Abstract
The genomes of eukaryotic cells predict the existence of multiple DNA polymerases, which are proposed to serve specialized roles in DNA replication and repair. We report here the isolation of the full-length human DNA POLQ gene, and an initial characterization of its gene product, DNA polymerase θ. POLQ is of particular interest as it is orthologous to Drosophila Mus308, a gene implicated in cellular resistance to interstrand DNA cross-linking agents. The POLQ cDNA encodes a polypeptide of 2592 amino acids with an ATPase-helicase domain in the N-terminal part of the protein, a central spacer domain, and a DNA polymerase domain in the C-terminal portion. This arrangement is conserved with Mus308. Expression of an mRNA of ∼8.5 kb was detected in human cell lines. In a survey of human and mouse tissues, expression was highest in testis. Immunoblotting with POLQ antibodies detected a protein of >250 kDa in extracts from HeLa cells. Prominent fragments of ∼100 kDa suggest that POLQ is readily proteolyzed. Full-length human POLQ was expressed from a baculovirus system. Purified POLQ showed DNA polymerase activity on nicked double-stranded DNA and on a singly primed DNA template. The enzyme activity was resistant to aphidicolin, consistent with its membership of the A family of DNA polymerases, and inhibited by dideoxynucleotides. POLQ further exhibited a single-stranded DNA-dependent ATPase activity.
INTRODUCTION
DNA interstrand cross-links (ICLs) cause mutations, chromosome rearrangements and inhibit DNA replication and transcription. The strong cytotoxicity exhibited by such cross-links is the basis of their use as chemotherapeutic drugs, including nitrogen mustards, mitomycin C, cisplatin and psoralens. It is consequently important to characterize enzymes involved in ICL repair. In Escherichia coli, one pathway of ICL repair has been well characterized biochemically. The UvrABC protein complex recognizes and incises one strand on both sides of a cross-link, and then RecA-dependent recombination provides a template for a second round of incision and nucleotide excision repair that also requires the UvrD DNA helicase and DNA polymerase I (Pol I) (1–3).
In eukaryotes, there appear to be several mechanisms for repair or tolerance of DNA cross-links (4–10) but, on the whole, the pathways are not yet well defined. An interesting lead comes from studies of the Drosophila melanogaster mus308 gene. Mutant alleles of this gene cause raised sensitivity of Drosophila cells to cross-linking agents without marked sensitivity to a monoalkylating agent, methyl methanesulfonate, suggesting a function specific for ICL repair (11). Some experiments have suggested that mus308 mutants have defects in repair after incision at an ICL (12). The mus308 gene encodes a protein with recognizable domains suggesting two functions. The N-terminal domain has seven motifs characteristic of DNA and RNA helicases, while the C-terminal domain shares a high sequence similarity with DNA polymerases in the A family, such as E.coli Pol I (13), but neither activity has been firmly demonstrated.
In this study, we sought a human ortholog of the mus308 gene. Human DNA sequences similar to a portion of mus308 have been reported (14) and were proposed to encode a gene product designated DNA polymerase θ. This sequence predicted a 1762 amino acid protein containing DNA polymerase motifs and a non-conserved N-terminal region but, unlike Mus308, no DNA helicase-like domain. The putative gene product was not studied and no DNA polymerase activity has been reported. By using RT–PCR and methods for rapid amplification of cDNA ends (RACE), we have isolated a considerably longer, full-length cDNA encoding human POLQ (Pol θ). This cDNA encodes a protein of 2592 amino acids with DNA helicase-like motifs at the N-terminus and DNA polymerase motifs at the C-terminus. The arrangement is similar to that of Mus308, although with a significantly longer domain connecting the two motifs. We also report here the production of recombinant POLQ protein, and a demonstration that its 290 kDa product is an A family DNA polymerase with unusual properties, and a DNA-dependent ATPase.
MATERIALS AND METHODS
Northern hybridization analysis
Northern hybridization was performed as described previously with Clontech blots (15). For human POLQ, a 300 bp cDNA C-terminal fragment derived by PCR amplification from expressed sequence tag (EST) W00829 (NCBI accession number) was used as described in the text. For mouse Polq, a C-terminal 1711 bp cDNA fragment containing motifs A, B and C of the DNA polymerase domain served as probe. The fragment was obtained by 3′ RACE, using primers designed from EST BB351500 (NCBI accession number), homologous to the D.melanogaster Mus308 polymerase domain (5′ end sequence GTGTGCTGGAATTCGGCTTGA; 3′end sequence TATATCTTTTTGCTGTTCAAATAA). After subcloning into the pT-AdV vector and sequencing, EcoRI sites in the multiple cloning sites of the vector were used to obtain the fragment used for hybridization.
Immunoblotting
DNA fragments encoding residues 844–1059 and 1634–1834 of human POLQ were inserted into pET15 expression plasmids (details available on request) and the resulting His-tagged polypeptides were expressed in E.coli. The insoluble products were purified on nickel resin under denaturing conditions, and used for immunization of rabbits.
HeLa S3 cells (5.0 × 107 per extract) were suspended in lysis buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 10 mM EDTA) and protease inhibitor cocktail (Calbiochem) was added. After incubation for 15 min on ice, the cells were lysed by passage through a 25 gauge needle. The supernatant after centrifugation was mixed with SDS loading buffer and applied to a 4–15% gradient polyacrylamide gel. Protein was electrophoretically transferred to PVDF membrane (Immobilon P, Millipore) in 10 mM CAPS, 10% methanol at 0.4 A for 45 min. After blocking treatment, membranes were incubated with primary antibodies for 1 h. Primary rabbit polyclonal antibodies were purified from sera on an antigen affinity column (Pierce). After incubation with secondary antibody (1:50 000 dilution of peroxidase-conjugated anti-rabbit IgG; Sigma), bands were visualized by chemiluminesence (Pierce).
Cloning of human POLQ cDNA
The C-terminal and central spacer portions of the POLQ cDNA were assembled starting from EST sequences W00829 and N69543 (NCBI accession numbers). From the ∼1110 bp EST sequence, primers were designed to isolate further 5′ sequences by 5′ RACE (Clontech SMART RACE cDNA Amplication Kit). Using total RNA prepared from 833K and MGH-U1 cells (15), a fragment of 6588 bp of the POLQ cDNA was isolated. After DNA sequencing, and comparison with human genome sequence information, several PCR errors were corrected by site-directed mutagenesis (Stratagene QuickChange™ Site-Directed Mutagenesis Kit). One sequence difference was not changed, a G found in place of C in the central domain which results in an R at POLQ amino acid residue 984 instead of a T. During the cloning process, a SmaI restriction site was introduced at the 3′ end of the open reading frame (ORF) to connect a FLAG tag. An XhoI site was engineered at the 3′ terminus of the FLAG tag. A SacI restriction site was introduced at the 5′ end of this clone to connect to the N-terminal portion of the gene.
To isolate the N-terminal part of the cDNA, RT–PCR was used to clone a region starting within exon 1 (where cDNA position +1 is the initiating ATG, Fig. 2A) and encompassing the helicase-like domain (to cDNA position 2526). Primers based on human genome sequence information were designed and used with total RNA from MOLT4 and HeLa cells, and poly (A)+ RNA from HeLa cells. RT–PCR used thermostable reverse transcriptase to help avoid problems with secondary structures (Thermoscript reverse transcriptase, Invitrogen), and RNase inhibitor (Invitrogen) was included in the reaction mixtures. For the 5′ RACE reactions (Fig. 2A), primary and nested second round PCR primers were: first strand primer, 5′-GCCTGCCATTCAAACATC-3′; and second nested primer, 5′-GGAAGTCCCCAGTTTGCCAATAGTAGC-3′. After DNA sequencing, both portions were connected and inserted between the BamHI and XhoI sites of pFastBac HTc (Invitrogen) for expression in baculovirus.
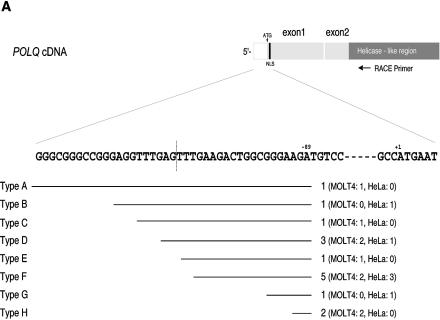
Figure 2.
Analysis of the 5′ end of the POLQ transcript and the POLQ coding sequence. (A) Diagram of the 5′ end of the cDNA determined by 5′ RACE product analysis. The dark gray rectangle is the helicase conserved region (which starts from exon 2). The 5′-non-coding region is shown in white. The black rectangle denotes the predicted nuclear localization signal (NLS). The first ATG in the POLQ ORF is indicated at position +1. The region in which the 5′ RACE primers annealed is shown by an arrow. The lower part of the diagram shows the transcribed DNA sequence around the 5′ end, as found by our 5′ RACE analysis. The dashed vertical line shows the position at which sequence NM_006596 currently predicts a junction between an exon and intron DNA, but in our analysis all of this sequence is part of the transcript. The starting site of each class of 5′ RACE products is indicated with a horizontal line. The numbers in parentheses give the number of products found in MOLT4 or HeLa S3 RNA. (B) Sequence alignment of the coding region of human POLQ (experimentally determined in this study) and mouse POLQ (18). Identical amino acids are indicated with dark shading and similar amino acids with light shading. The alignment was carried out using the Clustal X program. The locations of the conserved helicase motifs I–VI and the conserved DNA polymerase motifs 1–6 are indicated. The predicted NLS is indicated near the beginning of the protein. A conserved Q residue upstream of helicase motif I is indicated by an asterisk, and the conserved Y residue predicting ddNTP sensitivity is indicated with a caret (∧).
Purification of the human POLQ protein
The POLQ ORF cloned into plasmid pFastBacHTc resulted in a protein tagged with six His residues at the N-terminus (contributed by the pFastBac vector), and a FLAG tag at the C-terminus. The Bac-to-Bac baculovirus expression system (Invitrogen) was used to obtain recombinant baculovirus. Sf9 cells (600 ml, 6.0 × 106 cells/ml) were infected with His6-POLQ-FLAG baculovirus for 48 h at 27°C. Cells were lysed in 40 ml of buffer A [100 mM sodium phosphate buffer pH 6.0, 10% glycerol, 0.5% NP-40, 10 mM EDTA, 5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), EDTA-free protease inhibitor mixture from Roche diagnostics] for 15 min on ice. After centrifugation (6500 g for 10 min), 6 ml of buffer B [100 mM Tris–HCl pH 8.0, 0.6 M (NH4)2 SO4, 10% glycerol, 0.5% NP-40, 10 mM EDTA, 5 mM DTT, 1 mM PMSF, EDTA-free protease inhibitor mix] was added to the pellet and incubated for 10 min after gentle sonication on ice (10 cycles of 0.2 s with a 0.8 s pause). To remove DNA, polyethyleneimine was added and incubated for 15 min at 4°C. Debris was removed by centrifugation and the supernatant incubated overnight at 4°C with 100 µl of FLAG resin (Sigma). The resin was washed with 10 vols of FLAG binding buffer (50 mM sodium phosphate buffer pH 8.0, 10% glycerol, 0.1% NP-40, 5 mM EDTA, 10 mM β-mercaptoethanol, 1 M NaCl, 1 mM PMSF, EDTA-free protease inhibitor mix) and then with the same buffer without EDTA. The protein was eluted with 200 µg/ml FLAG peptide in the latter buffer. The eluate was incubated with 100 µl of Ni2+ -nitrilotriacetate superflow resin (Qiagen). The resin was washed with 10 vols of wash buffer (50 mM sodium phosphate buffer pH 8.0, 10% glycerol, 0.1% NP-40, 10 mM β-mercaptoethanol, 1 M NaCl, 1 mM PMSF, 25 mM imidazole) and the protein was eluted with the same buffer containing 250 mM imidazole.
DNA polymerase assay
An oligonucleotide forming a hairpin primer–template (Fig. 4A) was used to detect polymerase activity. The oligonucleotide was heat denatured for 5 min at 65°C and cooled down slowly for self-annealing. Standard reaction mixtures (25 µl) contained 20 mM Tris–HCl pH 7.5, 4% glycerol, 80 µg/ml bovine serum albumin (BSA), 8 mM MgCl2, 16 fmol of 5′-[32P]hairpin DNA and 1.25 ng of POLQ. dATP, dTTP, dGTP and dCTP (100 µM) each were also present unless otherwise indicated. After incubation for 30 min at 30°C, reactions were terminated by adding gel loading buffer (formamide, 0.1% xylene cyanol, 0.1% bromophenol blue, 20 mM EDTA) and boiling. Products were electrophoresed on a denaturing 10% polyacrylamide gel and analyzed with a Fuji phosphorimaging device. For assaying sensitivity to ddNTPs, 1 µg of a poly(dA)–oligo(dT) 2:1 template was used instead of the hairpin DNA. Escherichia coli DNA Pol I (Klenow fragment) was from Amersham Biosciences. To assay sensitivity to aphidicolin, 4 µg of activated calf thymus DNA (Amersham Biosciences) was used. Calf thymus DNA Pol δ was kindly provided by U. Hübscher. In both cases, reactions were stopped by adding 25 µl of 40 mM EDTA and placed on ice. A 10 µl aliquot of each mixture was spotted onto DE81 paper (Whatman), and washed three times with 0.5 M Na2HPO4 for 5 min and twice with ethanol. The paper was dried and radioactivity was quantified with a Fuji Phosphor Imager.
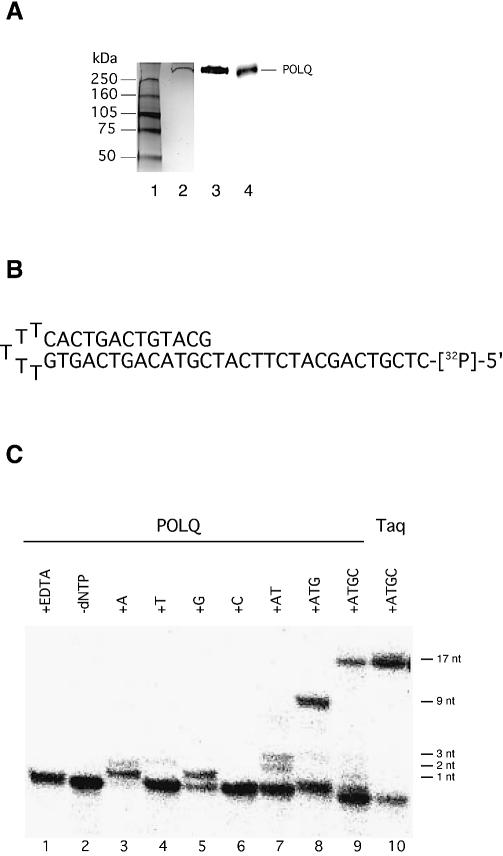
Figure 4.
DNA polymerase activity of POLQ. (A) Purified recombinant POLQ. Silver-stained gel showing molecular weight markers (lane 1) and 30 ng of purified POLQ (lane 2). Proteins were separated by electrophoresis on an SDS 4–15% polyacrylamide gradient gel. Two separate samples of 5 ng of POLQ were loaded onto similar gels, immunoblotted with a 1:1000 dilution of POLQ antibody 1 (lane 3) or antibody 2 (lane 4), and detected with secondary reagents as described in Materials and Methods. (B) The hairpin primer–template for the primer extension assay. The self- complementary oligonucleotide was labeled with 32P at the 5′ end. (C) The primer–template (16 fmol) was incubated with 1.25 ng of POLQ in the presence of a single dNTP (A, T, G or C, lanes 3–6), two dNTPs (lane 7), three dNTPs (lane 8) or all four dNTPs (lane 9). EDTA was added before incubation for the reaction in lane 1, and dNTPs were omitted in lane 2. The reaction mixture in lane 10 included all four dNTPs and 6 × 10 –4 U of Taq DNA polymerase (Promega), incubated for 30 min at 74°C.
Single-stranded DNA-dependent ATPase activity and DNA helicase activity
Reaction mixtures (10 µl) contained 50 mM KCl, 20 mM Tris–HCl pH 7.4, 8 mM MgCl2, 1 mM DTT, 50 µg/ml BSA, 0.25 µCi of [γ-32P]ATP, 1 ng of POLQ and 250 ng of M13GTGx single-stranded DNA. After incubation for 180 min at 37°C, reactions were terminated by the addition of 5 µl of 0.5 M EDTA. Released phosphate was separated from ATP by thin-layer chromatography on polyethyleneimine cellulose using 0.75 M KH2PO4 as the running buffer. Hydrolysis was quantified with the use of a Fuji phosphorimaging device. Helicase activity was tested as described (16), except that the helicase assay used M13-GTGx single-stranded phage DNA (17), annealed to a 17mer primer, TAAAACGACGGCCAGTG.
RESULTS
Identification of a mammalian gene transcript related to Pol I
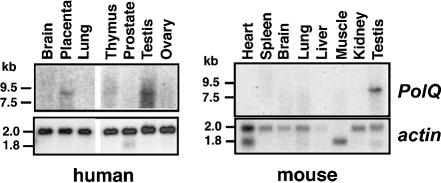
We initially searched in public databases for human sequences related to bacterial Pol I. The EST W00829 (NCBI EST accession number) was found and sequenced. A 300 bp C-terminal cDNA fragment from this EST was derived by PCR amplification and used as a probe in northern hybridization against poly(A)+ mRNAs from various human tissues. An mRNA ∼8.5 kb long was detected, albeit weakly, in human placental and testis tissue (Fig. 1, left). The expression pattern in mouse tissues was also investigated, using a 1711 bp probe generated by PCR from a mouse EST in the DNA polymerase homology region. A strong signal was detected only in mouse testis tissue, also showing a transcript of ∼8.5 kb (Fig. 1, right). A 4 kb band possibly representing a short alternatively spliced transcript was seen in human brain tissue but not observed in any mouse tissue (not shown). The human probe also detected an mRNA of ∼8.5 kb in HeLa S3 and MOLT4 (lymphoblastic leukemia) cells, with weaker signals in HL60, K562 and SW480 cells (not shown). This transcript is designated as that of POLQ, based on the experimental evidence summarized below.
Figure 1.
Detection of POLQ expression. Left: northern blot analysis of human POLQ expression in different human tissues. Right: northern blot analysis of human POLQ expression in different mouse tissues. Blots with POLQ probes were exposed to film for 15 days (human) or 8 days (mouse), while the same blots with the control actin probe were exposed for 30 min. The actin probe detected expression of non-muscle γ- and β-actin as well as the α-actin prominent in skeletal and heart muscle.
Isolation of the full-length human POLQ cDNA
After performing the northern hybridization with the human EST sequence, we noted that the EST had sequence identity with part of a DNA sequence predicted by Sharief et al. (14) to encode a protein designated as Pol θ. The 8.5 kb mRNA detected in our experiments was, however, considerably larger than the 5.7 kb that would be needed to encode the 1762 amino acid protein proposed by Sharief et al. (14). The sequence in the latter study was predicted to encode DNA polymerase motifs, but no helicase-like region. We examined accumulating human genome sequence information and found that additional potential coding sequences were present 5′ to the putative DNA polymerase-encoding sequences, and that these sequences included homology to the helicase similarity region of Drosophila Mus308. It therefore appeared that POLQ, like Mus308, contained both DNA polymerase and helicase similarity regions. We confirmed unification of the helicase and polymerase domains in one coding sequence by preliminary experiments, finding that RT–PCR using primers in the polymerase region and in the helicase homology region could recover single fragments.
A complete POLQ cDNA corresponding to the 8.5 kb transcript was assembled in several stages. The C-terminal part of POLQ was isolated by assembling sequences from ESTs and by RT–PCR as outlined in Materials and Methods. Molecular cloning of the N-terminal part of the cDNA utilized NCBI sequence information and gene prediction software. A full cDNA was cloned by RT–PCR of RNA prepared from HeLa S3 and MOLT4 cells.
To experimentally locate the 5′ end of the coding sequence, 5′ RACE-PCR was used. Primers for the nested PCRs were set in the area where helicase sequence conservation begins (Fig. 2A). Fifteen RACE-PCR products were obtained from HeLa S3 and MOLT4 total RNA, and the 5′ ends of these sequences were within 38 bp of one another (Fig. 2A). These fell into eight sequence categories, of which one appears to be derived from a splicing variant (type H) with a 56 bp deletion at the end of exon 1 that would cause early termination of the polypeptide. RACE-PCRs were not derived from contaminating genomic DNA, as no clones contained intron sequence between the first two exons.
The results indicate that the major transcripts in human cell lines start in the region indicated in Figure 2A. The first ATG in this region (position –89, Fig. 2A) is in a sequence context with a poor match to the Kozak translation initiation consensus and would give rise to a truncated polypeptide of 22 residues. The ATG start codon marked +1 in Figure 2A begins an ORF and would encode a protein of 2592 amino acids with a predicted Mr of 290 kDa. Consequently, the complete POLQ coding sequence is 7776 bp long, a size consistent with the ∼8.5 kb transcript after taking account of the 5′- and 3′-untranslated regions.
The sequence of human POLQ protein as deduced from this analysis is shown in Figure 2B. Overall, human POLQ is derived from 30 exons (Fig. 3A). The unusually large exon 16 comprises 3107 bp encoding 1036 amino acids. Exons 2–9 encode the conserved motifs found in superfamily II DNA and RNA helicases (Figs 2B and 3A). The coding sequences starting from exon 20 have significant homology with the DNA polymerase region of family A DNA polymerases. For example, the region between residues 2060 and 2592 of human POLQ is 41% similar (28% identical) to the homologous region of E.coli DNA Pol I. Analysis of the cDNA with P-SORT and other nuclear localization prediction programs strongly predicts a nuclear localization for POLQ, with a putative nuclear localization signal, KRRR, at residues 9–12. Residues 1–1028 and 1826–2592 of human POLQ are well conserved with Drosophila Mus308 (Figs 3A and 6). A poorly conserved area is encoded by the central part of the large exon 16.
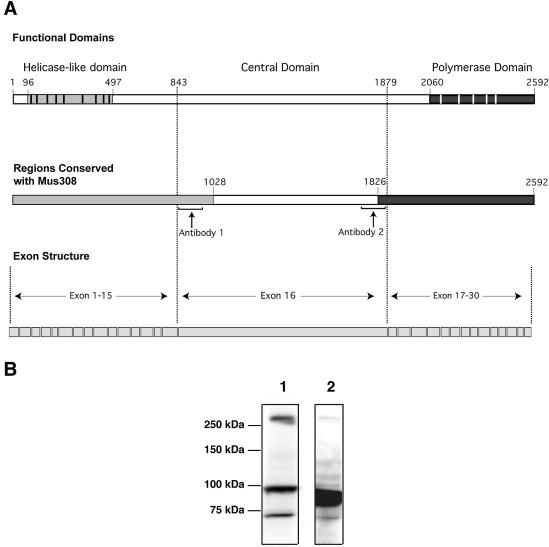
Figure 3.
POLQ domain structure, conservation and exon arrangement. (A) In the top line of the diagram, the position of a conserved helicase-like domain near the N-terminus of POLQ is shown with gray shading (black vertical bars show the positions of conserved motifs), and the position of a DNA polymerase domain is shown with black shading (white vertical bars show the positions of conserved motifs). Shaded regions in the middle line of the diagram show regions of the protein that are most highly conserved (see Fig. 6 for further detail). At the bottom, the exon structure of the protein is diagrammed. Exon 16 encodes residues 843–1879 of POLQ. Polyclonal antibodies 1 and 2 were raised against regions indicated by brackets. (B) HeLa S3 extract was analyzed by immunoblotting with polyclonal antibodies raised against the central domain of POLQ (positions indicated in the middle line in A). Lane 1, antibody 1; lane 2, antibody 2.
Figure 6.
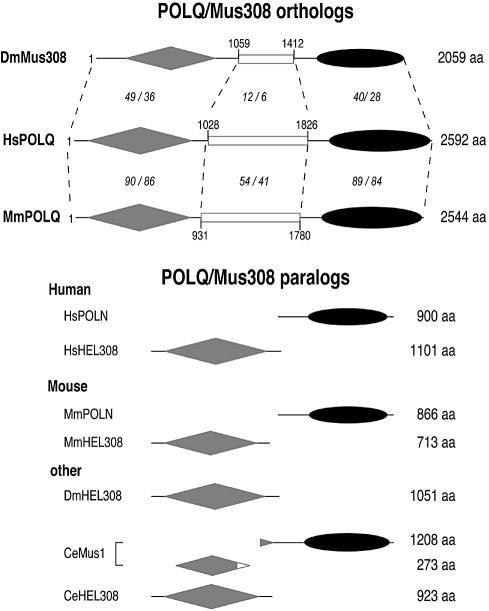
Domain structure of POLQ family homologs. The domain structure of D.melanogaster (Dm) Mus308 (13), human (Hs) POLQ (accession no. AY338826) and mouse (Mm) POLQ (accession no. AY074936) is schematized in the upper portion of the diagram. Gray diamonds indicate helicase-like domains, white rectangles the central non-conserved region, and black ovals the polymerase domain. Dotted lines separate the proteins into regions of highest conservation, with the least conserved region in the center. The percentage of sequence similarity/identity between each pair, as determined by a Clustal alignment and MacBoxShade analysis, is indicated by italic numerals. For example, the region between residues 1 and 1028 of human POLQ is 49% similar and 36% identical to a homologous region of Drosophila Mus308 between residues 1 and 1059. Paralogs showing genes closely related to POLQ encode only the helicase-like domain or the polymerase domain and are schematized in the lower portion of the diagram. References or accession numbers: DmHEL308 (NP_648178), Caenorhabditis elegans (Ce) HEL308 (AAF59587), Ce Mus1 (13). Hs and Mm POLN (15), Hs and Mm HEL308 (16).
Most of the cDNA sequence that we determined is consistent with a recent human genome database prediction for human POLQ (NM_006596). This entry, apparently a computational prediction, suggests 31 exons. The coding portion of exon 1 in our sequence corresponds to exon 2 of the previously predicted database sequence. We have found no experimental evidence for a further exon located 5′ of the sequences shown in Figure 2A. The 5′-untranslated region of the POLQ cDNA includes a site predicted in the current genome database to be a splice acceptor sequence for a putative upstream intron (Fig. 2A, vertical dotted line). As we have experimentally determined that this sequence is part of the cDNA, it can be concluded that the first exon currently proposed in NM_006596 does not exist, and that the coding region for POLQ starts as indicated in Figures 2A and 3. Shima and co-workers recently analyzed murine transcripts similar to POLQ (18), and predicted a mouse coding sequence homologous to the one that we have determined. This predicted mouse POLQ sequence is aligned with that of human POLQ in Figure 2B. The human and murine coding sequences share 71.5% identity (76.9% similarity) over their entire lengths.
Comparison of the human POLQ sequence with that proposed by Sharief et al. (14) for the putative Pol θ protein shows that the latter sequence largely corresponds to the C-terminal region of full-length POLQ, with amino acids 13–1762 of the sequence of Sharief et al. corresponding to amino acids 843–2592 of our determination and thus to the region encoded by exons 16–30 of POLQ as defined here. Amino acids 1–12 predicted by the sequence of Sharief et al. are not present in POLQ; their suggested occurrence corresponds to computational translation of a pyrimidine-rich intronic sequence just before exon 16.
Detection of POLQ protein in HeLa cell extract
Two POLQ-specific polyclonal antibodies were raised in rabbits against peptide antigens derived from regions of the central domain of POLQ (Fig. 3A). These antibodies were purified on columns to which their respective antigens had been cross-linked. Both antibodies recognize purified, recombinant POLQ (see Fig. 4). Upon immunoblotting of HeLa S3 cell extract, both antibodies detected a band of molecular size >250 kDa (Fig. 3B), consistent with the predicted size of full-length POLQ. Several faster migrating bands that cross-reacted with the antibodies were also observed, usually with greater intensity than the slowest migrating band. The >250 kDa band and slower migrating bands were also detected by immunoblotting of MOLT4 cell extract (not shown). In both cases, the slowest migrating protein was reproducibly observed, although its intensity changed in different experiments. To avoid protein degradation during extraction, some cell samples were lysed in SDS sample buffer and immediately boiled. However, this treatment did not prevent protein degradation, and it appears that POLQ protein degradation can occur easily in vivo. It is possible that junctions between globular domains of POLQ may be easily attacked by proteases as, for example, E.coli Pol I is readily degraded into the Klenow fragment and a 5′–3′ exonuclease domain.
DNA polymerase activity of purified human POLQ
A complete POLQ ORF was assembled, verified by DNA sequencing, and cloned into a baculovirus vector for protein expression in insect cells. Initial experiments with a construct containing an N-terminal His tag showed that the protein was easily degraded during extraction. A FLAG tag was added at the C-terminus to facilitate purification of intact protein. POLQ protein was purified by lysis of baculovirus-infected cells in the presence of protease inhibitors, extraction of protein from the pellet with ammonium sulfate, removal of DNA with polyethyleneimine, and then sequential purification on FLAG resin and Ni2+ resin. The purity was checked by silver staining (Fig. 4A).
Initial experiments showed that POLQ could incorporate dNTPs into activated calf thymus DNA and a poly(dA)–oligo(dT) substrate (e.g. see Fig. 5). For experiments at nucleotide resolution, a singly primed hairpin template was used (Fig. 4B). This substrate was chosen to simplify the analysis in case POLQ had significant DNA helicase activity that might displace a short oligonucleotide primer annealed to a template. Elongation by 17 nt to the end of the template occurred only when all four dNTPs were present in reaction mixtures (Fig. 4C, lane 9). The reaction was inhibited by EDTA, consistent with dependence of activity on a divalent cation (Mg2+ in this case, Fig. 4C, lane 1). The DNA polymerase activity was largely template dependent. Omission of dCTP from the reaction interrupted extension after 9 nt were incorporated, at the point where the first template G was encountered (Fig. 4C, lane 8). When only one dNTP was present, extension stopped after one or two bases were incorporated (Fig. 4C, lanes 3–6). The first template base is T, and extension of the primer occurred efficiently when the complementary deoxynucleotide dATP was present (lane 3). Interestingly, a less efficient extension by one base with dGTP was also observed (lane 5). A faint band was observed when dTTP was used, suggesting some incorporation of T opposite T, and then T opposite A at the following template position (lane 4). dCTP was not detectably incorporated (lane 6). When only dATP and dTTP were added (complementary to the first two template nucleotides), incorporation of both two and three nucleotides was observed with similar efficiency (lane 7). These initial results suggest that POLQ might have lower fidelity in some contexts, a possibility to be explored in future experiments.
Figure 5.
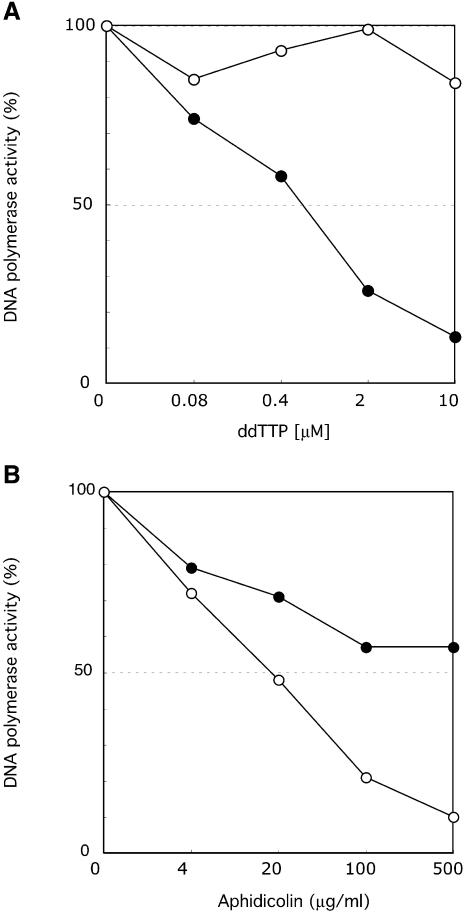
Sensitivity of POLQ DNA polymerase to inhibitors. (A) ddNTP sensitivity of POLQ on a poly(dA)–oligo(dT)2:1 template. The indicated amounts of ddTTP were added to reaction mixtures containing 10 µM dTTP. Activities for POLQ (filled circles) and E.coli Klenow fragment (open circles) are expressed as percentages relative to control reactions in the absence of ddTTP. (B) Aphidicolin resistance of POLQ. The reaction conditions were as described in Materials and Methods except that each dNTP was present at 20 µM. Activities for POLQ (filled circles) and calf thymus Pol δ (open circles) are expressed as percentages relative to control reactions in the absence of aphidicolin.
In the middle of motif 4 of family A DNA polymerases is a highly conserved aromatic residue, either F or Y, that is involved in binding of incoming nucleotide. DNA polymerases with an F residue, such as E.coli Pol I, are resistant to dideoxynucleotides (ddNTPs), while those with a Y residue at this position are ddNTP sensitive. POLQ has a Y residue at this position (amino acid 2389, Fig. 2B), as does Mus308, and both are predicted to be ddNTP sensitive (13,14). A poly(dA)–oligo(dT) template was used for these experiments. The DNA polymerase activity of POLQ was inhibited by ddTTP. At 10 µM ddTTP, the highest concentration tested, incorporation was reduced to 13% of the control (Fig. 5A). In contrast, E.coli Pol I Klenow fragment retained >80% polymerase activity under the same conditions. Aphidicolin sensitivity was also tested. Eukaryotic DNA polymerases involved in chromosomal DNA replication, such as Pol α, Pol δ and Pol ε, are sensitive to aphidicolin. In the presence of 500 µg/ml aphidicolin, only 10% of the activity of Pol δ remained, whereas POLQ polymerase activity was relatively resistant, with 60% polymerase activity under the same conditions (Fig. 5B).
In this study, we have concentrated on the DNA polymerase activity of POLQ. Nevertheless, because of the presence of conserved helicase family motifs in the N-terminal domain of the protein, several experiments to search for an activity relevant to this domain have been carried out. Single-stranded DNA-dependent ATPase activity was tested on the purified fraction. Released radiolabeled phosphate was separated from non-hydrolyzed ATP by thin-layer chromatography, and the extent of hydrolysis was quantified. POLQ showed ATPase activity upon addition of single-stranded DNA (Table 1), which could be inhibited by EDTA. The ATPase activity was 30% of that exhibited by the HEL308 ATPase-DNA helicase. We have also tested for DNA helicase activity in several ways. Under the same conditions that HEL308 is able to displace a 17 nt oligonucleotide from M13 viral single-stranded DNA (16), we found no significant oligonucleotide displacement (data not shown). We have also tested POLQ for its ability to displace other types of oligonucleotides, including those with 5′- and 3′-unpaired tails as well as RNA oligonucleotides, but so far without detecting helicase activity.
Table 1. Single-stranded DNA-dependent ATPase activity of POLQa.
| ssDNA | EDTA | Pi/Pi + ATP (%) | |
|---|---|---|---|
| HEL308 | – | – | 10.2 ± 1.1% |
| + | – | 35.7 ± 1.7% | |
| POLQ | – | – | 4.9 ± 0.3% |
| + | – | 10.8 ± 0.5% | |
| + | + | 4.9 ± 0.6% |
a0.25 µCi of [γ-32P]ATP was incubated with 1.25 ng of HEL308 or POLQ for 180 min at 37°C. Values shown in the last column are averages of three experiments ± SEM.
DISCUSSION
Multidomain structure of POLQ
We have reported here the experimental definition of full-length human POLQ and the initial demonstration of the DNA polymerase and DNA-dependent ATPase activity of the gene product. Human POLQ, like Mus308 of Drosophila, consists of three distinct regions: a helicase-like domain, a central spacer domain and a DNA polymerase domain. The N-terminal part of the protein includes nine conserved motifs (Fig. 2B) closely related to those in the helicase-like domains of Drosophila Mus308 and in the mammalian protein designated HEL308 (13,16). A recent analysis classified helicase sequences in yeast into several families, based on the local DNA sequence around the ATP-binding motif I (19). Using this scheme, motif I of the POLQ/HEL308/Mus308 helicase domain is most closely related to the Ski2 family of RNA helicases. As in many DNA and RNA helicases (19), there is a conserved glutamine residue about 20 amino acids upstream of motif I in POLQ and its homologs (Fig. 2B).
It is remarkable that most of the central spacer region between the two conserved functional domains is encoded by a single large exon (Fig. 3A). Mouse POLQ has a similar arrangement of exons. The sequence of the central domain of POLQ has no significant homology with other sequences that might suggest a specific function for this region of the protein. Interestingly, the first and last segments of the part of the POLQ protein encoded by exon 16 have significant similarity to Mus308 (Figs 3A and 6). These portions of POLQ and Mus308 fall outside the helicase-like and DNA polymerase domains, but their conservation suggests functional importance.
The POLQ family of polymerases and helicases
Mus308 homologs can be detected in the genome sequences of various multicellular eukaryotes, but not in yeast. These mus308 homologs fall into two classes based on their domain arrangements. Mammalian and Drosophila POLQ/Mus308 orthologs are arranged similarly, with helicase-like domains, a spacer and a polymerase domain (Fig. 6). A gene predicted in Arabidopsis thaliana (CAA18591) also appears to be a POLQ/Mus308 ortholog. In addition, the nematode gene mus1 on Caenorhabditis elegans chromosome 3 predicts a DNA polymerase domain with an N-terminal extension to give a 1208 amino acid protein of unknown function. It is noteworthy that 3.2 kb away on the same C.elegans chromosome, with another gene intervening, there is a predicted ORF (accession no. AAB93324) with sequences similar to part of the helicase domain of Mus308 and POLQ, and transcribed in the same direction.
As previously reported, there are also mammalian mus308 paralogs (Fig. 6). HEL308 is a 3′–5′ helicase showing homology to the Mus308 helicase domain (16). POLN encodes an A family DNA polymerase (15).
The possibility of functional diversity amongst this enzyme family is suggested by the different expression patterns of human and mouse mus308 homologs. In northern blots from multiple human tissues, HEL308 and POLN express in heart, muscle and testis, and HEL308 in ovary (15). In contrast, a full-length transcript for POLQ is most apparent in testis, which may suggest a specialized function in this organ (Fig. 1A and B). Many repair and recombination proteins are well expressed in testis, perhaps due to the high repair and recombination activity in this tissue. As Mus308 has a function in maintaining resistance to DNA-damaging agents, it is possible that human POLQ also has such a function. Recently, a mutant mouse strain was isolated, called chaos1 because of the high spontaneous and chemically induced levels of micronuclei in reticulocytes (18). Genetic mapping on mouse chromosome 16 suggested that mouse Polq is a candidate gene for the chaos1 defect (18).
Maga et al. recently reported partial purification of a human DNA polymerase activity from nuclear extracts of HeLa cells (20). Because the purified fraction cross-reacted with an antibody raised against a portion of the Pol θ polypeptide predicted by Sharief et al. (14), the responsible DNA polymerase activity was tentatively identified as DNA polymerase θ. However, the molecular mass of the protein isolated by Maga et al. was ∼100 kDa (20), and it thus cannot be POLQ. It is possible that the activity detected by Maga et al. could be derived from a proteolytic fragment of full-length POLQ, similar to fragments that we have detected in HeLa cell extract (Fig. 1C). It is also possible that the enzyme isolated by Maga and co-workers could be POLN, a Pol A family polymerase of ∼100 kDa (15). POLN has high homology (43% similarity) to POLQ in the conserved DNA polymerase region which could permit cross-reaction with a polyclonal antibody. The activity isolated by Maga and co-workers was ddNTP sensitive, and POLN is also predicted to be ddNTP sensitive based on the presence of a Y residue in polymerase motif 4. In any case, the identify of the activity studied by Maga et al. cannot be determined without further protein sequence information.
Oshige and co-workers partially purified a DNA activity from fly embryos from a fraction that was not present in mus308 embryos (21). The activity was resistant to aphidicolin, sensitive to ddTTP and had an Mr of 200– 300 kDa as estimated by gel filtration (21). This polymerase species was also not defined by protein sequencing. It appeared to co-purify with ATPase and 3′–5′ exonuclease activities. We have so far found no exonuclease activities associated with POLQ (e.g. POLQ does not degrade the primer–template, Fig. 4C). Motifs that would suggest such an activity are not apparent in either the POLQ or Drosophila Mus308 protein sequences.
POLQ has single-stranded DNA-dependent ATPase activity, consistent with the presence of seven conserved helicase family motifs in the protein. However, we have not detected helicase activity under the same conditions in which the closely related protein HEL308 works. HEL308 can displace 20–44mer oligonucleotides efficiently, without requiring forked structures with 5′ or 3′ tails. Some helicases working at the DNA replication fork such as E.coli DnaB helicase, T7 gene 4 helicase/primase and T4 helicase require tails for activity (22). For E.coli Dna B helicase, 20 bases are enough to load the enzyme (23,24), wherease T7 gene 4 helicase/primase (with two activities in one polypeptide) prefers a tail of 55 bases (25). With POLQ, we have tested 30 and 60mer tailed oligonucleotides in both the 5′ and 3′ orientations, without detecting significant strand displacement. It is possible that helicase activity, if it exists, may involve a more specialized substrate. During repair of ICLs in E.coli, the UvrD helicase unwinds DNA 3′ to an incision nick to form a single-stranded DNA region for initiation of recombination. Pol I cannot extend the primer terminus because the remaining arm of the cross-link blocks primer extension (3). A special substrate that mimics the cross-link repair intermediate might be required for the helicase activity of POLQ.
The fidelity of human POLQ requires further investigation. Under the conditions tested, it was able to incorporate G opposite template T with about half the yield of A opposite T (Fig. 4). This is not typical of A family DNA polymerases. For example, a proofreading-defective mutant of E.coli DNA Pol I incorporates G opposite template T with a misinsertion frequency of 6.5 × 10–4 (26). It is also conceivable that POLQ is a type of polymerase that can bypass DNA adducts, and future analyses will determine whether this is the case.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to our laboratory colleagues for discussions, and to Anthony Schuffert for assistance with DNA sequence analysis. This work was supported by an NIH grant (number R01 CA101980), the University of Pittsburgh Cancer Institute and the Imperial Cancer Research Fund (now Cancer Research UK).
REFERENCES
- 1.Cole R.S. and Sinden,R.R. (1975) Repair of cross-linked DNA in Escherichia coli. Basic Life Sci., 5B, 487–495. [DOI] [PubMed] [Google Scholar]
- 2.Van Houten B., Gamper,H., Hearst,J.E. and Sancar,A. (1988) Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem., 263, 16553–16560. [PubMed] [Google Scholar]
- 3.Sladek F.M., Munn,M.M., Rupp,W.D. and Howard-Flanders,P. (1989) In vitro repair of psoralen–DNA cross-links by RecA, UvrABC and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem., 264, 6755–6765. [PubMed] [Google Scholar]
- 4.Dronkert M.L. and Kanaar,R. (2001) Repair of DNA interstrand cross-links. Mutat. Res., 486, 217–247. [DOI] [PubMed] [Google Scholar]
- 5.Magana-Schwencke N., Henriques,J.A., Chanet,R. and Moustacchi,E. (1982) The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl Acad. Sci. USA, 79, 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McHugh P.J., Sones,W.R. and Hartley,J.A. (2000) Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossmann K.F., Ward,A.M., Matkovic,M.E., Folias,A.E. and Moses,R.E. (2001) S.cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat. Res., 487, 73–83. [DOI] [PubMed] [Google Scholar]
- 8.De Silva I.U., McHugh,P.J., Clingen,P.H. and Hartley,J.A. (2000) Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol., 20, 7980–7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng H., Wang,X., Warren,A.J., Legerski,R.J., Nairn,R.S., Hamilton,J.W. and Li,L. (2003) Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell. Biol., 23, 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Andrea A.D. and Grompe,M. (2003) The Fanconi anaemia/BRCA pathway. Nature Rev. Cancer, 3, 23–34. [DOI] [PubMed] [Google Scholar]
- 11.Aguirrezabalaga I., Sierra,L.M. and Comendador,M.A. (1995) The hypermutability conferred by the mus308 mutation of Drosophila is not specific for cross-linking agents. Mutat. Res., 336, 243–250. [DOI] [PubMed] [Google Scholar]
- 12.Boyd J.B., Sakaguchi,K. and Harris,P.V. (1990) mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics, 125, 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris P.V., Mazina,O.M., Leonhardt,E.A., Case,R.B., Boyd,J.B. and Burtis,K.C. (1996) Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 16, 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharief F.S., Vojta,P.J., Ropp,P.A. and Copeland,W.C. (1999) Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics, 59, 90–96. [DOI] [PubMed] [Google Scholar]
- 15.Marini F., Kim,N., Schuffert,A. and Wood,R.D. (2003) POLN, a nuclear Pol A family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem., 278, 32014–32019. [DOI] [PubMed] [Google Scholar]
- 16.Marini F. and Wood,R.D. (2002) A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem., 277, 8716–8723. [DOI] [PubMed] [Google Scholar]
- 17.Moggs J.G., Yarema,K.J., Essigmann,J.M. and Wood,R.D. (1996) Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J. Biol. Chem., 271, 7177–7186. [DOI] [PubMed] [Google Scholar]
- 18.Shima N., Hartford,S.A., Duffy,T., Wilson,L.A., Schimenti,K.J. and Schimenti,J.C. (2003) Phenotype-based identification of mouse chromosome instability mutants. Genetics, 163, 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner N.K., Cordin,O., Banroques,J., Doere,M. and Linder,P. (2003) The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell, 11, 127–138. [DOI] [PubMed] [Google Scholar]
- 20.Maga G., Shevelev,I., Ramadan,K., Spadari,S. and Hubscher,U. (2002) DNA polymerase theta purified from human cells is a high-fidelity enzyme. J. Mol. Biol., 319, 359–369. [DOI] [PubMed] [Google Scholar]
- 21.Oshige M., Aoyagi,N., Harris,P.V., Burtis,K.C. and Sakaguchi,K. (1999) A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product. Mutat. Res., 433, 183–192. [DOI] [PubMed] [Google Scholar]
- 22.Ahnert P. and Patel,S.S. (1997) Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem., 272, 32267–32273. [DOI] [PubMed] [Google Scholar]
- 23.Jezewska M.J., Kim,U.S. and Bujalowski,W. (1996) Binding of Escherichia coli primary replicative helicase DnaB protein to single-stranded DNA. Long-range allosteric conformational changes within the protein hexamer. Biochemistry, 35, 2129–2145. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan D.L. (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol., 301, 285–299. [DOI] [PubMed] [Google Scholar]
- 25.Hacker K.J. and Johnson,K.A. (1997) A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry, 36, 14080–14087. [DOI] [PubMed] [Google Scholar]
- 26.Bebenek K., Joyce,C.M., Fitzgerald,M.P. and Kunkel,T.A. (1990) The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem., 265, 13878–13887. [PubMed] [Google Scholar]