Abstract
Guanyl radicals, the product of the removal of a single electron from guanine, are produced in DNA by the direct effect of ionizing radiation. We have produced guanyl radicals in DNA by using the single electron oxidizing agent (SCN)2–, itself derived from the indirect effect of ionizing radiation via thiocyanate scavenging of OH. We have examined the reactivity of guanyl radicals in plasmid DNA with the six most easily oxidized amino acids cysteine, cystine, histidine, methionine, tryptophan and tyrosine and also simple ester and amide derivatives of them. Cystine and histidine derivatives are unreactive. Cysteine, methionine, tyrosine and particularly tryptophan derivatives react to repair guanyl radicals in plasmid DNA with rate constants in the region of ∼105, 105, 106 and 107 dm3 mol–1 s–1, respectively. The implication is that amino acid residues in DNA binding proteins such as histones might be able to repair by an electron transfer reaction the DNA damage produced by the direct effect of ionizing radiation or by other oxidative insults.
INTRODUCTION
Guanyl radicals, the product of the removal of a single electron from guanine, are produced in DNA by the direct effect of ionizing radiation (ionization of the DNA itself) (1–3) and also by related electron removal products such as photoionization, chemical oxidation and photosensitization (4–11). The site of electron loss tends to migrate to guanine (11,12) because this is the most stable location available in DNA (13,14). However, guanyl radicals are still strongly oxidizing species (15,16) and the opportunity exists for their repair by electron transfer even from quite mild reducing agents. The usual examples of naturally occurring mild reducing agents are compounds such as ascorbate and glutathione (17). However, in mammalian cells it is possible that DNA embedded in densely packed chromatin is not accessible to these antioxidants because it is protected by a close association with histone proteins (18). Although these proteins interfere with access of even small diffusible species, it is possible that they are themselves capable of repairing guanyl radicals by electron transfer. This is because of the presence of residues of amino acids such as tryptophan and tyrosine. These amino acids behave in this way because their side chains contain functional groups (indole and phenol, respectively) with appropriate redox properties (19).
The redox reaction of amino acids, peptides or proteins with guanyl radicals in monomeric nucleosides, oligonucleotides or DNA has been observed in several different experimental systems. Examples are electron transfer from an amino acid or peptide to a monomeric purine in aqueous solution (20), from a peptide to a polynucleotide in aqueous solution (21) and from a histone to DNA in irradiated chromatin at cryogenic temperatures (22). Endonuclease binding has been reported to decrease charge migration in DNA (23).
Further support is provided by the observation of amino acid radicals as intermediates in normal biochemistry (24). In addition to cofactors and/or prosthetic groups, some redox enzyme systems employ radicals located on their own amino acid residues as intermediates. Examples are ribonucleotide reductase (25) and DNA photolyase (26). Proteins whose regular function does not involve a redox reaction (such as lysozyme) may also undergo electron transfer reactions in response to an artificial stimulus (oxidative electron removal from a kinetically reactive site) (27).
The fundamental repeating structural unit of eukaryotic chromatin is the nucleosome (18), and there are numerous close contacts between amino acid residues in histones and the nucleotide residues in DNA. It therefore seems reasonable to suggest that electron transfer from amino acid to DNA may take place in response to oxidative or ionizing DNA damage. In these reactions, the amino acid residue is the electron donor and an ionized or oxidized guanine is the acceptor. It would be desirable to obtain estimates of the rate constants that characterize these reactions. To address this issue, we have used a single electron oxidizing agent generated by γ-radiation to produce guanyl radicals in plasmid DNA and then quantified their reactivity with the free amino acids and simple ester and amide derivatives of cysteine, cystine, histidine, methionine, tryptophan and tyrosine.
MATERIALS AND METHODS
Plasmid substrate
A sample of plasmid pHAZE (28) was generously provided by Dr W. F. Morgan (Department of Radiation Oncology, University of Maryland). It was grown to large scale, isolated and purified as described previously (29).
Base excision repair endonuclease
An expression vector containing formamidopyrimidine-DNA N-glycosylase (FPG) from Escherichia coli was generously provided by Dr Y. W. Kow (Department of Radiation Oncology, Emory University). The enzyme was overexpressed, isolated and purified as described previously (30).
Irradiation
The plasmid substrate was γ-irradiated in aerobic aqueous solution. These solutions contained pHAZE (25 µg ml–1), sodium phosphate (5 × 10–3 mol dm–3, pH 7.0), sodium thiocyanate (10–3 mol dm–3), sodium perchlorate (1.1 × 10–1 mol dm–3) and an amino acid or simple derivative thereof (10–7–10–4 mol dm–3). The amino acid was one of the following: cysteine, cystine, histidine, methionine, tryptophan or tyrosine. The derivatives were either the acetylated amine or the carboxylic acid methyl or ethyl ester. Each aliquot was 27 µl in volume. The dose rate of 5.58 × 10–2 Gy s–1 was quantified by means of the Fricke method (31).
Enzyme incubation
After irradiation, each 27 µl aliquot was mixed with 3 µl of a solution containing FPG (50 µg ml–1). The resulting solutions were incubated at 37°C for 30 min and assayed by gel electrophoresis.
Determination of strand break yields
After incubation, the yield of single-strand breaks (SSB) was quantified after agarose gel electrophoresis. The procedures for digital video imaging of ethidium fluorescence and for calculating the radiation chemical yield (G value) for SSB formation have been described previously (29).
RESULTS
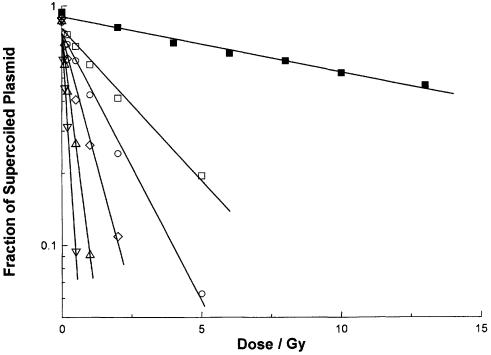
A typical experimental result is reproduced in Figure 1. An aerobic solution of plasmid pHAZE (25 µg ml–1) was irradiated by 137Ce γ-rays in the presence of sodium phosphate (5 × 10–3 mol dm–3, pH 7.0), sodium perchlorate (1.1 × 10–1 mol dm–3), sodium thiocyanate (10–3 mol dm–3) and the methyl ester of tyrosine (3 × 10–7–5 × 10–6 mol dm–3). After irradiation, the plasmid solutions were incubated for 30 min at 37°C under one of two conditions: with or without the enzyme FPG. For each radiation dose and incubation, the fraction of the plasmid remaining in the supercoiled conformation was determined using agarose gel electrophoresis. In Figure 1 this fraction is plotted against the radiation dose. For irradiation in the presence of 5 × 10–6 mol dm–3 tyrosine methyl ester and incubation in the absence of FPG, the fraction of supercoiled plasmid decreases to ∼0.5 at a dose of 10 Gy. After incubation in the presence of FPG, the fraction of supercoiled plasmid decreases to ∼0.2 at 5 Gy. Decreasing concentrations of tyrosine methyl ester result in an increasingly rapid loss of the supercoiled form.
Figure 1.
Loss of supercoiled plasmid with increasing dose of γ-radiation. Aliquots of a solution containing plasmid pHAZE (25 µg ml–1), sodium phosphate (5 × 10–3 mol dm–3, pH 7.0), sodium thiocyanate (10–3 mol dm–3), sodium perchlorate (1.1 × 10–1 mol dm–3) and tyrosine methyl ester [3 × 10–7 (open inverted triangle), 7.5 × 10–7 (open triangle), 1.5 × 10–6 (open diamond), 3 × 10–6 (open circle) or 5 × 10–6 (open and closed squares) mol dm–3] were irradiated under aerobic conditions with 137Ce γ-rays (662 keV) at dose rates between 1.7 × 10–3 and 9.0 × 10–2 Gy s–1. After irradiation, the solutions were incubated for 30 min at 37°C with FPG at a concentration of 0 (closed square) or 5 µg ml–1 (open symbols). The fraction of supercoiled plasmid remaining after each radiation dose and incubation was determined using agarose gel electrophoresis. These six data sets are plotted together. Each is fitted to least mean squares straight lines of the form y = ce–mx. From the slopes m of these fitted straight lines, the doses D0 and SSB yields for the six irradiation and incubation conditions are: open inverted triangle, 0.234 Gy, 1.59 × 10–2 µmol J–1; open triangle, 0.463 Gy, 8.04 × 10–3 µmol J–1; open diamond, 0.971 Gy, 3.84 × 10–3 µmol J–1; open circle, 1.94 Gy, 1.92 × 10–3 µmol J–1; open square, 3.41 Gy, 1.09 × 10–3 µmol J–1; closed square, 22.5 Gy, 1.66 × 10–4 µmol J–1.
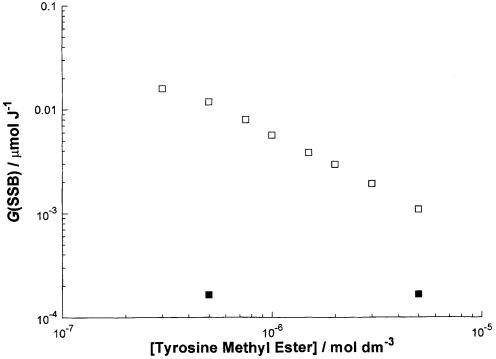
The loss of the supercoiled form of the plasmid results from the formation of SSB. The radiation chemical yield (G value) of these SSBs [G(SSB)] can be calculated from the slopes of straight lines fitted to yield–dose plots such as Figure 1. We have determined G(SSB) values for additional concentrations of the methyl ester of tyrosine (not shown in Fig. 1).
These G(SSB) values are plotted in Figure 2 against the concentration of the methyl ester of tyrosine. In the absence of FPG, the SSB yield is insensitive to the concentration of tyrosine methyl ester. This observation also applies to the other amino acids and their derivatives for concentrations in the range 10–7–10–4 mol dm–3 (data not shown). For incubation in the presence of FPG, the SSB yield decreases from ∼10–2 to ∼10–3 µmol J–1 (factor of 10) as the concentration of tyrosine methyl ester increases from 0.5 to 5 µmol dm–3 (factor of 10).
Figure 2.
Effect of tyrosine methyl ester concentration during irradiation on the SSB yield after incubation with or without FPG. Plasmid pHAZE was γ-irradiated in the presence of various concentrations of tyrosine methyl ester and the resulting SSB yield was determined (see Fig. 1) after incubation in the presence (open square) or absence (closed square) of FPG.
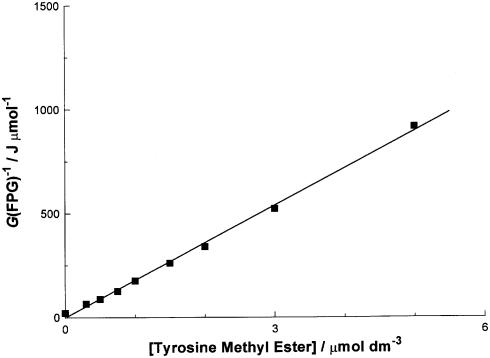
The data in Figure 2 are re-plotted according to competition kinetics in Figure 3 (see Discussion). The yield of FPG-sensitive sites [hereafter G(FPG)] was calculated by subtracting the SSB yield after incubation in the absence of FPG from the SSB yield determined after incubation in its presence. In principle, both yields should be determined at an identical concentration of the tyrosine derivative, but the dependence of G(SSB) on the amino acid concentration is very small when FPG is absent. Therefore, a constant value of 1.5 × 10–4 µmol J–1 was assumed. We have reported previously on the value for G(FPG) in the absence of any added amino acids (32). Under the conditions we have used here, the value of G(FPG) is 4.84 × 10–2 µmol J–1. From these values, and the slope of the straight line fitted to the data in Figure 3, it is possible to estimate the rate constant for reaction of the methyl ester of tyrosine with a DNA guanyl radical. The procedure is explained in Discussion.
Figure 3.
Reciprocal plot of the attenuation of the yield of FPG-sensitive sites, G(FPG), against the concentration of tyrosine methyl ester according to competition kinetics (see Discussion). The data set is fitted to a least mean squares straight line of the form y = mx + c. The slope m of the line is 1.8 × 108 MJ dm3 mol–2. See Discussion for an evaluation of the intercept c.
Rate constants were quantified for the six amino acids cysteine, cystine, histidine, methionine, tryptophan and tyrosine and simple derivatives of them (esters and amides) by constructing plots similar to Figure 3 (not shown). The derived rate constants are listed in Table 1. These data reveal that cystine and histidine derivatives are unreactive. Cysteine, methionine, tyrosine and tryptophan derivatives react to repair guanyl radicals in plasmid DNA with rate constants in the region of ∼105, 105, 106 and 107 dm3 mol–1 s–1, respectively. The combination of the abundance of these amino acids in the nucleosome (18) and their reactivity (Table 1) points to the importance of tyrosine.
Table 1. Rate constants for repair of DNA guanyl radicals by amino acids and their methyl or ethyl esters.
| Parent amino acid | k5 (dm3 mol–1 s–1) | ||
|---|---|---|---|
| Free acid | Methyl or ethyl estera | N-acetyl amide | |
| Cysteine | 2.0 × 106 | 8.8 × 105 | 2.7 × 105 |
| Cystine | 3.4 × 105 | <2.0 × 104 | |
| Histidine | <2.0 × 104 | <2.0 × 104 | |
| Methionine | 7.7 × 105 | 8.3 × 104 | <2.0 × 104 |
| Tryptophan | 4.4 × 107 | 1.0 × 107 | 7.7 × 106 |
| Tyrosine | 1.7 × 106 | 4.0 × 105 | 1.9 × 107 |
aMethyl ester in all cases except cystine, which was the diethyl ester.
DISCUSSION
Description of the experimental system
We have reported previously on the use of aqueous thiocyanate to mimic the direct effect of ionizing radiation (32). In the direct effect, the DNA itself is ionized. The site of the electron vacancy tends to migrate to guanine (equation 1) (1–3). The same electron-deficient guanine species (identified spectrophotometrically) can also be produced by several different single electron oxidizing agents (12,33–36). In the system we employ here, radiolysis of water produces the hydroxyl radical (OH), the hydrogen atom (H) and the hydrated electron (e–aq) (equation 2) (31). The latter two species are strongly reducing and are rapidly scavenged by the oxygen present in solution to form superoxide. The ineffectiveness of superoxide dismutase in this system suggests that subsequent reactions of superoxide can be safely ignored (32). The principal scavenger for OH present in solution is thiocyanate [rate constant k3 = 1.1 × 1010 dm3 mol–1 s–1 (37)], leading to the formation of the dimeric radical anion (SCN)2– (equation 3). This species is a strong single electron oxidizing agent {E°[(SCN)2–/2SCN–] = +1.33 V (38,39)} and reacts mainly with the highest concentration of oxidizable targets available, which under our conditions are guanine residues in plasmid DNA (equation 4). The product of this reaction is a single electron oxidized guanine radical cation (or more briefly a DNA guanyl radical), symbolized by DNA-G+. The value of the rate constant k4 is unknown for (SCN)2–, but has been determined as 3 × 107 dm3 mol–1 s–1 for the similarly reactive species SeO3– (35), which is also a single electron oxidant. The resulting guanyl radicals are relatively long lived by the standards of radicals (millisecond to second timescale; 4) in the absence of reducing agents. However, they accept electrons from even quite mild reducing agents R if any are present in solution (equation 5). The significance of equation 5 is that it reverses or repairs the DNA damage. Unrepaired guanyl radicals eventually produce the stable product 7,8-dihydro-8-oxoguanine (8-oxoG) (equation 6). It is generally agreed that 8-oxoG is the major product derived from a guanyl radical in double-stranded DNA under aerobic conditions (40–43). This modified base is detected by converting it to a SSB with the base excision repair endonuclease FPG (equation 7). Such SSBs in plasmid DNA are easily detected and quantified by electrophoretic methods.
DNA → DNA-G+ + e– 1
H2O → OH + H + e–aq 2
OH + 2SCN– → (SCN)2– + OH– 3
(SCN)2– + DNA → DNA-G+ + 2SCN– 4
DNA-G+ + R → DNA +R+ 5
DNA-G+ → DNA-8oxoG 6
DNA-8oxoG + FPG → SSB 7
We have previously reported that the amino acids methionine and tyrosine are able to repair guanyl radicals in plasmid DNA (44) and here we have extended the observation to additional amino acids and simple derivatives of them.
Derivation of rate constants
Figures 1 and 2 show that the presence during irradiation of tyrosine methyl ester protects against the formation of FPG-sensitive sites. We have argued above that these FPG-sensitive sites are probably 8-oxoG residues. The mechanism of the attenuation of their yield (Fig. 2) is very probably a reduction in the precursor guanyl radicals by the tyrosine compound (equation 5, R = tyrosine methyl ester). This is because equal attenuations are produced by a single reducing agent after DNA damage by different oxidizing agents (44). The competition between repair of the guanyl radical (equation 5) and its trapping to form a stable oxidized product (equation 6) can be analyzed quantitatively by competition kinetics (equation 8).
[1/G(FPG)] = [1/G0(FPG)] × {1 + (k5[R]/k6)} 8
In equation 8, the rate constants for equations 5 and 6 are symbolized by k5 and k6. G(FPG) and G0(FPG) represent the yields of FPG-sensitive sites in the presence and absence of the reducing agent R, respectively. The value of G0(FPG) is derived from the SSB yields observed in the absence of any added reducing agent and G(FPG) refers to the yield of FPG-sensitive sites observed in the presence of a reducing agent R. The yield of FPG-sensitive sites was estimated from the experimental data by subtracting the SSB yield determined in the absence of FPG (closed symbols in Fig. 2, assumed to have a constant value of 1.5 × 10–4 µmol J–1) from the SSB yield determined in its presence (open symbols in Fig. 2). Figure 3 is an example of a plot of 1/G(FPG) against the concentration of R, for the case where R is the methyl ester of tyrosine. The data fall on a straight line, as predicted by equation 8. Comparison of equation 8 with that of a straight line (y = mx + c) reveals that the value of k5 is found from k5 = mk6/c, where m and c are the slope and intercept of the straight line fitted to Figure 3, respectively, and k6 = 0.2 s–1 (4). In practice, the value for c was calculated as the reciprocal of the value of G(FPG) determined in the absence of any added amino acid (4.84 × 10–2 µmol J–1). Therefore, the value of k5 for tyrosine methyl ester is k5 = 1.8 × 108 × 0.2 × 4.84 × 10–2 = 1.7 × 106 dm3 mol–1 s–1 (Table 1). The other entries in Table 1 were derived in the same manner (not shown).
Reduction potentials of amino acids
Six amino acids are relatively easily oxidized. These are cysteine, cystine, histidine, methionine, tryptophan and tyrosine. Values for the reduction potentials of their single electron oxidation products at pH 7 (a close approximation to physiological conditions) are available in the literature. These are: E7 = +0.92 V (cysteine) (17,45); +1.1 V (cystine) (46,47); +1.17 V (histidine) (48); +1.5 V (methionine) (49); +1.03 V (tryptophan) (19,38,50); +0.93 V (tyrosine) (19,50). Values for guanine are: E7 = +1.29 V (guanosine) (15); +1.39 V (plasmid DNA) (16). This leads to a stability series of single oxidation products increasing in the order methionine ≈ guanine < histidine < cystine < tryptophan < cysteine < tyrosine. Therefore, all six of these amino acids are in principle capable of reacting with guanyl radicals. In contrast to the situation for the monomers, reduction potentials for amino acid radicals in proteins are not well characterized. Differences of –0.2 and +0.35 V have been reported for tyrosine and tryptophan, respectively (51), larger than the differences between adjacent amino acids. Therefore, the order of stability under cellular conditions may be quite different from that expected on the basis of model compounds.
Estimation of intramolecular rate constants
The bimolecular rate constants we have measured here for amino acids and plasmid DNA apply to an intermolecular reaction. For nucleosomal DNA in cells, the reaction is better described as intramolecular. A better experimental system would model this with DNA binding proteins or with an authentic nucleosomal structure such as the SV40 minichromosome. However, for the present it is possible to estimate values of intramolecular rate constants from our intermolecular measurements.
This estimation is simplified by existing literature data for electron transfer from tyrosine to tryptophan. Rate constants have been determined for several examples of intermolecular and intramolecular reactions. Intermolecular rate constants lie in the range 5 × 105–2 × 106 dm3 mol–1 s–1 for the amino acids or dipeptides containing them bound to glycine or alanine (52,53). Intramolecular rate constants for di- or tripeptides lie in the range 2 × 104–8 × 104 s–1 (53). In addition, a distance dependence has been observed with the rate constant decreasing consistently by ∼3-fold with each additional proline residue (from none to five prolines) (54,55). Intramolecular rate constants for the proteins DNA photolyase, concanavalin A and α-lactalbumin all fall in the range 4 × 103–2 × 104 s–1 (27,56). Therefore, extrapolating from the measurements using plasmid DNA and simple amino acid derivatives (Table 1), we would estimate expected intramolecular rate constants in the nucleosome of the order of 25- to 100-fold lower than the bimolecular rate constants.
The combination of abundance in histones (18) and reactivity (Table 1) identifies tyrosine as the most important amino acid. Bimolecular rate constants for tyrosine derivatives lie between 4 × 105 and 2 × 107 dm3 mol–1 s–1 (Table 1). As described above, this leads to an estimate for the intramolecular rate constant of 4 × 103–1 × 106 s–1, which is equivalent to a lifetime in the range 2.5 × 10–4–1 × 10–6 s. This microsecond timescale is significantly shorter than the lifetime of accessible guanyl radicals in the presence of ascorbate and glutathione (at their typical physiological concentrations), which has been estimated to be of the order of 2–5 ms (16). The conclusion is that amino acid residues, particularly tyrosine, may well play a significant role in the chemical repair of DNA guanyl radicals.
Possible biological effects
In nucleosomal DNA it is possible that histone proteins limit access of small diffusible compounds such as the reducing agents ascorbate and glutathione. We show here that some amino acids are able to reduce guanyl radicals (Table 1), and presumably this reactivity is also to be expected from proteins containing them. A potentially large number of subsequent reactions may then occur within the protein. One important product derived from amino acid radicals may be DNA–protein cross-links (57), so that reduction of DNA guanyl radicals is not necessarily equivalent to DNA repair.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NIH grant CA46295 and by the UCSD Cancer Center.
REFERENCES
- 1.Becker D. and Sevilla,M.D. (1993) The chemical consequences of radiation damage to DNA. Adv. Radiat. Biol., 17, 121–180. [Google Scholar]
- 2.O'Neill P. and Fielden,E.M. (1993) Primary free radical processes in DNA. Adv. Radiat. Biol., 17, 53–120. [Google Scholar]
- 3.Debije M.G. and Bernhard,W.A. (1999) Free radical yields in crystalline DNA X-irradiated at 4K. Radiat. Res., 152, 583–589. [PMC free article] [PubMed] [Google Scholar]
- 4.Hildenbrand K. and Schulte-Frohlinde,D. (1990) ESR spectra of radicals of single and double stranded DNA in aqueous solution. Implications for OH induced strand breakage. Free Radic. Res. Commun., 11, 195–206. [DOI] [PubMed] [Google Scholar]
- 5.Wolf P., Jones,G.D.D., Candeias,L.P. and O’Neill,P. (1993) Induction of strand breaks in polyribonucleotides and DNA by the sulphate radical anion. Role of electron loss centers as precursors of strand breakage. Int. J. Radiat. Biol., 64, 7–18. [DOI] [PubMed] [Google Scholar]
- 6.Melvin T., Botchway,S.W., Parker,A.W. and O’Neill,P. (1996) Induction of strand breaks in single stranded polyribonucleotides and DNA by photoionization: one electron oxidized nucleobase radicals as precursors. J. Am. Chem. Soc., 118, 10031–10036. [Google Scholar]
- 7.Melvin T., Bothe,E. and Schulte-Frohlinde,D. (1996) The reaction of triplet 2-methyl-1,4-naphthoquinone (menadione) with DNA and polynucleotides. Photochem. Photobiol., 64, 769–776. [DOI] [PubMed] [Google Scholar]
- 8.Cadet J., Berger,M., Douki,T. and Ravanat,J.-L. (1997) Oxidative damage to DNA. Formation, measurement and biological significance. Rev. Physiol. Biochem. Pharmacol., 131, 1–87. [DOI] [PubMed] [Google Scholar]
- 9.Spassky A. and Angelov,D. (1997) Influence of local helical conformation on the guanine modifications generated from one electron DNA oxidation. Biochemistry, 36, 6571–6576. [DOI] [PubMed] [Google Scholar]
- 10.Burrows C.J. and Muller,J.G. (1998) Oxidative nucleobase modifications leading to strand scission. Chem. Rev., 98, 1109–1152. [DOI] [PubMed] [Google Scholar]
- 11.Melvin T., Cunniffe,S.M.T., O’Neill,P., Parker,A.W. and Roldan-Arjona,T. (1998) Guanine is the target for direct ionization damage in DNA, as detected using excision enzymes. Nucleic Acids Res., 26, 4935–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Candeias L.P. and Steenken,S. (1993) Electron transfer in di(deoxy)nucleoside phosphates in aqueous solution: rapid migration of oxidative damage (via adenine) to guanine. J. Am. Chem. Soc., 115, 2437–2440. [Google Scholar]
- 13.Colson A.-O., Besler,B., Close,D.M. and Sevilla,M.D. (1992) Ab initio molecular orbital calculations of DNA bases and their radical ions in various protonation states: evidence for proton transfer in GC base pair radical anions. J. Phys. Chem., 96, 661–668. [Google Scholar]
- 14.Seidel C.A.M., Schulz,A. and Sauer,M.H.M. (1996) Nucleobase specific quenching of fluorescent dyes. 1. Nucleobase one electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem., 100, 5541–5553. [Google Scholar]
- 15.Steenken S. and Jovanovic,S.V. (1997) How easily oxidizable is DNA? One electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc., 119, 617–618. [Google Scholar]
- 16.Milligan J.R., Aguilera,J.A. and Ward,J.F. (2001) Redox equilibrium between guanyl radicals and thiocyanate influences base damage yields in gamma irradiated plasmid DNA. Estimation of the reduction potential of guanyl radicals in plasmid DNA in aqueous solution at physiological ionic strength. Int. J. Radiat. Biol., 77, 1195–1205. [DOI] [PubMed] [Google Scholar]
- 17.Buettner G.R. (1993) The pecking order of free radicals and antioxidants: lipid peroxidation, α-tocopherol and ascorbate. Arch. Biochem. Biophys., 300, 535–543. [DOI] [PubMed] [Google Scholar]
- 18.van Holde K.E. (1988) Chromatin. Springer-Verlag, New York, NY. [Google Scholar]
- 19.Harriman A. (1987) Further comments on the redox potentials of tryptophan and tyrosine. J. Phys. Chem., 91, 6102–6104. [Google Scholar]
- 20.Pan J., Lin,W., Wang,W., Han,Z., Lu,C., Yao,S., Lin,N. and Zhu,D. (2001) A kinetic study on the interaction of deprotonated purine radical cations with amino acids and model peptides. Biophys. Chem., 89, 193–199. [DOI] [PubMed] [Google Scholar]
- 21.Wagenknecht H.-A., Stemp,E.D.A. and Barton,J.K. (2000) DNA bound peptide radicals generated through DNA mediated electron transport. Biochemistry, 39, 5483–5491. [DOI] [PubMed] [Google Scholar]
- 22.Cullis P.M., Jones,G.D.D., Symons,M.C.R. and Lea,J.S. (1987) Electron transfer from a protein to DNA in irradiated chromatin. Nature, 330, 773–774. [DOI] [PubMed] [Google Scholar]
- 23.Nakatani K., Dohno,C., Ogawa,A. and Saito,I. (2002) Suppression of DNA mediated charge transport by BamHI binding. Chem. Biol., 9, 361–366. [DOI] [PubMed] [Google Scholar]
- 24.Stubbe J. and van der Donk,W.A. (1998) Protein radicals in enzyme catalysis. Chem. Rev., 98, 705–762. [DOI] [PubMed] [Google Scholar]
- 25.Gräslund A. and Sahlin,M. (1996) Electron paramagnetic resonance and nuclear magnetic resonance studies of class I ribonucleotide reductase. Annu. Rev. Biophys. Biomol. Struct., 25, 259–286. [DOI] [PubMed] [Google Scholar]
- 26.Kim S., Sancar,A., Essenmacher,C. and Babcock,G.T. (1993) Time resolved EPR studies with DNA photolyase. Excited state FADH0 abstracts an electron from trp-306 to generate FADH–, the catalytically active form of the cofactor. Proc. Natl Acad. Sci. USA, 90, 8023–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prütz W.A., Butler,J., Land,E.J. and Swallow,A.J. (1980) Direct demonstration of electron transfer between tryptophan and tyrosine in proteins. Biochem. Biophys. Res. Commun., 96, 408–414. [DOI] [PubMed] [Google Scholar]
- 28.Lutze L.H. and Winegar,R.A. (1990) pHAZE: a shuttle vector system for the detection and analysis of ionizing radiation induced mutations. Mutat. Res., 245, 305–310. [DOI] [PubMed] [Google Scholar]
- 29.Milligan J.R., Aguilera,J.A. and Ward,J.F. (1993) Variation of single strand break yield with scavenger concentration for plasmid DNA irradiated in aqueous solution. Radiat. Res., 133, 151–157. [PubMed] [Google Scholar]
- 30.Zharkov D.O., Rieger,R.A., Iden,C.R. and Grollman,A.P. (1997) NH2 terminal proline acts as a nucleophile in the glycosylase/AP lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem., 272, 5335–5341. [DOI] [PubMed] [Google Scholar]
- 31.Spinks J.W.T. and Woods,R.J. (1976) An Introduction to Radiation Chemistry, 2nd Edn. Wiley, New York, NY. [Google Scholar]
- 32.Milligan J.R., Aguilera,J.A., Paglinawan,R.A. and Ward,J.F. (2000) Mechanism of DNA damage by thiocyanate radicals. Int. J. Radiat. Biol., 76, 1305–1314. [DOI] [PubMed] [Google Scholar]
- 33.Jovanovic S.V. and Simic,M.G. (1989) The DNA guanyl radical: kinetics and mechanism of generation and repair. Biochim. Biophys. Acta, 1008, 39–44. [DOI] [PubMed] [Google Scholar]
- 34.Candeias L.P. and Steenken,S. (1989) Structure and acid base properties of one electron oxidized deoxyguanosine, guanosine and 1-methylguanosine. J. Am. Chem. Soc., 111, 1094–1099. [Google Scholar]
- 35.Martin R.F. and Anderson,R.F. (1998) Pulse radiolysis studies indicate that electron transfer is involved in radioprotection by Hoechst 33342 and methylproamine. Int. J. Radiat. Oncol. Biol. Phys., 42, 827–831. [DOI] [PubMed] [Google Scholar]
- 36.Faraggi M., Broitman,F., Trent,J.B. and Klapper,M.H. (1996) One electron oxidation reactions of some purine and pyrimidine bases in aqueous solutions. Electrochemical and pulse radiolysis studies. J. Phys. Chem., 100, 14751–14761. [Google Scholar]
- 37.Buxton G.V., Greenstock,C.L., Helman,W.P. and Ross,A.B. (1988) Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals (OH/O–) in aqueous solution. J. Phys. Chem. Ref. Data, 17, 513–887. [Google Scholar]
- 38.Wardman P. (1989) Reduction potentials of one electron couples involving free radicals in aqueous solution. J. Phys. Chem. Ref. Data, 18, 1637–1755. [Google Scholar]
- 39.Stanbury D.M. (1989) Reduction potentials involving inorganic free radicals in aqueous solution. Adv. Inorg. Chem., 33, 69–138. [Google Scholar]
- 40.Swarts S.G., Becker,D., Sevilla,M. and Wheeler,K. (1996) Radiation induced DNA damage as a function of hydration. II. Base damage from electron loss centers. Radiat. Res., 145, 304–314. [PubMed] [Google Scholar]
- 41.Angelov D., Spassky,A., Berger,M. and Cadet,J. (1997) High intensity UV laser photolysis of DNA and purine 2′-deoxyribonucleotides: formation of 8-oxopurine damage and oligonucleotide strand cleavage as revealed by HPLC and gel electrophoresis studies. J. Am. Chem. Soc., 119, 11373–11380. [Google Scholar]
- 42.Douki T. and Cadet,J. (1999) Modification of DNA bases by photosensitized one electron oxidation. Int. J. Radiat. Biol., 75, 571–581. [DOI] [PubMed] [Google Scholar]
- 43.Kan Y. and Schuster,G.B. (1999) Long range guanine damage in single stranded DNA: charge transport through a duplex bridge and in a single stranded overhang. J. Am. Chem. Soc., 121, 10857–10864. [Google Scholar]
- 44.Milligan J.R., Aguilera,J.A., Paglinawan,R.A. and Ward,J.F. (2002) One electron oxidation of plasmid DNA by selenium(V) species. Int. J. Radiat. Biol., 78, 359–374. [DOI] [PubMed] [Google Scholar]
- 45.Wardman P. and von Sonntag,C. (1995) Kinetic factors that control the fate of thiyl radicals in cells. Methods Enzymol., 251, 31–45. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad R. and Armstrong,D.A. (1984) The effect of pH and complexation on redox reactions between RS radicals and flavins. Can. J. Chem., 62, 171–177. [Google Scholar]
- 47.Koppenol W.H. (1993) A thermodynamic appraisal of the radical sink hypothesis. Free Radic. Biol. Med., 14, 91–94. [DOI] [PubMed] [Google Scholar]
- 48.Navaratnam S. and Parsons,B.J. (1998) Reduction potential of histidine free radicals. A pulse radiolysis study. J. Chem. Soc. Faraday Trans., 94, 2577–2581. [Google Scholar]
- 49.Schöneich C. (2002) Redox processes of methionine relevant to β-amyloid oxidation and Alzheimer’s disease. Arch. Biochem. Biophys., 397, 370–376. [DOI] [PubMed] [Google Scholar]
- 50.DeFilippis M.R., Murthy,C.P., Faraggi,M. and Klapper,M.H. (1989) Pulse radiolytic measurement of redox potentials: the tyrosine and tryptophan radicals. Biochemistry, 28, 4847–4853. [DOI] [PubMed] [Google Scholar]
- 51.Tommos C., Skalicky,J.J., Pilloud,D.L., Wand,A.J. and Dutton,P.L. (1999) De novo proteins as models of radical enzymes. Biochemistry, 38, 9495–9507. [DOI] [PubMed] [Google Scholar]
- 52.Jovanovic S.V., Harriman,A. and Simic,M.G. (1986) Electron transfer reactions of tryptophan and tyrosine derivatives. J. Phys. Chem., 90, 1935–1939. [Google Scholar]
- 53.Faraggi M., DeFilippis,M.R. and Klapper,M.H. (1989) Long range electron transfer between tyrosine and tryptophan in peptides. J. Am. Chem. Soc., 111, 5141–5145. [Google Scholar]
- 54.Mishra A.K., Chandrasekar,R., Faraggi,M. and Klapper,M.H. (1994) Long range electron transfer in peptides. Tyrosine reduction of the indolyl radical. Reaction mechanism, modulation of reaction rate and physiological considerations. J. Am. Chem. Soc., 116, 1414–1422. [Google Scholar]
- 55.DeFilippis M.R., Faraggi,M. and Klapper,M.H. (1990) Evidence for through bond long range electron transfer in peptides. J. Am. Chem. Soc., 112, 5640–5642. [Google Scholar]
- 56.Aubert C., Vos,M.H., Mathis,P., Eker,A.P.M. and Brettel,K. (2000) Intraprotein radical transfer during photoreactivation of DNA photylase. Nature, 405, 586–590. [DOI] [PubMed] [Google Scholar]
- 57.Gebicki S. and Gebicki,J.M. (1999) Cross linking of DNA and proteins induced by protein hydroperoxides. Biochem. J., 338, 629–636. [PMC free article] [PubMed] [Google Scholar]