Summary
Familial amyotrophic lateral sclerosis (ALS) has been linked to mutations in the copper/zinc superoxide dismutase (SOD1) gene. The mutant SOD1 protein exhibits a toxic gain-of-function that adversely affects the function of neurons. However, the mechanism of how mutant SOD1 initiates ALS is unclear. Lipid rafts are specialized microdomains of the plasma membrane that act as platforms for the organization and interaction of proteins involved in multiple functions including vesicular trafficking, neurotransmitter signaling and cytoskeletal rearrangements. In this study, we report a proteomic analysis using a widely used ALS mouse model to identify differences in spinal cord lipid raft proteomes between mice over-expressing wild-type (WT) and G93A mutant SOD1. A total of 413 and 421 proteins were identified in the lipid rafts isolated from WT and G93A mice, respectively. Further quantitative analysis revealed a consortium of proteins with altered levels between the WT and G93A samples. Functional classification of the 67 altered proteins revealed that the three most impacted subsets of proteins were involved in vesicular transport, neurotransmitter synthesis and release; cytoskeleton organization and linkage to plasma membrane; and metabolism. Other protein changes are correlated with alterations in microglia activation and inflammation; astrocyte and oligodendrocyte function; cell signaling; cellular stress response and apoptosis; and neuronal ion channels and neurotransmitter receptor functions. Changes of selected proteins were independently validated by immunoblotting and immunohistochemistry. The significance of the lipid raft protein changes in motor neuron function and degeneration in ALS is discussed, particularly those involved in vesicular trafficking and neurotransmitter signaling, and the dynamics and regulation of plasma membrane-anchored cytoskeleton.
Keywords: amyotrophic lateral sclerosis, proteomics, lipid rafts, vesicular trafficking, cytoskeleton dynamics
Introduction
Amyotrophic lateral sclerosis (ALS) is a chronic progressive neuromuscular disorder characterized by weakness, muscle wasting, fasciculation, and increased reflexes, with conserved intellect and higher functions [1]. The neuropathology of ALS is mostly confined to motor neurons in the cerebral cortex, some motor nuclei of the brainstem, and anterior horns of the spinal cord. An important discovery in the study of the disease was the identification of mutations in the copper/zinc superoxide dismutase (SOD1) gene in some families with hereditary ALS [2, 3]. To date, more than 100 mutations scattered throughout the SOD1 protein have been identified and it has been established that mutant SOD1 cause ALS through a gain-of-function mechanism(s) [4]. Many hypotheses of how mutant SOD1 could cause neurodegeneration, including aberrant redox chemistry, mitochondrial damage, excitotoxicity, microglial activation and inflammation, as well as SOD1 aggregation, have been proposed [4–6].
Lipid rafts are specialized microdomains of the plasma membrane enriched in cholesterol and sphingolipids. These rafts act as platforms for the organization and interaction of proteins involved in multiple functions including vesicular trafficking, signaling mechanisms and cytoskeletal rearrangements [7, 8]. In neurons, lipid rafts have been implicated in organizing and compartmentalizing proteins involved in many aspects of neurotransmitter signaling. These aspects include transport of neurotransmitters to the axon terminal and regulated exocytosis of neurotransmitters at the synapse, as well as organization of neurotransmitter receptors and other transduction molecules [7]. Lipid rafts and associated scaffold proteins have been implicated in the pathogenesis of several neurological disorders including Alzheimer's and Parkinson's diseases [7]. Several recent studies have shown that ALS is not an autonomous disease, i.e. various non-neuronal cells including astrocytes and microglia can contribute to the disease progression [9–11]. As plasma membrane microdomains enriched with signaling molecules, lipid rafts and alterations of lipid raft proteins may contribute to the neuron-glia interactions in ALS etiology. Despite several proteomic studies in ALS [12–18], no studies regarding alterations in lipid raft associated proteins have been reported.
In this study, we isolated and profiled lipid rafts from spinal cords of symptomatic G93A SOD1 transgenic mice and age-matched WT SOD1 transgenic mice. The G93A transgenic mice were chosen since it is the most extensively studied ALS model [19] and the findings from this proteomic study can be correlated to other studies. A one-dimensional SDS-PAGE combined with nano-HPLC-MS/MS approach was exploited to identify lipid raft proteins. A label-free quantitative analysis was then performed to distinguish protein changes in the lipid rafts of G93A and WT SOD1 transgenic mice. Functional classification of the altered proteins revealed that the affected proteins are mostly involved in the following: (i) vesicular transport, neurotransmitter synthesis and release, (ii) cytoskeleton organization and linkage to plasma membrane, (iii) metabolism, (iv) microglia activation and inflammation, (v) astrocyte and oligodendrocyte function, (vi) cell signaling, (vii) cellular stress responses and apoptosis, and (viii) neuronal ion channels and neurotransmitter receptor functions. Alterations of selected lipid rafts proteins were independently validated by immunoblotting and immunohistochemistry. The potential role of these lipid raft protein changes in ALS disease pathology is discussed.
Results
Lipid raft fraction isolation and purity analysis
Lipid rafts are specialized areas on the plasma membrane and act as platforms for spatiotemporal co-coordination of multiple cellular functions including vesicular transport and receptor signaling pathways. In this study, lipid rafts from spinal cord extracts of transgenic mice over-expressing human mutant G93A SOD1 and age-matched control mice over-expressing human WT SOD1, were isolated by OptiPrep gradient centrifugation [20]. The detergent free method is routinely used in the laboratory as the methods based on insolubility of lipid rafts in cold solutions containing Triton X-100 have been reported to be extensively contaminated by intracellular organelles and non-lipid rafts components [21, 22]. In addition to lipid rafts, cytoplasm and plasma membrane fractions were also collected. To evaluate the purity of the fractions, Western blotting using antibodies against the neuronal lipid raft marker flotillin-1 [23–26], the mitochondrial protein MnSOD, and the cytoplasmic protein triosephosphate isomerase (TIM) were performed. As seen in Figure 1, strong flotillin-1 signal was observed in the lipid rafts fractions. Weak Flotillin-1 signal was also observed in the plasma membrane fraction with prolonged exposure time (data not shown). No flotillin-1 signal could be detected in the cytoplasm fraction. In contrast, TIM and MnSOD were only detected in the cytoplasm but not the lipid rafts fraction. SOD1 protein was detected in all fractions including the plasma membrane and lipid raft fractions although SOD1 has been known as a highly soluble protein. These data show effective enrichment of lipid raft proteins using the centrifugation protocol.
Figure 1. Evaluation of the isolation of lipid raft proteins.
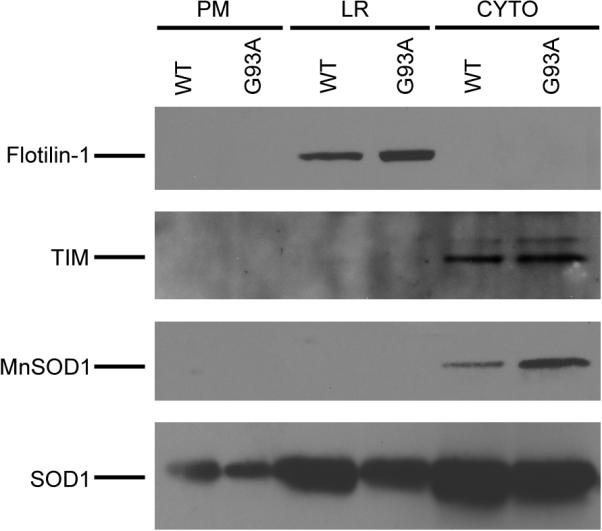
Lipid raft proteins were isolated from spinal cords of symptomatic G93A SOD1 transgenic mice and age-matched control WT SOD1 transgenic mice. The lipid raft (LR), cytoplasm (CYTO) and plasma membranes (PM) fractions (25 μg proteins from each fraction) were analyzed by Western blotting using antibodies against flotillin-1, TIM, MnSOD and SOD1.
Proteomic analysis of mouse spinal cord lipid rafts
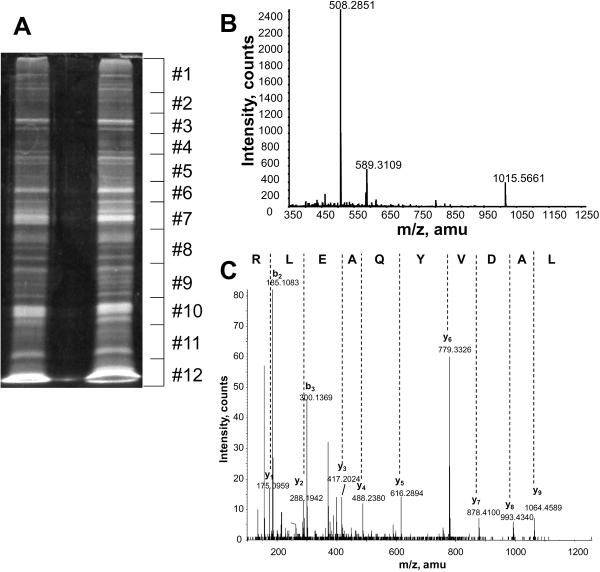
To identify proteins in lipid rafts, purified LR fractions were subjected to SDS-PAGE separation, in-gel digestion and nano-LC-MS/MS analysis. Figure 2A shows a representative image of SyproRuby stained SDS-PAGE of a set of G93A and WT lipid raft samples. Twelve equal bands were excised and each band was subjected to trypsin in-gel digestion and the tryptic peptides from each gel band were subjected to nano-LC-MS/MS analysis. Figure 2B shows a representative MS spectrum of tryptic peptides that were eluted at retention time 26.5 min during the LC-MS/MS analysis of band #6 of the G93A sample. Figure 2C shows the tandem MS/MS spectrum of the m/z 589.31 peptide in Figure 2B. A complete series of y ions were detected in the tandem MS/MS spectrum in Figure 2C, thus the identification of the peptide LADVYQAELR by subsequent MASCOT MS/MS ion search was unambiguous.
Figure 2. SDS-PAGE and mass spectrometry analysis of the lipid rafts samples.
(A). SyproRuby staining of the SDS-PAGE of the lipid raft samples from both WT and G93A transgenic mouse spinal cords. (B). MS of all peptides eluted at 26.5 min during the LC-MS/MS analysis of tryptic peptides from band #6 of the G93A lipid rafts.(C). Tandem MS/MS of the peptide with m/z 589.31 from (B). The complete series of y ions were detected from the collision induced dissociation of the peptide, thus yielding unambiguous identification of the peptide sequence as noted.
The MS/MS data generated from individual bands of each sample were submitted to a local Mascot server for protein identification using a merged search mode. Rigorous identification criteria were used to eliminate potential ambiguous protein identifications. All peptides were required to have an ion score greater than 30 (P < 0.05). Proteins with two or more unique peptides, each of which had a score greater than 30, were considered unambiguous identifications. Proteins with single-peptide identification were considered positive only if (i) the MS/MS ion score was consistently greater than 30 in multiple analyses of the lipid rafts sample isolated from the same mouse; and (ii) the protein was consistently identified in the independent analysis of lipid rafts isolated from at least two different mice. Otherwise, the proteins identified by a single peptide were discarded. The number of proteins, which were identified based on a single peptide but met the two criteria discussed above, was 70 and 73 in the lipid rafts of WT and G93A SOD1 mice, respectively. All LC-MS/MS data were also submitted to decoy MASCOT search against a randomized Sprot database [27], and the false discovery rates (FDR) in all MASCOT searches were in the range between 0.5% and 1.5% for each independent LC-MS/MS experiment. In total, we identified 413 and 421 proteins in the lipid rafts isolated from WT and G93A SOD1 mice, respectively. The complete list of proteins identified in the lipid rafts fractions is provided as supplementary tables S1 and S2.
Quantitative analysis of lipid raft proteins from WT and G93A mouse spinal cords
Quantitative analysis of protein changes between the WT and G93A lipid raft samples was performed and the results are presented in Table 1. First, 17 proteins were consistently identified in G93A samples but absent in WT samples; 9 proteins were identified in WT samples but absent in G93A samples. These proteins were considered as G93A and WT unique proteins, respectively. They represent a group of lipid rafts proteins that changed significantly between WT and G93A transgenic mice.
Table 1.
Quantitative Analysis of Proteins Changes between the Lipid Rafts Isolated from WT and G93A Mouse Spinal Cords
| Accession Number | Protein name | Function Category |
|---|---|---|
| Proteins uniquely identified in the lipid rafts isolated from G93A mouse spinal cord | ||
| ARPC3_MOUSE | Actin-related protein 2/3 complex subunit 3 | Cytoskeletal regulation |
| ANXA3_MOUSE | Annexin A3, Annexin III | Microglia/inflammation |
| CAPS1_MOUSE | Calcium-dependent secretion activator 1 | Vesicular trafficking, neurotransmitter synthesis and release |
| CLCA_MOUSE | Clathrin light chain A | Vesicular trafficking, neurotransmitter synthesis and release |
| DHRS1_MOUSE | Dehydrogenase/reductase SDR family member 1 | Other or unknown/uncharacterized |
| DEST_MOUSE | Destrin (Actin-depolymerizing factor) (ADF) | Cytoskeletal regulation |
| EFHD2_MOUSE | EF-hand domain containing protein 2, Swiprosin-1 | Microglia/inflammation |
| EZR1_MOUSE | Ezrin (p81) (Cytovillin) (Villin-2) | Cytoskeletal regulation |
| HSPB1_MOUSE | Heat shock 27 kDa protein (HSP 27) | Cellular stress/apoptosis |
| ITAM_MOUSE | Integrin alpha-M precursor | Microglia/inflammation |
| LAMP1_MOUSE | Lysosome-associated membrane glycoprotein 1 precursor (LAMP-1) | Protein degradation |
| MTCH2_MOUSE | Mitochondrial carrier homolog 2 | Cellular stress/apoptosis |
| NCKP1_MOUSE | Nck-associated protein 1 (NAP 1) (Membrane-associated protein HEM-2) | Cytoskeletal regulation |
| S10A1_MOUSE | Protein S100-A1 (S100 calcium-binding protein A1) | Cytoskeletal regulation |
| RRAS_MOUSE | Ras-related protein R-Ras | Microglia/inflammation |
| SCAM1_MOUSE | Secretory carrier-associated membrane protein 1 | Vesicular trafficking, neurotransmitter synthesis and release |
| UCR10_MOUSE | Ubiquinol-cytochrome c reductase complex 7.2 kDa protein | Metabolism |
| Proteins uniquely identified in the lipid rafts isolated from WT mouse spinal cord | ||
| THIL_MOUSE | Acetyl-CoA acetyltransferase, mitochondria precursor | Metabolism |
| SYUA_MOUSE | Alpha-synuclein | Vesicular trafficking, neurotransmitter synthesis and release |
| OST48_MOUSE | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 48 kDa subunit | Other or unknown/uncharacterized |
| NEGR1_MOUSE | Neuronal growth regulator 1 precursor (Kilon) | Neurite outgrowth |
| SPRE_MOUSE | Sepeiapterin reductase | Vesicular trafficking, neurotransmitter synthesis and release |
| DHSA_MOUSE | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor | Metabolism |
| PP2AA_MOUSE | Serine/threonine-protein phosphatase 2A catalytic subunit | Cell signaling |
| SPN90_MOUSE | SH3 adapter protein SPIN90 (NCK-interacting protein with SH3 domain) | Cytoskeletal regulation |
| SUSD2_MOUSE | Sushi domain-containing protein 2 precursor | Other or unknown/uncharacterized |
| Accession Number | Protein name | WT/G93A Ratio(Mean ± S.D.) | Function Category |
|---|---|---|---|
| Proteins with higher abundance in the lipid rafts isolated from G93A mouse spinal cord | |||
| AFG32_MOUSE | AFG3-like protein 2 | 0.63 ± 0.11 ** | Other or unknown/uncharacterized |
| ANXA2_MOUSE | Annexin A2, Annexin II | 0.52 ± 0.09 ** | Cytoskeletal regulation |
| ANXA5_MOUSE | Annexin A5 (Annexin V) | 0.37 ± 0.10 ** | Microglia/inflammation |
| BASI_MOUSE | Basigin precursor (Membrane glycoprotein gp42) | 0.69 ± 0.10 ** | Neuronal ion channel/pumps and neurotransmitter receptors |
| FLOT1_MOUSE | Flotillin-1 | 0.54 ± 0.08 ** | Cytoskeletal regulation |
| GFAP_MOUSE | Glial fibrillary acidic protein (GFAP) | 0.48 ± 0.12 ** | Astrocyte/oligodendrocyte function |
| NCB5R_MOUSE | NADH-cytochrome b5 reductase | 0.57 ± 0.16 * | Metabolism |
| SATT_MOUSE | Neutral amino acid transporter A (SATT) | 0.63 ± 0.10 ** | Metabolism |
| Proteins with lower abundance in the lipid rafts isolated from G93A mouse spinal cord | |||
| 1433G_MOUSE | 14-3-3 gamma | 2.56 ± 0.54 ** | Cell signalling |
| 1433T_MOUSE | 14-3-3 theta | 2.45 ± 0.65 * | Cell signalling |
| 1433Z_MOUSE | 14-3-3 zeta/delta | 2.08 ± 0.41 * | Cell signalling |
| ADT1_MOUSE | ADP/ATP translocase 1 (ANT 1) | 2.06 ± 0.31 ** | Cellular stress/apoptosis, Mitochondria |
| ARF1_MOUSE | ADP-ribosylation factor 1 | 1.39 ± 0.08 ** | Vesicular trafficking, neurotransmitter synthesis and release |
| AATC_MOUSE | Aspartate aminotransferase, cytoplasmic | 1.87 ± 0.45 * | Metabolism |
| AATM_MOUSE | Aspartate aminotransferase, mitochondrial precursor | 2.10 ± 0.52 * | Metabolism |
| CHP1_MOUSE | Calcium-binding protein p22, calcium binding protein CHP | 2.01 ± 0.32 ** | Vesicular trafficking, neurotransmitter synthesis and release |
| CD81_MOUSE | CD81 antigen, 26 kDa cell surface protein TAPA-1 | 1.91 ± 0.34 * | Astrocyte/oligodendrocyte function |
| CDC42_MOUSE | Cell division control protein 42 homolog precursor | 2.31 ± 0.31 ** | Cytoskeletal regulation |
| CAH2_MOUSE | Carbonic anhydrase 2 | 1.91 ± 0.35 * | Metabolism |
| DHPR_MOUSE | Dihydropteridine reductase (HDHPR) | 1.72 ± 0.24 ** | Vesicular trafficking, neurotransmitter synthesis and release |
| ALDOC_MOUSE | Fructose-biphosphate aldolase C (aldolase 3) | 1.88 ± 0.46 ** | Metabolism |
| LDHB_MOUSE | L-lactate dehydrogenase B chain (LDH-B) | 1.52 ± 0.32 * | Metabolism |
| MDHM_MOUSE | Malate dehydrogenase, mitochondria precursor | 1.67 ± 0.29 * | Metabolism |
| MBP_MOUSE | Myelin basic protein (MBP) | 1.90 ± 0.27 ** | Astrocyte/oligodendrocyte function |
| MYP0_MOUSE | Myelin P0 protein precursor, Myelin peripheral protein (MPP) | 2.17 ± 0.25 ** | Astrocyte/oligodendrocyte function |
| SIRT2_MOUSE | NAD-dependent deacetylase sirtuin-2 | 1.94 ± 0.23 ** | Cellular stress/apoptosis |
| NFL_MOUSE | Neurofilament triplet L protein, neurofilament light chain (NF-L) | 1.80 ± 0.17 ** | Cytoskeletal regulation |
| NPTN_MOUSE | Neuroplastin precursor, stromal cell-derived receptor 1 (SDR-1) | 2.44 ± 0.51 ** | Neurite outgrowth |
| PRDX5_MOUSE | Peroxidoxin-5, mitochondria precursor | 1.56 ± 0.07 ** | Cellular stress/apoptosis |
| MPCP_MOUSE | Phosphate carrier protein, mitochondria precursor | 1.98 ± 0.26 ** | Other or unknown/uncharacterized |
| PGAM1_MOUSE | Phosphoglycerate mutase 1 | 2.60 ± 0.39 ** | Metabolism |
| PA1B2_MOUSE | Platelet-activated factor acetylhydrolase IB subunit Beta | 2.32 ± 0.36 ** | Microglia/inflammation |
| RAB3A_MOUSE | Ras-related protein Rab-3A | 1.61 ± 0.25 * | Vesicular trafficking, neurotransmitter synthesis and release |
| RALA_MOUSE | Ras-related protein Ral-A | 1.82 ± 0.24 ** | Vesicular trafficking, neurotransmitter synthesis and release |
| RTN1_MOUSE | Reticulon-1 | 1.59 ± 0.21 ** | Vesicular trafficking, neurotransmitter synthesis and release |
| AT1A1_MOUSE | Sodium/potassium-transporting ATPase alpha-1 chain precursor | 1.48 ± 0.18 * | Neuronal ion channel/pumps and neurotransmitter receptors |
| AT1A3_MOUSE | Sodium/potassium-transporting ATPase alpha-3 chain | 1.52 ± 0.07 ** | Neuronal ion channel/pumps and neurotransmitter receptors |
| SNP25_MOUSE | Synaptosomal-associated protein 25 (SNAP-25) | 2.09 ± 0.25 ** | Vesicular trafficking, neurotransmitter synthesis and release |
| THY1_MOUSE | Thy-1 membrane glycoprotein precursor, Thy-1 antigen, CD90 antigen | 2.28 ± 0.30 ** | Neurite outgrowth |
| UBE2N_MOUSE | Ubiquitin-conjugating enzyme E2N | 1.95 ± 0.19 ** | Protein degradation |
| VAMP1_MOUSE | Vesicle-associated membrane protein 1 (VAMP-1), synaptobrevin-1 | 2.36 ± 0.43 ** | Vesicular trafficking, neurotransmitter synthesis and release |
P-values were calculated using t-Test.
p < 0.05
p < 0.01.
A total of 154 proteins were identified in all lipid rafts samples isolated from three WT and three G93A transgenic mice. These proteins were subjected to quantitative analysis using the label-free quantitative method described in the Materials and Methods. A ratio was calculated for each peptide identified in WT and G93A samples, an average ratio of all peptides for every protein was then obtained as the protein ratio in each pair of lipid rafts samples isolated from WT and G93A mice. The protein ratios from three independent pairs of WT and G93A mice were obtained and the average ratios and standard deviations were calculated. A P value for each protein in three independent sets of quantification data was obtained using Student t-test. Significant changes were recognized as the ratios between WT and G93A samples with P values less than 0.05. The quantification data in Table 1 are presented as the mean ratio ± S.D. In addition, the changes with P < 0.05 are indicated by * and those with P < 0.01 are indicated by ** in Table 1.
Out of the 154 proteins, 41 proteins showed changes with statistical significance (P < 0.05). Out of these 41 proteins, 8 proteins showed higher abundance in G93A samples than in WT, while 33 proteins showed lower abundance in G93A samples. These proteins with differential abundances in WT and G93A lipid rafts samples as well as the alteration ratios are listed in Table 1. The remaining 113 proteins, including actin, tubulin, cofilin and SOD1, showed either no changes in the lipid rafts of G93A versus WT, or an ratio with P > 0.05 among the three independent sets of WT and G93A samples. These proteins were all grouped as unchanged between WT and G93A mice.
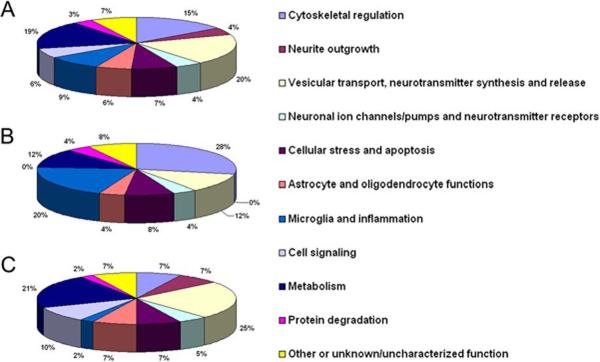
Functional classification of the 26 uniquely identified and 41 altered proteins is shown in Figure 3. Many of these 67 differential proteins are involved in: vesicular transport, neurotransmitter synthesis and release (13 proteins); metabolism (12 proteins); cytoskeleton organization and linkage to plasma membrane (10 proteins); microglia activation and inflammation (6 proteins); cellular stress responses and apoptosis (5 proteins); astrocyte and oligodendrocyte function (4 proteins); cell signaling (4 proteins); and neuronal ion channels and neurotransmitter receptor functions (3 proteins), see Figure 3A. Figure 3B shows that the 25 proteins over-represented in G93A (17 uniquely found in the G93A samples and 8 with higher abundance in the G93A samples) are mostly involved in cytoskeleton organization (7 proteins, 28%) and microglia activation/inflammation (5 proteins, 20%). It is noted that the majority of the proteins in the above two functional categories, i.e. 7 out of 10 proteins involved in cytoskeletal regulation and 5 out of 6 proteins involved in microglia activation, showed higher abundance in the G93A lipid rafts. Figure 3C shows that, among the 42 proteins under-represented in G93A (9 uniquely found in the WT samples and 33 with lower abundance in the G93A samples), the most impacted functional groups are vesicular transport/neurotransmitter synthesis and release (10 proteins, 24%) and metabolism (9 proteins, 21%). In addition, it is noted that all four altered proteins involved in cell signaling were found to have lower abundance in the G93A lipid rafts. Similarly, all three proteins involved in neurite outgrowth showed lower levels in the G93A lipid rafts.
Figure 3. Functional characterization of proteins with altered lipid raft association in G93A transgenic mouse spinal cord.
(A). Functional classification of all 67 proteins with altered lipid raft association in the G93A SOD1 transgenic mouse. The percentage of each functional category of the altered proteins is indicated. (B). Functional classification of the 25 proteins with increased lipid raft association in the G93A mouse. (C). Functional classification of the 42 proteins showing decreased lipid raft association in the G93A mouse.
Validation of lipid raft protein changes
We performed Western blotting to confirm the changes of a selected subset of lipid rafts proteins. Each protein change was examined using lipid rafts isolated from multiple sets of separate WT and G93A mice. As seen in Figure 4A, Western blotting of lipid raft fractions showed elevated levels of Flotillin-1, annexin II and GFAP in the G93A lipid rafts compared to the WT samples. Western blotting also demonstrated a reduced level of SNAP-25 in the G93A lipid rafts (Figure 4B), and an unaltered level of cofilin (Figure 4C). The Western blotting results support the quantitative proteomic data. For instance, quantitative analysis of scanned Western blots using the ImageJ program showed the ratio of SNAP in WT versus G93A samples was 2.4, consistent with that determined in the proteomic analysis (2.09 ± 0.25). In addition, Western blot of GFAP showed a ratio of 0.7 between WT and G93A samples, consistent with the ratio of 0.48 ± 0.12 determined by the proteomic analysis.
Figure 4. Validation of quantitative proteomic results.
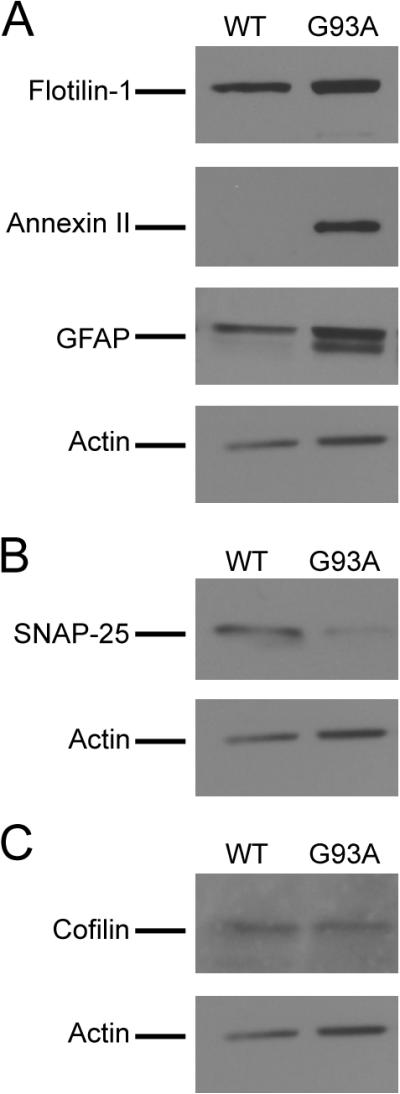
Selected proteins from the increased, decreased and unchanged categories as determined by the proteomic analysis were evaluated by Western blotting. (A) Increased levels of Flotillin-1, annexin II and GFAP in the lipid rafts fractions isolated from the G93A mice. (B) Decreased level of SNAP-25 in the G93A mouse lipid rafts. (C) Unchanged level of cofilin in the G93A mouse lipid rafts. Lipid rafts samples isolated from three pairs of WT and G93A transgenic mice were analyzed by Western blotting and representative images are shown. Twenty-five micrograms of lipid raft proteins were loaded in each lane for analysis.
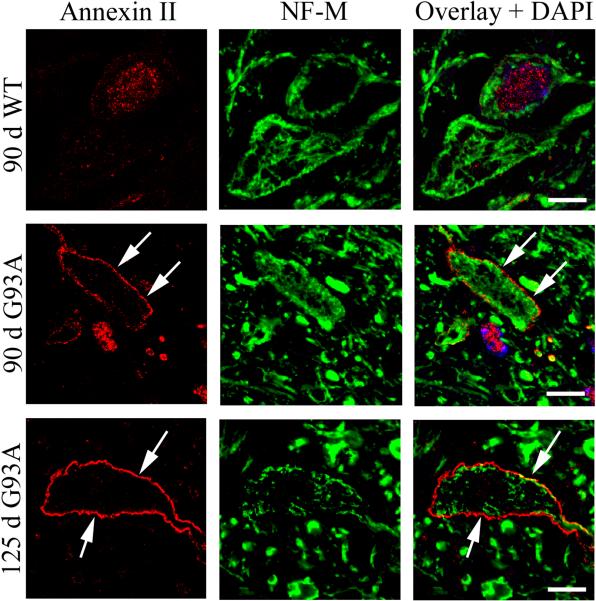
The up-regulation of annexin II in G93A lipid rafts were further analyzed by immunofluorescent stainings of spinal cords from WT and G93A SOD1 transgenic mice. As seen in Figure 5, anti-annexin II antibodies strongly stained the plasma membrane in motor neurons in the lumbar spinal cord of G93A mice, while mostly weak nuclear and cytoplasmic staining was observed in WT mice. The immunohistology clearly demonstrated the recruitment of annexin II to the plasma membrane of motor neurons in the diseased G93A transgenic mice.
Figure 5. Increased plasma membrane staining of annexin II in G93A motor neurons.
Immunofluorescent staining of annexin II and the neuronal marker NF-M (neurofilament M) in spinal cord motor neurons in 90 and 125 day old G93A SOD1 transgenic mice. Strong annexin II membrane staining was observed in a subset of motor neurons in G93A mice. Four pairs of WT and G93A transgenic mice (2 pairs of 90 and 125 days old, respectively) were analyzed in the immunohistochemical experiments and representative images are shown. Scale bars are 10 μm.
Discussion
In this paper we performed proteomic profiling of lipid raft proteins in G93A SOD1 ALS transgenic mice and age-matched controls. Alterations of selected proteins were validated by immunoblotting and immunohistochemistry. Functional analysis of the altered proteins revealed that these proteins are involved in multiple functions that are important to motor neuron health, thus their alterations may contribute to the ALS pathology.
Many of the identified proteins have previously been shown to localize to lipid rafts, including lipid raft markers flotillin-1 and flotillin-2 [26, 28]. This suggests that the lipid rafts purification protocol [20] is valid. This is further supported by Western blotting showing no signal for the cytoplasmic marker TIM or the mitochondria protein MnSOD in the lipid raft fraction (Figure 1). Many proteins identified in this study were also found in other published lipid rafts proteomics studies. For instance, a total of 106 proteins were identified in lipid rafts isolated from neutrophils [29] and 63 of them (60%) were also identified in this study. Another study of lipid rafts isolated from neonatal mouse brain identified 216 proteins [30], and 147 of them (68%) were also identified in this study. Given that these studies independently characterized the lipid rafts proteins isolated from different cell types using various mass spectrometers, differences are expected. The mouse spinal cord lipid rafts proteomic data obtained in this study are reasonably consistent with the literature.
We identified both endogenous mouse SOD1 and transgenically over-expressed human WT and G93A mutant SOD1 in lipid rafts in this study. SOD1 is conventionally believed to be a highly soluble protein, but have previously been identified in lipid rafts [31]. Western blotting revealed higher SOD1 levels in the lipid raft fraction than in the other areas of the plasma membrane (Figure 1). Interaction with lipids or biological membranes have been suggested to play a role in mutant SOD1 aggregation [32, 33]. Moreover, the ALS-linked SOD1 mutants have been shown to form pore-like aggregates in vitro [34, 35]. It is interesting to speculate that localization and subsequent aggregation of mutant SOD1 in lipid rafts could affect cellular functions as well as the interplay between different cell types, as lipid rafts are enriched in receptors and signaling molecules necessary for cell-cell communication.
The proteomics analysis identified 17 unique proteins in the G93A lipid rafts and 6 unique proteins in the WT lipid rafts. Only proteins that were positively identified in the lipid rafts samples in all six transgenic mice (3 WT and 3 G93A mice) were subjected to quantitative analysis. If a protein were not identified in all six samples, statistical analysis was unable to be performed, thus the protein was not included in the quantitative analysis. A total of 154 proteins met this criterion and their quantitative ratios from three independent experiments (using three separate pairs of WT and G93A mice) were averaged and subjected to statistical analysis. Out of the 154 proteins, 41 of them showed changes between the WT and G93A samples with statistical significance (P < 0.05). Among them, 8 and 33 proteins showed higher or lower levels in G93A lipid rafts, respectively. The remaining 113 proteins were considered unchanged since the ratios from three independent experiments were statistically insignificant (P > 0.05). Thus, a total of 25 proteins were over-represented in the G93A lipid rafts and 42 proteins that were under-represented in the G93A lipid rafts compared to WT (Table 1).
Western blotting analyses of lipid rafts samples isolated from multiple separate sets of WT and G93A mice were performed to validate the proteomics data. Seven proteins in all three categories (i.e. 1 unchanged, 3 with higher abundance and 3 with lower abundance in G93A) were selected for Western blotting. For the five proteins whose Western blotting showed clear results, the mass spectrometry-based quantification results were all confirmed by Western blotting (Figure 4). Two other proteins produced either high background or no signal in Western blotting (data not shown), likely due to the technical issues of the antibodies used. Additional sets of WT and G93A mice were used for immunohistochemical studies to confirm the increased lipid rafts association of annexin II in 90-day and 125-day old mice (Figure 5). The validation of protein changes in separate animals using both Western blotting and immunohistochemcial technique further supported the quantitative proteomic data.
We identified changes in neuronal as well as glial specific proteins (Table 1), supporting the involvement of motor neurons as well as different glial cells in the ALS pathology. The results are consistent with recent studies showing that various cell types including astrocytes and microglia can affect the survival of spinal motor neurons in ALS [9–11]. Although the G93A mice used in this study were symptomatic and some loss of neurons had occurred, we could identify neuronal proteins that showed decreased, unchanged and increased association with lipid rafts (Table 1). For instance, the increased plasma membrane localization of Annexin II was demonstrated in motor neurons in the 90 and 125 days G93A mice (Figure 5). For 6 altered lipid rafts proteins involved in microglia and neuroinflammation, 5 of them showed higher levels in the G93A lipid rafts, supporting that microglia activation plays a role in the ALS etiology [10]. In contrast, 3 out of 4 proteins involved in the astrocyte and oligodendrocyte functions actually showed decreased abundance in the G93A lipid rafts. Thus, the lipid rafts protein changes identified in this study are likely to reflect protein changes in multiple cell types involved in the disease, rather than simply the loss of neurons.
Changes in proteins involved in cellular stress response and apoptosis are expected in ALS. We detected an increase in the lipid raft association of HSP27, MTCH2 and carbonyl reductase. HSP27 up-regulation was reported in different ALS mouse models before [36] and HSP27 over-expression in transgenic mice may provide protective benefits to the ALS mice [37]. MTCH2 (mitochondrial carrier homolog 2) was reported to interact with pro-apoptotic protein BID to initiate apoptosis in response to TNF-α and Fas death receptor activation [38]. Carbonyl reductase clears harmful products formed by lipid peroxidation and has been suggested to be neuroprotective [39]. In addition, decreased levels of an antioxidant protein peroxiredoxin-5 detected in this study are consistent with previous studies implicating the peroxiredoxin family proteins in ALS [40] and Parkinson's disease [41].
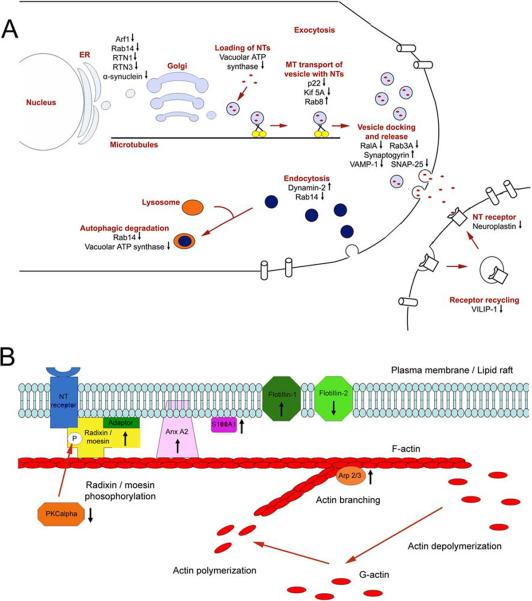
Approximately 20% (13 out of 67) of the altered proteins in G93A lipid rafts are involved in vesicular trafficking, neurotransmitter synthesis and release (Figure 3). The alterations of these proteins and their functionality in vesicular and neurotransmitter trafficking and release are illustrated in Figure 6A. Most proteins in this category (10 out of 13) showed reduced levels in lipid rafts of G93A mouse spinal cords. Alterations observed in this functional group include reduction of several Ras superfamily GTPases involved in trafficking of vesicles to plasma membrane (Arf-1) [42], and vesicle storage, docking and release at synapse (Ral-A, Rab3A) [42, 43]. Reduction of SNARE proteins VAMP-1 and SNAP-25 [44], involved in vesicle fusion and neurotransmitter release was also observed.
Figure 6. Schematic illustration of pathways with multiple altered lipid raft proteins in the G93A transgenic mouse.
(A) Proteins involved in axonal transport, vesicular trafficking, neurotransmitter release, endocytosis and exocytosis are mostly decreased in the spinal cord lipid rafts of the G93A mouse. (B) Proteins with altered levels involved in cytoskeletal organization and linkage of cytoskeleton to the plasma membrane. Arrows beside the proteins indicate increased or decreased levels in the G93A mouse lipid rafts.
Among the 13 proteins in the category of vesicular trafficking, several proteins involved in endocytosis and membrane recycling (clathrin light chain A, CLCA and secretory carrier associated membrane protein 1, SCAM1) showed increased levels in G93A (Figure 6A). Increased endocytosis could contribute to the activation of the macroautophagy-lysosome pathway, which is a major pathway responsible for degrading protein aggregates. Two proteins involved in protein degradation pathways were also found to show changes between WT and G93A samples in this study: LAMP1 (lysosome-associated membrane glycoprotein 1) abundance was increased in G93A whereas UBE2N (ubiquitin-conjugating enzyme E2N) level was decreased in G93A. The polyubiquitin-proteasome and macroautophagy-lysosome pathways are two major mechanisms for protein degradation. When proteasome function is compromised, the macroautophagy-lysosome pathway can be activated as an alternative [45]. A lower level of UBE2N in G93A could potentially contribute to the impairment of the polyubiquitin-proteasome system. A higher level of LAMP1 (a well recognized lysosome marker) in G93A would be reasonably expected, suggesting the induction of the macroautophagy-lysosome pathway. In fact, proteasome impairment [46, 47] and autuphagosome accumulation [48, 49] have been reported in ALS. The increased level of LAMP1 found in this study provides new insights into the activation of lysosomes downstream of autuphagosomes.
Another major functional group of the altered proteins (10 out of 67, 15%) are involved in cytoskeletal regulation and linkage of the cytoskeleton to lipid rafts (Figure 3). Figure 6B illustrates how these proteins are changed in G93A mice as determined in this study and how they may interact with other relevant proteins to regulate the cytoskeleton. For instance, we observed increased levels of annexin II, ezrin and flotillin-1 in the G93A mice, all of which are known actin-lipid raft adaptors [50, 51]. Increased association of annexin II with G93A lipid rafts was confirmed by Western blotting (Figure 4) and immunoflourescent microscopic analysis (Figure 5). Annexin II has previously been reported to be up-regulated in motor cortex of sporadic ALS and frontotemporal lobar degeneration patients [52, 53]. Altered levels of a subunit of the ARP 2/3 actin branching regulator [54] and a CDC42 homolog (a known Ras GTPaase controlling actin polymerization) [55] were identified in G93A lipid rafts. Taken together these changes suggest that the actin cytoskeleton is undergoing increased remodeling in spinal cords of G93A mice. This remodeling could take place either in neurons as an indication of neurodegeneration or attempts at neuroregeneration, or in various glial populations.
A large number of metabolic enzymes were altered in the G93A lipid rafts (12 proteins, 18%, Figure 3) and their functional relevance rather suggested the involvement of astrocytes in ALS. Aspartate aminotransferase, which is involved in oxidizing glutamate to 2-oxo-glutarate [56], showed decreased levels in the G93A lipid rafts. It has been reported that excitotoxicity induced by excess amount of glutamate can contribute to motor neuron degeneration in ALS [4–6]. Decreased levels of aspartate aminotransferase can potentially contribute to excess glutamate and excitotoxicity in ALS. In addition, this study showed altered levels of enzymes involved in metabolism of monocarboxylates such as lactate and ketone bodies. Monocarboxylates, especially lactate, are produced and exported by astrocytes and subsequently taken up by neurons as an alternative fuel source to glucose [57]. We observed a decreased level of L-lactate dehydrogenase, an enzyme converting pyruvate to lactate, in the G93A lipid rafts. The data suggest that altered or defective metabolic support by astrocytes could play a role in ALS neuronal degeneration.
In conclusion, we have carried out a proteomic analysis to profile alterations of lipid raft associated proteins in the spinal cord of the G93A SOD1 transgenic mouse model of ALS. Alterations of a consortium of 67 lipid raft associated proteins in the G93A mouse sample were detected, some of which were independently validated. The altered proteins are involved in multiple functions such as vesicular transport, neurotransmitter synthesis and release; cytoskeleton organization and linkage to plasma membrane; metabolism; microglia activation and inflammation. Many of the protein changes are consistent with the disease etiology hypotheses in the field. The comprehensive study of lipid rafts proteins in the transgenic mouse models support that multiple types of cells in the spinal cord participate in the disease pathogenesis and progression, suggesting that multiple pathways are affected in ALS and contribute to motor neuron degeneration.
Materials and Methods
Materials
Acrylamide (40%) was purchased from Bio-Rad (Hercules, CA). Trypsin (modified, sequence grade, lypholized) was from Promega (Madison, WI). Ammonium bicarbonate (AMBIC), dithiothreitol(DTT), iodoacetamide (IAA), formic acid and maltopentaose were form Sigma-Aldrich (St. Louis, MO). Acetonitrile and HPLC water were obtained from Fisher Scientific (Hampton, NJ). Antibodies used were: cofilin (#3312) from Cell Signaling, flotillin-1 (#610820) and annexin II (#610068) from BD Bioscience, MnSOD (06-984) from Upstate, annexin II (sc-9061), TIM (sc-22031), Neurofilament M (sc-51683), SNAP-25 (sc-20038), GFAP (sc-33673) and SOD1 (sc-11407) from Santa Cruz.
Animals
Transgenic mice strains over-expressing human WT or G93A mutant SOD1 [19] were maintained as hemizygotes at the University of Kentucky animal facility. Transgenic positives were identified using PCR as previously published [19, 58]. G93A SOD1 transgenic mice and the age-matched WT SOD1 transgenic mice were sacrificed and perfused with PBS before spinal cords were dissected. All animal procedures were approved by the University IACUC committee.
Isolation of lipid raft protein from spinal cord extracts
Spinal cords were lysed by douncing in buffer A (0.25 M sucrose, 20 mM Tricine and 1 mM EDTA, pH 7.8) and centrifuged at 4°C for 10 min at 1000 × g. The supernatant, called the post-nuclear supernatant (PNS) was collected, added onto 30 % percoll and centrifuged at 61,884 × g for 30 min at 4°C. After centrifugation three fractions were collected: the cytoplasm (CYTO), intracellular membranes (IM) and plasma membranes (PM). The PM fraction was sonicated and lipid rafts (LR) were isolated from the plasma membrane fraction by a detergent-free method utilizing the unique buoyant density of lipid rafts in OptiPrep gradient as previously described [20]. The detergent free method is routinely used in the laboratory as the methods based on insolubility of lipid rafts in cold solutions containing Triton X-100 have been reported to be extensively contaminated by intracellular organelles and non-lipid rafts components [21, 22]. Similar detergent-free gradient centrifugation methods have also been used in other lipid rafts proteomics studies [59–61]. After protein concentration determination by the Bradford assay (Biorad), CYTO, PM and LR fractions were TCA precipitated and proteins were re-dissolved in 2 × SDS running buffer. The purity of the lipid raft fractions were examined by Western blotting, probing for the neuronal lipid raft marker protein flotillin-1, the cytosolic protein triosephosphate isomerase (TIM), the mitochondria protein MnSOD, and SOD1.
Preparation of protein digests
Approximately 80 to 100 μg lipid rafts proteins were obtained from each mouse spinal cord. Equal amount of lipid raft proteins (32 μg) from G93A and WT control mice were loaded to SDS-PAGE followed by Sypro Ruby staining. Individual WT and G93A lanes were sliced into 12 pieces and each piece then underwent dithiothreitol (DTT) reduction, iodoacetamide (IAA) alkylation, and in-gel trypsin digestion using a standard protocol as previously reported [18]. The resulting tryptic peptides were extracted, concentrated to 20μl each using a SpeedVac, and subjected to nano-LC-MS/MS analysis.
Mass spectrometry and proteomics
LC-MS/MS data were acquired on a QSTAR XL quadrupole time-of-flight mass spectrometer (ABI/MDS Sciex) coupled with a nano-flow HPLC system (Eksigent) through a nano-electrospray ionization source (Protana). Desired volume of sample solution (typically 5 μl out of the 20 μl extracted tryptic peptides from each gel band) was injected by an autosampler, desalted on a trap column (LC Packings 300 μm i.d. × 5 mm), and subsequently separated by reverse phase C18 column (Vydac 75 μm i.d. × 150 mm) at a flow rate of 200 nL/min. The HPLC gradient was linear from 5% to 80% mobile phase B in 70 min using mobile phase A (H2O, 0.1% formic acid) and mobile B (80% acetonitrile, 0.1% formic acid). Peptides eluted out of the reverse phase column were analyzed online by mass spectrometry (MS) and selected peptides were subjected to tandem mass spectrometry (MS/MS) sequencing. The automated data acquisition using information-dependent mode was performed on QSTAR XL under control by Analyst QS software. Each cycle typically consisted of one 1-sec MS survey scan from 350 to 1600 (m/z) and two 2-sec MS/MS scans with mass range of 100 to 1600 (m/z).
The LC-MS/MS data were submitted to a local MASCOT server for MS/MS protein identification search. Mascot Daemon mode was used to combine the MS/MS data from 12 gel bands of G93A-SOD1 and WT-SOD1 samples to perform a single merged search. The typical parameters were: Mus, Sprot database (51.0), maximum of two trypsin missed cleavages, cysteine carbamidomethylation, methionine oxidation, a maximum of 100 ppm MS error tolerance, and a 0.5 Da MS/MS error tolerance. All peptides were required to have an ion score greater than 30 (P < 0.05). Protein identification is considered positive if two unique peptides were matched. For protein identified based on single peptide match, the same protein was identified in two independent LC-MS/MS analysis of lipid rafts isolated from two or more mice in order to consider the protein a positive identification. All LC-MS/MS data were also submitted to decoy MASCOT search against a randomized Sprot decoy database [27], and the false discovery rate (FDR) in each LC-MS/MS experiment was determined.
The quantitative analysis was done by using a label-free quantitative approach that is similar to other published label-free quantification protocols [62–64]. A minor modification of the protocol is to include an internal control to provide additional assurance for accuracy. Briefly, a monosaccharide (maltopentaose, C30H52O26, monoisotopic m/z 829.27) that does not bind to the stationary phase of the reverse phase C18 column was added to mobile phases A and B at equal concentration as an internal standard. Since the monosaccharide did not bind to the C18 column due to its high hydrophilicity and its concentration in mobile phases A and B was equal, the concentration of the internal standard remained constant throughout the HPLC gradient. Since the concentration of the internal standard remained constant in all LC-MS experiments, it serves as a common denominator to calculate the ratio of a peptide in different samples. The internal standard also serves as a real-time reference for normalizing peptide peak intensities during LC-MS analysis, thus eliminating potential experimental variation among different LC-MS experiments. All peptide intensities were normalized with the intensity of the internal standard in the same MS scan; then compared to the normalized intensity of the same peptide in another sample to calculate the ratio of the peptides between two samples. The monosaccharide was observed to have no effect on elution time of peptides.
Western blotting
Lipid rafts isolated from the spinal cords of three separate pairs of WT and G93A SOD1 transgenic mice were used for Western blotting analysis. Twenty-five μg of lipid rafts protein were subjected to SDS-PAGE and transferred onto nitrocellulose membranes, 35 V, over night at 4°C in 25 mM Tris-HCl, 192 mM Glycine, 20% (v/v) methanol. Membranes were blocked in 5% milk or 5% BSA in TBST (100 mM Tris-buffered saline, pH 7.4, 0.1% Tween-20) for 1h hour at room temperature, followed by incubation with the indicated primary antibodies in TBST for 5 hours at room temperature. After 3 washes with TBST membranes were incubated with the indicated secondary antibodies for 1 hour at room temperature. Again, membranes were washed 3 times with TBST and the protein of interest was visualized using Super West Pico Enhanced Chemiluminescent Substrate (ECL) or Supersignal West Dura extended duration substrate (ECL) kit (Pierce, Rockford, IL). Western blotting results were quantified by scanning the images and analyzing with the ImageJ program.
Immunohistochemistry
For immunofluorescent stainings lumbar spinal cords from 90 and 125 day old mice were dissected, post-fixed over night in 4% paraformaldehyde (PFA) in 0.1 M PBS, dehydrated and embedded in Paraplast X-tra (Tyco Healthcare). Sections (6 μm) were deparaffinized, rehydrated and boiled in 0.01 M citrate buffer (pH 6.0) at a high power setting for 15 min to retrieve antigens. Sections were then blocked in 10% heat-inactivated fetal bovine serum (FBS) in 0.1 M PBS with 0.1% Triton X-100 (PBST) for 30 min before incubated with primary antibodies (rabbit anti-annexin II and mouse anti-neurofilament M) diluted in 2% FBS-PBST over night at room temperature. Following primary antibody incubation sections were washed with PBST and incubated with 4', 6-diamidino-2-phenylindole dihydrochoride (DAPI, Sigma) at 1:7500 and Alexa Fluor 488 anti-mouse (Molecular Probes) at 1:350 in 10% FBS-PBST at RT for 1h. Sections were then washed with PBST, incubated with Alexa Fluor 594 anti-rabbit (Molecular Probes), washed and then mounted using vectashield-mounting medium. Fluorescence microscopy was carried out using a Leica DM IRBE laser scanning confocal microscope with a 100 x objective.
Supplementary Material
Acknowledgment
We are grateful to Dr. Sidney Whiteheart for providing SNAP-25 antibody. This study was in part supported by the NIH grants R01-NS049126 (to H.Z.), R21-DK075473 (to E.J.S and H.Z.), R01-HL078976 and R01-DK077632 (to E.J.S.). The Proteomics Core directed by H.Z. is in part supported by the NIH/NCRR Center of Biomedical Research Excellence in the Molecular Basis of Human Disease (P20-RR020171) and NIH/NIEHS Superfund Basic Research Program (P42-ES007380). The NIH Shared Instrumentation Grant S10RR023684 (to H.Z.) is acknowledged for purchasing the 4800 Plus MALDI-TOF-TOF mass spectrometer.
Abbreviations
- ALS
amyotrophic lateral sclerosis
- HPLC
high performance liquid chromatography
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- SOD1
cooper-zinc superoxide dismutase
- WT
wild-type (WT)
References
- 1.Hughes JT. Pathology of amyotrophic lateral sclerosis. Adv Neurol. 1982;36:61–74. [PubMed] [Google Scholar]
- 2.Deng HX, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, Getzoff ED, Hu P, Herzfeldt B, Roos RP, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 3.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 4.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 5.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 6.Shaw BF, Valentine JS. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Benarroch EE. Lipid rafts, protein scaffolds, and neurologic disease. Neurology. 2007;69:1635–1639. doi: 10.1212/01.wnl.0000279590.22544.c3. [DOI] [PubMed] [Google Scholar]
- 8.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 11.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen S, Heath PR, Kirby J, Wharton SB, Cookson MR, Menzies FM, Banks RE, Shaw PJ. Analysis of the cytosolic proteome in a cell culture model of familial amyotrophic lateral sclerosis reveals alterations to the proteasome, antioxidant defenses, and nitric oxide synthetic pathways. J Biol Chem. 2003;278:6371–6383. doi: 10.1074/jbc.M209915200. [DOI] [PubMed] [Google Scholar]
- 13.Cutler RG, Pedersen WA, Camandola S, Rothstein JD, Mattson MP. Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann Neurol. 2002;52:448–457. doi: 10.1002/ana.10312. [DOI] [PubMed] [Google Scholar]
- 14.Massignan T, Casoni F, Basso M, Stefanazzi P, Biasini E, Tortarolo M, Salmona M, Gianazza E, Bendotti C, Bonetto V. Proteomic analysis of spinal cord of presymptomatic amyotrophic lateral sclerosis G93A SOD1 mouse. Biochem Biophys Res Commun. 2007;353:719–725. doi: 10.1016/j.bbrc.2006.12.075. [DOI] [PubMed] [Google Scholar]
- 15.Pasinetti GM, Ungar LH, Lange DJ, Yemul S, Deng H, Yuan X, Brown RH, Cudkowicz ME, Newhall K, Peskind E, Marcus S, Ho L. Identification of potential CSF biomarkers in ALS. Neurology. 2006;66:1218–1222. doi: 10.1212/01.wnl.0000203129.82104.07. [DOI] [PubMed] [Google Scholar]
- 16.Ramstrom M, Ivonin I, Johansson A, Askmark H, Markides KE, Zubarev R, Hakansson P, Aquilonius SM, Bergquist J. Cerebrospinal fluid protein patterns in neurodegenerative disease revealed by liquid chromatography-Fourier transform ion cyclotron resonance mass spectrometry. Proteomics. 2004;4:4010–4018. doi: 10.1002/pmic.200400871. [DOI] [PubMed] [Google Scholar]
- 17.Ranganathan S, Williams E, Ganchev P, Gopalakrishnan V, Lacomis D, Urbinelli L, Newhall K, Cudkowicz ME, Brown RH, Jr., Bowser R. Proteomic profiling of cerebrospinal fluid identifies biomarkers for amyotrophic lateral sclerosis. J Neurochem. 2005;95:1461–1471. doi: 10.1111/j.1471-4159.2005.03478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukada K, Zhang F, Vien A, Cashman NR, Zhu H. Mitochondrial proteomic analysis of a cell line model of familial amyotrophic lateral sclerosis. Mol Cell Proteomics. 2004;3:1211–1223. doi: 10.1074/mcp.M400094-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 20.Smart EJ, Ying YS, Mineo C, Anderson RG. A detergent-free method for purifying caveolae membrane from tissue culture cells. Proc Natl Acad Sci U S A. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Oh P, Horner T, Rogers RA, Schnitzer JE. Organized endothelial cell surface signal transduction in caveolae distinct from glycosylphosphatidylinositol-anchored protein microdomains. J Biol Chem. 1997;272:7211–7222. doi: 10.1074/jbc.272.11.7211. [DOI] [PubMed] [Google Scholar]
- 22.Gaus K, Rodriguez M, Ruberu KR, Gelissen I, Sloane TM, Kritharides L, Jessup W. Domain-specific lipid distribution in macrophage plasma membranes. J Lipid Res. 2005;46:1526–1538. doi: 10.1194/jlr.M500103-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Parkin ET, Turner AJ, Hooper NM. Amyloid precursor protein, although partially detergent-insoluble in mouse cerebral cortex, behaves as an atypical lipid raft protein. Biochem J. 1999;344(Pt 1):23–30. [PMC free article] [PubMed] [Google Scholar]
- 24.Schulte T, Paschke KA, Laessing U, Lottspeich F, Stuermer CA. Reggie-1 and reggie-2, two cell surface proteins expressed by retinal ganglion cells during axon regeneration. Development. 1997;124:577–587. doi: 10.1242/dev.124.2.577. [DOI] [PubMed] [Google Scholar]
- 25.Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 26.Foster LJ, De Hoog CL, Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc Natl Acad Sci U S A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 28.Babuke T, Tikkanen R. Dissecting the molecular function of reggie/flotillin proteins. Eur J Cell Biol. 2007;86:525–532. doi: 10.1016/j.ejcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Feuk-Lagerstedt E, Movitz C, Pellme S, Dahlgren C, Karlsson A. Lipid raft proteome of the human neutrophil azurophil granule. Proteomics. 2007;7:194–205. doi: 10.1002/pmic.200600482. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Wakim B, Li M, Halligan B, Tint GS, Patel SB. Quantifying raft proteins in neonatal mouse brain by `tube-gel' protein digestion label-free shotgun proteomics. Proteome Sci. 2007;5:17. doi: 10.1186/1477-5956-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell. 2006;5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YJ, Nakatomi R, Akagi T, Hashikawa T, Takahashi R. Unsaturated fatty acids induce cytotoxic aggregate formation of amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutants. J Biol Chem. 2005;280:21515–21521. doi: 10.1074/jbc.M502230200. [DOI] [PubMed] [Google Scholar]
- 33.Aisenbrey C, Borowik T, Bystrom R, Bokvist M, Lindstrom F, Misiak H, Sani MA, Grobner G. How is protein aggregation in amyloidogenic diseases modulated by biological membranes? Eur Biophys J. 2008;37:247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 34.Chung J, Yang H, de Beus MD, Ryu CY, Cho K, Colon W. Cu/Zn superoxide dismutase can form pore-like structures. Biochem Biophys Res Commun. 2003;312:873–876. doi: 10.1016/j.bbrc.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Ray SS, Nowak RJ, Strokovich K, Brown RH, Jr., Walz T, Lansbury PT., Jr. An intersubunit disulfide bond prevents in vitro aggregation of a superoxide dismutase-1 mutant linked to familial amytrophic lateral sclerosis. Biochemistry. 2004;43:4899–4905. doi: 10.1021/bi030246r. [DOI] [PubMed] [Google Scholar]
- 36.Vleminckx V, Van Damme P, Goffin K, Delye H, Van Den Bosch L, Robberecht W. Upregulation of HSP27 in a transgenic model of ALS. J Neuropathol Exp Neurol. 2002;61:968–974. doi: 10.1093/jnen/61.11.968. [DOI] [PubMed] [Google Scholar]
- 37.Sharp PS, Akbar MT, Bouri S, Senda A, Joshi K, Chen HJ, Latchman DS, Wells DJ, de Belleroche J. Protective effects of heat shock protein 27 in a model of ALS occur in the early stages of disease progression. Neurobiol Dis. 2008;30:42–55. doi: 10.1016/j.nbd.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Grinberg M, Schwarz M, Zaltsman Y, Eini T, Niv H, Pietrokovski S, Gross A. Mitochondrial Carrier Homolog 2 Is a Target of tBID in Cells Signaled To Die by Tumor Necrosis Factor Alpha. Mol Cell Biol. 2005;25:4579–4590. doi: 10.1128/MCB.25.11.4579-4590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maser E. Neuroprotective role for carbonyl reductase? Biochem Biophys Res Commun. 2006;340:1019–1022. doi: 10.1016/j.bbrc.2005.12.113. [DOI] [PubMed] [Google Scholar]
- 40.Kato S, Kato M, Abe Y, Matsumura T, Nishino T, Aoki M, Itoyama Y, Asayama K, Awaya A, Hirano A, Ohama E. Redox system expression in the motor neurons in amyotrophic lateral sclerosis (ALS): immunohistochemical studies on sporadic ALS, superoxide dismutase 1 (SOD1)-mutated familial ALS, and SOD1-mutated ALS animal models. Acta Neuropathol. 2005;110:101–112. doi: 10.1007/s00401-005-1019-3. [DOI] [PubMed] [Google Scholar]
- 41.Lee YM, Park SH, Shin D-I, Hwang J-Y, Park B, Park Y-J, Lee TH, Chae HZ, Jin BK, Oh TH, Oh YJ. Oxidative Modification of Peroxiredoxin Is Associated with Drug-induced Apoptotic Signaling in Experimental Models of Parkinson Disease. J Biol Chem. 2008;283:9986–9998. doi: 10.1074/jbc.M800426200. [DOI] [PubMed] [Google Scholar]
- 42.Collins RN. Rab and ARF GTPase regulation of exocytosis. Mol Membr Biol. 2003;20:105–115. doi: 10.1080/0968768031000085892. [DOI] [PubMed] [Google Scholar]
- 43.Li G, Han L, Chou TC, Fujita Y, Arunachalam L, Xu A, Wong A, Chiew SK, Wan Q, Wang L, Sugita S. RalA and RalB function as the critical GTP sensors for GTP-dependent exocytosis. J Neurosci. 2007;27:190–202. doi: 10.1523/JNEUROSCI.2537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahn R. Principles of exocytosis and membrane fusion. Ann N Y Acad Sci. 2004;1014:170–178. doi: 10.1196/annals.1294.018. [DOI] [PubMed] [Google Scholar]
- 45.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 46.Kabashi E, Agar JN, Taylor DM, Minotti S, Durham HD. Focal dysfunction of the proteasome: a pathogenic factor in a mouse model of amyotrophic lateral sclerosis. J Neurochem. 2004;89:1325–1335. doi: 10.1111/j.1471-4159.2004.02453.x. [DOI] [PubMed] [Google Scholar]
- 47.Kabashi E, Agar JN, Hong Y, Taylor DM, Minotti S, Figlewicz DA, Durham HD. Proteasomes remain intact, but show early focal alteration in their composition in a mouse model of amyotrophic lateral sclerosis. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05317.x. in press. [DOI] [PubMed] [Google Scholar]
- 48.Morimoto N, Nagai M, Ohta Y, Miyazaki K, Kurata T, Morimoto M, Murakami T, Takehisa Y, Ikeda Y, Kamiya T, Abe K. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 49.Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–293. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- 50.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 51.Niggli V, Rossy J. Ezrin/radixin/moesin: Versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol. 2007 doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 52.Lederer CW, Torrisi A, Pantelidou M, Santama N, Cavallaro S. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics. 2007;8:26. doi: 10.1186/1471-2164-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mishra M, Paunesku T, Woloschak GE, Siddique T, Zhu LJ, Lin S, Greco K, Bigio EH. Gene expression analysis of frontotemporal lobar degeneration of the motor neuron disease type with ubiquitinated inclusions. Acta Neuropathol (Berl) 2007;114:81–94. doi: 10.1007/s00401-007-0240-7. [DOI] [PubMed] [Google Scholar]
- 54.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 55.Sarmiere PD, Bamburg JR. Regulation of the neuronal actin cytoskeleton by ADF/cofilin. J Neurobiol. 2004;58:103–117. doi: 10.1002/neu.10267. [DOI] [PubMed] [Google Scholar]
- 56.Daikhin Y, Yudkoff M. Compartmentation of brain glutamate metabolism in neurons and glia. J Nutr. 2000;130:1026S–1031S. doi: 10.1093/jn/130.4.1026S. [DOI] [PubMed] [Google Scholar]
- 57.Pellerin L. Lactate as a pivotal element in neuron-glia metabolic cooperation. Neurochem Int. 2003;43:331–338. doi: 10.1016/s0197-0186(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 58.Zhang F, Strom AL, Fukada K, Lee S, Hayward LJ, Zhu H. Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J Biol Chem. 2007;282:16691–16699. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- 59.Li N, Shaw AR, Zhang N, Mak A, Li L. Lipid raft proteomics: analysis of in-solution digest of sodium dodecyl sulfate-solubilized lipid raft proteins by liquid chromatography-matrix-assisted laser desorption/ionization tandem mass spectrometry. Proteomics. 2004;4:3156–3166. doi: 10.1002/pmic.200400832. [DOI] [PubMed] [Google Scholar]
- 60.Paradela A, Bravo SB, Henriquez M, Riquelme G, Gavilanes F, Gonzalez-Ros JM, Albar JP. Proteomic analysis of apical microvillous membranes of syncytiotrophoblast cells reveals a high degree of similarity with lipid rafts. J Proteome Res. 2005;4:2435–2441. doi: 10.1021/pr050308v. [DOI] [PubMed] [Google Scholar]
- 61.Gupta N, Wollscheid B, Watts JD, Scheer B, Aebersold R, DeFranco AL. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nat Immunol. 2006;7:625–633. doi: 10.1038/ni1337. [DOI] [PubMed] [Google Scholar]
- 62.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 63.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 64.Rinner O, Mueller LN, Hubalek M, Muller M, Gstaiger M, Aebersold R. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.