Abstract
Despite the progress in understanding the base excision repair (BER) pathway it is still unclear why known mutants deficient in DNA glycosylases that remove oxidised bases are not sensitive to oxidising agents. One of the back-up repair pathways for oxidative DNA damage is the nucleotide incision repair (NIR) pathway initiated by two homologous AP endonucleases: the Nfo protein from Escherichia coli and Apn1 protein from Saccharomyces cerevisiae. These endonucleases nick oxidatively damaged DNA in a DNA glycosylase-independent manner, providing the correct ends for DNA synthesis coupled to repair of the remaining 5′-dangling nucleotide. NIR provides an advantage compared to DNA glycosylase-mediated BER, because AP sites, very toxic DNA glycosylase products, do not form. Here, for the first time, we have characterised the substrate specificity of the Apn1 protein towards 5,6-dihydropyrimidine, 5-hydroxy-2′-deoxyuridine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine deoxynucleotide. Detailed kinetic comparisons of Nfo, Apn1 and various DNA glycosylases using different DNA substrates were made. The apparent Km and kcat/Km values of the reactions suggest that in vitro DNA glycosylase/AP lyase is somewhat more efficient than the AP endonuclease. However, in vivo, using cell-free extracts from paraquat-induced E.coli and from S.cerevisiae, we show that NIR is one of the major pathways for repair of oxidative DNA base damage.
INTRODUCTION
In organisms with aerobic respiration reactive oxygen species (ROS) such as O2·–, H2O2 and OH· are formed as by-products of oxidative metabolism. Exogenous factors such as ionising radiation and chemical carcinogens also generate ROS. DNA is a critical cellular target of oxygen radicals. ROS mostly produce non-bulky DNA lesions which are thought to be substrates for the base excision repair (BER) pathway (1–4).
The key components in BER are DNA glycosylases that recognise and remove damaged or mispaired bases from DNA by cleavage of their N-glycosidic bond, leaving either an abasic site or a single-stranded break in DNA (5,6). The DNA glycosylases are damage-specific and can excise modified bases of various types with high efficiency. Nevertheless, the BER pathway, which requires the sequential action of two enzymes for proper incision of DNA (7), raises theoretical problems for the efficient repair of oxidative DNA damage because it generates genotoxic intermediates such as apurinic/apyrimidinic (AP) sites and/or blocking 3′-terminal groups that must be eliminated by additional steps before initiating DNA repair synthesis. Genetic data indicate that DNA glycosylase mutants are not sensitive to oxidising agents and ionising radiation (8–11). Furthermore, a recent update of the database for mouse mutant strains shows that no single DNA glycosylase-deficient strain is sensitive to oxidising agents (12). This is in striking contrast to the highly sensitive phenotypes of AP endonuclease-deficient bacteria, yeast and mammals towards oxidising agents and ionising radiation (13–16). Recent findings that the AP endonucleases Nfo from Escherichia coli and Apn1 from Saccharomyces cerevisiae can initiate repair by nicking DNA on the 5′ side of 5,6-dihydrothymidine (DHT), 5,6-dihydro-2′-deoxyuridine (DHU), 5-hydroxy-2′-deoxyuridine (5ohU) and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine (me-FapyGua) deoxynucleotides, generating a 3′-hydroxyl and a 5′-dangling damaged nucleotide, provide an alternative nucleotide incision repair (NIR) pathway to the classic BER (17). Subsequently, it has been shown that removal of the 5′-dangling base can be catalysed by either E.coli DNA polymerase I or human flap endonuclease 1 (FEN1). Biological relevance of the NIR pathway is supported by the fact that it is evolutionarily conserved from E.coli to humans (17).
Endonuclease IV (Nfo) is an EDTA-resistant AP endonuclease that represents only 5% of the total AP endonuclease activity in wild-type E.coli. Nfo is a monomer of 30 kDa possessing several activities: AP endonuclease, 3′-phosphatase and 3′-phosphoglycoaldehyde diesterase (18). In E.coli, the nfo gene is a part of the soxRS system inducible by oxidative stress (19–22). Escherichia coli nfo mutants are extremely sensitive to the lethal effects of oxidative agents such as bleomycin and t-butylhydroperoxide. In addition, double nfo xth mutants are more sensitive to γ-radiation, H2O2 and alkylating agents (13). Overproduction of the Nfo protein fully restores drug resistance to xth mutants, but the opposite has not been observed, suggesting that the Nfo protein is able to process a broad spectrum of DNA lesions generated by oxidative stress (23). Indeed, it was shown that Nfo hydrolytically incises DNA containing α-deoxyadenosine (24) and a benzene-derived exocyclic adduct (25), which are not substrates for known DNA glycosylases. In these cases, Nfo leaves a dangling nucleotide on the 5′-terminus of the DNA single-strand break (17,25).
Homologues of E.coli Nfo have been identified in eukaryotes. The yeast APN1 gene encodes AP endonuclease 1 (Apn1), homologous to E.coli Nfo protein. This 41.4 kDa protein is the major AP endonuclease in yeast, accounting for >90% of total activity towards abasic sites (26). Apn1 has AP endonuclease, 3′-diesterase and 3′-phosphatase activities and, similarly to Nfo, is a metalloenzyme (27,28). Yeast mutants lacking Apn1 (apn1Δ) are viable but hypersensitive to oxidative (H2O2 and t-butylhydroperoxide) and alkylating (methyl- and ethylmethane sulfonate) agents and have 6- to 12-fold higher rates of spontaneous mutations than wild-type (14). This mutator phenotype is mainly characterised by a 60-fold increase in A·T→C·G transversion rate (29).
The eukaryotic nucleus is a very poorly oxygenated cellular compartment (30,31), which could greatly affect the formation of certain types of oxidative DNA damage in vivo. Indeed, under anoxic conditions in vitro, ring-saturated pyrimidines are formed in γ-irradiated DNA faster than 8-hydroxypurines (32). Among lesions produced by γ-irradiation of DNA, 5,6-dihydropyrimidines (DHPy) such as DHT and DHU are unique due to their strict requirement for anoxia during irradiation (33,34). DHPy, having lost their aromatic character, are susceptible to base loss and ring fragmentation via hydrolysis (35–37). The fragmentation products of DHPy strongly inhibit DNA synthesis and have been shown to be lethal in vivo (38,39). 5ohU arises by deamination and dehydration from cytosine glycol, an unstable DNA adduct (40,41). DHU, another cytosine-derived product, is produced by ionising radiation under anoxic conditions (42). 5ohU and DHU are miscoding lesions generating a C→T transition (43).
Major oxidative DNA damage products, such as DHT, DHU, 5ohU, 4,6-diamino-5-formamidopyrimidine (FapyAde) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) residues, are substrates for both DNA glycosylases and AP endonucleases (44). Therefore, in vivo these lesions could be repaired via alternative BER and NIR pathways. In this report, we evaluate the biological relevance of BER and NIR pathways by comparing the repair efficiency of Nfo and Apn1 with that of DNA glycosylases/β-lyases such as Nth and Fpg proteins on the same DNA substrates. Comparison of kinetic parameters for purified enzymes suggests that Nth cleaves the duplex oligonucleotides containing a single oxidative DNA base damage somewhat more efficiently than do Nfo and Apn1. However, a comparison of BER and NIR activities, in E.coli and S.cerevisiae cell-free extracts, suggests that ∼50% of oxidative DNA base damage can be processed in a DNA glycosylase-independent manner.
MATERIALS AND METHODS
Oligonucleotides and strains
All oligodeoxyribonucleotides were purchased from Eurogentec (Seraing, Belgium), including modified 30mer oligonucleotides d(TGACTGCATA-X-GCATGTAGACGA TGTGCAT), where X is a DHU, DHT, 5ohU or tetrahydrofuranyl (THF) residue, and complementary oligonucleotides, containing either dA, dG, dC or T opposite a modified base. The resulting duplex oligonucleotides are referred to as X·C(G,A,T) respectively, where X is the modified residue. The oligonucleotides were labelled at the 3′-end by terminal transferase (New England Biolabs, OZYME, France) in the presence of [α-32P]ddATP (3000 Ci/mmol) (Amersham Biosciences Europe GmbH, Orsay, France) and/or 5′-end-labelled by T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP (4500 Ci/mmol) (ICN Pharmaceuticals France SA, Orsay, France) as recommended by the manufacturers. The labelled oligonucleotides were annealed to their appropriate complementary oligonucleotides in a buffer containing 50 mM NaCl, 10 mM HEPES–KOH (pH 7.5) at 65°C for 3 min as previously described (45).
Escherichia coli strain AB1157 (wild-type) and its isogenic derivatives BH20 (fpg::kanR), BH150 (nth::kanR), BH130 (nfo::kanR) and BH160 (nth::kanR, [fpg::kanR, X::tetR]) were from laboratory stock. SW2-38 [isogenic to BW35 (KL16) except nth::kanR, nei::kanR] was a gift from Dr S. Wallace (University of Vermont, Burlington, VT). Saccharomyces cerevisiae strains isogenic to FF18733 (MATa his7-3 leu2-1,112 lys1-1 trp1-289 ura3-52), BG1 (apn1Δ::HIS3) and CD186 (ntg1Δ::LEU2; ntg2Δ::TRP1, ogg1Δ::URA3), were kindly provided by Dr S. Boiteux (CEA, Fontenay aux Roses, France).
DNA modifications
Oxidatively damaged DNA was produced by treatment of the supercoiled plasmid DNA with H2O2 or KMnO4. To generate DNA containing oxidised bases, 2 mg/ml pUC19 plasmid DNA in 10 mM degassed phosphate buffer, pH 7.5, was treated with 10 mM H2O2 in the presence of 0.1 mM FeCl2 and 0.4 mM EDTA at 24°C for 30 min under N2 as described (46). FapyAde, FapyGua, thymine glycol and 7,8-dihydro-8-oxoguanine (8oxoG) residues in plasmid DNA are expected to appear in plasmid DNA under these conditions (46). To generate mostly thymine glycol residues, the DNA was treated with 40 mM KMnO4 at 0°C for 5 min as described (47). The DNA samples were desalted on a Sephadex G50 column equilibrated in 10 mM Tris–HCl (pH 7.5) and 1 mM EDTA.
In DNA repair studies the me-FapyGua residue was used as a methylated analogue of hydroxyl radical-induced formamidopyrimidines (48,49). Although, the nature and formation of me-FapyGua and FapyGua are different, it has been demonstrated that me-Fapy and oxidised FapyGua are good substrates for the E.coli and mammalian oxidative damage-specific DNA glycosylases (48–51). To generate me-FapyGua residues, the 24mer oligonucleotide d(CCTCTTCTTTT GTTTTCTTCTCCC) or plasmid DNA was alkylated in buffer containing 0.05% dimethylsulfate (DMS), 50 mM sodium cacodylate (pH 7.0) and 1 mM EDTA at 23°C for 10 min. The reaction was stopped by the addition of 2-mercaptoethanol up to 0.5 M and the reaction mixture was immediately desalted on a Sephadex G50 column as described above. Under these conditions, the guanine residues are methylated, yielding 98% N7-methylguanine and 2% N3-methylguanine (52). The N7-methylguanine residues were converted to the ring-opened (me-FapyGua) form by incubating alkylated DNA in 60 mM phosphate buffer (pH 11.4) for 48 h at 23°C as described (52).
Enzymes
The Xth protein was purchased from Roche Diagnostics (Meylan Cedex, France). Human Nth1 protein was generously provided by Dr R. Roy (American Health Foundation, Valhalla, NY). Purification of the AlkA, Fpg and HAP1 proteins were performed as described (53,54). Purification of E.coli Nth protein was performed as described (33). The cDNAs encoding Nfo and Apn1 were cloned into plasmid pET11a (Novagen, Madison, WI) and the proteins overexpressed in E.coli BL21 Star cells (Invitrogen SARL, Gergy Pontoise, France). Nfo protein was purified using four chromatographic steps. Cells (4 g) were lysed using a French press at 18 000 p.s.i. in buffer A (20 mM HEPES–KOH pH 7.6, 1 mM dithiothreitol, 5% glycerol) supple mented with 500 mM KCl, 2 mM EDTA and Complete™ protease inhibitor cocktail (Roche, Switzerland). The homogenate was centrifuged at 40 000 g for 20 min and the supernatant (Fraction I) was adjusted to 200 mM KCl in buffer A and passed through a column packed with 40 ml QMA Anion Exchange (Waters-Accell) pre-equilibrated in the same buffer. The flow-through fraction was diluted with buffer A to 50 mM KCl and was applied to a 5 ml HiTrap™ Q-Sepharose column (Amersham Biosciences, Uppsala, Sweden). Proteins bound to the column were eluted by a 50–200 mM NaCl gradient in buffer A; Nfo eluted at 150–200 mM NaCl. Fractions containing Nfo were pooled, adjusted to 100 mM NaCl in buffer A and loaded onto a 1 ml HiTrap-Heparin™ column. Bound proteins were eluted in a 0.1–1.0 M NaCl gradient; Nfo eluted at 400–500 mM NaCl. Finally, an FPLC Superose 12 gel filtration column (Amersham Biosciences) was used to obtain a homogeneous preparation of the protein. Samples of Nfo (1.2 mg/ml) were stored at –20°C in buffer containing 50% glycerol, 250 mM NaCl, 20 mM HEPES–KOH pH 7.6, 2 mM ZnSO4 and 1 mM dithiothreitol.
Purification of Apn1 was similar to Nfo except that Fraction I was adjusted to 30 mM KCl in buffer A and loaded onto a QMA Anion Exchange column. The flow-through fraction was collected and purified further using HiTrap-Heparin™ as described above. Apn1-containing fractions were pooled and passed through a 1 ml HiTrap-Ni2+ ion-chelating column (Amersham Biosciences) and further purified using gel filtration step as described above.
The homogeneity of protein preparations was verified by SDS–PAGE. To exclude contamination of Nfo by DNA glycosylases and non-specific nucleases, the enzyme was heat treated at 65°C for 10 min. After the treatment Nfo retained most of its endonuclease activity towards THF- and DHU-containing oligonucleotides (data not shown). The activities of the various proteins were tested on appropriate DNA substrates immediately before use.
Incision assays
The standard assay mixture for damage-specific incision activity (20 µl final volume) contained 0.2 pmol of the [5′-32P]- or [3′-32P]ddAMP-end-labelled 30mer oligonucleotide duplex in 20 mM HEPES–KOH (pH 7.5), 50 mM KCl, 0.1 mM EDTA (or 2 mM EDTA for the cell-free extracts, 5 mM MgCl2 for Apn1 and HAP1 or 5 mM CaCl2 for Xth), 1 mM DTT, 100 µg/ml bovine serum albumin and either limiting amounts of purified enzyme or 6 µg of cell-free extract, unless otherwise stated. Incubations were carried out at 37°C for 5 min or 30 min for cell-free extracts, unless stated otherwise. Reaction products were analysed by electrophoresis in denaturing 20% (w/v) polyacrylamide gels (PAGE) (20:1, 7 M urea, 0.5× TBE), visualised with a Storm 840 PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and quantified using ImageQuant software.
RESULTS
Comparison of activities of various DNA repair proteins on plasmid DNA or oligonucleotides treated with DNA-damaging agents
AP endonucleases (Xth, Nfo, HAP1 and Apn1) as well as DNA glycosylases/AP lyases (Nth and Fpg) generate single-strand breaks at the site of damage in duplex DNA. Upon introduction of a single-strand nick, supercoiled plasmid DNA converts to an open circular form, which can be separated by electrophoresis in an agarose gel. This assay was used to compare the incision activity of the Xth, Nfo, HAP1, Nth and Fpg proteins towards supercoiled plasmid DNA treated with H2O2, KMnO4 or DMS/alkali. It should be emphasised that the absence of contaminating activities in the Nfo preparation was rigorously tested by confirming its activities on THF- and DHU-containing oligonucleotides after heat treatment for 10 min at 65°C. Nfo retained about 70% activity on all substrates tested (see Materials and Methods).
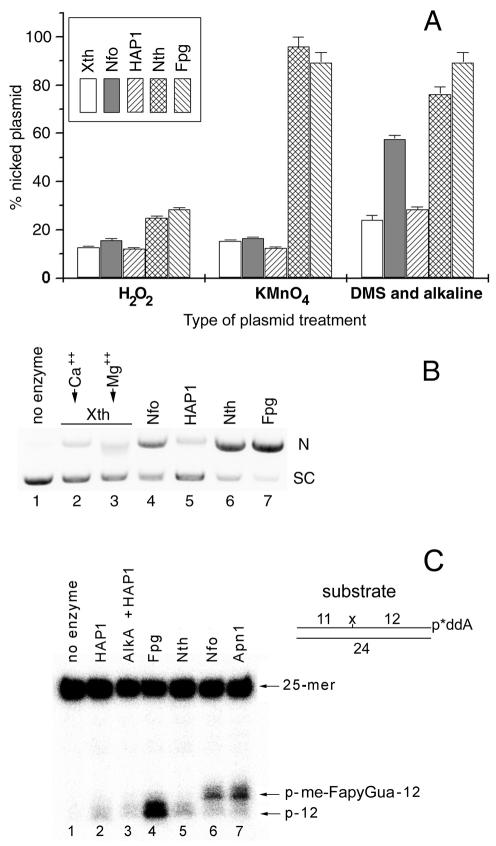
As shown in Figure 1A, the DNA glycosylases/AP lyases were more efficient than AP endonucleases on all plasmid DNA substrates tested. For all proteins tested, low incision of H2O2-treated plasmid DNA was detected as compared to KMnO4 and DMS/alkali treatments (Fig. 1A). Interestingly, the incision efficiencies of Nth and Fpg on all damaged plasmids were similar. In contrast, among AP endonucleases, the Nfo protein was more efficient than Xth and HAP1 on DMS/alkali-treated plasmid DNA, and to a lesser extent on DNA treated with H2O2 or KMnO4 (Fig. 1A). Figure 1B shows a typical agarose gel electrophoresis to measure the incision activity of the proteins on plasmid DNA substrates. As shown in Figure 1B, Nfo, Nth and Fpg all efficiently incise supercoiled plasmid DNA containing me-FapyGua residues (lanes 4, 6 and 7) whereas little incision was observed with Xth and HAP1 (lanes 2, 3 and 5). This result indicates that Nfo is able to incise me-FapyGua-containing DNA directly, although less efficiently than Nth and Fpg do.
Figure 1.
Incision of a supercoiled plasmid DNA by various DNA repair proteins. For the incision assay, 0.8 µg of chemically modified pUC19 and 10 ng of protein were incubated at 37°C for 10 min. The reaction products were separated by electrophoresis on a 0.8% agarose gel in the presence of ethidium bromide (0.5 µg/ml). The gel was photographed and quantified using Bio-Profil software. (A) Relative nicking activity of various enzymes. (B) DMS/alkali-treated plasmid DNA was incubated with different enzymes and analysed by agarose gel electrophoresis. (C) An aliquot of 1 pmol of [3′-32P]ddAMP-labelled me-FapyGua·C duplex oligonucleotide was incubated with 6 ng of protein at 37°C for 10 min.
To study the mechanism of DNA glycosylase-independent cleavage catalysed by AP endonucleases the [3′-32P]ddAMP-labelled me-FapyGua·C duplex oligonucleotide was incubated in the presence of HAP1, AlkA/HAP1, Fpg, Nth, Nfo and Apn1 proteins. As shown in Figure 1C, little incision was detected when using HAP1, AlkA/HAP1 and Nth proteins, suggesting that the oligonucleotide contains negligible numbers of AP sites or 7-methylguanine residues. In contrast and as expected, the Fpg, Nfo and Apn1 proteins very efficiently incise me-FapyGua·C (Fig. 1B, lanes 4 and 7). Essentially, Fpg-catalysed incision produced a 12mer DNA fragment lacking a me-FapyGua residue (Fig. 1C, lane 4), while Nfo and Apn1 generated mainly 13mer DNA fragments (Fig. 1C, lanes 6 and 7). This result indicates that the Nfo and Apn1 proteins do not excise me-FapyGua residues, but instead hydrolyse the phosphodiester bond 5′ to the lesion, leaving a dangling me-Fapy residue on the 5′-terminus of the downstream fragment.
Comparison of substrate specificities of Nfo, Apn1, Nth and Fpg proteins using defined oligonucleotide duplexes
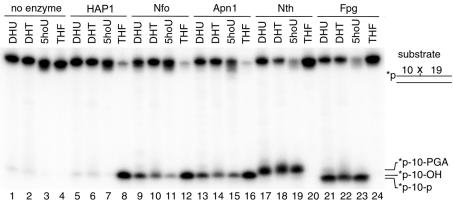
The incision activities of HAP1, Nfo, Apn1, Nth and Fpg proteins towards oxidatively damaged DNA were tested using 5′-32P-labelled duplex oligonucleotides containing DHU·G, DHT·A, 5ohU·G and THF·G base pairs (Fig. 2). The duplex oligonucleotides were incubated with the proteins and the products of the reaction were analysed by denaturing PAGE. As shown in Figure 2, all tested proteins, except for HAP1 (lanes 5–7), recognised the oxidatively damaged pyrimidines. However, the Nfo and Apn1 proteins (lanes 9–11 and 13–15) incised DHU·G, DHT·A and 5ohU·G less efficiently as compared to Nth and Fpg (lanes 17–19 and 21–23). As expected, HAP1, Nfo and Apn1 incised the oligonucleotide containing THF, a synthetic analogue of the AP site, with high efficiency (lanes 8, 12 and 16) and that Nth and Fpg were not able to incise THF·G (lanes 20 and 24).
Figure 2.
Analysis of the substrate specificities of various DNA repair proteins. Activities of Nfo, Apn1, Nth, Fpg and HAP1 proteins towards duplex oligonucleotides containing single DHU, DHT, 5ohU or THF residues. An aliquot of 0.2 pmol of the 5′-32P-labelled 30mer duplex oligonucleotide was incubated with 3 ng of the given repair proteins at 37°C for 20 min. The reaction products were analysed as described in Materials and Methods.
The products of enzymatic incision of the DHU·G, DHT·G and 5ohU·G duplex oligonucleotides migrate differently depending on the protein involved. Fpg generates, by β-δ elimination, a 10mer 32P-labelled fragment carrying a phosphate at the 3′-terminus (Fig. 2, lanes 21–23) (55). Nfo and Apn1 hydrolytically incise on the 5′ side of the lesion producing 10mer fragments with a 3′-hydroxyl terminus (Fig. 2, lanes 8–16) that migrate more slowly than products containing a 3′-phosphate (lanes 13–15) (17,55). Nth incises at the damaged site by β-elimination, generating a 10mer with a 3′-α,β-unsaturated aldehyde residue (Fig. 2, lanes 17–19) which migrates more slowly than products containing a 3′-hydroxyl (lanes 8–16) (55). Taken together, these results demonstrate that purified Apn1, similar to its prokaryotic homologue Nfo, directly recognises oxidatively damaged pyrimidines that are substrates for DNA glycosylases/AP lyases.
The effect of sequence context on Nfo and Apn1 activity was investigated. Both enzymes efficiently incised four different DHU·G oligonucleotides with random sequence contexts (data not shown), indicating that Nfo and Apn1 can initiate repair of the lesions randomly located in the genome.
Base pair specificity of the Nfo and Apn1 proteins
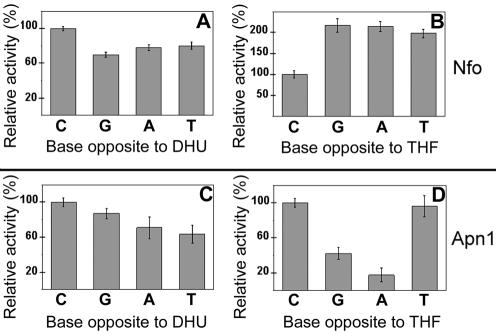
Nfo and Apn1 base pair specificity was investigated using duplex oligonucleotides containing each of the four naturally occurring bases opposite DHU and THF. The initial velocity of the incision was measured, presented in Figure 3 as relative activity of the two enzymes when acting upon the four different mismatches. Both enzymes incised DHU placed opposite any base with quite similar efficiency (Fig. 3A and C). In contrast, the repair of THF residues exhibited a marked preference for the opposite base. Nfo incised THF·dC with 2-fold lower efficiency as compared to THF·dG, THF·dA and THF·T mismatches (Fig. 3B). Apn1 strongly preferred THF·dC and THF·T mismatches whereas THF·dG and THF·dA were incised 2.5- and 7-fold less efficiently, respectively, as compared to THF·dC (Fig. 3D). Importantly, Nfo and Apn1 recognised DHU and THF only when present in duplex DNA since no detectable incision was observed on single-stranded oligonucleotide (data not shown).
Figure 3.
Base pair specificity of the Nfo (A and B) and Apn1 (C and D) proteins. The initial velocities of cleavage were measured under standard assay conditions and plotted as relative activities. For details see Materials and Methods.
Kinetic parameters for the incision of duplex DNA containing a single modified residue by the Nth, Fpg, Nfo and Apn1 proteins
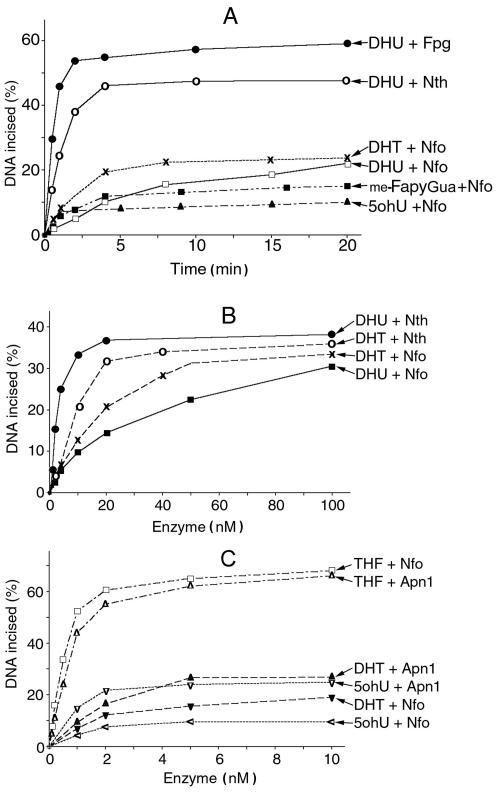
For quantitative evaluation of the substrate specificities of Apn1, Nfo, Fpg and Nth, we measured the amount of incised oligonucleotide containing a single DHU, DHT, 5ohU or me-FapyGua residue. Product formation time courses show dramatic differences between the DNA glycosylases/AP lyases and Nfo, with Fpg and Nth being most efficient (Fig. 4A). For all proteins tested, the amount of product increased linearly during the first 5 min of the reaction. Figure 4B shows protein concentration dependence of the incision of DHU·G and DHT·A by Nth and Nfo. It appears that Nth is more efficient as compared to Nfo at all concentrations tested. Up to 20 nM Nth protein is 3- to 5-fold more efficient than Nfo, although at higher enzyme concentrations the difference was very small (Fig. 4B). We have also compared the incision efficiencies of Nfo and Apn1 towards DHT·A, 5ohU·G and THF·G duplex oligonucleotides. Figure 4C shows that THF is the preferred substrate for Nfo and Apn1, whereas DHT and 5ohU are poor substrates. Interestingly, Apn1 was more efficient than Nfo when acting upon DHT·A and 5ohU·G.
Figure 4.
Activities of Nfo, Apn1, Nth and Fpg proteins towards oligonucleotides containing THF, DHU, DHT, me-FapyGua and 5ohU, as a function of time and protein concentration. (A) Time–course plot. Aliquots of 0.6 pmol of the 5′-32P-labelled oligonucleotides were incubated with 6 ng of a given repair protein at 37°C. (B and C) Enzyme concentration dependence. An aliquot of 1 pmol of the oligonucletide was incubated with the indicated amounts of the repair protein at 37°C for 5 min.
To further compare substrate specificities of the Nth, Nfo and Apn1 proteins, we measured the Km, kcat and kcat/Km values using duplex oligonucleotides containing a single modified residue. As shown in Table 1, the preferred substrate for the Nfo and Apn1 proteins was THF·G, whereas DHU·G, DHT·A, 5ohU·G and me-FapyGua·C were repaired 50- to 400-fold less efficiently. The apparent Km and kcat/Km values for Nth and hNth1 when acting upon DHU·G indicate that in vitro the DNA glycosylases/AP lyases have ∼4- to 7-fold higher catalytic efficiency as compared to Nfo and Apn1 (Table 1).
Table 1. Kinetic constants of the E.coli Nth and Nfo, S.cerevisiae Apn1 and human Nth1 proteins for the incision of duplex oligonucleotide containing a single modified residue.
| Protein | Substratea | Km (nM) | kcat (min–1) | kcat/Km (min–1 µM–1) |
|---|---|---|---|---|
| Nth | DHU·Gb | 15 ± 3 | 0.34 ± 0.02 | 23 |
| hNth1 | DHU·Gc | 47 | 0.6 ± 0.08 | 13 |
| Nfo | DHU·Gb | 74 ± 7 | 0.24 ± 0.02 | 3.2 |
| DHT·A | 38 ± 8 | 0.25 ± 0.01 | 6.6 | |
| 5ohU·G | 316 ± 44 | 1.6 ± 0.08 | 5.1 | |
| me-FapyGua·C | 250 ± 70 | 0.95 ± 0.1 | 3.8 | |
| THF·G | 1.3 ± 0.2 | 1.7 ± 0.1 | 1300 | |
| Apn1 | DHU·G | 103 ± 10 | 0.96 ± 0.03 | 9.3 |
| DHT·A | 23 ± 3 | 0.2 ± 0.01 | 8.6 | |
| 5ohU·G | 26 ± 3 | 0.34 ± 0.01 | 13 | |
| me-FapyGua·C | 50 ± 10 | 0.7 ± 0.1 | 14 | |
| THF·G | 0.9 ± 0.2 | 0.6 ± 0.04 | 670 |
a0.1–3000 nM of a 30mer duplex oligonucleotide substrate was incubated under standard reaction conditions (see Materials and Methods). For Km and kcat determination, the linear velocity was measured and the constants were determined from Lineweaver–Burk plots.
bData taken from Ischenko and Saparbaev (17).
cData taken from Ikeda et al. (70).
Nucleotide incision activity in cell-free extracts from E.coli and S.cerevisiae
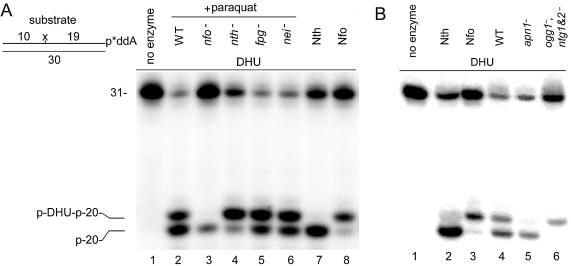
Analysis the of enzyme kinetics suggests that in vivo DNA glycosylases would recognise oxidative DNA base damage more efficiently than AP endonucleases. Therefore, we decided to compare endonuclease and DNA glycosylase activities in cell-free extracts. Previously we had shown that in cell-free extracts DNA glycosylase and AP endonuclease activities can be distinguished by the size of the cleavage product (17). As shown in Figure 5, when cleaving the 3′-labelled DHU·G oligonucleotide, DNA glycosylases/AP lyases generated a 20mer fragment (Fig. 5A, lane 7 and Fig. 5B, lane 2). In contrast, AP endonucleases generated a 21mer fragment (Fig. 5A, lane 8 and Fig. 5B, lane 3). Using this approach, we investigated the efficiency of the NIR and BER pathways in cell-free extracts from various E.coli and S.cerevisiae strains. In agreement with the kinetic data, we detected mainly DNA glycosylase and very little Nfo activity in extracts from wild-type E.coli (data not shown). Previously it was shown that in vivo, upon induction of the soxRS regulon by oxidative stress, the cellular concentration of Nfo protein increases up to 40 times. As shown in Figure 5A, addition of paraquat to growing cultures of wild-type E.coli (lane 2) greatly increased the amount of the 21mer fragment (lane 2) up to the level of the 20mer fragment generated by the DNA glycosylases/AP lyases. As expected from the known induction of Nfo protein by this oxidising agent, we detected no Nfo activity in paraquat-induced nfo E.coli (lane 3). However, the paraquat-induced Nfo activity is a dominant activity in all single DNA glycosylase/AP lyase mutants such as fpg–, nth– and nei– strains (lanes 4–6).
Figure 5.
NIR and BER activities in cell-free extracts of E.coli and S.cerevisiae. The [3′-32P]ddAMP-labelled DHU·G duplex oligonucleotides were incubated with cell-free extract of wild-type and mutant strains of paraquat-induced E.coli (A) or from S.cerevisiae (B) under standard reaction conditions. For details see Materials and Methods.
In contrast to its bacterial homologue, Apn1 is a major AP endonuclease in budding yeast. Previously we have shown that nucleotide incision is the main activity in crude extracts from S.cerevisiae strain MKp-o. However, depending on the genetic background, the level of DNA glycosylase and AP endonuclease activities could vary in different strains. Indeed, Figure 5B shows that in crude extracts from S.cerevisiae strain FF18733, Apn1 activity is a major, but not the main, activity, since about half of the cleavage is generated by DNA glycosylase-dependent incision (lane 4). Similar results were obtained when yeast cell extract was supplemented with 5 mM Mg2+ (data not shown). In agreement with the previous results, the NIR pathway in yeast was absolutely dependent on the APN1 gene product (Fig. 5B, lane 5), whereas the BER pathway required the NTG1 and NTG2 gene products (Fig. 5B, lane 6). These results suggest that AP endonuclease and DNA glycosylase activities in cell-free extracts can vary depending on the strain used. Nevertheless, in all yeast strains tested, nucleotide incision was a major activity on oxidative DNA base damage.
DISCUSSION
In the NIR pathway, an endonuclease nicks oxidatively damaged DNA in a DNA glycosylase-independent manner, providing the correct ends for DNA synthesis coupled to repair of the remaining 5′-dangling damaged nucleotide (17). This mechanistic feature provides an advantage over DNA glycosylase-mediated BER. It was shown earlier that E.coli DNA polymerase I (pol I) in the presence of dNTPs and/or human FEN1 in the presence of proliferating cell nuclear antigen efficiently eliminates the 5′-terminal dangling DHU residue from the nicked duplex oligonucleotide as a mononucleotide (17). Thus completion of NIR in eukaryotes would occur through the FEN1-dependent long patch repair pathway described in human cells (56). Obviously, the post-incision repair events would compete with simple re-ligation without repair. Nevertheless, a full DNA repair cycle seems to be a well-coordinated process, in which the first enzyme involved recruits the second, etc., and some enzymes modulate each other’s activities, as shown for the coordination of FEN1 and DNA ligase I by HAP1 in long patch BER (57). Furthermore, we have shown that distortion of a nicked duplex at the place of the damaged nucleotide, such as a me-Fapy or DHU residue, strongly inhibits the re-ligation process (data not shown). The existence of a human damage-specific endonuclease that is involved in the NIR pathway has been demonstrated, however, its identity has so far not been established (17) and searches of the human genome database have not revealed any homologue of Nfo or Apn1. Substrate specificities of the BER and NIR pathways overlap, therefore, the oxidative DNA damage could be repaired in vivo in either a DNA glycosylase-dependent or -independent manner. To evaluate the biological role of these repair pathways, we have investigated the substrate specificities of the homogeneous Nth, Fpg, Nfo and Apn1 proteins using various DNA substrates.
Supercoiled plasmid DNA treated with H2O2, KMnO4 and DMS/alkali was used to compare the incision efficiency of the Xth, Nfo, HAP1, Nth and Fpg proteins. In the presence of metal ions, hydrogen peroxide (H2O2) generates strand breaks (58) and base damage (59) in DNA. All enzymes tested generated little incision on H2O2-treated DNA, suggesting that H2O2 is inefficient, at least under the experimental conditions used (Fig. 1A). The relative efficiency of incision of H2O2-treated DNA was Fpg ≥ Nth > Nfo > Xth = HAP1. The major product of base oxidation in an oligonucleotide treated with KMnO4 is a 5R,6S-cis-thymine glycol (47), although amounts of 5-hydroxy-5-methylhydantoin, cytosine glycol, 5,6-dihydroxycytosine, 5-hydroxyhydantoin, 8-hydroxyadenine and Fapy derivatives of purines were also detected in plasmid DNA (60,61). In contrast to H2O2, KMnO4-treated DNA was a good substrate for DNA glycosylases/AP lyases, and the relative order of efficiency was Nth ≥ Fpg >> Nfo ≥ Xth > HAP1 (Fig. 1A). Alkali treatment of methylated DNA generates me-FapyGua, which closely mimics FapyGua residues generated in DNA by ionising radiation (48). Previously we have shown that Nfo incises duplex oligonucleotides containing a single me-FapyGua residue. In the present study, we show that plasmid DNA and oligonucleotides containing me-FapyGua residues are good substrates for Nfo and Apn1, but not for Xth and HAP1 (Fig. 1). As expected, Nfo and Apn1 proteins hydrolyse the phosphodiester bond 5′ to a me-FapyGua residue, leaving the lesion attached to the 5′-terminus of the downstream fragment and an OH group on the 3′-terminus of the upstream fragment (Fig. 1C, lanes 6 and 7). In agreement with previous data, DMS/alkali-treated DNA was efficiently incised by Fpg (49,62), and the relative order of efficiency was Fpg > Nth > Nfo >> HAP1 ≥ Xth (Fig. 1A). It has been demonstrated that Nth can excise me-FapyGua residues, however, this activity is low on me-FapyGua·C base pairs (63). In agreement, little activity was detected on me-FapyGua·C oligonucleotides (Fig. 1C, lane 5). However, in plasmid DNA Nth efficiently recognised me-FapyGua·C (Fig. 1B, lane 6), suggesting that the sequence context, rather than the opposite base, might have a dramatic effect on the enzyme activity. Overall, the results obtained with plasmid DNA demonstrate that: (i) in vitro the DNA glycosylases/AP lyases are more efficient as compared to AP endonucleases; (ii) Nfo has a broader substrate specificity than Xth and HAP1.
Using defined oligonucleotide substrates we characterised the substrate specificity of the homogeneous Apn1 protein and compared it with other enzymes. As shown in Figure 2, and in agreement with previous observations, Nfo, Apn1, Nth and Fpg proteins recognise DHU, DHT and 5-ohU residues when present in DNA. Nfo and Apn1 were somewhat less efficient as compared to Nth and Fpg proteins (Fig. 2). Furthermore, an abasic site analog (THF) was the preferred substrate for AP endonucleases as compared to modified bases (Fig. 2). Importantly, Nfo and Apn1 proteins were able to efficiently incise all four DHU·G oligonucleotides with different sequence contexts, suggesting that these enzymes can initiate repair of randomly located lesions in genomic DNA (data not shown).
The preferential recognition of a modified residue paired with one of the four natural bases in duplex DNA is an important feature of DNA repair enzymes. We investigated the influence of the base opposite DHU or THF residues on the incision of duplex oligonucleotides by Nfo and Apn1 (Fig. 3). The Nfo and Apn1 proteins incised DHU in all four mismatches with a similar efficiency (Fig. 3A and C). In contrast, incision of a duplex oligonucleotide containing THF varied dramatically depending on the mismatch (Fig. 3B and D). Nfo was twice as active on a THF residue opposite dG, dA or T as compared to dC, whereas Apn1 was 2- to 5-fold more active on THF opposite dC or T than opposite dG or dA. The apparently different activity profiles towards DHU and THF substrates may reflect the fact that the structure and thermodynamic stability of various DHU mismatches are quite similar, whereas for THF these parameters strongly depend on the opposite base. The data obtained with Nfo acting on THF mismatches in the present study are in agreement with previous observations (64).
The crystal structure of the Nfo·THF–DNA complex has been solved (65). Nfo is a single domain α8β8-barrel fold protein. The protein structure, well suited for binding to large molecules such as DNA, contains three Zn2+ ions at the active site which are critical for activity. The Zn2+ ions are bound in the protein by conserved residues that cluster at the centre of a crescent-shaped cavity. The Nfo protein detects AP sites by inserting its side chains into the minor groove of DNA. It then flips the target AP site and the opposite nucleotide out of the DNA base stack to produce a 90° bend in the DNA backbone. Importantly, the Nfo·THF–DNA complex reveals the basis of Nfo cleavage specificity, as normal β configuration nucleotides are sterically excluded from binding in the enzyme active pocket (65). Therefore, a nucleotide in the α configuration, such as α-deoxyadenosine, can be accommodated by placing the adduct in a solvent-accessible pocket on the enzyme surface. Consistent with this, Nfo efficiently binds and cleaves α-deoxyadenosine, one of the major products of ionising radiation under anoxic conditions (24). It has been shown that the NIR activity and AP site cleavage activity share the same active site (66,67). However, it is still unclear how an AP endonuclease can accommodate the abasic residue and bulky modified bases such as DHU, me-FapyGua (17) and 3,N4-benzetheno-dC (25) within the same active site.
The fact that DHU is a substrate for several different DNA repair enzymes in vitro does not necessarily indicate that all of these enzymes play equal roles in the processing of this lesion in vivo. In view of this problem, we measured linear velocities and kinetic parameters of Nth, Nfo and Apn1 towards various types of oxidative DNA damage. Analysis of the data presented in Figure 4 and Table 1 indicates that the primary substrate for the Nfo and Apn1 proteins is an AP site. Furthermore, as shown in Table 1, the kcat/Km value of Nth acting upon DHU is 7-fold higher than the value for Nfo, suggesting that in E.coli the majority of DHU residues may be processed by the BER pathway, whereas human Nth1 is only 1.4-fold more efficient than yeast Apn1, indicating that in eukaryotes DHU might be processed equally well by both the BER and NIR pathways (Table 1). Overall, a comparison of the kinetic parameters for AP endonucleases and DNA glycosylases/AP lyases demonstrates that Nfo and Apn1 are less efficient when acting upon oxidative DNA base damage as compared to Nth, Fpg and hNth1.
Data obtained in vitro using purified enzymes and defined oligonucleotide substrates cannot be simply extrapolated to an in vivo situation without taking into account the complex cellular response to DNA damage and integration of a given enzymatic reaction in the repair network. In E.coli, Nfo expression is induced by superoxide anion generators (19,68), whereas in S.cerevisiae Apn1 is the predominant constitutive AP endonuclease (26). Therefore, we used cell-free extracts from S.cerevisiae and paraquat-induced E.coli to compare NIR and BER activities towards DHU·G. Figure 5 clearly demonstrates that incision of DHU·G in bacteria and yeast cell extracts has an absolute requirement for both AP endonucleases (Fig. 5A, lane 3 and Fig. 5B, lane 5) and DNA glycosylases/AP lyases (Fig. 5A, lane 4 and Fig. 5B, lane 6). Furthermore, the level of NIR activity in extracts from wild-type strains was about the same as the level of BER activity. These results indicate that in vivo the choice between two alternative repair pathways does not solely depend on kinetic efficiency of a single enzyme, but also on other factors, such as the physiological concentration of the enzyme in the cell and oxidative stress conditions.
In conclusion, the severe phenotype of AP endonuclease-deficient mutants towards oxidising agents (13,15,16,69) supports the important role of the NIR pathway in vivo. Indeed, NIR activity is perfectly integrated in the repair reaction chain since it directly generates proper ends for DNA synthesis on one side of the nick and for elimination of the lesion by specific nucleases on the other side. The absence of genotoxic intermediates in the NIR pathway might create an advantage as compared to BER. The substrate specificities of Nfo-like AP endonucleases provide the biological relevance of the NIR pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Dr J. Laval for stimulating this work, Drs S. Boiteux and S. Wallace for yeast and bacterial strains, Dr R. Roy for human Nth1 protein and Drs B. Mullan and D. Zharkov for critical reading of the manuscript. This research was supported by European Community Grant QLK4-2000-00286 and l’Association pour la Recherche sur le Cancer (M.S.). A.A.I. is a recipient of grants from the Russian Foundation for Basic Research (01-04-48892) and is a Chercheur Associé in the CNRS.
REFERENCES
- 1.Seeberg E., Eide,L. and Bjoras,M. (1995) The base excision repair pathway. Trends Biochem. Sci., 20, 391–397. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. (2001) Keynote: past, present and future aspects of base excision repair. Prog. Nucleic Acid Res. Mol. Biol., 68, xvii–xxx. [DOI] [PubMed] [Google Scholar]
- 3.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scharer O.D. and Jiricny,J. (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays, 23, 270–281. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl T. (1976) New class of enzymes acting on damaged DNA. Nature, 259, 64–66. [DOI] [PubMed] [Google Scholar]
- 6.McCullough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 7.Laval J. (1977) Two enzymes are required from strand incision in repair of alkylated DNA. Nature, 269, 829–832. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham R.P. and Weiss,B. (1985) Endonuclease III (nth) mutants of Escherichia coli. Proc. Natl Acad. Sci. USA, 82, 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaisdell J.O. and Wallace,S.S. (2001) Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl Acad. Sci. USA, 98, 7426–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas D., Scot,A.D., Barbey,R., Padula,M. and Boiteux,S. (1997) Inactivation of OGG1 increases the incidence of G.C→T.A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet., 254, 171–178. [DOI] [PubMed] [Google Scholar]
- 11.Alseth I., Eide,L., Pirovano,M., Rognes,T., Seeberg,E. and Bjoras,M. (1999) The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol., 19, 3779–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedberg E.C. and Meira,L.B. (2003) Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage. Version 5. DNA Repair (Amst.), 2, 501–530. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham R.P., Saporito,S.M., Spitzer,S.G. and Weiss,B. (1986) Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol., 168, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramotar D., Popoff,S.C., Gralla,E.B. and Demple,B. (1991) Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol. Cell. Biol., 11, 4537–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett R.A. (1999) The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol. Cell. Biol., 19, 1800–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig D.L., MacInnes,M.A., Takiguchi,Y., Purtymun,P.E., Henrie,M., Flannery,M., Meneses,J., Pedersen,R.A. and Chen,D.J. (1998) A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res., 409, 17–29. [DOI] [PubMed] [Google Scholar]
- 17.Ischenko A.A. and Saparbaev,M.K. (2002) Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature, 415, 183–187. [DOI] [PubMed] [Google Scholar]
- 18.Ljungquist S. (1977) A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J. Biol. Chem., 252, 2808–2814. [PubMed] [Google Scholar]
- 19.Chan E. and Weiss,B. (1987) Endonuclease IV of Escherichia coli is induced by paraquat. Proc. Natl Acad. Sci. USA, 84, 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunoshiba T., Hidalgo,E., Li,Z. and Demple,B. (1993) Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J. Bacteriol., 175, 7492–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsaneva I.R. and Weiss,B. (1990) soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol., 172, 4197–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demple B. and Amabile-Cuevas,C.F. (1991) Redox redux: the control of oxidative stress responses. Cell, 67, 837–839. [DOI] [PubMed] [Google Scholar]
- 23.Ramotar D., Popoff,S.C. and Demple,B. (1991) Complementation of DNA repair-deficient Escherichia coli by the yeast Apn1 apurinic/apyrimidinic endonuclease gene. Mol. Microbiol., 5, 149–155. [DOI] [PubMed] [Google Scholar]
- 24.Ide H., Tedzuka,K., Shimzu,H., Kimura,Y., Purmal,A.A., Wallace,S.S. and Kow,Y.W. (1994) Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV. Biochemistry, 33, 7842–7847. [DOI] [PubMed] [Google Scholar]
- 25.Hang B., Chenna,A., Fraenkel-Conrat,H. and Singer,B. (1996) An unusual mechanism for the major human apurinic/apyrimidinic (AP) endonuclease involving 5′ cleavage of DNA containing a benzene-derived exocyclic adduct in the absence of an AP site. Proc. Natl Acad. Sci. USA, 93, 13737–13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popoff S.C., Spira,A.I., Johnson,A.W. and Demple,B. (1990) Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc. Natl Acad. Sci. USA, 87, 4193–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson A.W. and Demple,B. (1988) Yeast DNA diesterase for 3′-fragments of deoxyribose: purification and physical properties of a repair enzyme for oxidative DNA damage. J. Biol. Chem., 263, 18009–18016. [PubMed] [Google Scholar]
- 28.Johnson A.W. and Demple,B. (1988) Yeast DNA 3′-repair diesterase is the major cellular apurinic/apyrimidinic endonuclease: substrate specificity and kinetics. J. Biol. Chem., 263, 18017–18022. [PubMed] [Google Scholar]
- 29.Kunz B.A., Henson,E.S., Roche,H., Ramotar,D., Nunoshiba,T. and Demple,B. (1994) Specificity of the mutator caused by deletion of the yeast structural gene (APN1) for the major apurinic endonuclease. Proc. Natl Acad. Sci. USA, 91, 8165–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joenje H. (1989) Genetic toxicology of oxygen. Mutat. Res., 219, 193–208. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 32.Fuciarelli A.F., Wegher,B.J., Blakely,W.F. and Dizdaroglu,M. (1990) Yields of radiation-induced base products in DNA: effects of DNA conformation and gassing conditions. Int. J. Radiat. Biol., 58, 397–415. [DOI] [PubMed] [Google Scholar]
- 33.Dizdaroglu M., Laval,J. and Boiteux,S. (1993) Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry, 32, 12105–12111. [DOI] [PubMed] [Google Scholar]
- 34.Furlong E.A., Jorgensen,T.J. and Henner,W.D. (1986) Production of dihydrothymidine stereoisomers in DNA by gamma-irradiation. Biochemistry, 25, 4344–4349. [DOI] [PubMed] [Google Scholar]
- 35.Duplaa A.M. and Teoule,R. (1985) Sites of gamma radiation-induced DNA strand breaks after alkali treatment. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med., 48, 19–32. [DOI] [PubMed] [Google Scholar]
- 36.Schulhof J.C., Molko,D. and Teoule,R. (1988) Synthesis of DNA fragments containing 5,6-dihydrothymine, a major product of thymine gamma radiolysis. Nucleic Acids Res., 16, 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo Y. and Witkop,B. (1968) The selective photolysis of dihydrothymidine. J. Am. Chem. Soc., 90, 3258–3259. [DOI] [PubMed] [Google Scholar]
- 38.Ide H., Petrullo,L.A., Hatahet,Z. and Wallace,S.S. (1991) Processing of DNA base damage by DNA polymerases. Dihydrothymine and beta-ureidoisobutyric acid as models for instructive and noninstructive lesions. J. Biol. Chem., 266, 1469–1477. [PubMed] [Google Scholar]
- 39.Evans J., Maccabee,M., Hatahet,Z., Courcelle,J., Bockrath,R., Ide,H. and Wallace,S. (1993) Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat. Res., 299, 147–156. [DOI] [PubMed] [Google Scholar]
- 40.Dizdaroglu M., Holwitt,E., Hagan,M.P. and Blakely,W.F. (1986) Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem. J., 235, 531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner J.R., Hu,C.C. and Ames,B.N. (1992) Endogenous oxidative damage of deoxycytidine in DNA. Proc. Natl Acad. Sci. USA, 89, 3380–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teoule R. (1987) Radiation-induced DNA damage and its repair. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med., 51, 573–589. [DOI] [PubMed] [Google Scholar]
- 43.Kunkel T.A. and Bebenek,K. (2000) DNA replication fidelity. Annu. Rev. Biochem., 69, 497–529. [DOI] [PubMed] [Google Scholar]
- 44.Gros L., Saparbaev,M.K. and Laval,J. (2002) Enzymology of the repair of free radicals-induced DNA damage. Oncogene, 21, 8905–8925. [DOI] [PubMed] [Google Scholar]
- 45.Saparbaev M. and Laval,J. (1994) Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat and human alkylpurine DNA glycosylases. Proc. Natl Acad. Sci. USA, 91, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karakaya A., Jaruga,P., Bohr,V.A., Grollman,A.P. and Dizdaroglu,M. (1997) Kinetics of excision of purine lesions from DNA by Escherichia coli Fpg protein. Nucleic Acids Res., 25, 474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kao J.Y., Goljer,I., Phan,T.A. and Bolton,P.H. (1993) Characterization of the effects of a thymine glycol residue on the structure, dynamics and stability of duplex DNA by NMR. J. Biol. Chem., 268, 17787–17793. [PubMed] [Google Scholar]
- 48.Boiteux S., Gajewski,E., Laval,J. and Dizdaroglu,M. (1992) Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry, 31, 106–110. [DOI] [PubMed] [Google Scholar]
- 49.Asagoshi K., Yamada,T., Terato,H., Ohyama,Y., Monden,Y., Arai,T., Nishimura,S., Aburatani,H., Lindahl,T. and Ide,H. (2000) Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J. Biol. Chem., 275, 4956–4964. [DOI] [PubMed] [Google Scholar]
- 50.Karahalil B., Girard,P.M., Boiteux,S. and Dizdaroglu,M. (1998) Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res., 26, 1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiederholt C.J., Delaney,M.O., Pope,M.A., David,S.S. and Greenberg,M.M. (2003) Repair of DNA containing Fapy.dG and its beta-C-nucleoside analogue by formamidopyrimidine DNA glycosylase and MutY. Biochemistry, 42, 9755–9760. [DOI] [PubMed] [Google Scholar]
- 52.Boiteux S., Belleney,J., Roques,B.P. and Laval,J. (1984) Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res., 12, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saparbaev M. and Laval,J. (1998) 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 95, 8508–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Privezentzev C.V., Saparbaev,M. and Laval,J. (2001) The HAP1 protein stimulates the turnover of human mismatch-specific thymine-DNA-glycosylase to process 3,N(4)-ethenocytosine residues. Mutat. Res., 480/481, 277–284. [DOI] [PubMed] [Google Scholar]
- 55.Bailly V., Verly,W.G., O’Connor,T. and Laval,J. (1989) Mechanism of DNA strand nicking at apurinic/apyrimidinic sites by Escherichia coli [formamidopyrimidine]DNA glycosylase. Biochem. J., 262, 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klungland A. and Lindahl,T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ranalli T.A., Tom,S. and Bambara,R.A. (2002) AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J. Biol. Chem., 277, 41715–41724. [DOI] [PubMed] [Google Scholar]
- 58.McDonald R.J., Pan,L.C., St George,J.A., Hyde,D.M. and Ducore,J.M. (1993) Hydrogen peroxide induces DNA single strand breaks in respiratory epithelial cells. Inflammation, 17, 715–722. [DOI] [PubMed] [Google Scholar]
- 59.Blakely W.F., Fuciarelli,A.F., Wegher,B.J. and Dizdaroglu,M. (1990) Hydrogen peroxide-induced base damage in deoxyribonucleic acid. Radiat. Res., 121, 338–343. [PubMed] [Google Scholar]
- 60.Akman S.A., Doroshow,J.H. and Dizdaroglu,M. (1990) Base modifications in plasmid DNA caused by potassium permanganate. Arch. Biochem. Biophys., 282, 202–205. [DOI] [PubMed] [Google Scholar]
- 61.Nawamura T., Negishi,K. and Hayatsu,H. (1994) 8-Hydroxyguanine is not produced by permanganate oxidation of DNA. Arch. Biochem. Biophys., 311, 523–524. [DOI] [PubMed] [Google Scholar]
- 62.Graves R., Laval,J. and Pegg,A.E. (1992) Sequence specificity of DNA repair by Escherichia coli Fpg protein. Carcinogenesis, 13, 1455–1459. [DOI] [PubMed] [Google Scholar]
- 63.Asagoshi K., Yamada,T., Okada,Y., Terato,H., Ohyama,Y., Seki,S. and Ide,H. (2000) Recognition of formamidopyrimidine by Escherichia coli and mammalian thymine glycol glycosylases. Distinctive paired base effects and biological and mechanistic implications. J. Biol. Chem., 275, 24781–24786. [DOI] [PubMed] [Google Scholar]
- 64.Takeuchi M., Lillis,R., Demple,B. and Takeshita,M. (1994) Interactions of Escherichia coli endonuclease IV and exonuclease III with abasic sites in DNA. J. Biol. Chem., 269, 21907–21914. [PubMed] [Google Scholar]
- 65.Hosfield D.J., Guan,Y., Haas,B.J., Cunningham,R.P. and Tainer,J.A. (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell, 98, 397–408. [DOI] [PubMed] [Google Scholar]
- 66.Hang B., Rothwell,D.G., Sagi,J., Hickson,I.D. and Singer,B. (1997) Evidence for a common active site for cleavage of an AP site and the benzene-derived exocyclic adduct, 3,N4-benzetheno-dC, in the major human AP endonuclease. Biochemistry, 36, 15411–15418. [DOI] [PubMed] [Google Scholar]
- 67.Jilani A., Vongsamphanh,R., Leduc,A., Gros,L., Saparbaev,M. and Ramotar,D. (2003) Characterization of two independent amino acid substitutions that disrupt the DNA repair functions of the yeast Apn1. Biochemistry, 42, 6436–6445. [DOI] [PubMed] [Google Scholar]
- 68.Walkup L.K. and Kogoma,T. (1989) Escherichia coli proteins inducible by oxidative stress mediated by the superoxide radical. J. Bacteriol., 171, 1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walker L.J., Craig,R.B., Harris,A.L. and Hickson,I.D. (1994) A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res., 22, 4884–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikeda S., Biswas,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]