Abstract
CDC5L and PLRG1 are both spliceosomal proteins that are highly conserved across species. They have both been shown to be part of sub- spliceosomal protein complexes that are essential for pre-mRNA splicing in yeast and humans. CDC5L and PLRG1 interact directly in vitro. This interaction is mediated by WD40 regions in PLRG1 and the C-terminal domain of CDC5L. In order to determine whether this interaction is important for the splicing mechanism, we have designed peptides corresponding to highly conserved sequences in the interaction domains of both proteins. These peptides were used in in vitro splicing experiments as competitors to the cognate sequences in the endogenous proteins. Certain peptides derived from the binding domains of both proteins were found to inhibit in vitro splicing. This splicing inhibition could be prevented by preincubating the peptides with the corresponding partner protein that had been expressed in Escherichia coli. The results from this study indicate that the interaction between CDC5L and PLRG1 is essential for pre-mRNA splicing and further demonstrate that small peptides can be used as effective splicing inhibitors.
INTRODUCTION
Pre-mRNA splicing is the nuclear process whereby the intervening sequences in nascent transcripts, called introns, are removed and the protein coding sequences joined together to produce mRNA that is subsequently exported to the cytoplasm for use in the translation of proteins. The splicing process is catalysed by a large complex containing RNA and protein factors called the spliceosome (1). The RNA components (snRNA) form four ribonucleoprotein (snRNP) particles (U1, U2, U5 and U4/U6), each of which contains the corresponding snRNAs and a set of specific and common proteins (2–4). The spliceosome complex also contains multiple non-snRNP associated proteins that are essential for complex assembly and splicing catalysis (1,4). The assembly of the splicing factors on pre-mRNA to form a spliceosome involves the sequential addition of the snRNP particles and other proteins onto the pre-mRNA substrate prior to catalysis (5).
CDC5L and PLRG1 are two nuclear proteins that are highly conserved across species that have been identified as components of the human spliceosome complex in several studies (6–8). Further studies have also shown that both proteins are found in sub-spliceosomal multiprotein complexes in yeast and humans that are essential for splicing. In yeast a 40S complex that contains the yeast homologues of CDC5L and PLRG1 has been purified and analysed by mass spectrometry (9,10). Others have also identified a similar complex, which they called either the Cef1p or the Prp19p complex, by using epitope tagging methods and affinity chromatography (10–12). We have previously purified and characterised the human CDC5L complex, containing PLRG1 and other known splicing factors as well as novel proteins, and showed that this complex is needed for splicing in vitro using HeLa nuclear extract (13).
Several other cellular functions have been attributed to both the CDC5L and PLRG1 proteins, apart from their role in splicing. CDC5L has been implicated in mitogen-activated signalling in mammalian cells because the CDC5L protein has been shown (using serum-starved cells) to be able to translocate from the cytoplasm to the nucleus upon serum stimulation (14). The fission yeast homologue of CDC5L (the CDC5+ gene) was identified in a genetic screen of Schizosaccharomyces pombe for cell division cycle mutants. It was observed that the CDC5+ gene encodes an essential protein with a possible role in the G2 phase of the cell cycle (15). In humans it was observed that overexpression of CDC5L in mammalian cells shortens the G2 phase and reduces cell size. These data are consistent with a role for the protein in regulating G2 progression and mitotic entry (14). The N-terminal domain of CDC5L binds specifically and with high affinity to a 12 bp DNA sequence and this interaction is capable of activating transcription (16). Thus, CDC5L may also function in gene transcription.
PLRG1 contains seven copies of the phylogenetically conserved WD repeat motifs. These repeat WD domains were first identified in the β subunit of heterotrimeric GTP-binding proteins that transduce signals across the cell membrane. Proteins containing these WD motifs, also called WD40, β transducin or GH-WD repeats, are found in all eukaryotes, but are absent in prokaryotes (17,18). The PLRG1 protein homologue in Arabidopsis thaliana is called PRL1. The PRL1 protein is involved in the pleiotropic control of glucose and hormone responses in A.thaliana (19). A mutation in the prl1 gene has pleiotropic phenotypes in A.thaliana. It has been shown that prl1 mutation causes transcriptional derepression of glucose-responsive genes, augments the sensitivity of the plants to growth hormones such as cytokinin, abscisic acid, ethylene and auxin, stimulates the accumulation of sugars and starch in the plant leaves and inhibits root elongation (19). In both mammalian and A.thaliana cells, PRL1 accumulates in the cell nucleus and interacts with ATHKAP2, an α-importin nuclear import receptor (19). The observation of a regulatory interaction between PRL1 and SNF1-like protein kinases in A.thaliana is consistent with a role for this protein in the regulation of glucose responses in the plant (20). All the observed roles (described above) for CDC5L and PLRG1 and their homologues in other species indicate either that both proteins are involved in multiple cellular pathways or that the observed functions may be indirect effects of the function(s) of both proteins in the splicing of pre-mRNAs encoding proteins involved in these processes.
We have previously shown that the proteins CDC5L and PLRG1 in HeLa nuclear extract are very strongly associated and have demonstrated a direct interaction between both proteins in vitro (13,21). The regions of both proteins involved in the direct interaction were identified as the C-terminus of CDC5L and the WD40 repeat region of PLRG1. We subsequently showed that this interaction is essential for splicing by using dominant negative mutants containing the interacting regions of both proteins. In this study, we have analysed the interaction domains of both proteins sequentially by using peptide sequences derived from sites surrounding phylogenetically highly conserved amino acids. By using this strategy, we have identified essential amino acid sequences in the interacting regions of CDC5L and PLRG1. We show that the interaction between the two proteins is essential for splicing but not spliceosome assembly and demonstrate that short peptides can act as potent splicing inhibitors.
MATERIALS AND METHODS
Peptide synthesis and purification
Peptides were designed from sequences in the CDC5L and PLRG1 interaction domains and their syntheses performed commercially by Sigma-Genosys Ltd, UK. The synthesised peptides were purified by HPLC and their integrity confirmed by mass spectrometry. All the peptides used in the analyses were prepared to a final purity of >95%. The peptides were dissolved in water (7 nmol/µl) and stocks stored in small aliquots at –80°C. Peptides to be used in pull-down assays were biotinylated at their N-termini. The peptides designed and synthesised for use in this study are listed in Table 1.
Table 1. List of peptides synthesised for use in this study.
| Peptide name | Sequence of peptide |
|---|---|
| PL30-1 | PYLFSCGEDKQVKCWDLEYNKVIRHYHGHL |
| PL30-2 | CSRDSTARIWDVRTKASVHTLSGHT |
| PL30-3 | PQIITGSHDTTIRLWDLVAGKTRVTLTNHK |
| PL15-1 | PQIITGSHDTTIRLW |
| PL15-2 | DTTIRLWDLVAGKTR |
| PL15-3 | DLVAGKTRVTLTNHK |
| HC-2 | CPEELQVSPGPRQQLPPRQ |
| PL-SB15 | TTHLNVKDVLARKGT |
| PL-SB30 | TWTRHTLDNSVTKQDIPVTLGAH |
| CD24-1 | HMTTEAKRAAKMEKKMKILLGGYQ |
| CD24-2 | ELKKHEDSAIPRRLECLKEDVQRQ |
| CD24-3 | EREKELQHRYADLLLEKETLKSKF |
| CD-R24 | QYGGLLIKMKKEMKAARKAETT |
| CD-R12 | QYGGLLIKMKKE |
| CD12-1 | HMTTEAKRAAKM |
| CD12-2 | KRAAKMEKKMKI |
| CD12-3 | EKKMKILLGGYQ |
| CD8 | KILLGGYQ |
The table shows the names and sequences of the peptides used in this study.
Multiple protein sequence alignments
Sequence alignments were performed with the MegAlign application in the Lasergene software package (DNAStar Inc.) using the Clustal algorithm.
In vitro transcription
The hnRNPA1 plasmid construct was digested with ScaI, pBSAL4 (AL4) was digested with PvuII and the RPL32 construct was digested with the restriction enzyme RsaI. The pBSAd1 plasmid was digested with Sau3AI. All the digested plasmids were used for in vitro transcription under similar conditions to pBSAd1 (22) using the enzyme T3 RNA polymerase (Promega), except for the RPL32 constructs, which were transcribed using T7 RNA polymerase (Promega). Where the transcripts were to be used for purification of splicing complexes, biotin was incorporated into the pre-mRNA using biotin-UTP (6,7). Preparation of antisense probes for snRNA and northern blot hybridisation experiments were performed as described previously (23).
Splicing assays
Splicing assays were done using uniformly labelled, capped pre-mRNAs incubated with nuclear extract as described previously (24). All the splicing reactions were done using the transcript from pBSAd1 except where it is specifically mentioned that transcripts from other templates were used. The nuclear extracts used in the splicing assays were obtained commercially from the Computer Cell Culture Centre (Mons, Belgium). When the reactions were to be used for the analysis of splicing complexes, heparin was added at a final concentration of 5 mg/ml to stop each reaction and facilitate gel separation of splicing complexes. The reactions were loaded onto a polyacrylamide/agarose composite gel and run for ∼5 h at 25 mA (22).
Streptavidin–agarose pull-down assays
Streptavidin–agarose beads (Sigma) were washed in wash buffer I containing 0.1% NP40, 0.1% sodium azide, 50 mM KCl and 20 mM Tris, pH 7.5. The beads were then washed with buffer II containing 0.1% NP40, 0.1% sodium azide, 250 mM KCl and 20 mM Tris, pH 7.5, before a final wash in buffer I. The washed beads were preblocked with constant mixing to saturate the non-specific binding sites for ∼1 h at 4°C in preblock buffer (i.e. buffer I containing 100 µg/ml glycogen and 1 mg/ml BSA). The streptavidin–agarose beads were then washed again with buffer I. HeLa nuclear extract to be used in the pull-down assay was pre-cleared using a portion of the washed beads. Each pull-down sample contained ∼200 µg nuclear extract and 7 nmol biotinylated peptide. The reactions were mixed at 4°C for ∼1–1.5 h before washing the beads three times using phosphate-buffered saline (PBS) containing 0.1% Triton X-100. The washed beads were resuspended in SDS–PAGE buffer before electrophoretic separation of protein bound to the peptide.
In order to purify splicing complexes, the splicing reactions were prepared using HeLa nuclear extract and biotinylated pre-mRNA (6,7) under the same conditions as for assays used for spliceosome native gel analyses. The bound complexes were purified using streptavidin–agarose beads (Sigma or Boehringer) (6,7) and the snRNAs present in the complexes were released by proteinase K digestion and analysed by northern blotting.
SDS–PAGE and western blotting
SDS–PAGE gel analysis was done as described previously (25). For immunoblotting, the washed beads were resuspended in an equal volume of 2× SDS–PAGE loading buffer and heated at 70°C for 10 min. Approximately 20 µl of the supernatant was loaded on a 4–12% pre-cast gradient gel (Invitrogen, UK). The separated proteins were transferred onto Hybond-C extra membrane (Amersham Biociences, UK) by electroblotting. The membranes carrying the transferred proteins were blocked with 5% non-fat milk powder in PBS containing 0.3% Tween-20 for ∼1–16 h. The membranes were then incubated with primary antibody for 1 h at room temperature, washed with blocking buffer and incubated with the appropriate secondary antibody. The primary antibodies anti-CDC5L and anti-PLRG1 were used in immunoblotting as described previously (21). After washing the blots in blocking buffer three or four times (5 min per wash at room temperature), the membranes were then incubated with a secondary antibody to which had been covalently coupled horseradish peroxidase. Protein bands were detected by developing blots with the ECL kit (Amersham Biociences, UK) according to the manufacturer’s instructions.
Expression of recombinant proteins in Escherichia coli
Expression of recombinant proteins in E.coli was done as described previously (21). CDC5L and PLRG1 cDNAs cloned into the pGEX-4T1 vector were used to transform E.coli BL21(DE3). Overnight cultures were grown from single colonies, then diluted 1:10 in fresh LB medium with ampicillin (100 µg/ml) and grown at 25–30°C to an OD600 of 0.7–1.0 before induction with 0.04 mM IPTG. Three hours post-induction, cells were pelleted and resuspended in 10 ml of PBS containing 0.5% Triton X-100 and protease inhibitor cocktail (Boehringer). Cell lysis was achieved by sonication. The cell debris was removed by centrifugation at 10 000 g for 10 min. Pre-swollen glutathione–Sepharose beads pre- equilibrated in PBS were added to the supernatant (1 ml/l culture). The beads were incubated with the crude protein extract for 2 h at 4°C with rocking. Beads were collected and washed three times in PBS containing 0.5% Triton X-100, followed by three washes in PBS. Proteins were eluted from the beads by incubating in 25 mM glutathione in 50 mM Tris–HCl, pH 8.0. The proteins were dialysed into either PBS or a buffer containing 20 mM HEPES, pH 8.0, 20% glycerol, 100 mM KCl, 0.2 mM EDTA and 1 mM dithiothreitol and stored at –80°C. Protein concentrations were determined either by measuring the OD280 of the purified protein or by using the Coomassie Plus Protein Assay reagent (Perbio Science, UK) according to the manufacturer’s instructions.
RESULTS
Inhibition of pre-mRNA splicing by PLRG1 peptides containing sequences in the CDC5L binding region of the protein
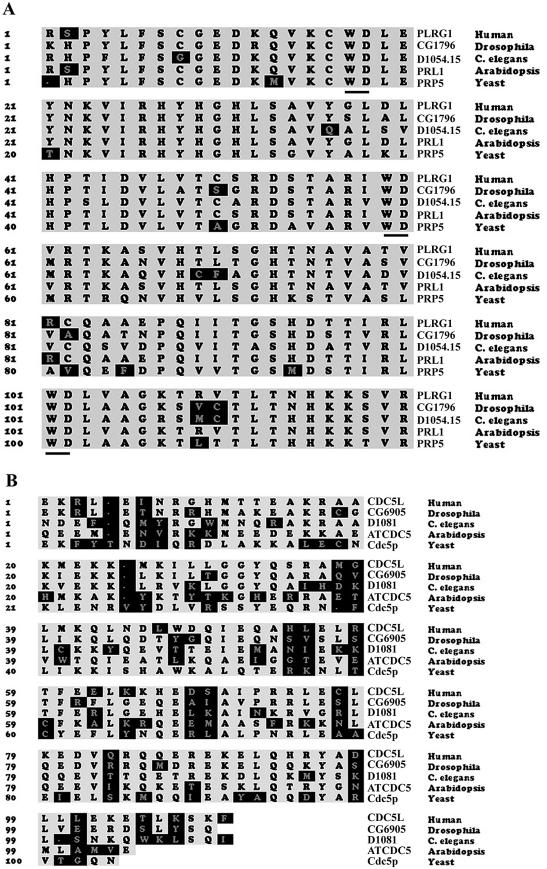
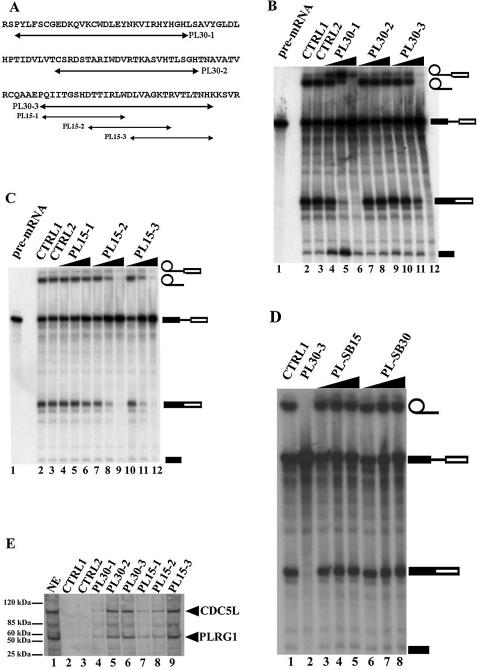
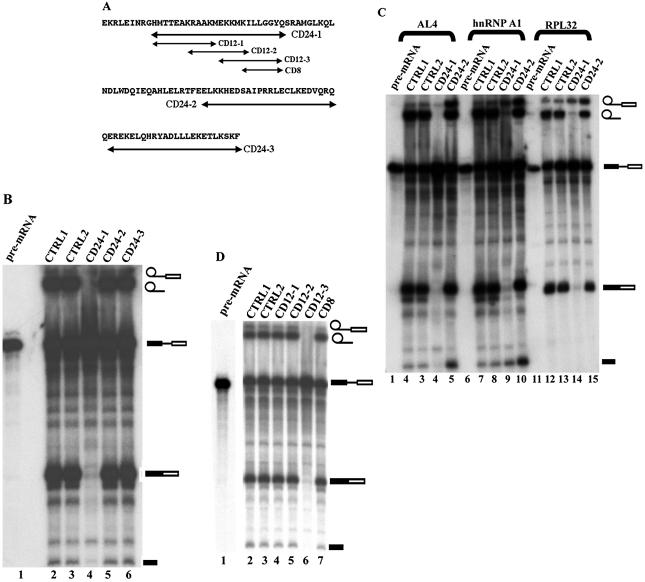
We have previously identified the CDC5L binding region in PLRG1 (21). Multiple sequence alignment of these regions in different species revealed highly conserved amino acid sequences (Fig. 1A). In order to determine the significance of these conserved sequences for pre-mRNA splicing, peptides from the binding region (Fig. 2A) were synthesised. Initially, 25mer–30mer peptides were made spanning the highly conserved WD40 sequences in the CDC5L binding domain. These peptides were then added to splicing reactions as competitors to the endogenous protein. The results obtained indicate that both PL30-1 and PL30-3 were able to inhibit splicing (Fig. 2B, lanes 6 and 12), whereas PL30-2 (Fig. 2B, lane 9) and the control non-specific peptide (Fig. 2B, lane 3) did not inhibit splicing. PL30-3 was found to be more efficient in blocking splicing than PL30-1. Because of the relatively stronger inhibitory activity of PL30-3 compared to PL30-1 at the highest peptide concentrations used (Fig. 2B, compare lanes 6 and 12), we decided to prepare three overlapping 15mer peptides spanning the PL30-3 sequence (Fig. 2A, PL15-1, PL15-2 and PL15-3). The 15mer peptides were added to splicing assays as above. The results from these experiments show that peptides PL15-2 and PL15-3 block splicing (Fig. 2C, lanes 9 and 12), whereas PL15-1 and the control peptide (Fig. 2C, lane 3) do not. The data from the above experiments indicate that PLRG1 sequences spanning or overlapping PL30-1, PL30-3, PL15-2 and PL15-3 are essential for the function of the protein in splicing.
Figure 1.
Multiple protein sequence alignments of the interacting regions in CDC5L and PLRG1. The highlighted regions in the figure represent amino acids that are not highly conserved across species. The names of the organisms from which the sequences were derived are on the right of the figure. Yeast sequence is from S.pombe. (A) Alignment of PLRG1 sequences. The WD40 motifs are underlined. (B) Alignment of CDC5L sequences.
Figure 2.
PLRG1 peptides will interact with CDC5L in nuclear extract and inhibit pre-mRNA splicing. (A) Design of peptides from sequences in the CDC5L binding region of PLRG1. The arrows indicate the sequences of the peptides synthesised. (B) Autoradiograph of a splicing gel from an experiment to determine the effect of 24mer–30mer peptides spanning the highly conserved WD40 sequences on splicing. Approximately 7–20 nmol peptide were added to the splicing reactions (lanes 4–12). Lane 1 contained the input pre-mRNA. CTRL1 is a control splicing reaction without peptide. CTRL2 is a control reaction containing 20 nmol control peptide HC-2 derived from another spliceosomal protein HCF-1 that has not been detected in complexes containing CDC5L and PLRG1. The symbols on the right of the panel represent the input RNA, splicing intermediates and products. (C) Autoradiograph of a splicing gel from an experiment to determine the effect of overlapping 15mer peptides spanning the PL30-3 sequence on splicing. Similar amounts of peptide were added (lanes 4–12) to the splicing reactions as in (B). The lanes marked CTRL1 and CTRL2 contained splicing reactions treated in a similar way to lanes with the same names in (B). (D) Peptides containing the same amino acids as PL15-3 and PL30-3 in a scrambled sequence do not inhibit splicing. Lane 1, CTRL1 is the control reaction without peptide; lane 2, ∼20 nmol PL30-3; lanes 3–5, 7–20 nmol PL-SB15; lanes 6–8, 7–20 nmol PL-SB30 peptide. (E) Pull-down of CDC5L onto streptavidin–agarose beads from HeLa nuclear extract using PLRG1 peptides. Lanes 4–9, pull-down assays with the corresponding peptides (used in marking each lane). CTRL1 did not contain any peptides whereas CTRL2 contained a control peptide that does not inhibit splicing. The blot was probed with a buffer containing both anti-CDC5L and anti-PLRG1 antibodies. The arrows on the right of the panel show the positions of the CDC5L and PLRG1 proteins.
In order to check that the splicing inhibition observed for the PLRG1 peptides is directly linked to the sequences of the peptides, rather than their amino acid composition, peptides were synthesised containing identical amino acids as in PL15-3 and PL30-3, respectively, except that the amino acid sequences in the new peptides (PL-SB15 and PLSB-30) were scrambled. The peptides were then added to splicing reactions as for PL15-3 and PL30-3. The results obtained from these experiments indicate that the peptides PL-SB15 and PLSB-30 do not inhibit splicing (Fig. 2D, lanes 3–8), whereas peptides containing amino acid sequences identical to the protein sequence, e.g. PL30-3 (Fig. 2D, lane 2), will inhibit splicing when added at a similar concentration. These results show that the splicing inhibition observed is linked to the sequence of the amino acids in the peptides.
PLRG1 peptides interact with CDC5L in HeLa nuclear extract without disrupting the CDC5L–PLRG1 complex
We have previously shown that a truncated protein containing the PLRG1 binding domain in CDC5L can disrupt the interaction between the two proteins in nuclear extract resulting in splicing inhibition (21). Because of the inhibitory effect of the PLRG1 peptides on splicing, we next decided to investigate whether these peptides are able to bind to the partner of PLRG1, i.e. CDC5L, in nuclear extract and disrupt the CDC5L–PLRG1 interaction. The peptides (containing an N-terminal biotin) were incubated with HeLa nuclear extract followed by a pull-down on streptavidin–agarose beads. After extensive washing of the beads, the bound proteins were separated by SDS–PAGE electrophoresis. The separated proteins in the gel were transferred to a nitrocellulose membrane by western blotting. The membrane was then probed using anti-CDC5L and anti-PLRG1 antibodies and the presence of the proteins revealed by ECL (Amersham Biosciences). The results obtained from these experiments show that all the PLRG1 peptides associate with the CDC5L complex in HeLa nuclear extract. Some of the peptides, e.g. PL30-2, PL30-3 and PL15-3, associated more stably with the complex as compared with the other PLRG1 peptides (Fig. 2E, compare lanes 4, 7 and 8 to lanes 5, 6 and 9). The control peptide (Fig. 2E, lane 3) did not associate stably with the CDC5L complex. We also observed that none of the peptides disrupted the endogenous CDC5L–PLRG1 interaction in nuclear extracts, because pull-down of CDC5L on the streptavidin–agarose beads also resulted in co-precipitation of the PLRG1 that was bound to CDC5L in the extract. These data indicate that the peptides associate with CDC5L in nuclear extract without disrupting the protein interaction with PLRG1. This may be because the peptides (due to their small size) do not completely interfere with the interaction interface between the two proteins.
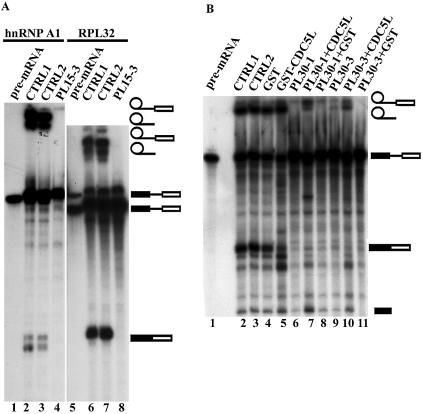
In order to determine whether the inhibitory effects of the PLRG1 peptides were template dependent, the PLRG1 peptide PL15-3 was added to splicing reactions using two other pre-mRNA templates, i.e. hnRNP A1 and RPL32 (26) (Fig. 3A). The results obtained show that the peptide will also inhibit splicing using the above templates (Fig. 3A, lanes 4 and 8), whereas the control peptide did not block splicing, as expected (Fig. 3A, lanes 3 and 7). These results indicate that the inhibitory effect of the peptides is not limited to a single template and suggest an essential role for the CDC5L–PLRG1 interaction in splicing.
Figure 3.
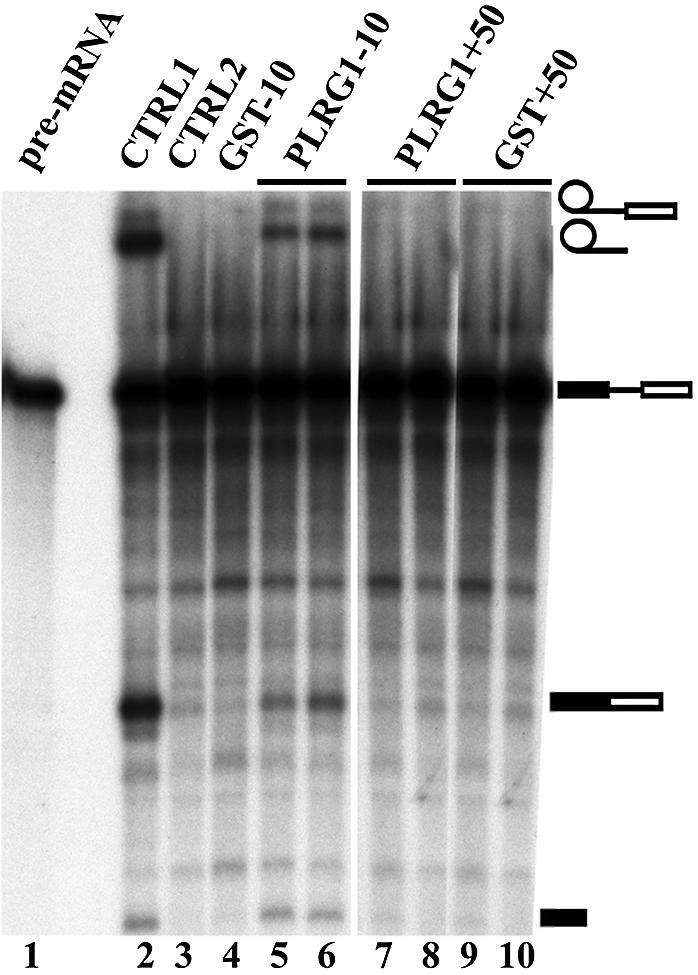
(A) Effect of the PLRG1 inhibitory peptide on splicing using other pre-mRNA templates (hnRNP A1 and RPL32). Lanes 1 and 5 contain the input pre-mRNA. All the lanes marked CTRL1 are splicing controls without peptide. The lanes marked CTRL2 contained the control peptide PL-SB15. Lanes 4 and 8 are splicing reactions containing the PLRG1 inhibitory peptide PL15-3. (B) Bacterially expressed CDC5L reduces the effect of PLRG1 peptides in splicing inhibition. Splicing assays were performed using peptides that had been preincubated with either GST–CDC5L (∼40 µg) or a similar molar amount of GST for 15 min at room temperature. Lane 1, input pre-mRNA; lane 2 (CTRL1), a control splicing reaction without peptide; lane 3 (CTRL2), a negative control splicing reaction containing PL30-2; lanes 4 and 5, splicing reactions without peptides but to which had been added GST and GST–CDC5L, respectively, at the same levels as the protein used in preincubation with peptides; lane 6, splicing reaction containing peptide without recombinant protein. The reactions in lanes 7 and 8 are similar to that in lane 6 except that the peptides were preincubated with GST–CDC5L and GST, respectively. The splicing reactions in lanes 9–11 are identical except that the reaction in lane 9 contained PL30-3 peptide alone whereas the reactions in lanes 10 and 11 contained peptides preincubated with GST–CDC5L and GST, respectively. The symbols on the right of the panel represent the input RNA, splicing intermediates and products. Other bands in the figure have been caused by partial degradation of the RNA in the reactions.
Bacterially expressed CDC5L will block the inhibitory activity of PLRG1 peptides
In order to test whether the inhibitory effect of the peptides on splicing is specific (i.e. is due to the peptides associating with CDC5L in HeLa nuclear extract), we next explored the possibility of blocking the activity of the peptides by using bacterially expressed CDC5L. CDC5L was expressed as a GST fusion and purified as described previously (22). The active PL30 peptides were pre-incubated with either GST–CDC5L or GST alone before addition to a splicing reaction. The results obtained from these experiments show that GST–CDC5L decreases the inhibitory activity of the peptides (Fig. 3B, lanes 7 and 10), whereas GST alone does not (Fig. 3B, lanes 8 and 11), as demonstrated by the presence of stable lariat and 3′-exon lariat bands in Figure 3B (lanes 7 and 10). The corresponding spliced exon bands have been partially degraded by RNase contamination from the bacterially expressed protein preparations. Consistent with this, some degradation of the spliced exons was observed when GST–CDC5L protein alone was added (Fig. 3B, lane 5). Similar results were obtained for the 15mer PLRG1 peptides (data not shown). Taken together, these data indicate that the inhibitory activity of the peptides is specific and likely due to the peptides interfering with the normal PLRG1–CDC5L interaction in nuclear extract.
Spliceosome assembly is not prevented by PLRG1 peptides in the splicing reaction
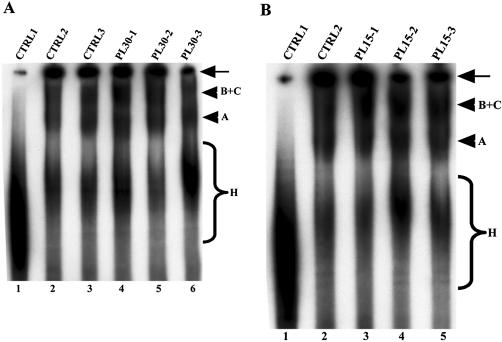
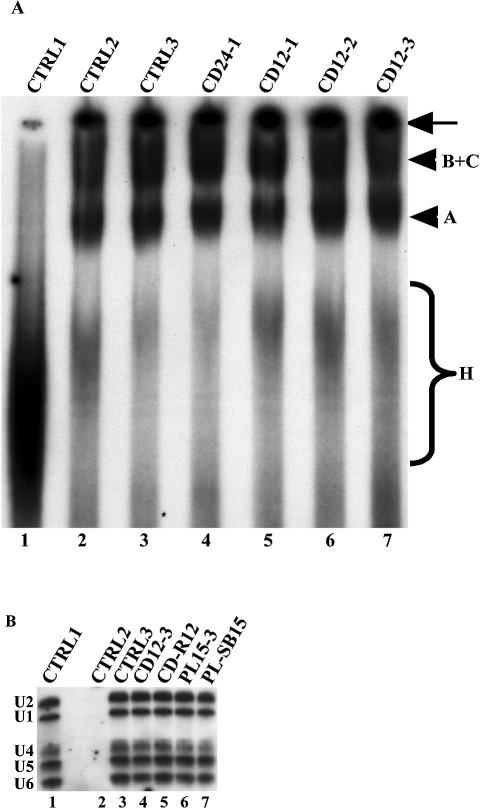
The above observations that PLRG1 peptides will inhibit splicing do not provide any information on the stage in the pathway where the inhibition occurs. We next investigated whether the inhibition of splicing by the peptides was due to a disruption of the spliceosome assembly pathway. Splicing reactions were set up as described in Materials and Methods and the PLRG1 peptides added to these reactions. After addition of the peptides, the reactions were allowed to proceed for up to 1 h. The reactions were then loaded onto native polyacrylamide/agarose composite gels and run for several hours. Bands corresponding to splicing complexes were revealed by autoradiography. The results obtained from these experiments show that neither inhibitory amounts of the PL30 peptides (Fig. 4A, lanes 4–6) nor the same amounts of the PL15 peptides (Fig. 4B, lanes 3–5) were able to block spliceosome assembly in HeLa nuclear extract. Taken together, our data suggest a possible role for the peptide regions analysed in either the catalytic steps of splicing or the rearrangements that occur in the spliceosome prior to catalysis.
Figure 4.
Effect of the PLRG1 peptides on spliceosome assembly. Splicing reactions were performed for 50–60 min at 30°C and the splicing complexes formed were separated on a polyacrylamide/agarose native gel. The bands representing splicing complexes were revealed by autoradiography. (A) Lane 1, a control splicing reaction incubated on ice; lane 2 (CTRL2), a control splicing reaction without peptide; lane 3, the HC-2 peptide; lanes 4–6, the PL30-1, PL30-2 and PL30-3 peptides, respectively. (B) Lane 1, similar to CTRL1 in (A); lane 2 (CTRL2), the HC-2 peptide; lanes 3–5, the PL15-1, PL15-5 and PL15-5 peptides, respectively. The arrows on the right of the panels indicate material that did not penetrate into the gel. The arrowheads represent the splicing complexes separated on the native gel. The braces (labeled H) show non-specific complexes assembled on the pre-mRNA.
Specific inhibition of pre-mRNA splicing by CDC5L peptides containing sequences in the PLRG1 binding region of the protein
Because of the effects of the PLRG1 peptides on splicing, we decided to perform a complementary analysis with the PLRG1 binding region of CDC5L. We carried out multiple sequence alignments of the binding region across species (Fig. 1B). Peptides (24mers) spanning highly conserved amino acids were designed (Fig. 5A), synthesised and purified as described in Materials and Methods. The 24mer peptides were added to in vitro splicing reactions to analyse their effect on splicing. The results obtained showed that one of these peptides (CD24-1) is able to block splicing efficiently (Fig. 5B, lane 4), whereas the other two peptides had little effect on splicing at similar concentrations.
Figure 5.
Splicing inhibition by peptides derived from the PLRG1 binding region of CDC5L. (A) Double-headed arrows indicate peptides synthesised from the PLRG1 binding region of CDC5L. (B) Autoradiograph of a splicing gel using CDC5L 24mer peptides. The peptides were added to splicing reactions to check the effect on splicing. Lane 1, the input pre-mRNA; lane 2 (CTRL1), a splicing reaction without peptides; lane 3 (CTRL2), a negative control reaction containing the peptide CD-R24 (CD-R24 contains the same amino acids as the peptide CD24-1 but in a different order); lanes 4–6, peptides CD24-1, CD24-2 and CD24-3, respectively. (C) Effect of the CDC5L inhibitory peptide on splicing using other pre-mRNA templates (AL4, hnRNP A1 and RPL32). Lanes 1, 6 and 11, the input pre-mRNA; lanes marked CTRL1, splicing controls without peptide; lanes marked CTRL2, control peptide CD-R24. The lanes with splicing reactions containing CD24-2 (i.e. lanes 5, 10 and 15) also served as internal controls because the peptide is also derived from the PLRG1 binding region of CDC5L. Lanes 4, 9 and 14 are splicing reactions that contained the CD24-1 peptide. Note that the size differences between the introns of these constructs have not been resolved by this gel because it was not run for the amount of time needed to resolve these size differences. (D) Effect of 8mer–12mer peptides derived from the CD24-1 sequence on splicing. Lane 1, input pre-mRNA; lane 2 (CTRL1), a splicing reaction without peptide; lane 3 (CTRL2), a splicing reaction containing the control peptide CD-R12 (CD-R12 contains the same amino acids as CD12-3 except that the order of the amino acids in the sequence is different); lanes 4–6, splicing reactions that included the 12mer peptides CD12-1, CD12-2 and CD12-3, respectively. The splicing reaction in lane 7 contained the 8mer peptide CD8. The symbols on the right of the panel represent the input RNA, splicing intermediates and products. The other bands in the figure have been caused by partial degradation of the RNA in the reactions.
We next investigated whether the splicing inhibition observed for CD24-1 was either a substrate-specific or a general characteristic of the peptide. Pre-mRNA was prepared by in vitro transcription from the globin AL4 plasmid, an hnRNP A1-533 series plasmid and plasmid pGEM-RPL32 (26 and references therein). Addition of the CD24-1 peptide to splicing reactions containing different pre-mRNA substrates resulted in inhibition of the formation of splicing products (Fig. 5C, lanes 4, 9 and 14). We also observed that at this CD24-1 concentration the first step of splicing was not completely blocked, resulting in the presence of some lariat exon intermediate on the gel, perhaps due to small variations in template pre-mRNA concentration in the splicing reaction. These data indicate that the activity of the peptides is not limited to a single pre-mRNA substrate and are consistent with a general function for the interaction between CDC5L and PLRG1 in splicing. We then designed three 12mer overlapping peptides spanning the CD24-1 sequence in order to narrow down the essential amino acids in this region. The 12mer peptides synthesised were then added to splicing reactions as described above. The results obtained from these experiments show that one of the 12mer peptides is active in blocking splicing (Fig. 5D, lane 6), whereas the other 12mer peptides, as well as an 8mer peptide (CD8) derived from a conserved motif in the CD12-3 sequence, were inactive in splicing inhibition (Fig. 5D, lanes 4, 5 and 7, respectively).
To test the specificity of the CDC5L peptides in splicing inhibition, we investigated whether E.coli-expressed PLRG1 blocks the splicing inhibitory activity of the CD12-3 peptide. The CD12-3 peptide was pre-incubated with either GST–PLRG1 or GST alone before addition to the splicing reaction. The results from these experiments show that GST–PLRG1 blocks the inhibitory activity of the peptide (Fig. 6, lanes 5 and 6), whereas GST alone does not block activity of the peptide (Fig. 6, lane 4). In order to check whether the peptide-blocked splicing reaction could be chased into products, either GST alone or GST–PLRG1 was added to the reactions containing the CD12-3 peptide 50 min after the start of the splicing assay. The reactions were incubated for a further 10–15 min at 30°C before separation of splicing complexes on a native gel. The results obtained show that the effect of the peptide cannot be reversed once the splicing reaction has been allowed to proceed through spliceosome assembly in the presence of the peptide inhibitor (Fig. 6, lanes 7 and 8). These data indicate that the effect of the peptide can only be attenuated if it is incubated with PLRG1 prior to the start of splicing.
Figure 6.
PLRG1 expressed in E.coli reduces the effect of CDC5L peptides in splicing inhibition. Lane 1, input pre-mRNA; lane 2 (CTRL1), a splicing reaction without peptides. The lane marked CTRL2 represents a splicing reaction containing the CDC5L peptide CD12-3. The splicing reaction in lane 4 contained CD12-3 peptide preincubated with GST (8 µg) for 10 min before addition to the reaction, whereas the samples in lanes 5 and 6 are duplicate splicing reactions containing CD12-3 peptide preincubated with an equivalent amount of GST–PLRG1 (i.e. for ∼10 min at room temperature prior to addition to the reaction). The contents of the splicing reactions in lanes 7 and 8 (duplicates) are similar to those of lanes 5 and 6 except that GST–PLRG1 was added after the splicing reactions had been incubated for 50 min at 30°C. The contents of the splicing reactions in lanes 9 and 10 (duplicates) are similar to that of lane 4 except that GST was added to the reactions after the splicing reaction had been incubated for 50 min at 30°C. The symbols on the right of the panel represent the input RNA, splicing intermediates and products. Other bands in the figure have been caused by partial degradation of the RNA in the reactions.
CDC5L peptides interact with PLRG1 in HeLa nuclear extract without disrupting the CDC5L–PLRG1 interaction
Because we had observed above that the PLRG1 peptides associated with CDC5L in HeLa nuclear extract without disrupting the CDC5L–PLRG1 interaction, we next decided to investigate whether the same happens for the CDC5L peptides. The CDC5L peptides were incubated with HeLa nuclear extract followed by a pull-down on streptavidin–agarose beads. After washing the beads, the bound proteins were separated by SDS–PAGE and the separated proteins in the gel were transferred to nitrocellulose membrane by western blotting. The membranes were then probed using either anti-PLRG1 antibodies (Fig. 7A and B) or combined anti-CDC5L and anti-PLRG1 antibodies (Fig. 7C). The protein bands were detected by ECL (Amersham Biosciences). The results for the CD24 peptides show that CD24-1 associates most strongly with PLRG1, whereas the other peptides showed relatively weaker associations with the protein (Fig. 7A, lanes 4–6). Results obtained using the CD12 peptides show that CD12-2 and CD12-3 bind to PLRG1 in HeLa nuclear extract (Fig. 7B, lanes 4 and 5), whereas the other peptide associated at background levels (Fig. 7B, lane 3). The CDC5L peptides that showed the strongest PLRG1 bands in the pull-down assays were then used to investigate whether the interactions between the peptides and PLRG1 caused disruption of the CDC5L–PLRG1 interaction. The peptides were incubated with HeLa nuclear extract followed by a pull-down on streptavidin–agarose beads and sample analysis as described above, except that the nitrocellulose membrane containing transferred proteins was probed using a combination of anti-CDC5L and anti-PLRG1 antibodies. The results obtained from these experiments (Fig. 7C) indicate that the CDC5L peptides bind to PLRG1 without disrupting the CDC5L–PLRG1 interaction in nuclear extract, as shown by the presence of both proteins in the pull-downs (Fig. 7C, lanes 4 and 5). Taken together, these results indicate that the peptides may block splicing by competing with CDC5L for essential contacts to PLRG1. These results are consistent with the existence of a large protein–protein interaction interface between CDC5L and PLRG1, involving multiple contacts between stretches of amino acids, perhaps mediated by the WD40 domains.
Figure 7.
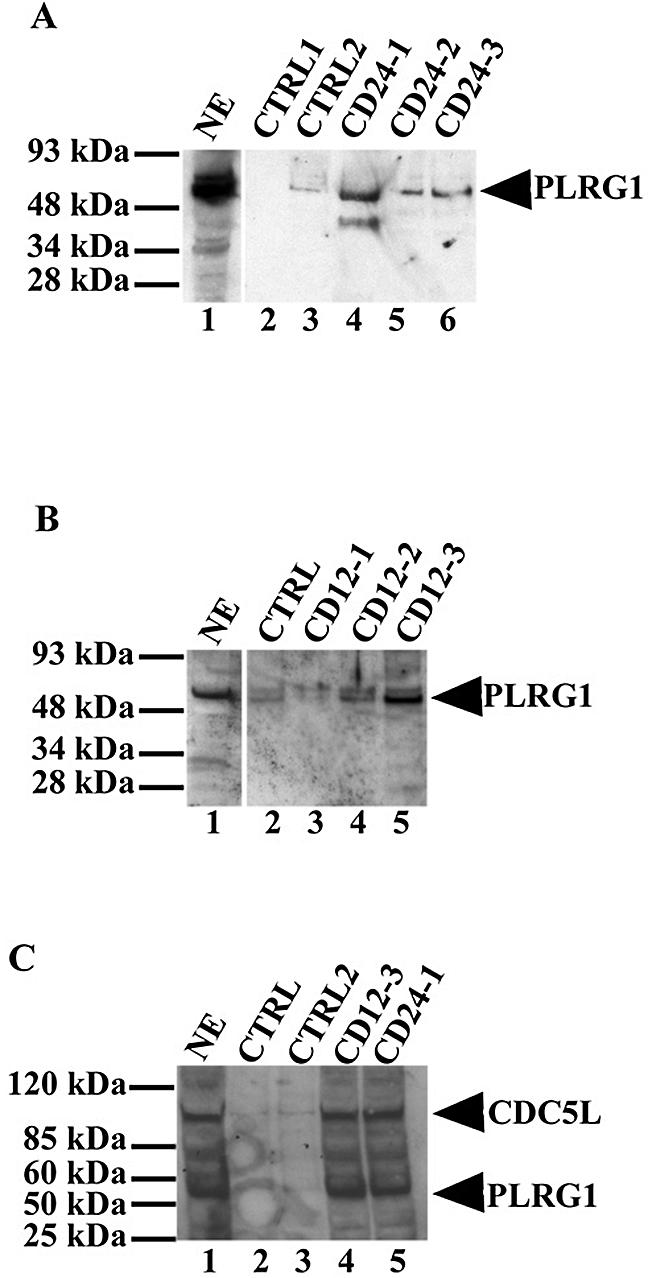
CDC5L peptides associate with PLRG1 in HeLa nuclear extract. CDC5L peptides were used in pull-down assays on streptavidin–agarose beads and the co-precipitated protein transferred to the nitrocellulose membranes was probed with anti-PLRG1 antibodies. Protein bands were then revealed by enhanced chemiluminescence (ECL). (A) Lane 1, a positive control containing HeLa nuclear extract; lanes 2 and 3, control pull-downs using beads only and CD-R24 peptide, respectively; lanes 4–6, protein from pull-down assays using the CD24-1, CD24-2 and CD24-3 peptides, respectively. (B) Pull-down assays using 12mer CDC5L peptides. Lane 1, a control containing HeLa nuclear extract; lane 2, a control pull-down assay using the CD-R12 peptide; lanes 3–5, protein from pull-down assays using the CD12-1, CD12-2 and CD12-3 peptides, respectively. (C) Binding of CDC5L peptides to PLRG1 in nuclear extract does not disrupt the CDC5L–PLRG1 interaction. Pull-down assays were performed as above using streptavidin–agarose beads except that the blots were probed with a buffer containing both anti-PLRG1 and anti-CDC5L antibodies. Lane 1, the positive control (HeLa nuclear extract); lanes 2 and 3, control pull-downs using the CD-R12 and CD-R24 peptides, respectively; lanes 4 and 5, pull-downs performed using the CD12-3 and CD24-1 peptides, respectively. The arrowheads on the right of the figure point to the bands representing PLRG1 or CDC5L on the nitrocellulose membrane.
Assembly of the spliceosome complex is not prevented by the presence of active CDC5L peptides in the splicing reaction
Because some of the CDC5L peptides inhibited splicing, we next investigated whether the observed inhibition was due to either disruption of the spliceosome assembly pathway or just the catalytic steps of splicing. Splicing reactions were performed in vitro in the presence of active CDC5L and control peptides. The reactions were then loaded onto a native agarose/polyacrylamide gel. The spliceosome complexes were separated by electrophoresis and bands corresponding to the major splicing complexes revealed by autoradiography (Fig. 8A). The results from these experiments show that the inhibitory CDC5L peptides (CD24-1 and CD12-3) do not prevent the formation of spliceosome complexes (Fig. 8A, lanes 4 and 7).
Figure 8.
Characterisation of splicing complexes formed in the presence of inhibitory peptides. (A) Splicing complexes are formed in the presence of CDC5L peptides that inhibit catalysis. CDC5L peptides were added to splicing reactions as described previously and the splicing complexes formed were separated on a polyacrylamide/agarose native gel. The bands representing splicing complexes were revealed by autoradiography. Lane 1, a control splicing reaction incubated on ice; lane 2 (CTRL2), a control splicing reaction without peptide; lane 3, a negative control containing the CD-R24 peptide that does not inhibit splicing; lanes 4–7, the CD24-1, CD12-1, CD12-2 and CD12-3 peptides, respectively. The arrow on the right indicates material that did not penetrate into the gel. The arrowheads represent the splicing complexes separated on the native gel. The brace labeled H indicates non-specific complexes assembled on the pre-mRNA. (B) Identification of snRNAs in splicing complexes formed in the presence of inhibitory peptides. Splicing complexes were formed (as above) using biotinylated pre-mRNA and as described in Materials and Methods. snRNAs in streptavidin–agarose pull-downs were analysed by northern blotting (26). The lane marked CTRL1 is the positive control showing all five snRNAs in HeLa nuclear extracts. CTRL2 contained a splicing reaction assembled on ice. CTRL3 contained a splicing reaction without peptide. The reaction in lane 4 contained the inhibitory CDC5L peptide CD12-3 and lane 5 contained the control CDC5L peptide that does not block splicing. The splicing reaction in lane 6 contained the inhibitory PLRG1 peptide, whereas the reaction in lane 7 contained the control PLRG1 peptide that does not inhibit splicing.
We next investigated the snRNA composition of the splicing complexes formed in the presence of inhibitory peptides. Splicing complexes were purified using streptavidin–agarose beads as described in Materials and Methods. The snRNA composition of the splicing complexes was determined by northern blotting using antisense snRNA probes. The results obtained from these experiments indicate that the splicing complexes formed upon inhibition of splicing using peptides contain all four snRNPs (Fig. 8B, lanes 4 and 6).
Taken together, these results suggest an essential role for the CDC5L–PLRG1 interaction in splicing and suggest that the peptides may block splicing by interfering either with splicing catalysis or the molecular rearrangements that occur in the spliceosome before catalysis.
DISCUSSION
We have previously shown that the C-terminal domain of CDC5L interacts with the central WD40 domains of PLRG1 (21). Others have confirmed the same interaction between the yeast homologues (Cef1p and Prp46) of both proteins (27). Multiple sequence alignments of the interaction domains reveal the presence of highly conserved amino acid residues. We have exploited the conserved sequences to design peptides for use in analysing the role of the CDC5L–PLRG1 interaction in the splicing mechanism. Several previous studies have analysed the CDC5L complex components using mass spectrometry and genetic methods (9–13). Some of these analyses have also involved the characterisation of protein–protein interactions within these complexes in yeast and humans (12,21,27,28). These studies all suggest that the CDC5L complex components are required for splicing. However, a direct role for any of the core components in specific steps of the splicing mechanism is still unclear. Some of the previous studies mentioned above have shown an accumulation of pre-mRNA either upon repression of the cdc5+ gene in S.pombe (9) or when temperature-sensitive mutants of components of the complex were grown at the non-permissive temperature (29). It is also not clear from these studies whether the effect of splicing inhibition occurs at the level of spliceosome assembly or catalysis. It has been suggested that one of the components of the CDC5L complex in yeast, i.e. Prp19, may function in mediating the conformational rearrangements that occur in the spliceosome because the protein is thought to associate with the complex, either concomitant with or just after dissociation of U4 from the spliceosome (30,31). In this study, although we observed formation of splicing complexes in the presence of the active peptides, it is not possible using these assays to ascertain if the active peptides inhibited splicing by interfering with conformational rearrangements in the spliceosome. However, it is more likely that the active peptides do not prevent spliceosome assembly but instead inhibit splicing by interfering with essential contacts between the CDC5L and PLRG1 proteins in nuclear extract and the splicing reaction. In our previous analysis of the human CDC5L complex, we observed that immunodepletion of CDC5L (13) results in splicing inhibition, with the second catalytic step being more sensitive to immunodepletion. However, immunodepletion of CDC5L from nuclear extract did not prevent spliceosome assembly. Subsequent analysis of the immunodepleted extracts also revealed co-depletion of PLRG1 upon immunodepletion of CDC5L from HeLa nuclear extract, suggesting a tight association between both proteins in these extracts (21). Although immunodepletion of CDC5L/PLRG1 from HeLa nuclear extract did not prevent spliceosome assembly, we have observed that disruption of the interaction between CDC5L and PLRG1 inhibits spliceosome assembly (21), presumably through a dominant negative effect.
In this study, we have used peptides derived from the interaction domains of PLRG1 and CDC5L in order to analyse the roles of both proteins in the pre-mRNA splicing mechanism. We observed that peptides derived from the interaction domains of both proteins were capable of inhibiting splicing catalysis without preventing spliceosome assembly. We also observed that these peptides could interact with their partner proteins in HeLa nuclear extract. The inhibitory peptides, however, did not disrupt the interaction between endogenous CDC5L and PLRG1 in nuclear extract, suggesting that the protein–protein interaction interface between both CDC5L and PLRG1 may be extensive and involve multiple amino acid contacts. Consistent with this view, we note that while the inhibitory peptides are unable to disrupt the CDC5L–PLRG1 interaction in HeLa nuclear extract, in contrast, a truncated protein fragment that contains the entire PLRG1 binding region in CDC5L can disrupt the CDC5L–PLRG1 interaction in nuclear extract (21). Although CDC5L and PLRG1 may not be strictly required for spliceosome assembly to occur (as observed from the immunodepletion data; 13), disruption of the CDC5L complex by the truncated protein may cause other proteins in the complex, such as hPrp19 (which may be involved in spliceosome assembly or the rearrangements that occur during the assembly step; 30,31), to be lost, thus destabilising the spliceosome. Alternatively, the presence of disrupted complexes in the nuclear extract may interfere with assembly. On the other hand, CDC5L and PLRG1 may be involved in a network of interactions that are essential for both spliceosome assembly and catalysis. Thus, disruption of the CDC5L–PLRG1 complex in HeLa nuclear extract blocks the formation of splicing complexes, whereas peptide inhibition of specific amino acid contacts in the complex, which does not disrupt the binding of CDC5L and PLRG1 proteins to each other, only inhibits the catalytic steps of splicing and not spliceosome assembly. This hypothesis is consistent with the observation that the active peptides are unable to disrupt the endogenous CDC5L–PLRG1 interaction in HeLa nuclear extracts. It is also possible that the CDC5L complex as a whole needs to be targeted to the assembling splicing complexes for efficient assembly and that following assembly the distinct components of the complex may function in the catalytic steps.
Using some pre-mRNA substrates, i.e. hnRNPA1 and RPL32, we observed that peptide concentrations that would normally inhibit the first step of splicing in other pre-mRNAs, e.g. adenovirus pre-mRNA, were less efficient in inhibiting this step of splicing in these templates. However, at higher peptide concentrations, both steps of splicing were blocked in all templates used (data not shown). These results are consistent with our previous observations (13) that the second step of splicing was more sensitive to immunodepletion of CDC5L from HeLa nuclear extract and may be due to the fact that it is not always possible to completely remove factors from nuclear extracts using antibodies. Thus, any remaining protein molecules may allow the first step of splicing to proceed, albeit at a lower efficiency. Here, the peptide concentrations are sufficient to interfere with the functions of both CDC5L and PLRG1 and suggest a function for the CDC5L complex proteins in splicing catalysis and/or the rearrangements that occur in the spliceosome complex prior to the first catalytic step of splicing.
The fact that the inhibitory activity of the peptides could be blocked by pre-incubation with their interaction partner expressed exogenously in E.coli indicates that the splicing inhibition observed is specific. This specificity is further supported by the evidence that peptides from the interaction domains of two different proteins that interact directly, i.e. CDC5L and PLRG1, have the same inhibitory effect on splicing. This means that the peptides are most likely inhibiting splicing by interfering with specific amino acid contacts between the two proteins in the complex. The splicing complexes formed in the presence of active peptides could not be chased into splicing products by the addition of bacterially expressed protein, suggesting that these peptides may be incorporated into the spliceosome complex and then are either inaccessible to recombinant proteins after assembly has occurred or else the splicing complexes formed in the presence of the peptides constitute complexes blocked in an inactive conformation. It is, however, more likely that the peptides in the splicing complexes are not accessible to the recombinant protein.
The work presented in this study therefore demonstrates for the first time that small peptides derived from regions involved in direct protein–protein interactions between pre-mRNA splicing factors can act as effective splicing inhibitors. This work also indicates that peptides could be potentially very useful tools in determining the functions of interacting proteins in the splicing mechanism and may facilitate the future development of splicing inhibitors that can be used to specifically block pre-mRNA splicing in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank all our colleagues in the Lamond laboratory for their helpful comments and discussions. A.I.L. is a Wellcome Trust Principal Research Fellow.
REFERENCES
- 1.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 15–26. [DOI] [PubMed] [Google Scholar]
- 2.Krämer A. (1995) The biochemistry of pre-mRNA splicing. In Lamond,A.I. (ed.), Pre-mRNA Processing. Springer-Verlag, Heidelberg, Germany, pp. 39–64. [Google Scholar]
- 3.Krämer A. (1996) The structure and function of proteins in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 357–409. [DOI] [PubMed] [Google Scholar]
- 4.Will C.L. and Lürhmann,R. (1997) snRNP structure and function. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 130–173. [Google Scholar]
- 5.Reed R. and Palandjian,L. (1997) Spliceosome assembly. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. Oxford University Press, New York, NY, pp. 103–129. [Google Scholar]
- 6.Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- 7.Rappsilber J., Ryder,U., Lamond,A.I. and Mann,M. (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res., 12, 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Z., Licklider,L.J., Gygi,S.P. and Reed,R. (2002) Comprehensive proteomic analysis of the human spliceosome. Nature, 419, 182–185. [DOI] [PubMed] [Google Scholar]
- 9.McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohi M.D., Link,A.J., Ren,L., Jennings,J.L., McDonald,W.H. and Gould,K.L. (2002) Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors and snRNAs. Mol. Cell. Biol., 22, 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarn W.Y., Hsu,C.H., Huang,K.T., Chen,H.R., Kao,H.Y., Lee,K.R. and Cheng,S.C. (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J., 13, 2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai W.Y., Chow,Y.T., Chen,H.R., Huang,K.T., Hong,R.I., Jan,S.P., Kuo,N.Y., Tsao,T.Y., Chen,C.H. and Cheng,S.C. (1999) Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem., 274, 9455–9462. [DOI] [PubMed] [Google Scholar]
- 13.Ajuh P., Kuster,B., Panov,K., Zomerdijk,J.C., Mann,M. and Lamond,A.I. (2000) Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J., 19, 6569–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein H.S. and Coughlin,S.R. (1997) Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J. Biol. Chem., 272, 5833–5837. [DOI] [PubMed] [Google Scholar]
- 15.Ohi R., McCollum,D., Hirani,B., Den Haese,G.J., Zhang,X., Burke,J.D., Turner,K. and Gould,K.L. (1994) The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J., 13, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei X.H., Shen,X., Xu,X.Q. and Bernstein,H.S. (2000) Human Cdc5, a regulator of mitotic entry, can act as a site-specific DNA binding protein. J. Cell Sci., 113, 4523–4531. [DOI] [PubMed] [Google Scholar]
- 17.Fong H.K., Hurley,J.B., Hopkins,R.S., Miake-Lye,R., Johnson,M.S., Doolittle,R.F. and Simon,M.I. (1986) Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl Acad. Sci. USA, 83, 2162–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neer E.J., Schmidt,C.J., Nambudripad,R. and Smith,T.F. (1994) The ancient regulatory-protein family of WD-repeat proteins. Nature, 371, 297–300. [DOI] [PubMed] [Google Scholar]
- 19.Nemeth K., Salchert,P., Putnoky,R., Bhalerao,Z., Konc-Kalman,B., Stankovic-Stangeland,L., Bako,J., Mathur,L., Okresz,S., Stabel,P. et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev., 12, 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhalerao R.P., Salchert,K., Bako,L., Okresz,L., Szabados,L., Muranaka,T., Machida,Y., Schell,J. and Koncz,C. (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl Acad. Sci. USA, 96, 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajuh P., Sleeman,J., Chusainow,J. and Lamond,A.I. (2001) A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem., 276, 42370–42381. [DOI] [PubMed] [Google Scholar]
- 22.Konarska M.M. and Sharp,P.A. (1987) Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell, 49, 763–774. [DOI] [PubMed] [Google Scholar]
- 23.Ryder U., Sproat,B.S. and Lamond,A.I. (1990) Sequence-specific affinity selection of mammalian splicing complexes. Nucleic Acids Res., 18, 7373–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamond A.I., Konarska,M.M. and Sharp,P.A. (1987) A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev., 1, 532–543. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 26.Ajuh P., Chusainow,J., Ryder,U. and Lamond,A.I. (2002) A novel function for human factor C1 (HCF-1), a host protein required for herpes simplex virus infection, in pre-mRNA splicing. EMBO J., 21, 6590–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohi M.D. and Gould,K.L. (2002) Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA, 8, 798–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H.R., Tsao,T.Y., Chen,C.H., Tsai,W.Y., Her,L.S., Hsu,M.M. and Cheng,S.C. (1999) Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 5406–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H.R., Jan,S.P., Tsao,T.Y., Sheu,Y.J., Banroques,J. and Cheng,S.C. (1998) Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol., 18, 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarn W.Y., Lee,K.R. and Cheng,S.C. (1993) Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl Acad. Sci. USA, 90, 10821–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarn W.Y., Lee,K.R. and Cheng,S.C. (1993) The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol., 13, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]