Abstract
Borrelia burgdorferi lipoprotein Lp6.6 is a differentially produced spirochete antigen. An assessment of lp6.6 expression covering representative stages of the infectious cycle of spirochetes demonstrates that the gene is solely expressed during pathogen persistence in ticks. Deletion of lp6.6 in infectious B. burgdorferi did not influence in vitro growth, or its ability to persist and induce inflammation in mice, migrate to larval or nymphal ticks or survive through the larval-nymphal molt. However, Lp6.6-deficient spirochetes displayed significant impairment in their ability to transmit from infected ticks to naïve mice, which was restored upon genetic complementation of the mutant with a wild-type copy of lp6.6, establishing that Lp6.6 plays a role in pathogen transmission from ticks to mammals. Lp6.6 is a subsurface, yet highly abundant, outer membrane antigen. Two-dimensional blue native/SDS-PAGE coupled with liquid chromatography-mass spectrometry (LC-MS/MS) analysis and protein cross-linking studies independently shows that Lp6.6 exists in multiple protein complexes in the outer membrane. We speculate that the function of Lp6.6 is connected to the physiological processes of these membrane complexes. Further characterization of differentially produced membrane antigens and associated protein complexes will likely aid in our understanding of the molecular details of B. burgdorferi persistence and transmission through a complex enzootic cycle.
Introduction
Lyme borreliosis is a highly prevalent tick-borne disease in the USA, Europe and many parts of Asia (Piesman and Eisen, 2008). Borrelia burgdorferi, the spirochetal agent of Lyme borreliosis, is maintained in nature via a complex enzootic life cycle, which involves wild rodents and ticks of the genus Ixodes. The pathogen contains a highly unique segmented and unstable genome (Fraser et al., 1997; Casjens et al., 2000) that differs significantly even from closely related pathogenic spirochetes (Porcella and Schwan, 2001). To persist in its natural transmission cycle, B. burgdorferi must adapt physiologically to markedly different host and vector tissue environments. A significant portion of the B. burgdorferi genome encodes hypothetical proteins of undefined functions (Fraser et al., 1997; Casjens et al., 2000), many of which are responsive to changes in the surrounding environment (Caimano et al., 2007) and differentially expressed during mammalian- or arthropod-specific phases of the spirochete life cycle (de Silva and Fikrig, 1997), contributing to microbial persistence or transmission (Grimm et al., 2004; Pal et al., 2004; Yang et al., 2004; Ramamoorthi et al., 2005; Seshu et al., 2006; Li et al., 2007; Neelakanta et al., 2007). As Lyme borreliosis may be difficult to diagnose (Aguero-Rosenfeld et al., 2005) and a human vaccine to prevent the incidence of the infection is not available, characterization of antigens that impact B. burgdorferi persistence through the vector–host transmission cycle is important for the development of preventative and therapeutic measures against the disease.
The gene product of the B. burgdorferi bba62 locus, annotated as a 6.6 kDa lipoprotein (Lp6.6), was originally described as an abundant, phenol-chloroform-petroleum ether-extractable low-molecular-weight lipoprotein (Katona et al., 1992). Subsequent studies further identified Lp6.6 as an outer membrane (OM)-associated antigen, which appeared to be downregulated during mammalian infection (Lahdenne et al., 1997). Lp6.6 is highly conserved among major B. burgdorferi isolates 297, N40 and B31. Lp6.6 production is regulated by alterations in the environment, such as changes in temperature (Ojaimi et al., 2002) and pH (Yang et al., 2001), exposure to blood in the culture medium (Tokarz et al., 2004), growth in host-implanted dialysis membrane (Akins et al., 1998; Brooks et al., 2003; Caimano et al., 2007), or in the murine host (Lahdenne et al., 1997). Collectively, these studies established that lp6.6 expression follows a prototypic ‘ospA’-like expression (Caimano et al., 2007) where alternative sigma factor RpoS is required for the repression of both ospA and lp6.6 in vivo (Caimano et al., 2005), and favours the notion that the function of Lp6.6 could be linked to the arthropod phases of the spirochete enzootic life cycle. Despite all these studies, the detailed expression profile of lp6.6 in the spirochete infection cycle has not previously been studied and its role in B. burgdorferi infectivity is unknown.
Although Lp6.6 is an abundant lipoprotein in cultured spirochetes and associated with the microbial OM (Katona et al., 1992; Lahdenne et al., 1997), the antigen is not likely exposed on the microbial surface (Lahdenne et al., 1997), and thus, may not directly participate in host–pathogen interaction. Certain cellular proteins, including small OM bacterial lipoproteins (Sklar et al., 2007; Lewis et al., 2008), often assemble into protein complexes (Alberts, 1998) that carry out specific roles in microbial biology including energy generation, protein assembly, lipoprotein trafficking and small molecule transport, which contributes to microbial pathogenesis and survival in the host environment (Stenberg et al., 2005; Pyndiah et al., 2007). As Lp6.6 is abundant in the spirochete membrane, we have assessed if Lp6.6 forms protein complexes, and determined whether Lp6.6 function is required for B. burgdorferi persistence through an experimental tick–mouse infection cycle. The characterization of membrane antigens that are differentially expressed during the host- or vector-specific pathogen life cycle is important for the development of novel strategies to interfere with B. burgdorferi transmission and prevention of Lyme borreliosis.
Results
Expression of lp6.6 throughout the mouse–tick infection cycle of B. burgdorferi
Borrelia burgdorferi bba62 encodes for a major membrane lipoprotein, annotated as Lp6.6, that appears to be downregulated during mammalian infection (Lahdenne et al., 1997). To understand the role of Lp6.6 in the spirochete enzootic life cycle, we assessed the temporal and spatial expression of lp6.6 throughout representative stages of the infectious cycle of B. burgdorferi using ticks and murine hosts. C3H/HeN mice were infected with B. burgdorferi and skin, joint, heart and bladder samples were collected following 2 weeks of infection. Larval and nymphal ticks were fed on parallel groups of mice following 2 weeks of infection (25 ticks per mouse) and engorged ticks were isolated at 3 days of feeding. One group of fed intermolt larvae were allowed to molt to nymphs and analysed as infected unfed nymphs. Another parallel group of unfed infected nymphs were allowed to feed on naïve mice (25 ticks per mice), and their gut and salivary glands were isolated at 2 days of feeding. Total RNA was prepared from murine and tick samples, and subjected to quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis to measure lp6.6 transcripts. As lp6.6 has been speculated to follow ‘ospA’-like expression (Caimano et al., 2007), the same set of tick samples were also assessed for ospA transcripts. The results supported a previous study (Lahdenne et al., 1997) showing that lp6.6 transcripts are undetectable in infected murine tissues (Fig. 1). lp6.6 expression is upregulated as soon as B. burgdorferi enters ticks, either larvae or nymphs, from infected mice and, similarly to ospA, remains highly expressed throughout the tested stages of the spirochete life cycle in the vector. However, unlike ospA, the levels of lp6.6 transcripts were abundant during transmission of B. burgdorferi from ticks to the murine host.
Fig. 1.
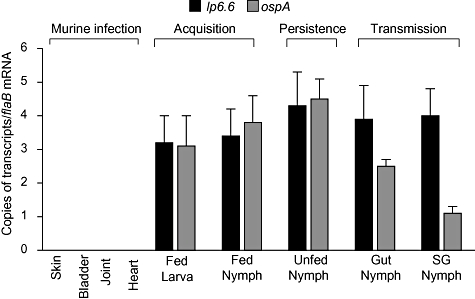
Expression of lp6.6 and ospA in representative stages of B. burgdorferi enzootic life cycle. The relative expression levels of lp6.6 during murine infectivity, acquisition and persistence in larval and nymphal ticks, and transmission through infected nymphs are analysed. Expression of ospA was also assessed in the tick samples. Transcript levels of lp6.6 and ospA were measured using quantitative RT-PCR and presented as copies of target transcripts per copy of the flaB transcript. RNA was isolated from mice at 2 weeks after B. burgdorferi infection, from larvae and nymphs at 3 days of feeding on B. burgdorferi-infected mice or from freshly molted unfed infected nymphs, and from gut and salivary glands (SG) of B. burgdorferi-infected nymphs feeding on naïve mice at 2 days of feeding. The bars represent the mean values and the error bars represent the SEM values from six quantitative RT-PCR analyses of two independent murine-tick infection experiments.
Generation and characterization of Lp6.6-deficient B. burgdorferi
To further study the role of Lp6.6 in the B. burgdorferi life cycle, we created Lp6.6-deficient B. burgdorferi. An infectious B. burgdorferi isolate was used to create an isogenic mutant by exchanging the lp6.6 (bba62) open reading frame with a kanamycin-resistance cassette via homologous recombination (Fig. 2A). Out of five transformed clones that grew in antibiotic-containing media, one clone was selected through polymerase chain reaction (PCR) analysis of the desired integration of the antibiotic cassette (Fig. 2B) and retention of all endogenous plasmids present in the parental isolate (data not shown). RT-PCR analysis showed that the mutant failed to produce lp6.6 mRNA, and that mutagenesis did not impose polar effects on the transcription of the immediate upstream gene bba61 (Fig. 2C). Transcription of the downstream gene, bba63, which has a small open reading frame (126 nucleotides) with a highly repetitive DNA sequence, could not be evaluated; however, a polar effect is unlikely due to its opposite transcriptional direction to that of lp6.6. The protein profile of the lp6.6 mutant was similar to that of the wild-type spirochete (Fig. 2D, left), and the mutant failed to produce Lp6.6 protein (Fig. 2D, right). Compared with parental isolates, the lp6.6 mutant displayed a similar growth rate when cultured in vitro at 33°C (Fig. 2E) or at 23°C (data not shown).
Fig. 2.
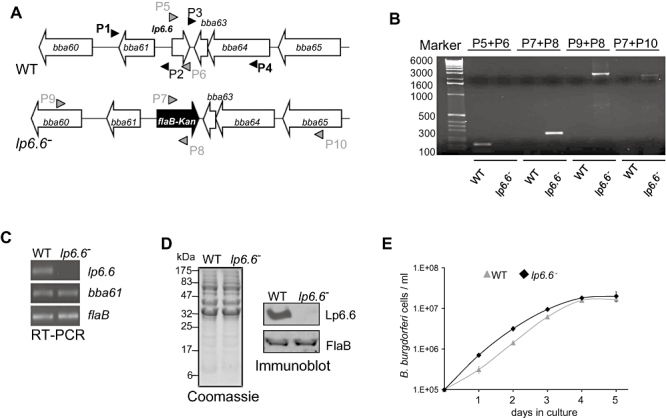
Construction and analysis of the lp6.6 mutant B. burgdorferi. A. Schematic drawings of the wild-type (WT) and the lp6.6 mutant (lp6.6−) isolates at the lp6.6 locus. Genes bba60–bba65 (white box arrows) and the kanamycin-resistance cassette driven by the B. burgdorferi flaB promoter (flaB-Kan, black box arrow) are indicated. Primers P1–P4 (black arrowheads) were used to amplify 5′ and 3′ arms for homologous recombination and ligated on either side of the flaB-Kan cassette as detailed in the text. B. Desired integration of the mutagenic construct, flaB-Kan, in lp6.6 genomic locus. Primers 5–10 (grey arrowheads, positions indicated in A) were used for DNA amplifications in wild type or lp6.6 mutant and subjected to gel electrophoresis. The combination of primers used for PCR is indicated at the top, and migration of the DNA ladder is shown on the left. C. RT-PCR analysis of lp6.6 deletion and the polar effect of mutagenesis. Isolated cDNA was used to amplify regions within lp6.6, flaB, and the gene downstream of lp6.6 locus (bba61) and visualized on a gel. D. Protein analysis of B. burgdorferi. Equal amounts of proteins from wild type and lp6.6 mutant were separated on a SDS-PAGE gel, and either stained with Coomassie blue (left) or transferred onto a nitrocellulose membrane and probed with Lp6.6 and FlaB antibodies (right). Migration of protein standards is shown to the left in kDa. E. Growth curves for the wild-type and lp6.6 mutant spirochetes. Spirochetes were diluted to a density of 105 cells ml−1, grown at 33°C in BSK-H medium, and counted under a dark-field microscope every 24 h using a Petroff–Hausser cell counter. Differences between wild type and lp6.6 mutant numbers were insignificant at all times of growth (P > 0.05).
lp6.6 mutants remain infectious in mice
To examine whether the lack of lp6.6 influences B. burgdorferi infectivity in mammals, C3H/HeN mice (five animals per group) were inoculated intradermally with equal numbers of wild-type or lp6.6 mutant B. burgdorferi (105 spirochetes per mouse). B. burgdorferi infection was assessed by qRT-PCR analysis of viable pathogen burden in murine skin, heart, bladder and joint samples isolated after 1, 2, 3 and 12 weeks of infection. Murine spleen samples were collected at the same time points and spirochete viability was further assessed by culture analysis. Results indicated insignificant differences between burdens of wild type and lp6.6 mutants in a diverse range of tissues (skin, heart, bladder and joint) and phases of infection, between 1 and 3 weeks (Fig. 3A) or even at 12 weeks of infection (data not shown). Similarly, both the mutant and wild-type spirochetes were isolated by culture of infected spleen (data not shown). As suggested by the similar pathogen burdens, mice infected with wild-type or lp6.6 mutant B. burgdorferi developed similar levels of disease, as evaluated by the development of ankle swelling (Fig. 3B) and histopathological signs of arthritis (data not shown).
Fig. 3.
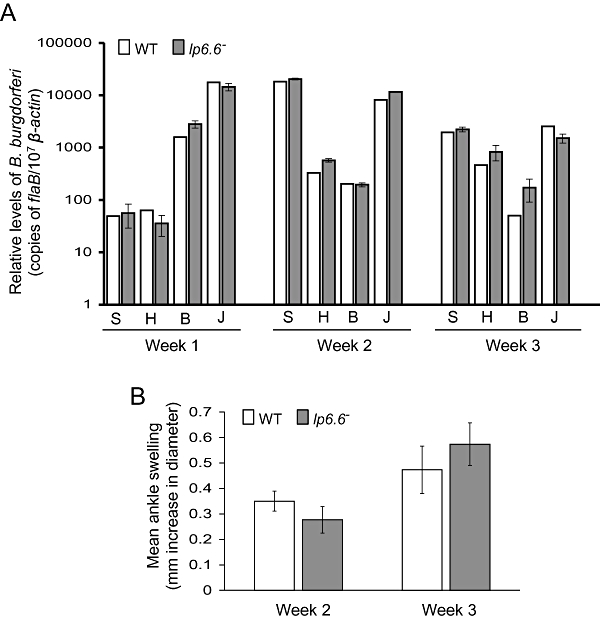
lp6.6 mutant B. burgdorferi retains murine infectivity. A. The B. burgdorferi burdens in multiple tissues of infected mice are shown. Mice (five animals per group) were infected with either the wild-type or the lp6.6 mutant B. burgdorferi and spirochete burdens were analysed in skin (S), heart (H), bladder (B) and joint (J) samples by measuring copies of B. burgdorferi flaB RNA at weeks 1, 2 and 3 following infection. Amounts of murine β-actin mRNA were determined in each sample and used to normalize the quantities of spirochete RNA. The bars represent the mean values and the error bars represent the SEM values from four quantitative RT-PCR analyses from two independent infection experiments. Differences between wild-type and lp6.6 mutant burdens were statistically insignificant in any tissue or time point measured (P > 0.05). B. Severity of joint swelling in B. burgdorferi-infected mice. Groups of mice (five animals per group) were separately infected with wild type or lp6.6 mutant and inflammation was evaluated by the assessment of joint swelling following weeks 2 and 3 of spirochete challenge using a digital caliper. The bars represent the mean values and the error bars represent the SEM values from two independent infection experiments. Wild type and lp6.6 mutant induced similar joint swelling (P > 0.05).
lp6.6 mutants display no defects in acquisition and persistence in ticks
We next determined whether Lp6.6-deficient B. burgdorferi could efficiently migrate from the infected murine host to the arthropod vector and then persist within Ixodes scapularis. To achieve this, Ixodes larval and nymphal ticks (25 ticks per group) were allowed to engorge on parallel groups of mice after 12 days of infection with wild-type or lp6.6 mutant B. burgdorferi. The ticks were collected following 1, 2 and 3 days of feeding and also during intermolt stages, 7 and 25 days after engorgement. The samples were subjected to qRT-PCR analyses to assess viable spirochete burdens. The results indicated no significant difference in the burdens of lp6.6 mutant and wild-type isolates in any stages of spirochete acquisition and persistence in larval or nymphal ticks (Fig. 4). Overall, these observations suggest that despite highly detectable lp6.6 expression (Fig. 1), Lp6.6 is not required for the acquisition or persistence of B. burgdorferi in ticks.
Fig. 4.
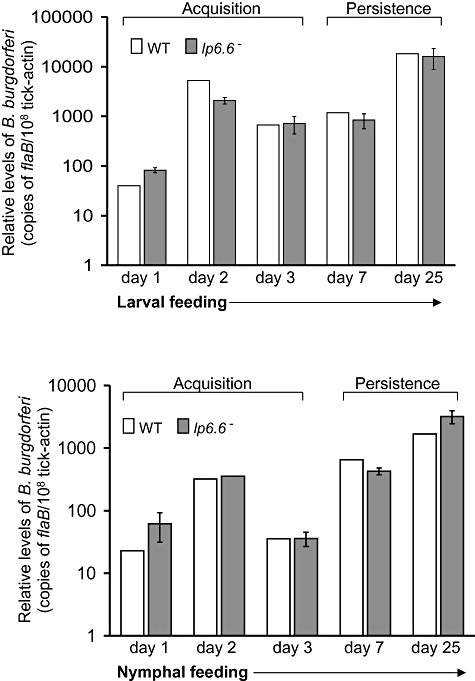
lp6.6 mutant B. burgdorferi displays no defects in its ability to enter and persist in ticks. Mice were infected with B. burgdorferi (two mice per group) and following 12 days of infection, naïve I. scapularis larvae or nymphs (25 ticks per group) were allowed to feed on mice, and B. burgdorferi burdens in ticks were analysed at the indicated time intervals following feeding by measuring copies of the B. burgdorferi flaB RNA. Amounts of tick β-actin mRNA were determined in each sample and used to normalize the quantities of spirochete RNA. The bars represent the mean values and the error bars represent the SEM values from two independent experiments. Burdens of wild type and lp6.6 mutant are similar at all time points (P > 0.05).
lp6.6 mutant displays a phenotypic defect in transmission from infected ticks to naïve mice
We finally assessed whether Lp6.6 is required for B. burgdorferi transmission from ticks to mice. To mimic the natural transmission process, we first generated B. burgdorferi-infected nymphs by allowing larvae to acquire spirochetes from infected mice and then molt into B. burgdorferi-infected nymphs in the laboratory. When infected nymphs were allowed to feed on naïve C3H mice, the burden of lp6.6 mutants declined in ticks at 2 days of feeding. To rule out the possibility that the potential phenotypic defects of the mutant during the transmission process was the result of anomalous effects of genetic manipulation, we sought to complement the lp6.6 mutant spirochetes with a wild-type copy of the lp6.6 gene, and use this isolate in tick–mouse transmission studies. For stable integration of the complemented construct, lp6.6, along with an antibiotic resistance cassette, was inserted into an intergenic chromosomal locus in B. burgdorferi. As our efforts for complementation of lp6.6 mutant involving the native promoter failed to yield any transformants, we sought to complement the mutant using the B. burgdorferi flaB promoter as described (Yang et al., 2009). To accomplish this, we first fused the open reading frame of lp6.6 with the flaB promoter and cloned it into the pKFSS1 vector (Frank et al., 2003), which carries an aadA streptomycin resistance cassette. A DNA element encompassing the flaB–lp6.6 fusion, along with the aadA cassette, was then assembled into the recombinant plasmid pXLF14301-plp6.6 (Fig. 5A) and integrated into B. burgdorferi genome via allelic exchange. Five transformants that grew in BSK medium containing both kanamycin and streptomycin were isolated. PCR analyses further identified one of the lp6.6-complemented clones, which retained the endogenous plasmids similar to those present in the parental isolate (data not shown). RT-PCR and immunoblotting showed that the lp6.6-complemented isolate produced both lp6.6 mRNA (Fig. 5B) and Lp6.6 protein (Fig. 5C).
Fig. 5.
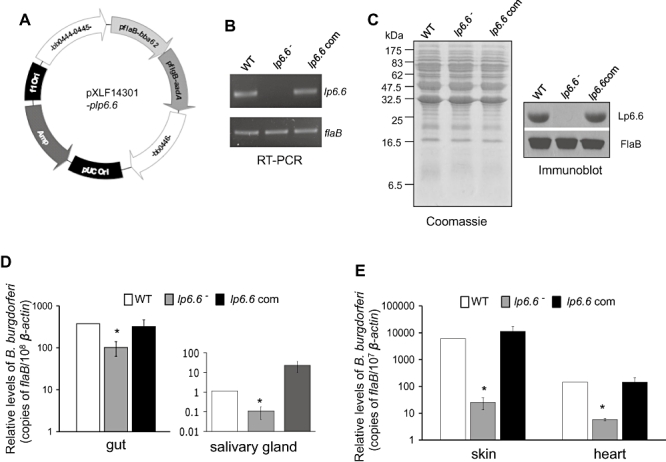
Complementation of lp6.6 mutant B. burgdorferi with lp6.6 restores the phenotypic defects of the mutant to transmit from ticks to mice. A. Construction of plasmid DNA pXLF14301-plp6.6 for cis-integration of the lp6.6 gene. The open reading frame of lp6.6 gene was fused with the flaB promoter and cloned into the shuttle vector pKFSS1 that houses the streptomycin resistance gene (aadA) under B. burgdorferi flgB promoter. The flaB promoter–lp6.6 gene fusion and aadA cassette was finally cloned into pXLF14301, which contains DNA fragments for homologous recombination and integration of the complemented gene in the B. burgdorferi genome. B. RT-PCR analysis of the lp6.6 transcripts. Total RNA was isolated from either the wild-type (WT), lp6.6 mutant (lp6.6−) or lp6.6-complemented B. burgdorferi (lp6.6 com), converted to cDNA, then subjected to PCR analysis with flaB and lp6.6 primers, and analysed on an agarose gel. C. Restoration of Lp6.6 protein by the complemented B. burgdorferi. Lysates of B. burgdorferi were separated on a SDS-PAGE gel, which was either stained with Coomassie blue or transferred to nitrocellulose membrane, and probed with antiserum against Lp6.6 or FlaB. D. B. burgdorferi transmission through infected ticks. B. burgdorferi-infected nymphs were generated by feeding larvae on mice infected with wild-type and genetically manipulated B. burgdorferi (two mice per group) as described in the text. Newly molted B. burgdorferi-infected nymphs were allowed to feed on naïve mice (five ticks per mouse, five animals per group). B. burgdorferi burdens were assessed in dissected tick guts (left) or salivary glands (right) at 2 days of feeding by measuring copies of the B. burgdorferi flaB RNA and normalized against tick β-actin RNA levels. E. B. burgdorferi transmission from infected ticks to mice. B. burgdorferi-infected nymphs were fed on mice as described in (D) and spirochete levels were assessed in the indicated murine tissues at 7 days of tick feeding. The bars represent the mean values and the error bars represent the SEM values from two independent animal infection experiments. Burdens of wild type and lp6.6-complemented mutants are significantly higher than corresponding levels of lp6.6 mutants (*P < 0.05).
We then compared the ability of the wild-type, lp6.6 mutant and complemented spirochetes to transmit from infected ticks to naïve mice. Naturally infected nymphs were generated by allowing larval ticks to acquire spirochetes from mice infected with wild-type or genetically manipulated B. burgdorferi. Infected ticks were allowed to feed on naïve C3H mice (five ticks per mouse), collected at 1, 2 and 4 days of feeding and viable spirochete burdens were assessed by qRT-PCR analysis. The results showed that, while differences between the burden of wild-type and mutant spirochetes were insignificant in ticks collected at 1 or 4 days of feeding (data not shown), the levels of Lp6.6-deficient B. burgdorferi, compared with wild-type and lp6.6-complemented spirochetes, were significantly lower in the gut (Fig. 5D, left) and salivary glands (Fig. 5D, right) collected at 2 days of feeding. Spirochete burdens in mice were assessed by qRT-PCR at an early time point of infection, 7 days of tick feeding, which indicated that the lp6.6 mutant is significantly impaired in transmission to mice (Fig. 5E), whereas lp6.6-complemented isolates were transmitted to the host at comparable levels to the wild type. In spite of impaired transmission, the lp6.6 mutant was recovered by culture analyses of infected murine tissues (data not shown) suggesting that while Lp6.6 is not essential, it facilitates B. burgdorferi transmission through feeding ticks to the murine host.
Lp6.6 is a member of multiple protein complexes in the OM
Although the above studies indicate that Lp6.6 plays a role in spirochete transmission, the function of Lp6.6 in the biology of B. burgdorferi is unknown. Lp6.6 expression in cultured spirochetes is highly sensitive to environmental alterations and is differentially expressed in vivo (Fig. 1); however, the antigen lacks surface exposure (Fig. 6A) and thus is unlikely to be directly involved in pathogen interaction with the surrounding environment. While Lp6.6 was previously identified as an OM lipoprotein (Katona et al., 1992; Lahdenne et al., 1997), we assessed the precise cellular distribution of Lp6.6. To accomplish this, cultured spirochetes were separated into two major fractions, outer membrane vesicle (OMV) and protoplasmic cylinder (PC), and subjected to immunoblot analyses with anti-Lp6.6, -OspA and -FlaB antibodies. While FlaB was almost undetectable in the OM, both OspA and Lp6.6 were abundant in the OM (Fig. 6B).
Fig. 6.
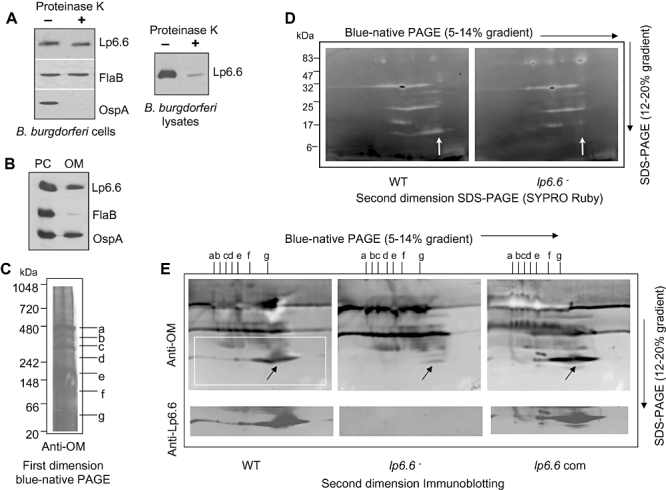
Lp6.6 is localized in the B. burgdorferi outer membrane (OM) with subsurface topology and is a member of multiple membrane protein complexes. A. Lp6.6 is insensitive to proteinase K-mediated degradation of B. burgdorferi surface proteins. Viable spirochetes were incubated with (+) or without (−) proteinase K for the removal of protease-sensitive surface proteins and processed for immunoblot analysis using Lp6.6 antibodies (left). B. burgdorferi OspA and FlaB antibodies were utilized as controls for surface-exposed and subsurface proteins respectively. Lp6.6 is highly sensitive to proteinase K digestion in lysed preparations of B. burgdorferi (right). B. Cellular Lp6.6 is found in the isolated OM of cultured spirochetes. B. burgdorferi protoplasmic cylinders (PC) and OM were separated by sucrose density gradient centrifugation and equal amounts of protein from two subcellular fractions were separated by SDS-PAGE, and immunoblotted with Lp6.6, OspA and FlaB antiserum. C. OM protein complexes separated by one-dimensional blue-native (BN) gel. OM protein complexes from wild-type B. burgdorferi were separated in 5–14% BN-PAGE as described in the text, transferred onto a nitrocellulose membrane and probed with OM antibodies. Identified protein complexes were labelled from a to g. D. Two-dimensional analysis of OM protein complexes in wild type and lp6.6 mutant. Strips of BN-PAGE were excised and placed on the top of the 12–20% denaturing gels and stained by SYPRO Ruby. The vertical arrow indicates the position of Lp6.6. E. Immunoblots of 2D-BN/SDS-PAGE from isolated OM from wild-type, lp6.6 mutant and lp6.6-complemented (com) isolates using antibodies specific for OM and Lp6.6. The arrow indicates the bands assigned to Lp6.6.
As Lp6.6 is abundant in the OM, we assessed whether the antigen participates in protein complexes, which contribute to membrane physiology and microbial pathogenesis. To explore this, we used two-dimensional blue native (2D-BN)/SDS-PAGE combined with mass spectrometry and protein cross-linking approaches to determine whether Lp6.6 is a member of membrane protein complexes. OM proteins from wild-type B. burgdorferi were separated using one-dimensional BN-PAGE and immunoblotting with anti-OM antibodies identified seven major protein complexes (Fig. 6C, labelled as a–g). The approximate molecular masses of the protein complexes ranged from 50 to 480 kDa. To identify if Lp6.6 participates in protein complex formation, OMV were isolated from both wild-type and genetically manipulated B. burgdorferi, separated in the first dimension through BN-PAGE and further analysed in the second dimension by SDS-PAGE, which resolved individual complexes into vertical ‘channels’, enabling visualization of the individual constituents of the complexes. Gels were either stained with SYPRO Ruby (Fig. 6D) or immunoblotted with OM or Lp6.6 antibodies (Fig. 6E), which indicated that Lp6.6 is a member of protein complexes. Liquid chromatography-mass spectrometry (LC-MS/MS) analysis further confirmed the presence of Lp6.6 in multiple protein complexes, where each complex (marked as a–g, Fig. 6C) contained matching Lp6.6 peptides with a minimum of 38% coverage of the protein. Unlike lp6.6 mutant, both wild-type and lp6.6-complemented B. burgdorferi displayed protein complexes containing Lp6.6 (Fig. 6E). To obtain independent experimental evidence that Lp6.6 forms membrane protein complexes, isolated OM from wild-type and Lp6.6-deficient B. burgdorferi was subjected to protein cross-linking analysis using dithiobis(succinmidylpropionate) (DSP), an amine-reactive, homobifunctional cross-linker with a spacer arm of 12 Å length, which contains a disulphide bond that can be cleaved with reducing agents, such as β-mercaptoethanol (βME). Immunoblot analysis with Lp6.6 indicated that DSP readily cross-linked Lp6.6 in OM isolated from wild-type spirochetes, where multiple high-molecular-weight cross-link products containing Lp6.6 were obtained (arrows, Fig. 7). Although the components of these Lp6.6-containing complexes, either different sizes of Lp6.6 polymers or multiprotein units, remain indistinguishable, DSP-mediated Lp6.6 cross-linking was specific as the products were completely dissociated into free proteins in the presence of βME (arrowhead, Fig. 7). Cross-linked products were also undetectable in OM isolated from lp6.6 mutant spirochetes (left lanes, Fig. 7).
Fig. 7.
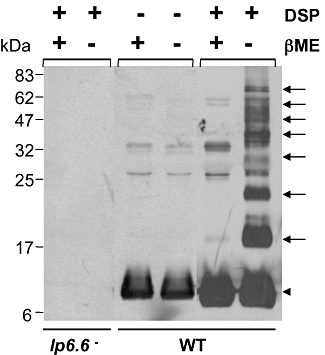
Outer membrane Lp6.6 forms high-molecular-weight complexes in the presence of a protein cross-linker. Outer membrane proteins were isolated from wild-type and Lp6.6-deficient B. burgdorferi and subjected to cross-linking with dithiobis(succinmidylpropionate) (DSP). DSP-treated samples were further incubated in the presence or absence of β-mercaptoethanol to cleave the DSP-mediated cross-linking, separated on SDS-PAGE gels and immunoblotted with Lp6.6 antibodies. Arrows indicated formation of multiple high-molecular-weight cross-linked protein complexes containing Lp6.6. Arrowhead donates non-cross-linked free Lp6.6 proteins.
Discussion
The present study establishes that in vivo expression of a small lipoprotein of B. burgdorferi, Lp6.6, is confined to the vector phases of the pathogen life cycle, and that while Lp6.6 is not essential for pathogen persistence, it facilitates the transmission of spirochetes from ticks to the murine host. We show that Lp6.6 is a constituent member of multiple protein complexes in the spirochete OM, indicating that the function of Lp6.6 may be related to as yet undefined aspects of membrane physiology.
Borrelia burgdorferi persists in a diverse range of tissue environments as it cycles between the arthropod vector and the mammalian host. Spirochetes alter their gene expression in response to alterations in in vitro growth conditions (Ojaimi et al., 2002; Revel et al., 2002; Brooks et al., 2003; Tokarz et al., 2004; Caimano et al., 2007) and in different phases of microbial persistence in mammalian hosts and ticks (Narasimhan et al., 2002; 2003). Many of these transcriptional modulations lead to changes in the antigenic composition of B. burgdorferi, including OM lipoproteins (Hefty et al., 2002), which directly face the surrounding environment and likely play important roles in microbial adaptation to a given microenvironment. A significant fraction of the B. burgdorferi genome encodes for proteins with export signals (Casjens et al., 2000; Schulze and Zückert, 2006), many of which are abundant in the membrane and produced in vivo (Nowalk et al., 2006; Barbour et al., 2008) and, thus, likely involved in spirochete adaptation. Lp6.6 is subsurface lipoprotein (Katona et al., 1992) that lacks a detectable antibody response in infected hosts (Lahdenne et al., 1997). Previous studies determined that overexpression of vector-specific borrelial genes in mice, such as ospA, by genetically manipulated isolates, induces robust and specific borreliacidal antibodies that severely impact B. burgdorferi persistence (Strother et al., 2007; Xu et al., 2008). However, continual lp6.6 expression by complemented mutants within the murine host did not significantly influence spirochete persistence (data not shown). Many of the borrelial membrane proteins that are differentially expressed in the vector or in hosts, such as OspA/B, BmpA/B, DbpA/B and CRASP-related proteins, are members of a common operon or paralogous gene family (Casjens et al., 2000), and in many instances are co-transcribed (Dobrikova et al., 2001; El-Hage and Stevenson, 2002). Although lp6.6 is a unique gene, its expression pattern mostly resembles that of ospA, and is likely regulated by similar environmental signals and genetic regulatory networks (Caimano et al., 2007). However, lp6.6 does not downregulate in parallel with ospA during nymphal feeding, raising the possibility that, at least during transmission, lp6.6 expression might be governed by an unknown regulatory pathway. Nevertheless, the consistent expression of lp6.6 within ticks, which is analogous to other borrelial genes important in the vector, such as ospA and ospB (Yang et al., 2004; Neelakanta et al., 2007), indicates that Lp6.6 function is likely important for the spirochete life cycle in ticks. While lp6.6 mutant did not display detectable growth deficiencies in vitro, transient defects in persistence within feeding-infected ticks were apparent (Fig. 5D). This is the phase at which spirochetes dramatically increase in number (de Silva and Fikrig, 1995) and disseminate from feeding ticks to the host (Piesman et al., 1987; Piesman, 1993; Ohnishi et al., 2001). The detection of lower numbers of lp6.6 mutants in the tick gut due to possibly more efficient transmission is unlikely, as burdens of lp6.6 mutants that exited the gut and migrated either to the salivary glands (Fig. 5D, right) or to the murine host (Fig. 5E) were significantly lower than wild-type and complemented isolates. Lp6.6 thus serves an unknown function during the transmission of spirochetes, which involves a series of carefully orchestrated yet poorly defined events including timely escape through gut epithelia and potential peritrophic barriers, dissemination via the haemolymph, invasion of the salivary gland and transfer to the mammalian dermis (Fikrig and Narasimhan, 2006). Spirochete proteins that are differentially produced in ticks, including Lp6.6 and other proteins regulated by the RpoN–RpoS pathway (Fisher et al., 2005), might play a role in B. burgdorferi transmission from ticks as well as in establishment of early host infection.
In cellular membranes, multiple proteins assemble into distinct protein complexes (Alberts, 1998), which perform specific roles in membrane physiology and contribute to microbial virulence (Stenberg et al., 2005; Pyndiah et al., 2007). We show that Lp6.6, although differentially expressed in the spirochete life cycle, is one of the prominent constituent members of all OM protein complexes. Both 2D-BN/SDS-PAGE and protein cross-linking analyses identified Lp6.6 as a participant in multiple protein complexes, with the highest abundance in low-molecular-weight complexes. However, we cannot rule out the possibility that the observed chemical cross-linking of Lp6.6 could be due to the abundance of antigen in the OM, and thus, opportunistic cross-linking to other membrane proteins. The appearance of Lp6.6-containing protein complexes in 2D-BN/SDS-PAGE as a continual smear is commonly observed in protein complexes of other bacteria (Krall et al., 2002; Stenberg et al., 2005; Pyndiah et al., 2007). The occurrence of protein complexes in the bacterial OM and their roles in pathogen biology vary among Gram-negative bacteria. For example, Helicobacter pylori OM contains only two multiprotein complexes (Pyndiah et al., 2007), fumarate reductase and urease enzyme complexes, composed of 10 individual proteins and primarily involved in the energy metabolism, which may not be essential for H. pylori survival in vitro but are required for pathogen colonization in the host gut (Ge et al., 2000). Escherichia coli OM, on the other hand, contains nine homo- or hetero-oligomeric complexes composed of a total of 12 participating proteins and each complex carries a specific function related to general diffusion, drug transport, stationary-phase adaptation and biogenesis or integrity of the OM (Stenberg et al., 2005). We have identified seven protein complexes in the OM of wild-type spirochetes, which are also maintained in lp6.6 mutants. Therefore, Lp6.6 is probably not a core member of the protein complexes required for their assembly, but may provide an auxiliary role, for example, by maintaining complex stability, as shown for a small OM lipoprotein in E. coli (Sklar et al., 2007) and Salmonella enterica (Lewis et al., 2008). Similarly, a non-essential yet supportive role of Lp6.6 in the spirochete infection cycle was also shown by our mutagenesis studies.
The B. burgdorferi membrane undergoes dramatic antigenic variation in vivo, due to sequence-specific recombination (Coutte et al., 2009) and differential gene expression (Schwan, 2003) as it cycles through the tick–rodent infection cycle. Here we show at least one of these differentially produced antigens, Lp6.6, to be a member of the membrane protein complexes of cultured organisms. We do not know the biogenesis, stability or contributions to spirochete infectivity of these complexes. Future characterization of the spirochete membrane protein complexome, identification of interacting proteins besides Lp6.6, and their roles in pathogen biology and infectivity may provide new insights into the molecular details of B. burgdorferi survival through a complex enzootic cycle.
Experimental procedures
B. burgdorferi, ticks and mice
A clonal and low-passage infectious isolate of B. burgdorferi, B31-A3 (Elias et al., 2002), was used throughout this study. Four- to six-week-old C3H/HeN mice were purchased from the National Institutes of Health. I. scapularis ticks used in this study originated from a colony that is maintained in the laboratory (Coleman et al., 2008). All animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee and Institutional Bio-safety Committee of the University of Maryland, College Park.
Polymerase chain reaction
The oligonucleotide sequences for each of the primers used in specific PCR reactions are indicated in Table S1. Total RNA was isolated using TRIzol reagent (Invitrogen), reverse transcribed to cDNA (AffinityScript, Stratagene), and RT-PCR or qRT-PCR analysis was performed as described (Coleman et al., 2008). Expression of lp6.6 was analysed in groups of five C3H/HeN mice (105 spirochetes per mouse) at 2 weeks after infection or in larval and nymphal ticks that fed on mice infected for 2 weeks (25 ticks per mouse) as detailed (Coleman et al., 2008). For lp6.6 and ospA expression analyses during spirochete acquisition and persistence in ticks, whole larva and nymph were analysed at 3 days of feeding or as freshly molted infected unfed nymphs. For expression analysis during transmission, B. burgdorferi-infected nymphs were fed on naïve mice (25 ticks per mice, two mice per group), and dissected guts and salivary glands were analysed at 2 days of feeding as described (Pal et al., 2004; Coleman et al., 2008). The levels of B. burgdorferi lp6.6 transcript in tick and mouse samples were normalized against flaB transcripts in the qRT-PCR reaction as detailed (Coleman et al., 2008).
Production of recombinant Lp6.6, generation and characterization of Lp6.6 antiserum
The lp6.6 gene was cloned into pGEX-4T-2 (Amersham-Pharmacia Biotech) using specific primers (Table S1) and the recombinant protein Lp6.6 without the N-terminal leader sequence was produced in E. coli. Expression and purification of the glutathione S-transferase (GST) fusion protein were performed as described previously (Pal et al., 2008a). Generation of polyclonal murine antiserum against recombinant Lp6.6–GST fusion protein and assessment of titre and specificity of the antisera, using ELISA and immunoblotting, were performed as described (Pal et al., 2008a).
Proteinase K accessibility assay
Proteinase K accessibility assays were performed as detailed (Coleman et al., 2008). Briefly, spirochetes (1 × 108) were suspended in phosphate-buffered saline (PBS) and split into two equal volumes. One aliquot received 200 mg ml−1 of proteinase K (Sigma) while the other aliquot received an equal volume of PBS without the enzyme. Both aliquots were incubated for 20 min, treated with phenylmethylsulphonylfluoride (Sigma), pelleted and re-suspended in PBS for immunoblot analysis using antibodies against Lp6.6, FlaB or OspA. To assess whether Lp6.6 is sensitive to protease digestion, additional batches of spirochetes (1 × 108) were sonicated, and equal amounts of solubilized proteins were incubated in the absence and presence of proteinase K, and immunoblotted with Lp6.6 antibodies.
Purification of OM, generation and characterization of anti-OM antiserum
Isolation of the OM of B. burgdorferi was performed as described (Skare et al., 1995). Briefly, 5 × 1010 to 1 × 1011B. burgdorferi cells were washed in PBS, pH 7.4, supplemented with 0.1% BSA. The cells were re-suspended in ice-cold 25 mM citrate buffer pH 3.2 containing 0.1% BSA and incubated on a rocker at room temperature for 2 h. OMV were released from whole cells and were isolated from protoplasmic cylinder (PC) by using sucrose density gradient centrifugation. The isolated OM was monitored for purity by immunoblotting using antibodies against OspA and FlaB. Immunoblotting results showed an enrichment of OspA in OM proteins and only with minor cross-contamination of FlaB. Generation of polyclonal OM antisera in rabbits, assessment of titre and specificity of the antisera using ELISA and immunoblotting were performed as described (Pal et al., 2008b).
2D-BN/SDS-PAGE and immunoblotting
Analysis of OM proteins was performed under native conditions by BN-PAGE as described (Schagger and von Jagow, 1991). Isolated membranes were solubilized with β-dodecyl maltoside (DM) (DM/protein = 20 w/w) and protein complexes were analysed at 4°C in 5–14% polyacrylamide gel. Native protein markers (Invitrogen) were used to estimate the size of protein complexes in BN gels. Subunit composition of the protein complexes was assessed by electrophoresis in denaturing 12–20% linear gradient polyacrylamide gels containing 7 M urea (Komenda et al., 2002). Pre-stained protein markers (New England BioLabs) were used for estimation of apparent molecular masses of proteins in SDS-PAGE gels. The whole lanes from the BN gel were excised, and either transferred onto nitrocellulose membrane for immunodetection with antibodies against OM or incubated for 30 min in 25 mM Tris/HCl, pH 7.5, containing 1% SDS (w/v) and 1% DTT (w/v), and placed on the top of the denaturing gel. Proteins separated in the gel were either stained by SYPRO Ruby (Invitrogen) or transferred onto nitrocellulose membrane for immunodetection with antibodies against OM and Lp6.6. Protein signals were visualized using ECL Western Blotting Detection Reagent (Amersham, UK).
Liquid chromatography-mass spectrometry (LC-MS/MS)
Samples for tandem mass spectrometry were prepared by tryptic in-gel digestion of excised protein bands as described (Williams and Stone, 1997). Briefly, protein complexes from one-dimensional BN-PAGE were excised and put into 1.5 ml tubes that were pre-washed with 500 μl of buffer containing 0.1% TFA and 60% acetonitrile (ACN). Gel bands were successively washed with 250 μl of 50% ACN, buffer containing 50% ACN, 50 mM NH4HCO3 and 10 mM NH4HCO3, dried with a speedvac and digested with sequencing grade trypsin (0.1 mg ml−1), further extracted with 50 μl of 50% ACN and 5% TFA and finally dried with a speedvac. LC-MS/MS analyses and protein identification were performed at the Proteomics Core Facility of College of Chemical and Life Sciences, University of Maryland. Briefly, tryptic digests were injected onto a Zorbax SB300 C18 column (1.0 × 100 mm, Agilent Technologies) connected to an Accela HPLC system (Thermo Electron) and interfaced to a Thermo Finnigan LTQOrbitrapXL mass spectrometer equipped with an Ion Max electrospray source. Separation of peptides was achieved by a gradient of 5–35% solvent B at 50 μl min−1 for 30 min. Solvent B contained 5% water in ACN with 0.1% formic acid. Precursor mass between m/z 350 and 3000 was scanned using the Orbitrap with resolution of 60 000 at m/z 400. LC-MS/MS data files were searched using Sequest search engine through Bioworks (Thermo Electron) and Mascot search engine through in-house Mascot Server (Matrix Science) against bacterial databases. Results from the two search engines were combined using Scaffold Distiller (Proteome Software) for identification of proteins.
Cross-linking
Chemical cross-linking analysis of Lp6.6-mediated protein complex formation was performed as described (Steiner et al., 2008) with minor modifications. Briefly, isolated OM preparations were incubated with dithiobis(succinimidylpropionate) (DSP) for 2 h on ice according to the instructions of the supplier (Pierce). Cross-linked proteins were incubated in the absence or presence of 5% β-mercaptoethanol for 10 min at 95°C and analysed by immunoblotting with the Lp6.6 antibodies.
Isolation and infection studies of lp6.6 mutant and complemented isolates of B. burgdorferi
Generation of B. burgdorferi mutants were performed using published procedures (Coleman et al., 2008). All primers used for genetic manipulation process are listed in Table S1. The lp6.6 mutant was constructed by exchanging the lp6.6 open reading frame with a kanamycin-resistance cassette via homologous recombination. The 5′ and 3′ arms flanking lp6.6 were amplified using primers P1–P4, then cloned into multiple-cloning sites flanking the kanAn cassette in plasmid pXLF10601 and electroporated into B. burgdorferi. Five clones that grew in antibiotic-containing media were further assessed using primers P5–P10 to confirm the desired integration of the flaBp-kan cassette into the B. burgdorferi genome. One of the lp6.6 mutants that retained the same set of plasmids as the wild-type isolate was selected for further experiments. Although the construct pXLF10601 (Li et al., 2006) contained an ampicillin resistance marker, no ampicillin-resistant transformants that might result from single-cross-over events were selected. PCR analysis further confirmed the absence of an ampicillin resistance marker in lp6.6 mutants and no differences in ampicillin sensitivity between the wild-type and mutant spirochetes were observed. For in vitro growth analysis, an equal number of wild type and lp6.6 mutants were diluted to a density of 105 cells ml−1 and grown at 23°C or 33°C in BSK-H medium until they reached the stationary phase (108 cells ml−1). Aliquots of spirochetes were enumerated using a Petroff–Hausser cell counter under a dark-field microscope.
Complementation of the lp6.6 mutant was achieved by re-insertion of a wild-type copy of the lp6.6 gene in the B. burgdorferi chromosome as detailed (Li et al., 2007; Yang et al., 2009). Briefly, two DNA inserts encompassing the lp6.6 open reading frame and the flaB promoter were PCR-amplified, fused and cloned into the BamHI and SalI sites of pKFSS1 (Frank et al., 2003) housing a streptomycin-resistance cassette (aadA). The flaB promoter–lp6.6 gene fusion construct along with aadA cassette was excised from the recombinant pKFSS1 plasmid and inserted into the corresponding restriction sites of the plasmid pXLF14301, which contains the required 5′ and 3′ arms for homologous recombination in the B. burgdorferi chromosomal locus bb0444–0446. The final construct was sequenced to confirm identity, and 25 μg of the recombinant plasmid was electroporated into the lp6.6 mutant. One of the five lp6.6-complemented clones that was able to grow in antibiotic medium was further selected based on the intended recombination event and the expression of lp6.6 mRNA and Lp6.6 protein. Analysis of wild-type, lp6.6 mutant and lp6.6-complemented isolates indicated the presence of the same set of endogenous plasmids.
For phenotypic analysis of wild-type and genetically manipulated spirochetes, burdens of viable pathogen in mice and ticks were assessed using qRT-PCR analysis of flaB mRNA and normalized against murine or tick β-actin genes as described (Coleman et al., 2008; Pal et al., 2008a). To assess whether flaB RNA- or flaB DNA-based quantitative analyses of spirochete burden produce similar results, qRT-PCR and qPCR analyses were performed using the same infected tissues. C3H mice (three animals per group) were infected with B. burgdorferi (105 spirochetes per mouse) and sacrificed at 3 weeks following inoculation, and skin, bladder, heart and joint samples were isolated. In parallel, nymphal ticks (20 ticks per mouse) were allowed to feed on mice infected for 12 days and collected as repleted ticks. Tissues were homogenized in liquid nitrogen, divided into two parts and separately processed for flaB DNA-based qPCR (Pal et al., 2004) and flaB RNA-based qRT-PCR (Coleman et al., 2008), which produced similar patterns in the differences of the tissue burdens of wild type and lp6.6 mutants (Fig. S1). Additionally, similar patterns in qPCR- and qRT-PCR-based detection of borrelial burdens were observed when spirochete levels were compared in multiples murine tissues at a later time point, 12 weeks after challenge (data not shown). As mRNA assessment has been shown to be a better surrogate for the detection of viable microbes (Sheridan et al., 1998; Bleve et al., 2003) and previous studies also indicated a general positive correlation between flaB mRNA levels and flaB DNA-based assessment of pathogen burdens (Hodzic et al., 2003), in subsequent studies, we measured flaB RNA using qRT-PCR analysis, rather than analysis of the DNA target. Furthermore, we have determined that deletion of lp6.6 did not affect flaB transcript levels, which are similar in wild-type and mutant B. burgdorferi (data not shown). To assess pathogen persistence in the murine host, C3H mice (five animals per group) were infected with B. burgdorferi (105 spirochetes per mouse) and sacrificed at 1, 2, 3 and 12 weeks following inoculation. The skin, bladder, heart and joint samples were isolated and stored in liquid nitrogen, and aliquots of blood and spleen tissues were cultured in BSK medium for further assessment of spirochete viability. Pathogen burdens were assessed in larval and nymphal ticks (five ticks per mouse) that fed on B. burgdorferi-infected mice after 12 days of infection. Naturally infected nymphs were also generated, allowed to feed on naïve mice (five ticks per mouse, five mice per group) and spirochete burdens in ticks were determined by qRT-PCR at 1, 2 and 4 days of feeding. B. burgdorferi burdens were assessed in intact salivary glands and the gut at day 2 of feeding, which is a biologically relevant time point of transmission (Piesman et al., 1987; Piesman, 1993) prior to the major blood meal when intact organs can be isolated with less possibility of cross-contamination (Sonenshine, 1993). For other time points, B. burgdorferi burdens were assessed in whole ticks without dissection. At 7 days of tick feeding, all the mice were sacrificed, and the skin and heart tissues were isolated and assessed for the spirochete burden by qRT-PCR and culture analysis. To assess if continual expression of lp6.6 by complemented isolates had any effect on borrelial persistence, parallel groups of C3H mice (six animals) were infected with wild-type or genetically manipulated isolates (105 spirochetes per mouse). After 2 weeks post infection, samples of skin, bladder, heart and joint were isolated and analysed for lp6.6 transcripts and pathogen burdens as described above.
Evaluation of disease
Borrelia burgdorferi-infected mice were examined for disease by the assessment of joint swelling and histological evaluation of inflammation as detailed earlier (Coleman et al., 2008; Pal et al., 2008a). Ankle joints of each mouse were measured using a precision metric caliper, and development of swelling was monitored weekly. For histology, at least five ankle joints from each group of mice (five animals per group) were collected and fixed in 10% formalin, decalcified and processed for Haematoxylin and Eosin staining. Sections were blindly examined for assessment of histological parameters of B. burgdorferi-induced inflammation as detailed (Pal et al., 2008a).
Statistical analysis
Results are expressed as the mean ± standard error (SEM). The significance of the difference between the mean values of the groups was evaluated by two-tailed Student's t-test.
Acknowledgments
This work was supported by funding from the National Institute Of Allergy and Infectious Diseases (Award No. R01AI080615) and by a Faculty Start-up Fund from the University of Maryland. We sincerely thank Adam Coleman, Xiuli Yang and Michael Vasil for their excellent assistance with this study.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aguero-Rosenfeld ME, Wang G, Schwartz I, Wormser GP. Diagnosis of Lyme borreliosis. Clin Microbiol Rev. 2005;18:484–509. doi: 10.1128/CMR.18.3.484-509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akins DR, Bourell KW, Caimano MJ, Norgard MV, Radolf JD. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Invest. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Jasinskas A, Kayala MA, Davies DH, Steere AC, Baldi P, et al. A genome-wide proteome array reveals a limited set of immunogens in natural infections of humans and white-footed mice with Borrelia burgdorferi. Infect Immun. 2008;76:3374–3389. doi: 10.1128/IAI.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleve G, Rizzotti L, Dellaglio F, Torriani S. Development of reverse transcription (RT)-PCR and real-time RT-PCR assays for rapid detection and quantification of viable yeasts and molds contaminating yogurts and pasteurized food products. Appl Environ Microbiol. 2003;69:4116–4122. doi: 10.1128/AEM.69.7.4116-4122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Gonzalez CA, Radolf JD. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J Bacteriol. 2005;187:7845–7852. doi: 10.1128/JB.187.22.7845-7852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Coleman AS, Yang X, Kumar M, Zhang X, Promnares K, Shroder D, et al. Borrelia burgdorferi complement regulator-acquiring surface protein 2 does not contribute to complement resistance or host infectivity. PLoS ONE. 2008;3:3010e. doi: 10.1371/journal.pone.0003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutte L, Botkin DJ, Gao L, Norris SJ. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 2009;5:e1000293. doi: 10.1371/journal.ppat.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova EY, Bugrysheva J, Cabello FC. Two independent transcriptional units control the complex and simultaneous expression of the bmp paralogous chromosomal gene family in Borrelia burgdorferi. Mol Microbiol. 2001;39:370–378. doi: 10.1046/j.1365-2958.2001.02220.x. [DOI] [PubMed] [Google Scholar]
- El-Hage N, Stevenson B. Simultaneous coexpression of Borrelia burgdorferi Erp proteins occurs through a specific, erp locus-directed regulatory mechanism. J Bacteriol. 2002;184:4536–4543. doi: 10.1128/JB.184.16.4536-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikrig E, Narasimhan S. Borrelia burgdorferi– traveling incognito? Microbes Infect. 2006;8:1390–1399. doi: 10.1016/j.micinf.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, et al. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci USA. 2005;102:5162–5167. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CE, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Ge Z, Feng Y, Dangler CA, Xu S, Taylor NS, Fox JG. Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb Pathog. 2000;29:279–287. doi: 10.1006/mpat.2000.0391. [DOI] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, et al. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci USA. 2004;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Akins DR. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 2002;70:3468–3478. doi: 10.1128/IAI.70.7.3468-3478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic E, Feng S, Freet KJ, Barthold SW. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect Immun. 2003;71:5042–5055. doi: 10.1128/IAI.71.9.5042-5055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona LI, Beck G, Habicht GS. Purification and immunological characterization of a major low-molecular-weight lipoprotein from Borrelia burgdorferi. Infect Immun. 1992;60:4995–5003. doi: 10.1128/iai.60.12.4995-5003.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Lupinkova L, Kopecky J. Absence of the psbH gene product destabilizes photosystem II complex and bicarbonate binding on its acceptor side in Synechocystis PCC 6803. Eur J Biochem. 2002;269:610–619. doi: 10.1046/j.0014-2956.2001.02693.x. [DOI] [PubMed] [Google Scholar]
- Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:11405–11410. doi: 10.1073/pnas.172390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahdenne P, Porcella SF, Hagman KE, Akins DR, Popova TG, Cox DL, et al. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect Immun. 1997;65:412–421. doi: 10.1128/iai.65.2.412-421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C, Skovierova H, Rowley G, Rezuchova B, Homerova D, Stevenson A, et al. Small outer-membrane lipoprotein, SmpA, is regulated by sigmaE and has a role in cell envelope integrity and virulence of Salmonella enterica serovar Typhimurium. Microbiology. 2008;154:979–988. doi: 10.1099/mic.0.2007/011999-0. [DOI] [PubMed] [Google Scholar]
- Li X, Liu X, Beck DS, Kantor FS, Fikrig E. Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect Immun. 2006;74:3305–3313. doi: 10.1128/IAI.02035-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, et al. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- Narasimhan S, Santiago F, Koski RA, Brei B, Anderson JF, Fish D, et al. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J Bacteriol. 2002;184:3122–3125. doi: 10.1128/JB.184.11.3122-3125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Caimano MJ, Liang FT, Santiago F, Laskowski M, Philipp MT, et al. Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc Natl Acad Sci USA. 2003;100:15953–15958. doi: 10.1073/pnas.2432412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelakanta G, Li X, Pal U, Liu X, Beck DS, Deponte K, et al. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog. 2007;3:e33. doi: 10.1371/journal.ppat.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowalk AJ, Gilmore RD, Carroll JA. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect Immun. 2006;74:3864–3873. doi: 10.1128/IAI.00189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi J, Piesman J, de Silva AM. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc Natl Acad Sci USA. 2001;98:670–675. doi: 10.1073/pnas.98.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Akins D, Casjens S, Rosa P, Elias A, et al. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 2002;358:165–177. doi: 10.1016/s0076-6879(02)58088-5. [DOI] [PubMed] [Google Scholar]
- Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, et al. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–230. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Dai J, Li X, Neelakanta G, Luo P, Kumar M, et al. A differential role for BB0365 in the persistence of Borrelia burgdorferi in mice and ticks. J Infect Dis. 2008a;197:148–155. doi: 10.1086/523764. [DOI] [PubMed] [Google Scholar]
- Pal U, Wang P, Bao F, Yang X, Samanta S, Schoen R, et al. Borrelia burgdorferi basic membrane proteins A and B participate in the genesis of Lyme arthritis. J Exp Med. 2008b;205:133–141. doi: 10.1084/jem.20070962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piesman J. Dynamics of Borrelia burgdorferi transmission by nymphal Ixodes dammini ticks. J Infect Dis. 1993;167:1082–1085. doi: 10.1093/infdis/167.5.1082. [DOI] [PubMed] [Google Scholar]
- Piesman J, Eisen L. Prevention of tick-borne diseases. Annu Rev Entomol. 2008;53:323–343. doi: 10.1146/annurev.ento.53.103106.093429. [DOI] [PubMed] [Google Scholar]
- Piesman J, Mather T, Sinsky R, Spielman A. Duration of tick attachment and Borrelia burgdorferi transmission. J Clin Microbiol. 1987;25:557–558. doi: 10.1128/jcm.25.3.557-558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcella SF, Schwan TG. Borrelia burgdorferi and Treponema pallidum: a comparison of functional genomics, environmental adaptations, and pathogenic mechanisms. J Clin Invest. 2001;107:651–656. doi: 10.1172/JCI12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyndiah S, Lasserre JP, Menard A, Claverol S, Prouzet-Mauleon V, Megraud F, et al. Two-dimensional blue native/SDS gel electrophoresis of multiprotein complexes from Helicobacter pylori. Mol Cell Proteomics. 2007;6:193–206. doi: 10.1074/mcp.M600363-MCP200. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, et al. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature. 2005;436:573–577. doi: 10.1038/nature03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA. 2002;99:1562–1567. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Zückert WR. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol. 2006;59:1473–1484. doi: 10.1111/j.1365-2958.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- Schwan TG. Temporal regulation of outer surface proteins of the Lyme-disease spirochaete Borrelia burgdorferi. Biochem Soc Trans. 2003;31:108–112. doi: 10.1042/bst0310108. [DOI] [PubMed] [Google Scholar]
- Seshu J, Esteve-Gassent MD, Labandeira-Rey M, Kim JH, Trzeciakowski JP, Hook M, et al. Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol Microbiol. 2006;59:1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x. [DOI] [PubMed] [Google Scholar]
- Sheridan GE, Masters CI, Shallcross JA, MacKey BM. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva AM, Fikrig E. Growth and migration of Borrelia burgdorferi. Ixodes ticks during blood feeding. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- de Silva AM, Fikrig E. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J Clin Invest. 1997;99:377–379. doi: 10.1172/JCI119169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare JT, Shang ES, Foley DM, Blanco DR, Champion CI, Mirzabekov T, et al. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J Clin Invest. 1995;96:2380–2392. doi: 10.1172/JCI118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of Ticks. New York: Oxford University Press; 1993. [Google Scholar]
- Steiner H, Winkler E, Haass C. Chemical cross-linking provides a model of the gamma-secretase complex subunit architecture and evidence for close proximity of the C-terminal fragment of presenilin with APH-1. J Biol Chem. 2008;283:34677–34686. doi: 10.1074/jbc.M709067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, von Heijne G, et al. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005;280:34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- Strother KO, Hodzic E, Barthold SW, de Silva AM. Infection of mice with Lyme disease spirochetes constitutively producing outer surface proteins A and B. Infect Immun. 2007;75:2786–2794. doi: 10.1128/IAI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KR, Stone KL. Enzymatic cleavage and HPLC peptide mapping of proteins. Mol Biotechnol. 1997;8:155–167. doi: 10.1007/BF02752260. [DOI] [PubMed] [Google Scholar]
- Xu Q, McShan K, Liang FT. Modification of Borrelia burgdorferi to overproduce OspA or VlsE alters its infectious behaviour. Microbiology. 2008;154:3420–3429. doi: 10.1099/mic.0.2008/019737-0. [DOI] [PubMed] [Google Scholar]
- Yang X, Popova TG, Goldberg MS, Norgard MV. Influence of cultivation media on genetic regulatory patterns in Borrelia burgdorferi. Infect Immun. 2001;69:4159–4163. doi: 10.1128/IAI.69.6.4159-4163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med. 2004;199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Coleman AS, Anguita J, Pal U. A chromosomally encoded virulence factor protects the Lyme disease pathogen against host-adaptive immunity. PLoS Pathog. 2009;5:e1000326. doi: 10.1371/journal.ppat.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


