Abstract
Insulin-like growth factor-1 (IGF-1) is a small protein that promotes cell survival and growth, often acting over long distances. Although for decades IGF-1 has been considered to have therapeutic potential, systemic side effects of IGF-1 are significant, and local delivery of IGF-1 for tissue repair has been a long-standing challenge. In this study, we designed and purified a novel protein, heparin-binding IGF-1 (Xp-HB-IGF-1), which is a fusion protein of native IGF-1 with the heparin-binding domain of heparin-binding epidermal growth factor-like growth factor. Xp-HB-IGF-1 bound selectively to heparin as well as the cell surfaces of 3T3 fibroblasts, neonatal cardiac myocytes and differentiating ES cells. Xp-HB-IGF-1 activated the IGF-1 receptor and Akt with identical kinetics and dose response, indicating no compromise of biological activity due to the heparin-binding domain. Because cartilage is a proteoglycan-rich environment and IGF-1 is a known stimulus for chondrocyte biosynthesis, we then studied the effectiveness of Xp-HB-IGF-1 in cartilage. Xp-HB-IGF-1 was selectively retained by cartilage explants and led to sustained chondrocyte proteoglycan biosynthesis compared to IGF-1. These data show that the strategy of engineering a “long-distance” growth factor like IGF-1 for local delivery may be useful for tissue repair and minimizing systemic effects.
Keywords: heparin binding domain, tissue repair, biosynthesis
Insulin-like growth factor-1 (IGF-1) is a growth factor well known as an important mediator of cell growth and differentiation. IGF-1 stimulates several signaling pathways through the tyrosine kinase IGF-1 receptor, including phosphatidylinositol (PI) 3-kinase and mitogen-activated protein kinases (MAPKs) (1). PI3-kinase has many downstream targets, including the kinase Akt, and activation of Akt promotes survival, proliferation, and growth (2).
IGF-1 has been extensively studied for its therapeutic potential in tissue repair and regeneration (3-5). IGF-1 is a small and highly diffusible protein that can act over long distances (6). However, systemic administration of IGF-1 has significant side effects (7) as well as the potential to promote diabetic retinopathy (8) and cancer (9). Therefore, local delivery of IGF-1 has been a longstanding challenge. Here, we describe the design of a new protein, formed by fusion of IGF-1 with the heparin-binding (HB) domain of heparin-binding epidermal growth factor-like growth factor (HB-EGF). HB-EGF binds selectively to glycosaminoglycans through its highly positively charged heparin-binding domain (10-12), and HB-EGF acts locally within tissues (13). Thus, we hypothesized that engineering IGF-1 to bind to glycosaminoglycans could provide selective delivery of IGF-1 to cell surfaces or to specific tissues. We demonstrate that this heparin-binding IGF-1 (Xp-HB-IGF-1) can bind to cell surfaces as well as the proteoglycan-rich tissue of cartilage; furthermore, Xp-HB-IGF-1 prolongs the stimulation of chondrocyte biosynthesis, demonstrating its potential for tissue specific repair.
MATERIALS AND METHODS
Vector construction
Rat IGF-1 cDNA was amplified by polymerase chain reaction (PCR) using the primers (5′ to 3′) GGACCAGAGGACCCTTTGCG (forward) and AGCTGACTTTGTAGGCTTCAGC (reverse). We used the sequence for the mature peptide IGF-1 (70 amino acids), encoded by exons 3 and 4 (14, 15). The PCR product was subcloned into the pTrcHis-TOPO vector (Invitrogen, Carlsbad, CA, USA) with the addition of a stop codon (TAG) at the C-terminus of IGF-1, thus encoding an Xpress-tagged IGF-1 (Xp-IGF-1). To encode Xp-HB-IGF-1, the heparin binding sequence (AA 93–113) of rat HB-EGF (AAAAAGAAGAGGAAAGGCAAGGGGTTAGGAAAGAAGAGAGATCCATGCCTTAAGAAATACAAG) was inserted between the Xpress tag and the IGF-1 sequence through mutagenesis (Fig. 1A—C). Amplification was performed with PfuUltra HF DNA Polymerase (Stratagene, Cedar Creek, TX, USA), and the template plasmid was digested with DpnI (New England Biolabs, Beverly, MA, USA) before transformation in Escherichia coli. All sequences were confirmed by DNA sequencing.
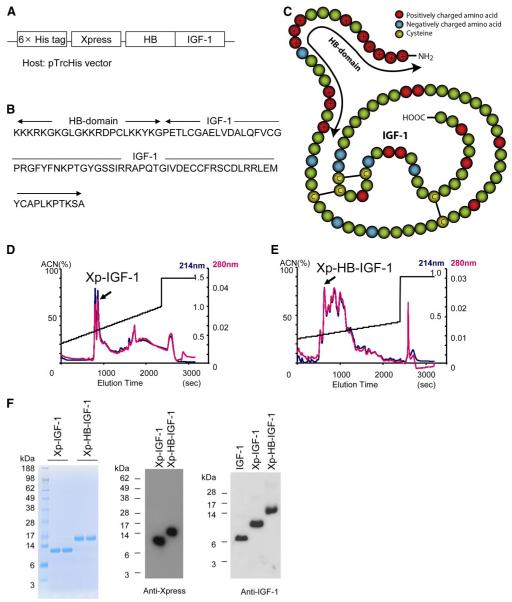
Figure 1.
Construction and purification of a new Xp-HB-IGF-1 fusion protein. A) The heparin binding domain of HB-EGF was inserted N-terminal to IGF-1 to generate the fusion protein. The construct included the hexahistidine and Xpress tags from the pTrcHis vector for purification and detection. B) The resulting amino acid sequence of HB-IGF-1. C) Schematic for the structure of HB-IGF-1. Red circles: positively charged amino acids; blue circles: negatively charged amino acids; yellow circles: cysteines. The arrow shows the HB domain. In this figure the epitope tags are not shown. D, E) Representative reverse-phase high-performance liquid chromatography (RP-HPLC) elution profiles with single peaks containing correctly folded protein. Readings of optical density at 214 nm are in blue; readings at 280 nm are in red; elution is by acetonitrile (ACN) gradient. F) After RP-HPLC, Coomassie blue staining and Western blot analysis demonstrate isolation of single bands containing Xpress-tagged protein. The right panel shows that the Western blot analysis of IGF-1, and the two engineered IGF-1 proteins yield similar results using an anti-IGF-1 antibody.
Purification of protein
Xp-IGF-1 and Xp-HB-IGF-1 were expressed in E. coli BL21 cells and grown in Luria-Bertani (LB) medium in 4 L batches. Protein synthesis was induced with 1 mM isopropyl β-D-thiogalactoside for 4 h, and cells were then harvested by centrifugation, lysed in lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate, 500 mM NaCl, pH 7.8), and homogenized. The first purification step consisted of affinity purification by the polyhistidine tag in fusion proteins with Ni-NTA (Invitrogen). Ni-NTA resin was washed with wash buffer (8 M urea, 500 mM NaCl, 20 mM phosphate, pH 6.2), and bound protein was eluted at pH 4. Eluted proteins were then subjected to oxidative refolding to restore biological activity. The proteins were incubated overnight at 4°C with refolding buffer (50 mM Tris, 75 mM NaCl, 100 μM glutathione disulfide, and 100 μM glutathione, pH 7.8). After refolding, the samples were adjusted to 0.1% trifluoroacetic acid and loaded on a C18 reverse-phase high-performance liquid chromatography column (Delta-Pak C18, Waters, Milford, MA, USA) as a final purification step. The column was subjected to a linear gradient from 25% to 40% acetonitrile in 0.1% trifluoroacetic acid (16, 17). The yields of purified Xp-IGF-1 and Xp-HB-IGF-1 were 10 μg and 50 μg from 8 LLB medium, respectively.
Cell culture
Primary cultures of cardiac myocytes were prepared from the ventricles of neonatal Sprague Dawley rats and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with 7% fetal bovine serum (Invitrogen); the medium was replaced after 24 h with serum-free medium. 3T3 fibroblast cells were cultured in DMEM with 10% newborn calf serum (Invitrogen) and the medium was replaced with serum-free medium 24 h before experiments. Mouse embryonic stem (ES) cells were grown on gelatin-coated dishes without feeder cells in Glasgow Minimum Essential Medium (Invitrogen) supplemented with 15% Knockout SR (Invitrogen) and leukemia inhibitory factor (Chemicon, Billerica, MA, USA). Cells were passaged every 3 days. To induce differentiation, cells were first enzymatically dissociated and cultured as hanging drops for embryoid body formation as described previously (18). Differentiation medium with 10% ES cell-qualified fetal bovine serum (Invitrogen) without leukemia inhibitory factor was added. These ES cells became green fluorescent protein (GFP) positive after differentiation into cardiac myocytes, because they were stably transfected with an α-myosin heavy chain promoter-driven enhanced GFP vector (18). After embryoid body formation (day 7), cells were plated on gelatin-coated dishes.
Harvest and culture of cartilage
Bovine articular cartilage explants (3-mm-diameter, 1-mm-thick disks) were harvested from the femoropatellar grooves of 1- to 2-wk-old calves and cultured in low-glucose DMEM with 10 mM HEPES, 0.1 mM nonessential amino acids, 0.4 mM L-proline, 20 μg/ml ascorbate, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 atmosphere (19).
Protein analysis
Neonatal cardiac myocytes and 3T3 fibroblasts were lysed using phosphate-buffered saline (PBS) with 1% Triton-X, 0.25% Na-deoxycholate, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM NaF, 1 mM Na3VO4, and 1:1000 protease inhibitor cocktail (Sigma, St. Louis, MD, USA). Cartilage disks were pulverized and lysed with 100 mM NaCl, 50 mM Tris, 0.5% Triton-X, 5 mM EDTA, 1 mM PMSF, and 1:1000 protease inhibitor cocktail. Protein concentration was measured by Bradford assay and 10 μg protein was loaded in each well for Western blot analysis. Similar GAG content was observed in all samples as measured by DMMB dye binding (20, 21). Anti-Xpress antibody (Invitrogen), polyclonal anti-IGF-1 anti-body (Abcam, Cambridge, MA, USA), antiphospho-IGF-1 receptor antibody (Cell Signaling, Danvers, MA, USA), antiphospho-Akt antibody (Cell Signaling), and anti-Actin antibody (Sigma) were used. Recombinant human IGF-1 was purchased from Sigma as a control protein.
To detect the fusion proteins by enzyme-linked immunosorbent assay (ELISA), 96-well plates were coated with an anti-Xpress antibody (10 μg/ml) overnight. Identical amounts of protein from cartilage extracts were added to each well. Polyclonal IGF-1 antibody was used as the primary antibody, and anti-rabbit-horseradish peroxidase (Bio-Rad, Hercules, CA, USA) was used as the secondary antibody. After addition of ABTS Peroxidase Substrate (KPL, Gaithersburg, MD, USA), plates were read at 405 nm.
Binding assays
Heparin agarose beads (Sigma) were incubated with 300 pmol Xp-HB-IGF-1 or Xp-IGF-1 for 2 h and washed 3× with PBS. Fusion proteins bound to the beads were extracted by boiling with SDS-PAGE sample buffer (Invitrogen). 3T3 fibroblast cells or neonatal rat cardiac myocytes were incubated with 100 nM Xp-HB-IGF-1 or control IGF-1 (Sigma) for 2 h and then washed with PBS 3×. The cells were lysed with lysis buffer and then subjected to Western blot analysis with an anti-IGF-1 antibody. Embryoid bodies (10 days after induction of differentiation) were incubated with fusion proteins for 2 h, washed with PBS 3×, and fixed with paraformaldehyde before immunohistochemistry with an anti-Xpress antibody. Cartilage disks were cultured in serum-free DMEM supplemented with either 500 nM Xp-HB-IGF-1 or 500 nM Xp-IGF-1. After 48 h (on day 0), disks were washed 3× with PBS, then incubated in DMEM with no IGF-1. Disks were collected on days 0, 1, 2, and 4. Protein remaining in cartilage extracts was detected by Western blot analysis and ELISA.
Chondroitin sulfate digestion from cartilage tissue
Chondroitin sulfate in 0.5-mm-thick cartilage disks was digested by protease-free chondroitinase ABC (Seikagaku Corporation, Tokyo, Japan). Each disk was incubated for 2 days at 37°C in 250 μl 0.4 U/ml chondroitinase ABC with 150 mM NaCl, 50 mM Tris, 2 mM EDTA, 1 mM PMSF, and proteinase inhibitor cocktail (Sigma) at pH 8.0. After 2 days digestion, 95.5% of GAG was digested, as measured by DMMB assay. The chondroitinase-treated disks vs. no treatment disks were incubated with 500 nM Xp-HB-IGF-1 or Xp-IGF-1 for 2 days and then IGF-1 binding was studied.
Cartilage biosynthesis assay
Chondrocyte proteoglycan synthesis was measured by incorporation of [35S]sulfate (PerkinElmer, Waltham, MA, USA), as described previously (21). Cartilage disks were equilibrated in serum-free medium for 1 day and incubated in medium containing 100 nM Xp-HB-IGF-1, Xp-IGF-1, or control IGF-1 (Sigma) for 2 days. The disks were then washed 3× with PBS and changed to IGF-1-free medium. Cultured disks were radiolabeled with 5 μCi/ml [35S]sulfate for the final 24 h of culture. After labeling, each disk was washed 3× in 1.0 ml of PBS with 0.8 mM proline and 1 mM Na2SO4 at 4°C to remove free radiolabel. Disks were digested in 1.0 ml of proteinase K (125 μg/ml in 0.1 M Na2SO4, 5 mM EDTA, and 5 mM cysteine at pH 6.0). The [35S]sulfate content of the digests was then measured in a scintillation counter (Wallac Micro-Beta TriLux, PerkinElmer, Waltham, MA, USA), with corrections for spillover and quenching.
Statistical analysis
Statistical analyses were performed by Student’s t test with acceptance level α = 0.05. t Tests were corrected for multiple comparisons using α = 1 − (1 − α0)1/n, where α0 = 0.05 and n = total number of comparisons. All data were expressed as means ± se.
RESULTS
Purification of Xp-HB-IGF-1
Figure 1A—C shows the constructs for Xp-HB-IGF-1 and the control Xp-IGF-1 fusion proteins. IGF-1 has 3 disulfide bonds and includes 70 amino acids. The IGF-1 fusion proteins both contain polyhistidine tags for protein purification and Xpress tags for protein detection. The expected molecular masses of Xp-HB-IGF-1 and Xp-IGF-1 are 14,018 and 11,548 Da, respectively. Xp-HB-IGF-1 has the HB domain on the N terminus of IGF-1. The HB domain has 21 amino acids and includes 12 positively charged amino acids. Final purification of the new fusion proteins after refolding was performed with RP-HPLC (Fig. 1D, E). Identification of the correctly folded protein was performed as described previously (16) and confirmed with bioactivity assays. Figure 1F shows the Coomassie blue staining and Western blot analysis with an anti-Xpress antibody of the refolded IGF-1 proteins after RP-HPLC, revealing a single band. To eliminate concerns about differential transfer rates for Xp-HB-IGF-1, Xp-IGF-1, and unmodified IGF-1 during Western blot analysis, we used the same amount of these proteins (1.5 pmol each) and performed Western blot analysis using IGF-1 antibody (Fig. 1F, right panel). These 3 IGF-1s yielded similar intensities, indicating that the transfer of these proteins is similar under the conditions used.
Xp-HB-IGF-1 binds to heparin and cell surfaces
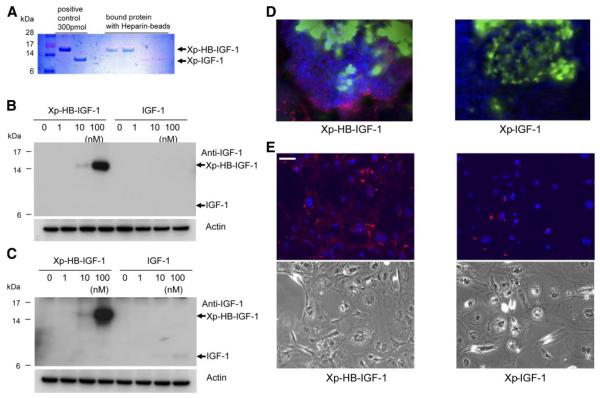
We first tested whether Xp-HB-IGF-1 binds selectively to heparin. After 2 h incubation of heparin agarose beads with 300 pmol Xp-HB-IGF-1 or Xp-IGF-1, bound proteins were extracted from beads by boiling. Coomassie blue staining showed that Xp-HB-IGF-1 binds selectively to heparin compared with Xp-IGF-1 (Fig. 2A). Next, we tested the ability of Xp-HB-IGF-1 to bind to cell surfaces, which have heparan sulfate proteoglycans, using 3T3 fibroblast cells (Fig. 2B) and neonatal rat cardiac myocytes (Fig. 2C). After pretreatment with 0–100 nM of Xp-HB-IGF-1 for 2 h, cells were washed with PBS 3×. For these experiments, commercial IGF-1 was used as control. Xp-HB-IGF-1 bound to 3T3 fibroblast cells when treated with 10 and 100 nM concentrations (Fig. 2B). Figure 2C shows Xp-HB-IGF-1 binding with neonatal cardiac myocytes, with clear selective binding of Xp-HB-IGF-1 at 10 and 100 nM and a very weak band of IGF-1 at 100 nM. These results are consistent with binding of this HB domain to heparan sulfate in the submicromolar range (11, 22). We also studied the ability of Xp-HB-IGF-1 to bind to ES cells in embryoid bodies, which contain multiple cell types. Xp-HB-IGF-1 was readily detected on the surfaces of cells in the embryoid bodies by immunofluorescence for the Xpress epitope tag, indicating that Xp-HB-IGF-1 can bind to multiple cell types (Fig. 2D). We studied the binding of Xp-HB-IGF-1 with ES cells after dissociation of embryoid bodies by trypsin and EDTA. Figure 2E shows a monolayer of differentiated ES cells, with more Xpress epitope tag in Xp-HB-IGF-1 group than the Xp-IGF-1 group. This result is consistent with the data shown in Fig. 2D, suggesting that Xp-HB-IGF-1 binds with heparan sulfate on the cell surface.
Figure 2.
Xp-HB-IGF-1 binds heparin and is retained more strongly in cell cultures. A) 300 pmol Xp-HB-IGF-1 or Xp-IGF-1 was incubated with heparin agarose beads for 2 h and washed 3× with PBS. Fusion proteins retained on the beads were detected by Coomasie blue staining. B, C) Doses of Xp-HB-IGF-1 or control IGF-1 were incubated with 3T3 fibroblast cells (B) or neonatal rat cardiac myocytes (C) for 2 h. The cells were collected after washing 3× with PBS, and the protein retained by the cell monolayer was detected by Western blot analysis using an anti-IGF-1 antibody. The results shown are representative of results from 3 independent experiments. D) Embryoid bodies were incubated with 100 nM Xp-HB-IGF-1 or Xp-IGF-1 for 2 h and then washed 3× with PBS. Retained protein was detected by immunofluorescent chemistry with an anti-Xpress antibody (red: anti-Xpress; green:α-myosin heavy chain; blue: DAPI). E) Embryoid bodies were dissociated by trypsin and EDTA, and monolayer differentiated ES cells were incubated with 100 nM Xp-HB-IGF-1 or Xp-IGF-1 for 2 h and then washed 3× with PBS. Retained protein was detected by immunofluorescence with an anti-Xpress antibody (red: anti-Xpress, blue: DAPI). Bottom panels show phase contrast. Scale bar = 50 μm.
Xp-HB-IGF-1 bioactivity
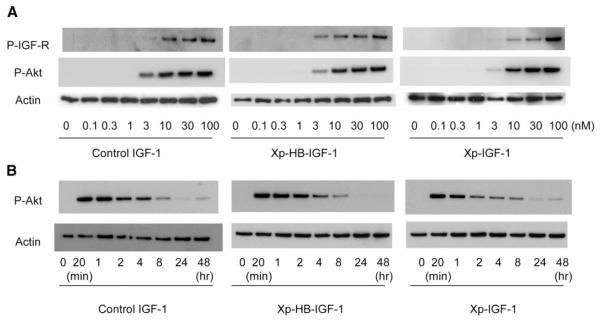
To determine whether the HB domain interferes with bioactivity, bioassays for IGF-1 receptor phosphorylation and Akt activation were performed. Control IGF-1, Xp-HB-IGF-1, and Xp-IGF-1 all activated the IGF-1 receptor of neonatal cardiac myocytes dose-dependently and induced Akt phosphorylation identically (Fig. 3A). Control IGF-1, Xp-HB-IGF-1, and Xp-IGF-1 all activated Akt with a similar time course (Fig. 3B). These data demonstrate that addition of the heparin-binding domain does not interfere with the bioactivity of IGF-1.
Figure 3.
Xp-HB-IGF-1 and Xp-IGF-1 activity is equivalent to IGF-1. A) Dose-response for phosphorylation of the IGF-1 receptor and Akt: neonatal rat cardiac myocytes were incubated with control IGF-1, Xp-HB-IGF-1, or Xp-IGF-1 at the indicated doses for 20 min. B) Time course of Akt phosphorylation: neonatal rat cardiac myocytes were incubated with 30 nM of control IGF-1, Xp-HB-IGF-1 or Xp-IGF-1 for the indicated time periods. The results shown are representative of results from 3 independent experiments. P-IGF-R and P-Akt represent phosphorylation of IGF-1 receptor and phosphorylation of Akt, respectively.
Xp-HB-IGF-1 transport in cartilage
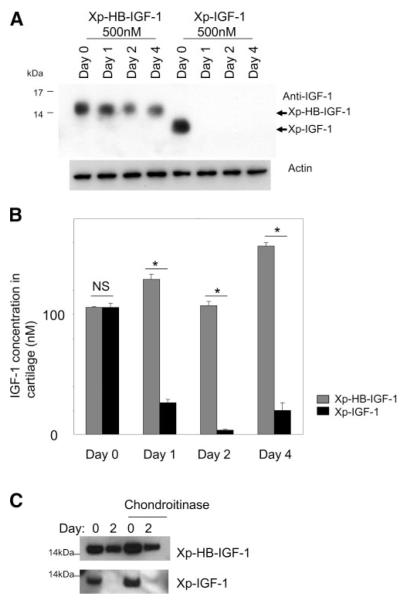
Cartilage is a proteoglycan-rich tissue, and chondrocytes respond to IGF-1 with increased extracellular matrix synthesis (19). Because prolonged local stimulation of IGF-1 signaling could thus be beneficial for cartilage repair, we studied the ability of Xp-HB-IGF-1 to bind to cartilage. Identically sized bovine articular cartilage disks were incubated with 500 nM Xp-HB-IGF-1 or Xp-IGF-1 for 1, 3, or 6 days, and no differences were found in the amount of IGF-1 protein that diffused into cartilage over this time period (data not shown). After preincubation with Xp-HB-IGF-1 or Xp-IGF-1 for 48 h, cartilage disks were washed with PBS at day 0 and similar amounts of IGF-1 were detected. However, on days 1, 2, and 4 after removal of the IGF-1 proteins, only Xp-HB-IGF-1 remained in the cartilage, suggesting that Xp-HB-IGF-1 bound to the proteoglycan-rich extracellular matrix (Fig. 4A). In contrast Xp-IGF-1 was undetectable even 1 day after washing. We also performed this selective binding experiment with cartilage extracts and ELISA measurements (Fig. 4B). These results confirmed that Xp-HB-IGF-1 is selectively retained by cartilage, while Xp-IGF-1 is rapidly lost.
Figure 4.
Xp-HB-IGF-1 is retained in cartilage tissue. A, B) Cartilage disks were incubated with 500 nM Xp-HB-IGF-1 or Xp-IGF-1 for 2 days, then washed 3× with PBS and changed to media with no IGF-1 (day 0). IGF-1 fusion proteins remaining in the cartilage were detected by Western blot analysis with an anti-IGF-1 antibody (A) or by an ELISA using anti-Xpress and anti-IGF-1 antibodies (B). Results are expressed as mean ± se; n = 4. *P < 0.01. NS, not significant. C) No treatment cartilage disks or chondroitinase-treated cartilage disks were incubated with 500 nM Xp-HB-IGF-1 or Xp-IGF-1 for 2 days, then washed 3× with PBS and changed to media with no IGF-1 (day 0). IGF-1 fusion proteins remaining in the cartilage were detected by Western blot analysis with an anti-IGF-1 antibody.
Xp-HB-IGF-1 can bind to cartilage after chondroitin sulfate digestion
To explore the possibility of nonspecific binding of Xp-HB-IGF-1 to glycosaminoglycans other than heparan sulfate, we studied the binding of Xp-HB-IGF-1 after chondroitinase ABC digestion. Figure 4C shows that Xp-IGF-1 was not retained from control or chondroitinase-treated cartilage disks after 2 days. On the other hand, Xp-HB-IGF-1 was retained after 2 days, and there was no significant difference between control and chondroitinase-treated disks. These data suggest that Xp-HB-IGF-1 retention is not mediated by the pool of chondroitin sulfated proteoglycans in the cartilage matrix.
Xp-HB-IGF-1 increases chondrocyte biosynthesis
The selective retention of Xp-HB-IGF-1 to cartilage suggests that this fusion protein could deliver a sustained stimulus for chondrocyte biosynthesis. Therefore, we measured chondrocyte biosynthesis of extracellular matrix proteoglycans by incorporation of [35S]sulfate. Cartilage disks were incubated with 100 nM Xp-HB-IGF-1, Xp-IGF-1, or control IGF-1 for 2 days and washed 3× with PBS, followed by culture in medium with no IGF-1 (Fig. 5A). [35S]sulfate incorporation was measured for 24 h beginning on day 0 (before washout), day 2 (just after washout), and days 4, 6, and 8. During incubation with the IGF-1 constructs on day 0, control IGF-1, Xp-IGF-1, and Xp-HB-IGF-1 groups all stimulated proteoglycan synthesis as expected (Fig. 5B, day 0). However, after washing, neither control IGF-1 nor Xp-IGF-1 stimulated proteoglycan synthesis at day 4 or beyond. In contrast, Xp-HB-IGF-1 led to sustained stimulation of proteoglycan synthesis for at least 8 days. Proteoglycan synthesis was significantly higher in cartilage incubated with Xp-HB-IGF-1 vs. Xp-IGF-1 on days 2, 4, 6, and 8. These data demonstrate that Xp-HB-IGF-1, which is selectively retained in the cartilage, stimulates chondrocyte biosynthesis over a more sustained period.
Figure 5.
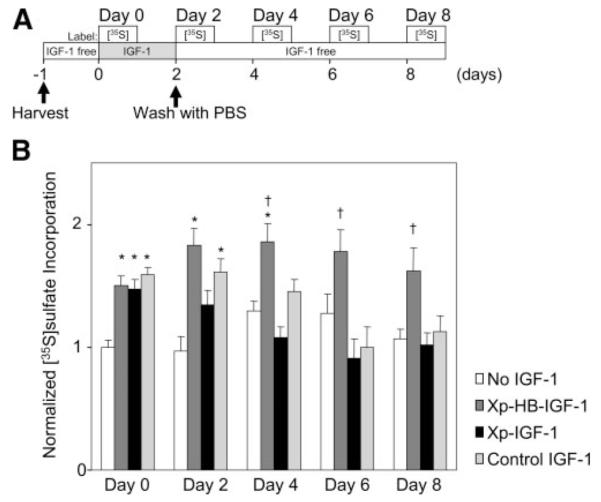
Stimulation of cartilage biosynthesis by Xp-HB-IGF-1 is sustained longer than by Xp-IGF-1. Starting on day 0, cartilage disks were incubated with 100 nM Xp-HB-IGF-1, Xp-IGF-1, control IGF-1 or IGF-1 free (No IGF-1) for 2 days. The disks were then washed 3× with PBS and changed to media with no IGF-1. Incorporation of [35S]sulfate over 24 h was measured starting at the indicated times. A) Schematic for the time course of the biosynthesis assay. B) Normalized [35S]sulfate incorporation. The data were normalized to the incorporation rate of the No IGF-1 group on day 0. Results are shown as mean ± se; n = 12. Asterisk (*) indicates significant differences vs. the No IGF-1 group; dagger (†) indicates significant differences between Xp-HB-IGF-1 and Xp-IGF-1.
DISCUSSION
Local delivery of IGF-1 has the potential for improving tissue repair and regeneration while minimizing systemic adverse effects. In this study, we describe a novel IGF-1 protein, Xp-HB-IGF-1, which binds to proteoglycan-rich tissue and cell surfaces but has the same bioactivity as IGF-1. Our data indicate that Xp-HB-IGF-1 can activate the IGF-1 receptor and Akt and thus that the heparin-binding domain does not interfere with interactions of IGF-1 and its receptor. IGF-1 has four domains (23, 24): B domain (aa 1–29), C domain (aa 30 – 41), A domain (aa 42–62) and D domain (aa 63–70), with the C domain playing the most important role in binding to the IGF-1 receptor (25). Replacement of the entire C domain causes a 30-fold decrease in affinity for the IGF-1 receptor (25). Thus, the addition of the heparin-binding domain to the N terminus of IGF-1 was not anticipated to interfere with interactions with the IGF-1 C domain.
Both extracellular matrix and cell surfaces are rich in proteoglycans and can serve as reservoirs for proteoglycan-binding growth factors. A classic example is the fibroblast growth factor-2 (FGF-2) system, where a low-affinity, high-capacity pool of proteoglycan receptors serves as a reservoir of FGF-2 for its high-affinity receptor (26). Our experiments suggest that Xp-HB-IGF-1 could function in some circumstances in a similar manner, since Xp-HB-IGF-1 is selectively retained on cell surfaces. Many growth factors are known to interact with heparan sulfate, including HB-EGF (10-12), FGF-2 (26), vascular endothelial growth factor-A (VEGF-A) (27), transforming growth factor beta (TGF-β) (28), platelet-derived growth factors (PDGFs) (29), and hepatocyte growth factor (HGF) (30). However, other proteins such as nerve growth factor (NGF), which induces differentiation and reduces apoptosis of neurons, does not have the heparin-binding domain (30). Thus, the strategy of engineering growth factors for selective matrix or cell surface binding could be used for other growth factors.
IGF-1 can also bind with extracellular matrix via IGF binding proteins (IGFBPs) (31); in the circulation, at least 99% of IGF-1 is bound to IGFBPs (IGFBP-1 to −6) (32). Further experiments are necessary to determine whether addition of a heparin-binding domain to IGF-1 changes interactions with IGFBPs and whether this changes its biological activity.
We used several types of cells, including 3T3 fibroblasts, cardiac myocytes, and ES cells, to demonstrate that Xp-HB-IGF-1 binds to cells. Xp-HB-IGF-1, therefore, has the potential to promote repair of injured myocardium or dermal wounds. Xp-HB-IGF-1 may also be useful for cell therapies using ES cells; for example, Xp-HB-IGF-1 can reduce apoptosis of ES cells (data not shown). Thus, the ability of Xp-HB-IGF-1 to bind to multiple cell types may enhance repair and regeneration in tissues other than cartilage.
IGF-1 can promote the synthesis of cartilage extracellular matrix and inhibit cartilage degradation (19); however, a practical mode of IGF-1 delivery to cartilage has yet to be developed (33). Heparan sulfate proteoglycans are prevalent in the pericellular matrix of cartilage, particularly as chains on perlecan and syndecan-2, and are known to bind other ligands such as FGF-2 (34). Our experiments suggest that Xp-HB-IGF-1 protein can bind with matrix and increase local, long-term bioavailability to chondrocytes and thus may improve cartilage repair. Previous studies have shown the IGF-1 concentrations of 100 ng/ml (∼13 nM) can induce chondrocyte biosynthesis (19). Based on ELISA we estimated that the concentration of Xp-HB-IGF-1 in the cartilage was ∼20 nM for our experiments with sulfate incorporation. Thus, the tissue dose of Xp-HB-IGF-1 here is in the same range as the previously described effective concentrations of IGF-1.
It is possible that the positively charged heparinbinding domain used in this fusion protein can bind nonspecifically to negatively charged sulfated proteoglycans other than heparin. Our experiments with chondroitin-depleted cartilage showed that Xp-HB-IGF-1 is retained by cartilage even after extensive digestion of chondroitin sulfates, suggesting that binding may be specific to heparan sulfated proteoglycans on the cell surface. However, these data do not exclude nonheparan sulfated proteoglycans as binding reservoirs, and further experiments are needed to determine the specificity of Xp-HB-IGF-1 binding.
In addition to cartilage, Xp-HB-IGF-1 has potential for use in other tissues. For example, IGF-1 induces the axon outgrowth of PC12 cells (35) and corticospinal motor neurons (36), and thus IGF-1 may benefit motor neuron degeneration diseases. In dermal wound healing, IGF-1 is also effective because IGF-1 stimulates collagen synthesis and mitogenicity of fibroblasts and keratinocytes (37, 38). The ability of Xp-HB-IGF-1 to bind to the surfaces of cells may enhance cell therapies and other regenerative strategies.
Acknowledgments
T.T. was supported by a fellowship of the Banyu Life Science Foundation International. R.M. was supported by National Defense Science and Engineering Graduate and National Science Foundation graduate student fellowships. M.D. was supported by a fellowship from the National Heart, Lung, and Blood Institute (F32 HL073574). V.S. was supported by a Ph.D. fellowship of the Research Foundation—Flanders (FWO) and by a Belgian American Educational Foundation research fellowship. This work was supported by U.S. National Institutes of Health grant EB003805 and the Center for Integration of Medicine and Innovative Technology.
REFERENCES
- 1.Delafontaine P, Song YH, Li Y. Expression, regulation, and function of IGF-1, IGF-1R, and IGF-1 binding proteins in blood vessels. Arterioscler. Thromb. Vasc. Biol. 2004;24:435–444. doi: 10.1161/01.ATV.0000105902.89459.09. [DOI] [PubMed] [Google Scholar]
- 2.Coffer PJ, Jin J, Woodgett JR. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem. J. 1998;335(Pt 1):1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nixon AJ, Brower-Toland BD, Bent SJ, Saxer RA, Wilke MJ, Robbins PD, Evans CH. Insulinlike growth factor-I gene therapy applications for cartilage repair. Clin. Orthop. Relat. Res. 2000:S201–S213. doi: 10.1097/00003086-200010001-00026. [DOI] [PubMed] [Google Scholar]
- 4.Mourkioti F, Rosenthal N. IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 2005;26:535–542. doi: 10.1016/j.it.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Capito RM, Spector M. Collagen scaffolds for nonviral IGF-1 gene delivery in articular cartilage tissue engineering. Gene. Ther. 2007;14:721–732. doi: 10.1038/sj.gt.3302918. [DOI] [PubMed] [Google Scholar]
- 6.Fraidenraich D, Stillwell E, Romero E, Wilkes D, Manova K, Basson CT, Benezra R. Rescue of cardiac defects in id knockout embryos by injection of embryonic stem cells. Science. 2004;306:247–252. doi: 10.1126/science.1102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jabri N, Schalch DS, Schwartz SL, Fischer JS, Kipnes MS, Radnik BJ, Turman NJ, Marcsisin VS, Guler HP. Adverse effects of recombinant human insulin-like growth factor I in obese insulin-resistant type II diabetic patients. Diabetes. 1994;43:369–374. doi: 10.2337/diab.43.3.369. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson-Berka JL, Wraight C, Werther G. The role of growth hormone, insulin-like growth factor and somatostatin in diabetic retinopathy. Curr. Med. Chem. 2006;13:3307–3317. doi: 10.2174/092986706778773086. [DOI] [PubMed] [Google Scholar]
- 9.Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, Hennekens CH, Pollak M. Plasma insulin-like growth factor-I and prostate cancer risk: a prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 10.Higashiyama S, Abraham JA, Miller J, Fiddes JC, Klagsbrun M. A heparin-binding growth factor secreted by macrophage-like cells that is related to EGF. Science. 1991;251:936–939. doi: 10.1126/science.1840698. [DOI] [PubMed] [Google Scholar]
- 11.Higashiyama S, Lau K, Besner GE, Abraham JA, Klagsbrun M. Structure of heparin-binding EGF-like growth factor. Multiple forms, primary structure, and glycosylation of the mature protein. J. Biol. Chem. 1992;267:6205–6212. [PubMed] [Google Scholar]
- 12.Thompson SA, Higashiyama S, Wood K, Pollitt NS, Damm D, McEnroe G, Garrick B, Ashton N, Lau K, Hancock N, Klagsbrun M, Abraham JA. Characterization of sequences within heparin-binding EGF-like growth factor that mediate interaction with heparin. J. Biol. Chem. 1994;269:2541–2549. [PubMed] [Google Scholar]
- 13.Yoshioka J, Prince RN, Huang H, Perkins SB, Cruz FU, MacGillivray C, Lauffenburger DA, Lee RT. Cardiomyocyte hypertrophy and degradation of connexin43 through spatially restricted autocrine/paracrine heparin-binding EGF. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10622–10627. doi: 10.1073/pnas.0501198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J. Physiol. 2003;547:247–254. doi: 10.1113/jphysiol.2002.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shavlakadze T, Winn N, Rosenthal N, Grounds MD. Reconciling data from transgenic mice that overexpress IGF-I specifically in skeletal muscle. Growth Horm. IGF Res. 2005;15:4–18. doi: 10.1016/j.ghir.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Milner SJ, Francis GL, Wallace JC, Magee BA, Ballard FJ. Mutations in the B-domain of insulin-like growth factor-I influence the oxidative folding to yield products with modified biological properties. Biochem. J. 1995;308(Pt 3):865–871. doi: 10.1042/bj3080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milner SJ, Carver JA, Ballard FJ, Francis GL. Probing the disulfide folding pathway of insulin-like growth factor-I. Biotechnol. Bioeng. 1999;62:693–703. [PubMed] [Google Scholar]
- 18.Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, Lee RT. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 19.Bonassar LJ, Grodzinsky AJ, Srinivasan A, Davila SG, Trippel SB. Mechanical and physicochemical regulation of the action of insulin-like growth factor-I on articular cartilage. Arch. Biochem. Biophys. 2000;379:57–63. doi: 10.1006/abbi.2000.1820. [DOI] [PubMed] [Google Scholar]
- 20.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 21.Sah RL, Kim YJ, Doong JY, Grodzinsky AJ, Plaas AH, Sandy JD. Biosynthetic response of cartilage explants to dynamic compression. J. Orthop. Res. 1989;7:619–636. doi: 10.1002/jor.1100070502. [DOI] [PubMed] [Google Scholar]
- 22.Besner G, Higashiyama S, Klagsbrun M. Isolation and characterization of a macrophage-derived heparin-binding growth factor. Cell Regul. 1990;1:811–819. doi: 10.1091/mbc.1.11.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denley A, Cosgrove LJ, Booker GW, Wallace JC, Forbes BE. Molecular interactions of the IGF system. Cytokine Growth Factor Rev. 2005;16:421–439. doi: 10.1016/j.cytogfr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Musaro A, Dobrowolny G, Rosenthal N. The neuroprotective effects of a locally acting IGF-1 isoform. Exp. Gerontol. 2007;42:76–80. doi: 10.1016/j.exger.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Bayne ML, Applebaum J, Underwood D, Chicchi GG, Green BG, Hayes NS, Cascieri MA. The C region of human insulin-like growth factor (IGF) I is required for high affinity binding to the type 1 IGF receptor. J. Biol. Chem. 1989;264:11004–11008. [PubMed] [Google Scholar]
- 26.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 27.Robinson CJ, Mulloy B, Gallagher JT, Stringer SE. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J. Biol. Chem. 2006;281:1731–1740. doi: 10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- 28.Ruoslahti E, Yamaguchi Y. Proteoglycans as modulators of growth factor activities. Cell. 1991;64:867–869. doi: 10.1016/0092-8674(91)90308-l. [DOI] [PubMed] [Google Scholar]
- 29.Raines EW, Ross R. Compartmentalization of PDGF on extracellular binding sites dependent on exon-6-encoded sequences. J. Cell Biol. 1992;116:533–543. doi: 10.1083/jcb.116.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harmer NJ, Chirgadze D, Kim K. Hyun, Pellegrini L, Blundell TL. The structural biology of growth factor receptor activation. Biophys. Chem. 2003;100:545–553. doi: 10.1016/s0301-4622(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 31.Bhakta NR, Garcia AM, Frank EH, Grodzinsky AJ, Morales TI. The insulin-like growth factors (IGFs) I and II bind to articular cartilage via the IGF-binding proteins. J. Biol. Chem. 2000;275:5860–5866. doi: 10.1074/jbc.275.8.5860. [DOI] [PubMed] [Google Scholar]
- 32.Frystyk J. Free insulin-like growth factors—measurements and relationships to growth hormone secretion and glucose homeostasis. Growth Horm. IGF Res. 2004;14:337–375. doi: 10.1016/j.ghir.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt MB, Chen EH, Lynch SE. A review of the effects of insulin-like growth factor and platelet derived growth factor on in vivo cartilage healing and repair. Osteoarthritis Cartilage. 2006;14:403–412. doi: 10.1016/j.joca.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Vincent TL, McLean CJ, Full LE, Peston D, Saklatvala J. FGF-2 is bound to perlecan in the pericellular matrix of articular cartilage, where it acts as a chondrocyte mechanotransducer. Osteoarthritis Cartilage. 2007;15:752–763. doi: 10.1016/j.joca.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 35.Topalli I, Etgen AM. Insulin-like growth factor-I receptor and estrogen receptor crosstalk mediates hormone-induced neurite outgrowth in PC12 cells. Brain Res. 2004;1030:116–124. doi: 10.1016/j.brainres.2004.09.057. [DOI] [PubMed] [Google Scholar]
- 36.Ozdinler PH, Macklis JD. IGF-I specifically enhances axon outgrowth of corticospinal motor neurons. Nat. Neurosci. 2006;9:1371–1381. doi: 10.1038/nn1789. [DOI] [PubMed] [Google Scholar]
- 37.Martin P. Wound healing—aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 38.Guler HP, Zapf J, Scheiwiller E, Froesch ER. Recombinant human insulin-like growth factor I stimulates growth and has distinct effects on organ size in hypophysectomized rats. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4889–4893. doi: 10.1073/pnas.85.13.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]