamine and antibiotics.
Budding yeast Ctf7p and Rad30p sequences and fission yeast Eso1p sequence were used to perform PSI-BLAST searches of non-redundant nucleic acid and protein sequence databases (GenBank CDS, PDB, SWISS-PROT, PIR and PRF), expressed sequence tag (EST) databases and recent submissions to the human genonome project (http://www.ncbi.nlm.nih.gov/BLAST). Clustal X and Clustal W were used to connect overlapping sequences to produce extended contiguous (contig) coding sequences. A full-length cDNA containing human EFO1 was identified from the HUGE (Human Unidentified Gene-Encoded Large Proteins) database and obtained from the Kazusa DNA Research Institute in Japan (37).
Cloning and molecular methods
The human EFO1 opening reading frame (ORF) was independently derived by the HUGE database. hEFO1 coding sequence, flanked by BamHI restriction sites, was generated by PCR using KIAA1911 as a template (obtained from Kazusa DNA Research Institute) and oligos 5′-CGCGGATCCATGTCCATTCAGGAGAAATCAAAAGAG-3′ and 5′-CGCGGATCCGGTTGTTGCCAGTCCTGAGTTCATTGT-3′. The resulting product contains hEFO1p in which the N-terminal methionine is deleted. The PCR product was then cloned into pCR 2.1 (Invitrogen) to generate pBS1088. The entire EFO1 region was sequenced, and two errors, presumably generated during PCR, were identified (Y93F and L384S). We termed this allele hEFO1-1. These two alterations are located well outside of the Ctf7p functional core domain (762–831) and did not adversely affect either acetyltransferase activity or nuclear localization (see Results). To generate wild-type hEFO1, a BplI–BsgI fragment obtained from KIAA1911 cDNA was used to replace the short DNA region containing both PCR-based alterations. The resulting clone was confirmed as correct by sequence analysis and used for further analyses.
To generate in-frame green fluorescent protein (GFP)–hEFO1p and GST–hEFO1p expression vectors, pCR 2.1-hEFO1 was digested with BamHI and the resulting drop-out fragment ligated into either pGEX 4T-3 linearized with BamHI (Amersham/Pharmacia) or pEGFP C.1 linearized with BglII (Clontech). For both GST and GFP constructs, plasmids harboring hEFO1p in the reverse orientation were also identified. To generate a C-terminal truncation of hEFO1p, the GST–hEFO1 construct was digested with BsmI to drop-out a 497 nucleotide fragment containing the CTF7 core domain coding region. The remaining construct was ligated back together, termed hEFO1-2, and encodes a protein harboring a C-terminal truncation.
Expression and detection of recombinant proteins
pGST–hEFO1p and control constructs were transformed into BL-21 bacterial cells and selected on LB plates containing carbenicillin. Overnight liquid cultures were diluted 10-fold and incubated for 2 h at 37°C to obtain log phase growth. The cultures were then brought to a final concentration of 100 µM isopropyl-β-d-thiogalactopyranoside (IPTG; Fisher) and protein expression induced for 3–4 h at 23°C. Cells were then spun for 10 min at 9500 r.p.m. (Sorvall SS-34) and the pellet resuspended in SDS/Laemmli buffer and boiled prior to resolving on polyacrylamide gels. Proteins were visualized by transferring onto PVDF membrane (Amersham/Pharmacia) and detected with a combination of a mouse antibody directed against GST (Santa Cruz), goat anti-mouse horseradish peroxidase (HRP; Bio-Rad) and ECL+ (Amersham/Pharmacia).
For acetylation assays, the bacterial cell pellet was suspended in 400 µl of 120 ELB buffer (38) supplemented with EDTA-free protease inhibitors (Roche) and phenylmethylsulfonyl fluoride (PMSF). De-acetylase inhibitors sodium butyrate (10 mM, Sigma) and nicotinamide (5 mM, Sigma) were added prior to cell lysis. The cell suspensions were lysed by sonication and brought to a final concentration of 50–100 µM acetyl-CoA (Sigma). After 5–10 min, the reaction was stopped by the addition of SDS/Laemmli buffer, boiled and resolved by polyacrylamide gel electrophoresis. Western blot detection of acetylated proteins was performed using a combination of mouse antibody directed against acetylated lysine residues (Cell Signaling Technology), goat anti-mouse HRP (Bio-Rad) and ECL+ (Amersham/Pharmacia). An additional step to enrich for GST-coupled proteins was optionally performed using glutathione–Sepharose 4B beads (Amersham-Pharmacia) as previously described (18).
Transfection of pEGFP–hEFO1p into human tissue culture HeLa cells was performed according to the manufacturer’s instructions (Roche). Briefly, cells were grown to ∼30% confluence on coverslips housed in 6-well culture dishes. pEGFP–hEFO1p and control plasmids were diluted into serum-free medium supplemented with Fugene (Roche) and the resulting mixture added drop-wise into each of the wells before returning the cells to 37°C. Transfection and recovery were assessed 24–48 h later.
For visualization of GFP–hEFO1 protein in vivo, coverslips were dipped in warm PEM buffer and then fixed by plunging into –20°C methanol for 2 min. The cells were rehydrated in phosphate-buffered saline (PBS) and the coverslip mounted onto a glass slide using an antifade reagent to reduce photobleaching (Vectastain). Images were collected on a Nikon microscope (Eclipse E800) coupled to a Photometrics cooled CCD digital camera (CoolSnap fx).
For detection of GFP proteins by western blot analysis, coverslips were rinsed twice in cold PBS and the cells immediately lysed in SDS/Laemmli buffer, boiled and resolved by acrylamide gel electrophoresis. GFP-tagged proteins were transferred onto PVDF membrane and detected with a combination of a mouse antibody directed against GFP (Cell Signaling Technology), goat anti-mouse HRP (Bio-Rad) and ECL+ (Amersham/Pharmacia).
Northern blot analyses
Expression of hEFO1p was assessed using a multi-tissue human northern blot (Clontech) following the manufacturer’s instructions. Two confirm expression, the membrane was independently probed twice using distinct probes: a 705 nt PCR product (oligos 5′-GACCCACCATTGGATAATCAG-3′ and 5′TGGATATGTTCCGCAATTAGG-3′) or a 690 bp fragment released via EcoRI–BamHI digestion. In both cases, the DNA probes were labeled with [32P]dCTP by random priming and separated from unincorporated dNTPs prior to hybridization (39).
RESULTS
Ctf7p/Eco1p is highly conserved but genetically mobile through evolution
In budding yeast, Ctf7p is a 281 amino acid protein essential for sister chromatid cohesion (7,16). In fission yeast, Eso1p is an 872 amino acid protein that also functions in sister chromatin cohesion but is comprised of two distinct domains: a Rad30p domain as the N-terminus and a Ctf7p domain as the C-terminus (29,30). To date, it is unknown whether higher order eukaryotes display the stand-alone Ctf7p of budding yeast, the bi-functional Rad30p–Ctf7p fusion of fission yeast or a novel genetic fusion. Resolving this issue thus remains an important first step in identifying the factors and elucidating the molecular activities required for cohesion establishment in human cells.
We used computer-assisted database searches to assess which form of Ctf7p predominates in higher eukaryotes. First, we identified Rad30p-like proteins and then determined whether the Rad30p domain was coupled to a downstream Ctf7p domain. The Rad30p domain of fission yeast Eso1p was used to query non-redundant databases, including ESTs and newly released human genome sequences (Materials and Methods). We obtained multiple hits, consistent with previous studies documenting the high degree of conservation of Rad30p proteins through evolution (40–42). Sequence analyses revealed that these Rad30p-like proteins failed to contain domains that exhibited significant homology to Ctf7p (data not shown). The inability to identify Rad30p proteins that also contain a Ctf7p domain in higher eukaryotes suggests that the genetic fusion comprising Eso1p may be limited to close relatives of fission yeast.
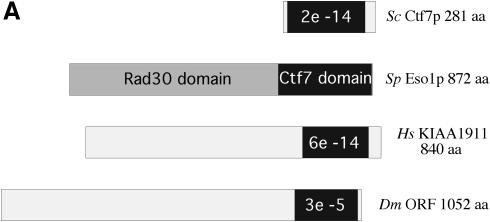
Second, we asked the reciprocal question: is the Ctf7p domain coupled to an upstream Rad30p domain in higher eukaryotes? The Ctf7p domain of fission yeast Eso1p was used to query non-redundant and EST databases as described above. We then ascertained if any of the putative Ctf7p homologs contain a Rad30p domain. Database analyses revealed that conceptual translations of putative Ctf7p orthologs exist in several model organisms including Drosophila, mice and humans (Fig. 1A). An amino acid alignment of the Ctf7p core domain indicates the high degree of conservation (Fig. 1B). We found full-length cDNAs for both Drosophila and human candidate orthologs that contain coding regions that extend well upstream of the Ctf7p domain (N-terminal extensions of 854 residues in Drosophila and 641 residues in humans). Neither of these N-terminal upstream domains exhibit similarity to Rad30p. These results show that, in both Drosophila and humans, the highly conserved Ctf7p domain is fused to a domain other than Rad30p, indicating that Ctf7p is a modular element that has been genetically recombined to other elements through evolution.
Figure 1.
Identification of Ctf7p-like proteins in higher eukaryotes. (A) Schematic illustrating Eso1p and conceptual translations of cDNAs that encode Ctf7p-like proteins. E values (a measurement of similarity) for Ctf7p domains are shown in black. The N-terminal region of Eso1p that exhibits homology to Rad30p is shown in dark gray. Regions that extend upstream of the Ctf7p region in human and Drosophila are shown in light gray. (B) Amino acid alignments reveal the high degree of conservation of the Ctf7p domain for a selection of coding sequences. Residues are numbered according to conceptual translations of cDNA sequences, except for contig sequences which were assigned a starting residue number of 1. Accession numbers are as follows: Schizosaccharomyces pombe Eso1p (BAA95122), Saccharomyces cerevisiae Ctf7p/Eco1p (NP 116683), Drosophila melanogaster (AAF50579.1), Mus musculus ORF (MGC:30637), Homo sapiens KIAA1191 (AB067498) and Homo sapiens contig (comprised of N94144, BG288675 and BE247544). Other mouse (AAH08220, BAB26905 and AA881675) and human orthologs were also identified.
Human EFO1p is one of four candidate proteins exhibiting homology to Ctf7p
A human EST that exhibits sequence homology to a single Ctf7p candidate ortholog in humans was previously reported (16). More recently, other short Ctf7p-like encoding sequences have been identified (http://mendel.imp.univie.ac.at). Here, we extend each of these sequences, all of which reside on different chromosomes and contain the highly conserved Ctf7p core domain (Fig. 2). At least one of these human ORFs contains an N-terminal extension that is absent from budding yeast Ctf7p and unique to the N-terminal region of fission yeast Eso1p (see below). To differentiate between these human sequences in an unbiased way (ScCtf7p/Eco1p versus SpEso1p) and to reflect that at least one of these human coding regions contains a domain unique to any Ctf7p protein thus far characterized, we name this family human Establishment Factor Orthologs (hEFO1s). In this report, we provide the full-length translation, expression pattern and characterization for one family member, hEFO1p (see below). hEFO1 (KIAA1911, accession No. AB067498) resides on chromosome XVIII. hEFO2–hEFO4 exhibit significant homology to the C-terminus of hEFO1, although their expression is currently unknown. hEFO2 resides on chromosome VIII and represents the first human coding sequence identified that exhibited significant homology to Ctf7p (16). We used database search methods to extend the hEFO2 coding region, identifying two new overlapping ESTs that produce a conceptual contiguous amino acid sequence of 334 residues (GenBank accession Nos N94144, BG288675 and BE247544). hEFO3 (accession No. NT 005334.10) and hEFO4 (accession No. NT 033232.2) reside on chromosomes II and XI, respectively. Both hEFO3 and hEFO4 coding sequences contain a 762 bp region that exhibits significant homology to positions 3139–3900 encoding hEFO1p.
Figure 2.
Amino acid alignments for conceptual translations of candidate human Ctf7p family members. See text for accession numbers and details.
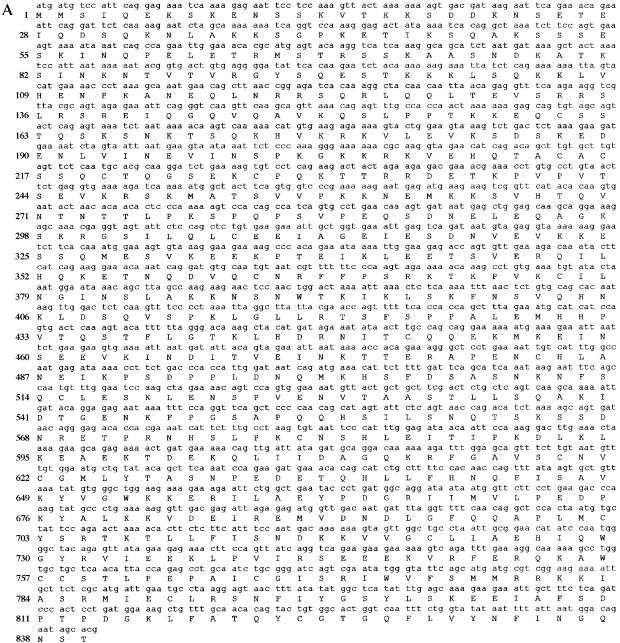
Conceptual translation of hEFO1 cDNA indicates that hEFO1p is comprised of 840 amino acid residues (Fig. 3A) encoded by a 4.6 kb message containing 5′- and 3′ untranslated regions (37). To both test for hEFO1 expression and assess its relative abundance among various cell types, we probed a multi-tissue northern blot containing RNA isolated from heart, brain, placenta, lung, liver, muscle, kidney and pancreas tissues. 32P-labeled probes from two regions of the hEFO1 ORF were independently used and identified a single transcript (Fig. 3B). The results show that hEFO1 is expressed in the majority of tissues tested, but elevated expression was detected in skeletal muscle.
Figure 3.
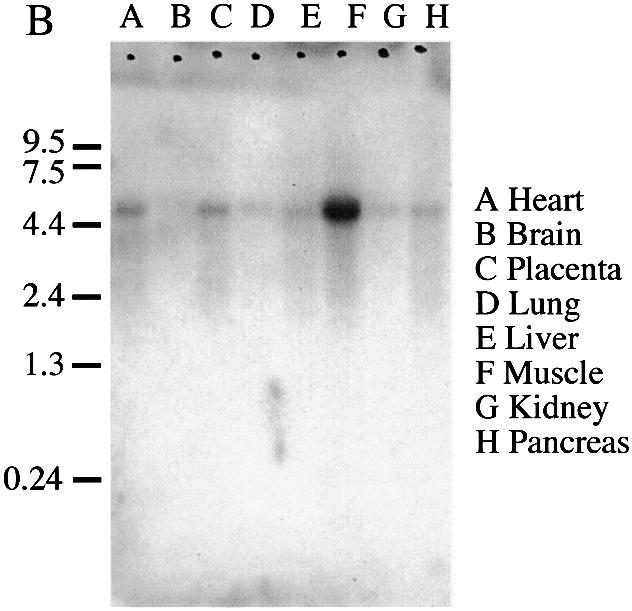
Expression and conceptual translation of hEFO1p. (A) Nucleotide and corresponding amino acid sequence for hEFO1p. (B) Northern blot analysis reveals that hEFO1p is expressed in most of the human tissues tested.
Human EFO1p is comprised of linker histone protein and Ctf7p domains
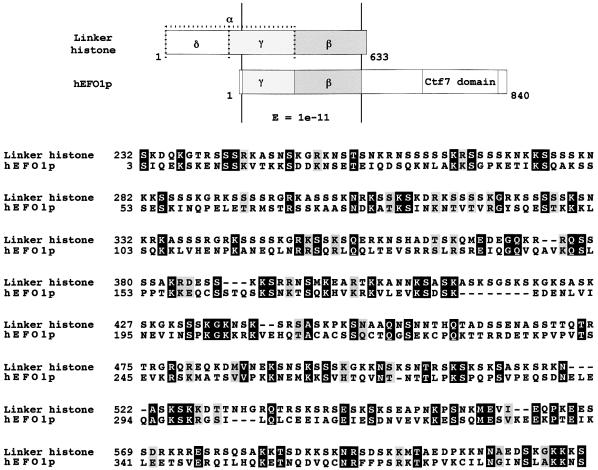
In both hEFO1 and in the Drosophila cDNA containing the Ctf7p core domain that we identified by computer-assisted database searches (Fig. 1), the C-terminal Ctf7p domain is fused to an N-terminal extension other than Rad30p. We next determined whether these N-terminal extensions are similar to each other or represent yet further novel genetic fusions. Database search methods reveal that both N-terminal extensions are conserved. Importantly, the N-terminal extensions of both hEFO1p and Drosophila cDNA translations exhibit significant homology to micronuclear linker histone proteins identified in Tetrahymena (Fig. 4). Based on similarities between these unique genetic fusions, we term the Drosophila homolog dEFO1. Micronuclear linker histone proteins are unique from macronulcear histone H1 and are critical for normal chromosome compaction (43,44). In Tetrahymena, the 71 kDa linker histone polyprotein is processed to produce independent α, β, γ and δ linker histone proteins (α linker histone is an early cleavage product, but later processed to generate γ and δ linker histone proteins). Sequence alignments reveal that the N-terminal extension in hEFO1p is comprised of δ and β linker histone proteins (Fig. 4). These findings document a unique genetic fusion between linker histone protein and a Ctf7p domain.
Figure 4.
hEFO1p and Tetrahymena linker histone alignments. Top: the schematic depicts the region of conservation between Tetrahymena linker histone proteins and hEFO1p. The MLH polyprotein cleavage products α, δ, γ and β linker histones are shaded (AAC18874). Bottom: amino acid alignment reveals the high degree of conservation (E of 1 × 10 –11) between hEFO1p and Tetrahymena γ and β linker histone proteins. Identical residues are boxed in black; conserved residues are boxed in gray.
EFO1p expressed in human cells is a nuclear protein
In budding yeast, Ctf7p is a nuclear protein required for proper chromosome segregation (7,16). It thus became important to test whether hEFO1p localizes to a specific region in the vertebrate cell. The hEFO1 coding cDNA was obtained from the Kazusa DNA Research Institute in Japan (37) and inserted in-frame behind GFP in a mammalian expression vector containing the cytomegalovirus (CMV) promotor (Materials and Methods). The resulting GFP–hEFO1p construct and GFP control plasmids were transfected into HeLa tissue culture cells. Within 24 h, GFP–hEFO1p expression in HeLa cell extracts was readily detected by western blot at the appropriate molecular weight. A protein band of similar molecular weight was absent in HeLa extracts expressing GFP alone (data not shown).
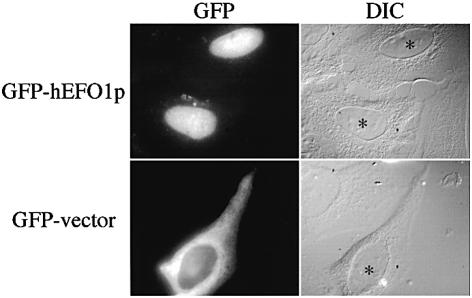
We then determined the localization of GFP–hEFO1p in human tissue culture cells. Transfected HeLa cells were grown on coverslips for 24–48 h. Images were then collected using both phase contrast microscopy to visualize cell morphology and epi-illumination to detect GFP-tagged proteins. GFP visualization revealed that hEFO1p localizes specifically to the human cell nucleus (Fig. 5). In contrast, GFP was predominantly cytoplasmic. Untransfected cells were devoid of GFP signal. These results reveal that hEFO1p localizes to the nucleus of human cells, consistent with a role in chromosome segregation.
Figure 5.
Localization of hEFO1p in mammalian cells. GFP-tagged hEFO1p versus GFP vector alone constructs were transfected into HeLa cells and localization assessed 24 h later. Cell images obtained using the GFP channel reveal that hEFO1p specifically localizes to the nucleus while GFP is uniformly distributed throughout the cell. Cell images obtained by differential interference contrast microscopy (DIC) are also shown (* denotes nuclei in transfected cells). GFP expression was not observed in untransfected cells.
Human EFO1p exhibits acetyltransferase activity
Bacterial expressed budding yeast Ctf7p was recently shown to possess acetyltransferase activity in vitro (15). Although physiologically relevant substrates have yet to be identified, Ctf7p is known to serve as its own substrate. We exploited this autoacetylation activity to determine whether hEFO1p also exhibits acetyltransferase activity. The hEFO1 coding sequence was inserted in-frame behind that of GST in an inducible bacterial expression vector. The resulting GST–hEFO1 construct was transformed into bacterial cells and protein expression induced using IPTG (Materials and Methods). As a positive control, GST–CTF7 was independently transformed and expressed in bacteria. Post-induction, the bacterial strains were lysed and assessed for protein expression. An optional protein enrichment step via glutathione–Sepharose bead pull-down was also used (Materials and Methods). Both GST–hEFO1p and GST–Ctf7p protein bands of the appropriate molecular weights were readily detectable by western blot analyses (Fig. 6).
Figure 6.
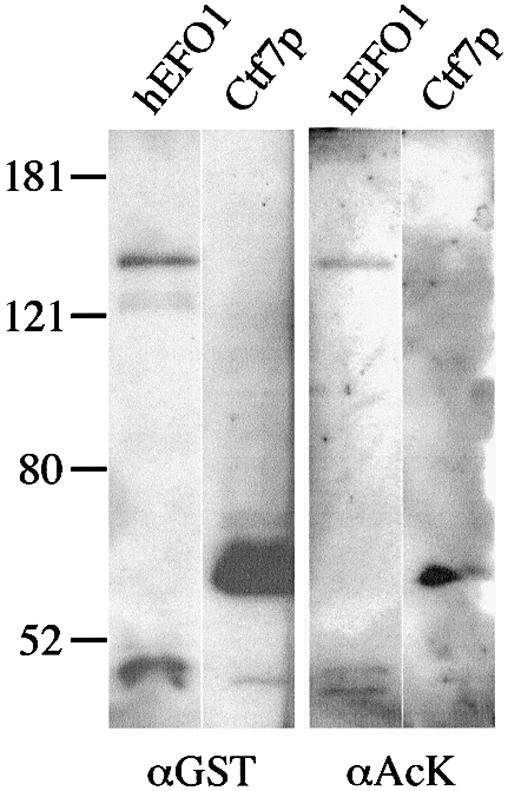
Expression and acetyltransferase activity of human EFO1p and budding yeast Ctf7p. (A) GST–EFO1p and GST–Ctf7p expressed in bacterial cells and visualized using an antibody directed against GST (α GST). Acetyltransferase activity of human EFO1p and budding yeast Ctf7p using an antibody directed against acetylated lysines (α Ac-K) previously described to detect Ctf7p acetylation reactions (15).
We then tested whether bacterially expressed hEFO1p was capable of acetyltransferase activity. As before, GST–EFO1p and GST–Ctf7p expression was induced, the bacterial cells lysed and GST-containing proteins enriched by glutathione–Sepharose bead pull-downs. The resulting protein fractions were incubated for 10 min in buffer containing a final concentration of 50 µM acetyl-CoA. The addition of acetyl moieties onto GST–hEFO1p or GST–Ctf7p was then visualized by western blot analysis using an acetylated lysine-specific antibody previously documented to detect Ctf7p acetylation reactions (15). As expected, GST–Ctf7p exhibited a robust autoacetylation activity. GST–hEFO1p also exhibited a similar autoacetylation activity (Fig. 6). No antibody cross-reactivity was observed for GST alone. These findings reveal that the acetyltransferase activity of Ctf7p-like proteins is conserved through evolution. We next tested for the specificity of the hEFO1p acetylation reaction by generating a loss-of-function allele in which the C-terminal region containing the Ctf7p core domain was deleted (hEFO1-2p). The resulting construct was expressed at levels similar to hEFO1p but was unable to catalyze the autoacetylation reaction (data not shown).
DISCUSSION
Sister chromatid cohesion plays a central role in chromosome transmission, cell cycle progression and genome maintenance (45,46). To date, Ctf7p-like proteins identified in yeast remain the only essential establishment factors identified: factors that couple sister chromatid cohesion to DNA replication without functioning in DNA replication per se (7,16,29). In this report, we identify and characterize the first candidate Ctf7p ortholog in humans, hEFO1p. hEFO1p is present in almost all tissues tested. It is unknown whether the inability to detect hEFO1 in brain tissue is due to detection limitations or to the terminally differentiated nature of this tissue, but BLAST searches of EST databases identify CTF7-like cDNAs from brain libraries (A.Bellows and R.V.Skibbens, unpublished observations). We also identified three other chromosomal loci within the human genome that encode Ctf7p-like proteins (hEFO2p–hEFO4p). One of the probes used to identify the hEFO1 transcript is co-incident with hEFO2–hEFO4 sequences. However, we did not detect novel sized transcripts, suggesting that either the reaction temperature precluded binding of similar but not identical sequences, that the other transcripts occur at very low levels, or that they are not expressed. Thus, while our analyses of hEFO2–hEFO4 greatly extend the limited sequences previously described (15,16), the full-length conceptual translations and expression patterns remain unknown. Given the essential function of Ctf7p in both budding and fission yeast and the apparent frequency through which CTF7 recombines through evolution, it would not be surprising that multiple editions exist within the human genome.
hEFO1p exhibits several characteristics consistent with a role in chromosome segregation and cohesion establishment. First, hEFO1p contains the highly conserved Ctf7p core domain found in budding yeast Ctf7p and fission yeast Eso1p (this study) (7,16,29). Secondly, hEFO1p localizes specifically to the nucleus. We were unable to identify mitotic cells in which hEFO1p associated with condensed chromosomes (M.Kenna and R.V.Skibbens, personal observation). Similarly, budding yeast Ctf7p is a nuclear protein that functions during S phase. Ctf7p is not required during mitosis, nor is Ctf7p detectable within the bulk of the chromatin mass (7,16) (R.V.Skibbens and D.Koshland, unpublished results). Instead, Ctf7p is thought to transiently associate with the DNA replication fork during cohesion establishment (18). In combination, these findings indicate that the cell cycle activity and chromatin association of Ctf7p-like factors are probably conserved through evolution. Thirdly, hEFO1p exhibits acetyltransferase activity and is capable of acting as its own substrate. A comparable activity has been reported for budding yeast Ctf7p (15). Currently, physiologically relevant substrates of budding yeast Ctf7p acetylation have yet to be identified (15); thus, characterization of hEFO1p acetylation targets in human cells is beyond the scope of this report. However, the finding that acetyltransferase activities are conserved from yeast to humans strongly suggests that this enzymatic activity will play a central role in establishing cohesion. Future endeavors in solving the mystery of sister chromatid pairing for both yeast and humans will probably entail identifying physiologically relevant substrates for Ctf7p acetylation.
Our findings are novel in that human EFO1p is comprised of a Ctf7p core domain genetically fused to a domain that exhibits significant homology to structural nucleosome compaction factors—the linker histones. In Tetrahymena, the micronuclear linker histone gene (MLH) is translated into a single 71 kDa polypeptide which is then cleaved to produce four distinct linker histones: α and β in which α is further processed to produce δ and γ linker histone proteins. These four linker histone proteins are distinct from one another and unique from macronuclear histone H1 protein (43). In humans, EFO1p is comprised of a C-terminal Ctf7p domain fused to an N-terminal domain comprised of contiguous γ and β linker histone proteins in which the predominant form appears uncleaved (Fig. 6). This genetic fusion is conserved through evolution in that a similar linker histone protein–Ctf7p chimera exists in Drosophila (this study). At present, efforts to complement yeast ctf7 mutant phenotypes with hEFO1p have been unsuccessful (A.Bellows, M.Kenna and R.V.Skibbens, personal observation), although even close orthologs often fail to rescue essential functions across model organisms.
Linker histones have been implicated in transcription regulation and chromosome compaction (47). For instance, in Tetrahymena MLH knockouts, loss of α, β, δ and γ linker histone proteins resulted in micronuclei that were over twice the size of wild-type cell micronuclei (44). Similarly, in budding yeast, mutations in many cohesion factors, including Mcd1p, Pds5p and Ctf7p, also exhibit defects in chromosome condensation (4,9,16) (R.V.Skibbens and D.Koshland, unpublished results). It is worth speculating that linker histone modifications may help target the acetyltransferase activity of hEFO1p to chromatin in a cell cycle-dependent manner. Previous studies revealed that Tetrahymena linker histones contain cAMP-dependent protein kinase (PKA) target sequences, with evidence that at least δ linker histone protein is a PKA substrate. Thus, the phosphorylation state of δ linker histone may regulate chromosome condensation/decondensation (48). Future directions will relate to the role of post-translational hEFO1p modifications in chromosome condensation and phenotypic analyses of cells lacking hEFO1p. These endeavors may be difficult given the possible functional redundancy of the hEFO family and the recent observations that a separate class of proteins, called the condensins, appears to perform the majority of chromosome condensation in humans (49,50). Similarly, there is an ever-growing number of cohesion proteins that, in addition to the structural and deposition cohesion factors, include RFC factors, DNA polymerases and an acetyltransferase (15,18–21,23–25). Deciphering the extent to which these players promote sister chromatid cohesion and chromosome condensation will continue to be an intriguing endeavor for cell biology.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Skibbens lab members Dr A. Brands, L. Antoniacci and B. Satish for their critical reading of this manuscript, and Steve Eastman for his technical expertise. This research was funded by a Howard Hughes Medical Institution undergraduate grant to A.M.B., Vice Provost funding to M.A.K., a National Institutes of Health grant to L.C. (GM 58025) and a National Science Foundation award to R.V.S. (MCB-0212323).
DDBJ/EMBL/GenBank accession no. AB067498
REFERENCES
- 1.Skibbens R.V. (2000) Holding your own: establishing sister chromatid cohesion. Genome Res., 10, 1664–1671. [DOI] [PubMed] [Google Scholar]
- 2.Koshland D.E. and Guacci,V. (2000) Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol., 12, 297–301. [DOI] [PubMed] [Google Scholar]
- 3.Nasmyth K., Peters,J.M. and Uhlmann,F. (2000) Splitting the chromosome: cutting the ties that bind sister chromatids. Science, 288, 1379–1385. [DOI] [PubMed] [Google Scholar]
- 4.Guacci V., Koshland,D. and Strunnikov,A. (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S.cerevisiae. Cell, 91, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaelis C., Ciosk,R. and Nasmyth,K. (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell, 91, 35–45. [DOI] [PubMed] [Google Scholar]
- 6.Kurlandzka A., Rytka,J., Gromadka,R. and Murawski,M. (1995) A new essential gene located on Saccharomyces cerevisiae chromosome IX. Yeast, 11, 885–890. [DOI] [PubMed] [Google Scholar]
- 7.Toth A., Ciosk,R., Uhlmann,F., Galova,M., Schleiffer,A. and Nasmyth,K. (1999) Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev., 13, 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strunnikov A.V., Larionov,V.L. and Koshland,D. (1993) SMC1: an essential yeast gene encoding a putative head–rod–tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol., 123, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartman T., Stead,K., Koshland,D. and Guacci,V. (2000) Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol., 151, 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panizza S., Tanaka,T., Hochwagen,A., Eisenhaber,F. and Nasmyth,K. (2000) Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol., 10, 1557–1564. [DOI] [PubMed] [Google Scholar]
- 11.Anderson D.E., Losada,A., Erickson,H.P. and Hirano,T. (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol., 156, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haering C.H., Lowe,J., Hochwagen,A. and Nasmyth,K. (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell, 9, 773–788. [DOI] [PubMed] [Google Scholar]
- 13.Ciosk R., Shirayama,M., Shevchenko,A., Tanaka,T., Toth,A., Shevchenko,A. and Nasmyth,K. (2000) Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4. Mol. Cell, 5, 243–254. [DOI] [PubMed] [Google Scholar]
- 14.Furuya K., Takahashi,K. and Yanagida,M. (1998) Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev., 12, 3408–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov D., Schleiffer,A., Eisenhaber,F., Mechtler,K., Haering,C.H. and Nasmyth,K. (2002) Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr. Biol., 12, 323–328. [DOI] [PubMed] [Google Scholar]
- 16.Skibbens R.V., Corson,L.B., Koshland,D. and Hieter,P. (1999) Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelman Z. (1997) PCNA: structure, functions and interactions. Oncogene, 14, 629–640. [DOI] [PubMed] [Google Scholar]
- 18.Kenna M.A. and Skibbens,R.V. (2003) Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol., 23, 2999–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer M.L., Gygi,S.P., Aebersold,R. and Hieter,P. (2001) Identification of RFC (Ctf18p, Ctf8p, Dcc1p): an alternate RFC complex required for sister chromatid cohesion in S.cerevisiae. Mol. Cell, 7, 959–970. [DOI] [PubMed] [Google Scholar]
- 20.Hanna J.S., Kroll,E.S., Lundblad,V. and Spencer,F.A. (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol., 21, 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause S.A., Loupart,M.L., Vass,S., Schoenfelder,S., Harrison,S. and Heck,M.M. (2001) Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol. Cell. Biol., 21, 5156–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen-Fix O. (2001) The making and breaking of sister chromatid cohesion. Cell, 106, 137–140. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., Castano,I.B., De Las Penas,A., Adams,C. and Christman,M.F. (2000) Pol κ: a DNA polymerase required for sister chromatid cohesion. Science, 289, 774–779. [DOI] [PubMed] [Google Scholar]
- 24.Carson D.R. and Christman,M.F. (2001) Evidence that replication fork components catalyze establishment of cohesion between sister chromatids. Proc. Natl Acad. Sci. USA, 98, 8270–8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards S., Li,C.M., Levy,D.L., Brown,J., Snow,P.M. and Campbell,J. (2003) Saccharomyces cerevisiae DNA polymerase varε and polymerase σ interact physically and functionally, suggesting a role for polymerase varε in sister chromatid cohesion. Mol. Cell. Biol., 23, 2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase eta. Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- 28.Washington M.T., Johnson,R.E., Prakash,S. and Prakash,L. (1999) Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase eta. J. Biol. Chem., 274, 36835–36848. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K., Yonekawa,T., Kawasaki,Y., Kai,M., Furuya,K., Iwasaki,M., Murakami,H., Yanagida,M. and Okayama,H. (2000) Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol., 20, 3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madril A.C., Johnson,R.E., Washington,M.T., Prakash,L. and Prakash,S. (2001) Fidelity and damage bypass ability of Schizosaccharomyces pombe Eso1 protein, comprised of DNA polymerase eta and sister chromatid cohesion protein Ctf7. J. Biol. Chem., 276, 42857–42862. [DOI] [PubMed] [Google Scholar]
- 31.Rose M.D., Winston,F. and Hieter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 32.Schiestl R.H. and Gietz,R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- 33.Ito H., Fukuda,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connelly C. and Hieter,P. (1996) Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell, 86, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doheny K.F., Sorger,P.K., Hyman,A.A., Tugendreich,S., Spencer,F. and Hieter,P. (1993) Identification of essential components of the S.cerevisiae kinetochore. Cell, 73, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuno R., Nagase,T., Waki,M. and Ohara,O. (2002) HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res., 30, 166–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb J.R., Michaud,W.A., Sikorski,R.S. and Hieter,P.A. (1994) Cdc16p, Cdc23p and Cdc27p form a complex for mitosis. EMBO J., 13, 4321–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Sutton M.D. and Walker,G.C. (2001) Managing DNA polymerases: coordinating DNA replication, DNA repair and DNA recombination. Proc. Natl Acad. Sci. USA, 98, 8342–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev., 13, 2191–2195. [DOI] [PubMed] [Google Scholar]
- 42.McDonald J.P., Tissier,A., Frank,E.G., Iwai,S., Hanaoka,F. and Woodgate,R. (2001) DNA polymerase iota and related Rad30-like enzymes. Philos. Trans. R. Soc. Lond. B Biol. Sci., 356, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu M., Allis,C.D., Sweet,M.T., Cook,R.G., Thatcher,T.H. and Gorovsky,M.A. (1994) Four distinct and unusual linker proteins in a mitotically dividing nucleus are derived from a 71-kilodalton polyprotein, lack p34cdc2 sites and contain protein kinase A sites. Mol. Cell. Biol., 14, 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen X., Yu,L., Weir,J.W. and Gorovsky,M.A. (1995) Linker histones are not essential and affect chromatin condensation in vivo. Cell, 82, 47–56. [DOI] [PubMed] [Google Scholar]
- 45.Cassimeris L. and Skibbens,R.V. (2003) Regulated assembly of the mitotic spindle: a perspective from two ends. Curr. Issues Mol. Biol., 5, 99–112. [PubMed] [Google Scholar]
- 46.Nasmyth K. (2001) Disseminating the genome: joining, resolving and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet., 35, 673–745. [DOI] [PubMed] [Google Scholar]
- 47.Wolffe A.P., Khochbin,S. and Dimitrov,S. (1997) What do linker histones do in chromatin? Bioessays, 19, 249–255. [DOI] [PubMed] [Google Scholar]
- 48.Sweet M.T., Carlson,G., Cook,R.G., Nelson,D. and Allis,C.D. (1997) Phosphorylation of linker histones by a protein kinase A-like activity in mitotic nuclei. J. Biol. Chem., 272, 916–923. [DOI] [PubMed] [Google Scholar]
- 49.Hirano T. (1998) SMC protein complexes and higher-order chromosome dynamics. Curr. Opin. Cell Biol., 10, 317–322. [DOI] [PubMed] [Google Scholar]
- 50.Koshland D. and Strunnikov,A. (1996) Mitotic chromosome condensation. Annu. Rev. Cell. Dev. Biol., 12, 305–333. [DOI] [PubMed] [Google Scholar]