Abstract
Osteocytes are the most abundant osteoblast lineage cells within the bone matrix. They respond to mechanical stimulation and can participate in the release of regulatory proteins that can modulate the activity of other bone cells. We hypothesize that neuropeptide Y (NPY), a neurotransmitter with regulatory functions in bone formation, is produced by osteocytes and can affect osteoblast activity. To study the expression of NPY by the osteoblast lineage cells, we utilized transgenic mouse models in which we can identify and isolate populations of osteoblasts and osteocytes. The Col2.3GFP transgene is active in osteoblasts and osteocytes, while the DMP1 promoter drives green fluorescent protein (GFP) expression in osteocytes. Real-time PCR analysis of RNA from the isolated populations of cells derived from neonatal calvaria showed higher NPY mRNA in the preosteocytes/osteocytes fraction compared to osteoblasts. NPY immunostaining confirmed the strong expression of NPY in osteocytes (DMP1GFP+), and lower levels in osteoblasts. In addition, the presence of NPY receptor Y1 mRNA was detected in cavaria and long bone, as well as in primary calvarial osteoblast cultures, whereas Y2 mRNA was restricted to the brain. Furthermore, NPY expression was reduced by 30–40% in primary calvarial cultures when subjected to fluid shear stress. In addition, treatment of mouse calvarial osteoblasts with exogenous NPY showed a reduction in the levels of intracellular cAMP and markers of osteoblast differentiation (osteocalcin, BSP, and DMP1). These results highlight the potential regulation of osteoblast lineage differentiation by local NPY signaling.
Keywords: Neuropeptide Y, Osteocytes, Osteoblasts, GFP, Bone
Neuropeptide Y (NPY) is a 36-amino acid polypeptide that belongs to the larger family of neuropeptides, which also includes the pancreatic polypeptide and the peptide YY [Tatemoto et al., 1982; Allen et al., 1992]. NPY signals through a class of receptors known as Y receptors, members of the G-protein-coupled receptors [Lemos et al., 1997; Raimondi et al., 2002]. The Y receptor system consists of five Y receptors; Y1, Y2, Y4, Y5, and Y6 (which is present only in the mouse). NPY is highly expressed in the hypothalamus, and its receptors Y1, Y2, and Y5 are found in the central nervous system, including the hypothalamus [Sar et al., 1990; Parker and Herzog, 1999].
Functionally, NPY is a potent orexigenic peptide; its expression is up-regulated in the hypothalamus of experimental diabetic rats [Sahu et al., 1990; la Fleur et al., 2003]. NPY acts through the Y2 receptor in the hypothalamus to centrally regulate bone mass [Baldock et al., 2002]. This observation was reinforced using germline deletion of Y2 or Y1 receptors in mice, resulting in a higher trabecular bone volume and elevated cortical bone mass [Baldock et al., 2005]. Moreover, targeted deletion of Y2 in the hypothalamus revealed an increased bone volume comparable to the effect of germ line deletion of Y1 or Y2 in mice, thus confirming the Y2-mediated central regulation of bone mass [Baldock et al., 2005]. In contrast to the germline Y1 deletion, Y1-targeted deletion in the hypothalamus did not affect the bone mass in mice indicating the possibility for a local NPY-Y1 signaling pathway in the bone tissue, especially since mouse stromal cells expresses Y1 [Baldock et al., 2007]. Thus, central NPY, acting through Y2 receptors, is known to affect bone mass. However, within the bone microenvironment, it is still unclear whether the actions of Y1 receptors result from local production of NPY, and if so, what cell type(s) are responsible. In adipose tissue, local production of NPY by adipocytes has been shown to regulate fat mass [Kuo et al., 2007]. Thus, we examined the production of NPY within the osteoblast lineage, and the effect of this production on cell function.
Studying the gene expression profile of the bone cells in mice has become easier due to the availability of transgenic mice in which promoters of bone-specific genes direct the green fluorescent protein (GFP) expression. For instance, transgenic mice in which GFP is under the control of either the 3.6 kb or the 2.3 kb of the rat col1a1 promoter fragment has enabled the isolation of relatively homogenous populations of preosteoblasts and mature osteoblasts, respectively [Kalajzic et al., 2002]. In the case of the osteocyte, it has been shown that dentin matrix protein 1 (DMP1) is preferentially expressed in cells embedded within the bone matrix (osteocytes) or in partially embedded cells (preosteocytes). This was the key observation leading to the generation of an osteocyte-specific GFP transgenic mouse (DMP1GFP) that can be used to selectively isolate this terminally differentiated population of osteoblast lineage cells [Kalajzic et al., 2004].
An intriguing observation was the expression of NPY in the Col2.3GFP population of mature osteoblasts with an increase in expression in DMP1GFP-positive population. In this study, we evaluated the expression of NPY in osteoblast lineage cells and studied the effects of NPY on osteoprogenitor lineage differentiation. This study defines NPY as a local regulator of osteoblast lineage activity that shows a response in relation to in vitro mechanical stimulation.
Materials and Methods
TRANSGENIC MICE MODELS
3 kb col1a1 GFP
To define cells as mature osteoblasts we utilized a transgenic mouse in which a 2.3-kb fragment of collagen type I promoter directs the expression of emerald or cyan variants of GFP to mature osteoblasts lineage cells (Col2.3GFPemd or Col2.3CFPc-yan) [Kalajzic et al., 2002].
DMP1 promoter-directed GFP expression
To define cells as osteocytes we used a previously developed transgenic mouse in which a DMP1 regulatory sequence directs the expression of the GFP transgene primarily to osteocytes (DMP1GFPtpz) [Kalajzic et al., 2004].
Transgenic lines were crossed to generate dual GFP-expressing Col2.3GFPcyan/DMP1GFPtopaz mice that were used for histological analysis and isolation of osteoblast and osteocytes by flow cytometry. All procedures involving the use of animals were approved by University of Connecticut Health Center Institutional Animal Care Committee (protocol # ACC 2007-344).
Separation of cell populations
Calvaria were isolated from 5-to 8-day-old double transgenic mice (pOBCol2.3-GFPcyan and DMP1GFPtopaz) sacrificed by CO2 asphyxiation. After the removal of the sutures, calvarial tissues were subjected to four sequential, 30-min long digestions in a mixture containing 0.05%/0.2 mM trypsin/EDTA and 1.5 U/ml collagenase-P (Roche) at 37°C. Cell fractions 2–4 were collected, pooled, and resuspended in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) containing 10% fetal calf serum (FBS). Cells were resuspended in PBS, filtered through a 70-μm cell strainer, centrifuged, resuspended in the PBS/2% FBS. Cell sorting was performed using a FACS-Vantage BD cell sorter with a 130-μm nozzle at a speed of 3–5K cells/s. Sorting was performed using appropriate lasers to distinguish GFPcyan+ from GFPtopaz+ expressing cells. Sorting allowed us to separate DMP1GFPtpz+ osteocytes from Col2.3GFPcyan+ osteoblasts.
Calvarial Osteoblast Culture
Calvarial cells were isolated from 7-day-old neonatal mice using a modified protocol developed by Wong and Cohn [Wong and Cohn, 1975; Kalajzic et al., 2002]. Calvaria were subjected to four sequential 30-min digestions in an enzyme mixture containing 0.05% trypsin and 1.5 U/ml collagenase P at 37°C. Cell fractions 2–4 were pooled and enzyme activity was terminated by the addition of media containing FBS. Cells were plated at a density of 1.5 × 105 cells/well in 6-well culture dishes in DMEM with 10% FBS and switched to differentiation medium (αMEM containing 10% FBS, 50 μg/ml ascorbic acid, 4mM β-glycerophosphate) when they reached confluence.
Preparation Of Cryosections
Bone tissues were fixed for 3–4 days in 4% paraformaldehyde/PBS (pH 7.4) at 4°C, decalcified for 3 days, placed in 30% sucrose/PBS overnight, and embedded (Cryomatrix, Thermo Shandon, Pittsburgh). Bones were cryosectioned at 5 μm sections using a CryoJane tape transfer system (Instrumedics, NJ). After rehydration in 1 mM MgCl2/physiological saline, GFP expression was observed and photographed (Zeiss Axiovert 200 M microscope and Axiocam digital camera; Zeiss) using GFP-variant specific filters (Chroma; Rockingham, VT) [Kalajzic et al., 2008]. A dual bandpass design was used to distinguish GFP signal from autofluorescence of bone and bone marrow.
Immunostaining
For immunofluorescent labeling primary calvaria cells were prepared as indicated above. Following activation of the DMP1GFP transgene, cells were sorted into DMP1GFP+ and DMPGFP1− populations. Briefly, cells were enzymatically digested using 0.2% collagenase/0.2% hyalouronidase in 2.5% trypsin at 37°C. Cells were collected and resuspended in DMEM (Life Technologies) containing 10% FBS (Hyclone) and centrifuged. Cells were resuspended in PBS, filtered through a 70-μm cell strainer, centrifuged, resuspended in the PBS/2% FBS, and filtered through a 45-μm filter. Cell sorting was performed using a FACS-Vantage BD cell sorter with a 130-μm nozzle at a speed of 3–5K cells/s. Sorting was performed using an argon laser to excite GFPtopaz-expressing cells at 488 nm employing a 550/30 emission filter. Following sorting, cells were replated on collagen-coated 2 mm2 cover slide chamber plates (0.15 mg/ml, rat type I collagen). Cells were fixed for 10 min in 4% PFA, and after washing in PBS, cells were blocked with 5% normal donkey serum, before incubating with rabbit anti-NPY (1:1,000 dilutions, Cat: ab10980, Abcam, Inc., MA) at 4°C overnight. Signal was detected with donkey anti-rabbit conjugated with CY3.
Histochemical Analysis Of Cell Cultures
Histochemical staining for ALP activity was performed using a commercially available kit (86-R ALP; Sigma Diagnostics, Inc., MO) according to the manufacturer's instruction. Mineralization was assessed using the von Kossa silver nitrate staining method. The results of staining procedures were recorded using a scanner (UMax Astra 4000U) and Adobe Photoshop.
Rna Extraction And Real-Time Pcr
Total RNA was isolated from tissues or cell cultures using TRIzol reagent (Invitrogen, USA). RNA samples were subjected to cDNA synthesis, and gene expression analysis was completed using semiquantitative and real-time PCR. A quantity of 3 μg of total RNA was reverse transcribed in a 20-μl reaction mixture containing 50 ng random hexamers mix together with 200 U of Superscript III reverse transcriptase according to the manufacturer's instructions for Superscript III first strand synthesis (Invitrogen). Real-time PCR was carried out using the TaqMan Gene Expression Assays (Applied Biosystems, CA) (assay ID:Dmp1, Mm01208365_m1, BSP, Mm00492555_M1). For RT-PCR, we used the following primers: NPY, forward 5′-GTTTCAGGGGATGAGATGAG-3′, reverse 5′-GCTCTGCGACACTACATCAA-3′; Y1, forward 5′-CTCGCTGGTTCTCATCGCTG TGGAACGG-3′, reverse 5′-gcgaatgtatatcttgaagtag-3; Y2, forward 5′-TCCTGGATTCCTCATCTGAG-3′, reverse 5′-ggtccagagcaatgactgtc-3′, 18s forward 5′-tcaagaacg aaagtcggagg-3′, reverse 5′-ggacatctc cgggccacaca-3′ [Lundberg et al., 2007].
Northern Blot Analysis
Total RNA (10μg) was separated on a 2.2 M formaldehyde/1% agarose gel and transferred onto a nylon membrane (Maximum Strength Nytran Plus, Schleicher & Schuell). Membranes were probed with the 900 bp PstI fragment of rat Col1a1 (pα1R2) [Genovese et al., 1984], and a 440-bp mouse OC fragment (p923) [Celeste et al., 1986]. Probes were radiolabeled by the random primer method using (α-32P) dCTP (New England Nuclear, 3,000 Ci/mmol) to obtain probes with specific activities of approximately 1 × 109 cpm/μg. Filters were hybridized with 3 × 106 cpm/ml 32P-labeled probe at 42°C in 50% formamide, 5× SSPE (1× SSPE = 0.149 M NaCl, 10 mM NaH2PO4, 1mM EDTA, pH 7.4), 1.2× Denhardt's, and 0.5% sodium dodecyl sulfate and washed according to published procedures.
In Vitro Shear Stress
Fluid shear stress (FSS) was performed using the previously described fluid flow chamber [Wadhwa et al., 2002]. The fluid flow system includes a flow chamber, a roller pump (Masterflex, IL), a tubing, and a water bath kept at 37°C. The fluid flow generates a uniform flow field in which the magnitude and direction of the velocity vector are constant with flow rate of 280 ml/min, generating FSS of 10 dynes/cm2, applied at a frequency of 5 Hz. Mouse calvarial osteoblasts were isolated by sequential digestion as described above. Cells were cultured for 7 days in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% nonessential amino acid, and then re-plated on slides coated with type I collagen (rat tail collagen, Collaborative Biomedical Products, Bedford, MA, 50 μg/ml) at a density of 100,000 cells/slide and cultured in a 4-well slide chamber plate. Twenty-four hours after re-plating, cells were switched to osteogenic media (see the Calvarial Osteoblast Culture Section) and cultured for 10 days additional until cells started to form mineralized nodules. The media were changed to low concentration FBS (1%) 1 h before subjecting cells to FSS for 30 min in the same media. After exposure to FSS, cells (including the controls) were cultured in the same media for 30 min before RNA was extracted from the cultures and prepared as described above for real-time PCR.
Intracellular Cyclic Amp Measurement
Following NPY treatment, cells were scraped into 500 μl of ice-cold ethanol. The ethanolic cell suspension was collected in tubes and centrifuged at 1,500g for 10 min at 4°C. Supernatants were collected and lyophilized. The dried residue was dissolved in EIA buffer and cAMP was measured according to the manufacturer's instructions (Cayman Chemicals, Ann Arbor, MI). The cAMP activity was normalized to the total protein in the sample measured by BCA protein assay (Pierce, USA).
Statistical Analysis
To compare differences in mean, we used unpaired t-test. ELISA data were normalized to protein concentration. P < 0.05 was considered statistically significant.
Results
Detection Of Npy Expression In Cells Of Osteoblasts Lineage
To investigate NPY expression in cells of the osteoblast lineage, we analyzed the RNA derived from different tissues of 2-month-old mice. NPY mRNA was detected in mouse calvaria, femur, and in the brain, which was used as a positive control (Fig. 1A). Lower expression of NPY was observed in heart, kidney, and adipose tissue. To further evaluate the expression of NPY during the different stages of osteoblast lineage differentiation, we utilized previously developed transgenic mice in which promoter-GFP transgenes were directed to specific stages of lineage differentiation. We generated primary neonatal calvarial cultures and separated cells based on GFP expression using flow cytometry cell sorting. We focused the analysis on sorted GFP+ and GFP− cells derived from Col2.3GFP and DMP1-GFP transgenic mice, as they direct GFP in areas of mineralization (Fig. 1B). RT-PCR analysis of RNA from the sorted cells shows strong NPY mRNA expression in the Col2.3GFP+ and DMP1-GFP+ cells (Fig. 1C). Since the Col2.3GFP-positive cells also contain the osteocyte population, we aimed to identify if the NPY expression is localized in the mature osteoblasts or within the preosteocyte/osteocyte population. In order to evaluate this, we utilized dual color transgenic mice obtained by breeding of Col2.3GFPcyan and DMP1-GFPtopaz mice to isolate mature osteoblasts (Col2.3GFPcyan+, DMP1GFPtpz−) and osteocytes (DMP1GFPtpz+). Col2.3GFP cyan is expressed in osteoblast lining bone surfaces and in some osteocytes, while DMP1GFP is active in osteocytes and preosteocytes (cells that are fully or partially embedded within the matrix) (Fig. 2A). Real-time PCR analysis of RNA from these fractions demonstrated that NPY expression is significantly higher in the preosteocyte/osteocyte fraction than in osteoblasts (Fig. 2B,D). Similar results on NPY gene expression in sorted cells were obtained by comprehensive microarray analysis of RNA derived from sorted osteoblasts and osteocytes (data not shown).
Fig. 1.
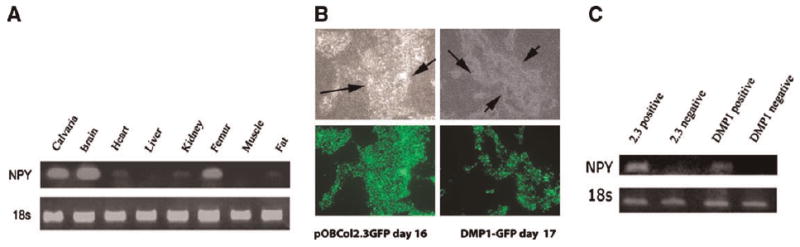
Detection of NPY mRNA in osteoblast lineage cells. A: Expression of NPY mRNA by RT-PCR was completed in selected tissues isolated from 2-month-old mice. NPY mRNA was detected in mouse calvaria, brain, and femur. B: GFP expression in mouse calvarial osteoblasts cultures. Cells were derived from neonatal GFP calvarial from pOBCol2.3GFP and DMP-1-GFP (areas undergoing mineralization are indicated by the arrows). The upper panel shows phase-contrast images, while epifluorescence images are shown in the lower panel. C: Expression of NPY in pOBCol2.3GFP+ and DMP-1-GFP+ sorted cells. No expression of NPY was detected in GFP− cells (images were taken at a magnification of 10× (B)).
Fig. 2.
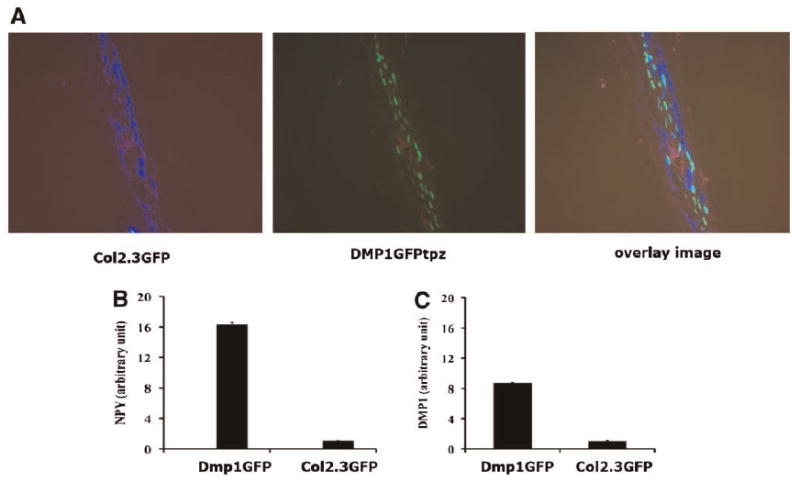
Detection of NPY in DMP1GFPtpz/pOBCol2.3GFPcyan dual transgenic mice. A: Expression of DMP1GFPtpz and pOBCol2.3GFPcyan in calvaria derived from dual GFP transgenic mice. DMP-1GFPtpz is detected in the cells within calvarial bone, while pOBCol2.3GFPcyan is detected in cells lining bone surfaces of the parietal bone. B: Real-time PCR analysis of RNA from freshly isolated calvarial cells obtained from dual transgenic mice (DMP1GFP/Col2.3GFP). Higher expression of NPY and DMP-1 was observed in DMP1GFPtopaz+ population than in Col2.3GFPcyan+ cells. A representative experiment out of three different biological experiments is presented (images were taken at a magnification of 20× (A)).
Detection Of Npy Protein In Cells Of Osteoblast Lineage
Having determined that NPY mRNA is expressed in cells of the osteoblast lineage, we investigated whether NPY protein could be detected in primary bone cell cultures of transgenic mice. Cells derived from DMP1-GFP mice were cultured for 7 days in basal media and then placed under osteogenic induction for another 10 days. We observed that DMP1-GFP was expressed in association with mineralized nodules (Fig. 3A,B). Cultures were enzymatically digested and cells were sorted into DMP1-GFP+ (Fig. 3C) and DMP1-GFP− (Fig. 3D). DMP1-GFP+ and DMP1-GFP− cells were immunostained for NPY expression. Consistent with the mRNA results, NPY immunoreactivity was observed in DMP1-GFP+ cells (Fig. 3E), while weaker but detectable NPY expression was observed in DMP1-GFP− cells (Fig. 3F).
Fig. 3.
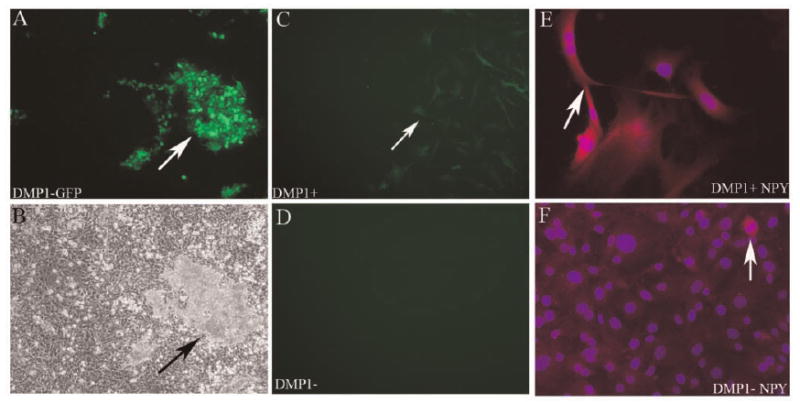
Detection of NPY protein by immunofluorescence. A,B: DMP1-GFP expression in mouse calvarial osteoblast cell cultures at day 16 prior to cell sorting under epifluorescence (A) and phase contrast (B). C: Epifluorescent image of DMP1+ cells after sorting. D: Epifluorescent imaging shows no GFP expression in DMP1− cells following sorting. E: DMP1+ cells and (F) DMP1− cells immunostained for NPY (red, TRITC+), and nuclear staining (blue, DAPI). A strong immunoreactivity was observed in DMP1+ cells (E), while a lower but detectable expression was observed in DMP1− cells (F).
Expression Of Npy Receptors In Osteoblasts
The Y1 and Y2 receptors are known to play a major role in NPY-dependent central regulation of bone formation. Therefore, we evaluated their expression in various tissues including calvaria and long bones. RNA analysis demonstrated the presence of Y1 mRNA in the calvarial and long bone tissues as well as in the other organs including heart, kidney, muscle, and adipose tissue (Fig. 4A). Y2 expression was observed in the brain, but was not detected in the calvaria or long bones (Fig. 4A), or in cultured mouse calvarial osteoblasts (data not shown). We evaluated the expression of NPY Y1 in mouse primary calvarial osteoblasts. Quantitative real-time PCR of RNA from cultured calvarial osteoblasts revealed an increase of NPY Y1 expression as the osteoprogenitors differentiate and mineralize in culture (Fig. 4B). The expression of alkaline phosphatase is observed by day 7 and increases at later time points (Fig. 4C). More mature stages of osteoblast lineage are detected by expression of bone sialoprotein (BSP) and DMP1 at days 14 and 21 along with mineralization (Fig. 4C–E).
Fig. 4.
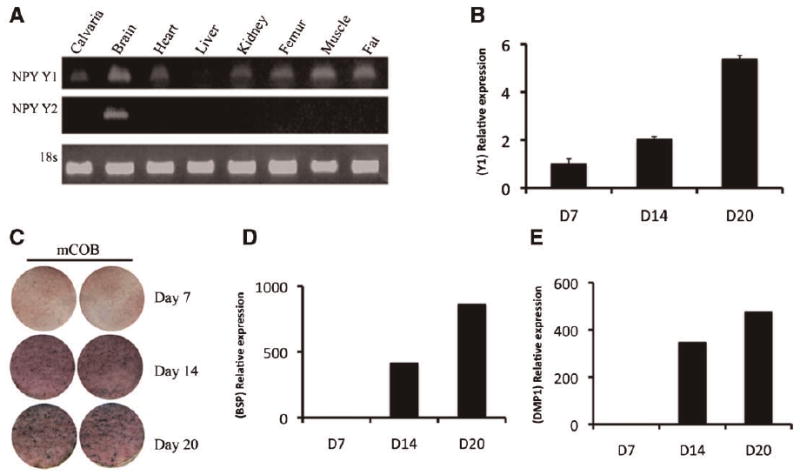
Detection of NPY-Y1 receptors. A: In vivo detection of NPY-Y1 and Y2 by RT-PCR in selected tissues derived from 2-month-old mice. B: mRNA expression of NPY-Y1 by real-time PCR in samples of cultured mouse calvarial osteoblasts. RNA was isolated on days 7, 14, and 20; values represent mean ± SE (n = 3) from a representative experiment. C: Alkaline phosphatase expression (days 7–20) and mineralization on days 14 and 20 by Von Kossa (black spots) indicate differentiation of the osteoprogenitor cells. D,E: Real-time PCR analysis for BSP and DMP1 gene expression. Data shown are obtained from one of the three representative experiments and are expressed as the relative gene expression ratio to day 7. Both BSP and DMP1 expression are very low or undetectable on day 7. Following osteogenic induction, an increase in both BSP and DMP1 is observed by days 14 and 21.
Fluid Shear Stress Negatively Regulates Npy Expression In The Osteoblast Lineage Cells
We have previously shown that DMP1 expression is responsive to in vivo and in vitro mechanical stimulation [Yang et al., 2005]. Here we evaluated whether the expression of NPY by the osteoblast lineage cells is regulated by in vitro FSS using DMP-1 as the positive readout for FSS. Following 3 weeks in culture, calvarial osteoblasts grown on rat type 1 collagen-coated slides were fully differentiated to a stage containing mineralizing nodules. The mineralized cultures were subjected to FSS for 30min at 10 dynes/cm2. After 30 min of post-shear stress culture, the RNA levels of DMP1 and NPY were assessed. As determined by quantitative real-time PCR, NPY mRNA levels were significantly decreased by 35% (P<0.05, n = 3) (Fig. 5A), while the mRNA level of DMP1 was increased twofold (Fig. 5B). These results suggest that NPY expression is negatively regulated by FSS.
Fig. 5.
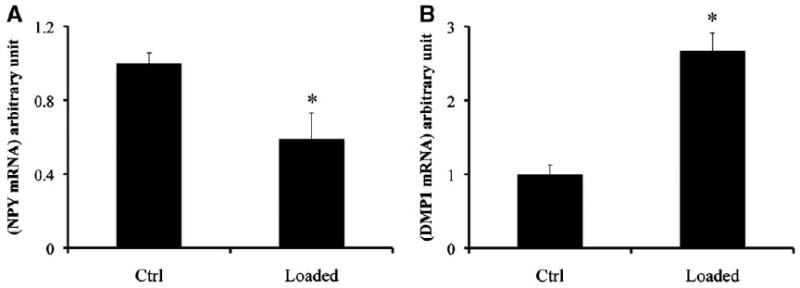
Regulation of NPY expression by FSS. A,B: The effect of in vitro fluid shear stress on the expression of NPY in primary mouse calvarial osteoblasts cultures. Fully differentiated mouse calvarial osteoblast were subjected to shear stress for 30min and then cultured without stress for 30min. A: Real-time PCR analysis of NPY and (B) DMP1 mRNA levels in loaded and control samples (values are mean ± SE of three experiments, *P-value = 0.03).
Role Of Npy On In Vitro Osteoblastogenesis
The observation that the expression of NPY-Y1 receptor in the osteoblast increases with time in culture (Fig. 4B,C) prompted us to evaluate whether NPY has the ability to regulate osteoblastic differentiation. In order to test this, we cultured mouse calvarial osteoblasts in the presence of 1 nM NPY for 20 days and then monitored the expression of markers of osteoblast differentiation. Primary calvarial osteoblasts cultured in the presence of NPY exhibited no detectable difference in the expression of ALP at days 7 and 14. However, decreased mineral deposition as detected by von Kossa staining (Fig. 6A) was observed by day 21 of culture. In addition, RNA analysis of NPY-treated cells demonstrated decreased expression of BSP, osteocalcin (OC) and DMP1 at days 14 and 21 of culture (Fig. 6B–D).
Fig. 6.
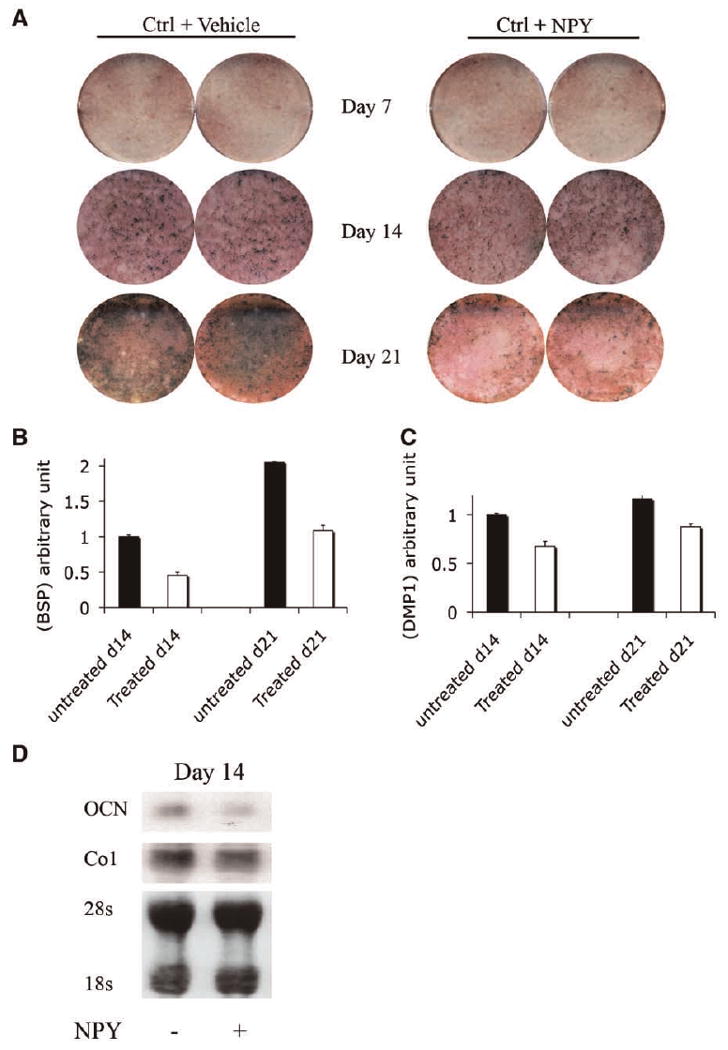
Effect of exogenous NPY on osteoblast cultures. Mouse calvarial osteoblasts were plated at a density of 1.5 × 105 cells/well in 6-well plates. NPY protein (1 nM) was added in culture media on day 2 and every second day when media were changed. Cells were switched to differentiation media on day 7. A: Alkaline phosphatase activity and von Kossa assay showing no major changes in the expression of ALP, while decreased mineral deposition (dark staining) in NPY-treated cultures for days 14 and 21 was observed. B,C: Real-time PCR showing mRNA levels of BSP and DMP1 expression on days 14 and 21, respectively. Values represent mean ± SE of three experiments. D: Northern blot analysis for osteocalcin and type I collagen mRNA expression.
Npy Treatment Decreased Intracellular Camp In Mouse Calvarial Osteoblasts
NPY-Y1 is classified as a G-protein-coupled receptor that can inhibit adenylate cyclase activity [Amano et al., 2007]. We aimed to evaluate whether the NPY treatment can modulate the levels of intracellular cAMP and explain the decrease in osteoblast differentiation markers observed in our NPY-treated osteoblasts. After treating calvarial osteoblasts with 10nM NPY for 1 h, we observed a 30% decrease in cAMP (data not shown). Because of the low level of cAMP in unstimulated osteoblasts, we validated this observation by evaluating the effect of NPY on forskolin-induced release of cAMP in calvarial osteoblast cultures. Treatment of calvarial osteoblasts with forskolin induced the release of intracellular cAMP at days 7 and 14 of culture (Fig. 7B,C). Consistent with the observation in Figure 7A, NPY significantly blunted forskolin-induced cAMP levels (Fig. 7B–D). In addition, this effect of NPY was mediated mostly via NPY-Y1 signaling since NPY-Y1 receptor blocker BIB03304 blunted this effect (Fig. 7D). These results indicate that NPY/NPY-1 signaling can alter the intracellular cAMP levels in osteoblast cells and modulate osteogenic differentiation.
Fig. 7.
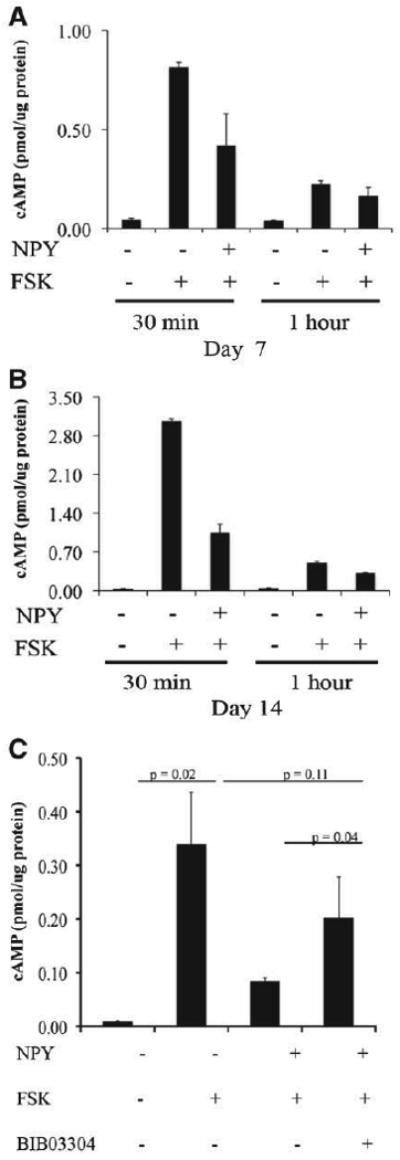
Effect of NPY on intracellular cAMP levels. A: Effect of treatment with NPY on the intracellular cAMP levels in primary mouse calvarial osteoblasts cultures. Cells were treated with 10nM NPY for 1 h while the control group received vehicle only. Values represent cAMP levels at days 7 and 14, as assessed by ELISA (n = 2). B,C: Effect of NPY on forskolin-induced intracellular cAMP levels primary calvarial osteoblasts cultures. D: The effect of NPY-Y1 blocker BIB03304 on the regulation of intracellular cAMP levels in primary calvarial osteoblast cultures. Cells were cultured for 7 days and then treated with forskolin alone or with either NPY or NPY-Y1 blocker BIB03304 (values are mean ± SD from three experiments).
Discussion
Over the last two decades, since the discovery of NPY, its expression has been thought be exclusive to the brain [Tatemoto, 1982]. However, NPY expression has been observed in other tissues. For instance, the white adipocytes were reported to synthesize and release NPY (Fig. 1A) [Kuo et al., 2007; Yang et al., 2008]. In this study, we present evidence for the first time of the expression of the NPY by the cells of the osteoblast lineage. Bone tissue is well innervated and it had been previously reported that NPY immunoreactivity was associated with the nerve endings along vascular elements within the bone [Hill and Elde, 1991]. Here, using DMP1-GFP, Col2.3-GFP, and DMP1-GFP/Col2.3-GFP dual transgenic mice, we were able to isolate population of cells that are highly enriched with mature osteoblasts (Col2.3-GFP) and osteocytes (DMP1-GFP and DMP1-GFP/Col2.3-GFP). This enabled us to perform gene expression analysis on RNA from these two different cell populations. We detected greater NPY mRNA expression in the fractions that were highly enriched with mature osteoblasts and osteocytes. Osteocytes are characterized by a specific morphology that distinguishes them from other bone cells; they also share some of these features with the neuronal cells, such as extensive network of processes that run through out the bone tissue. Much like the neuronal processes, osteocytes processes may actively participate in the release of regulatory proteins that can modulate the activity of other bone cells. One of such proteins is NPY, a neurotransmitter with functions in energy and bone regulation. The possibility of NPY expression by the bone cells is not surprising as other cells within the bone marrow niche such as the megakaryocytes and the leukocytes has been shown to express NPY [Holler et al., 2008].
Local production of NPY in the bone could indicate a novel alternative pathway to the previously described hypothalamic-mediated central regulation of bone, which requires Y2 expression on the pre-synaptic nerve ending to maintain the feedback inhibition of NPY release [King et al., 2000; Sainsbury et al., 2002]. This is plausible since we did not detect Y2 in any of the bone cells including the osteocytes and therefore a non-Y2-regulated source of NPY may represent a potential alternate peripheral source of NPY in the bone tissue. NPY-Y2 signaling is supposedly a part of the hypothalamo-pitutary-corticotropic axis, which is also involved in the regulation of NPY. The complexity of NPY regulation has been examined at different levels. Mice lacking either Y2 or leptin receptor demonstrated increased hypothalamic NPY levels [Wilding et al., 1993; Sainsbury et al., 2002]. In addition, stress and insulin have also been implicated in the regulation of NPY in the brain [Wilding et al., 1993; Zhou et al., 2008]. We investigated whether mechanical stimulation regulates NPY expression, since osteocytes are known to respond to mechanical stress [Aarden et al., 1994; Weinbaum et al., 1994]. DMP1 expression can be utilized as a marker for studying the response of osteocytes to mechanical loading [Yang et al., 2005; Harris et al., 2007]. The DMP1 protein is found within the lacunae and along the processes of mature osteocytes, where it is believed to contribute to the mechanosensory properties of the osteocytes. In a model of shears stressed cells, DMP1 response was observed after 30 min. In contrast to the increase in DMP1 mRNA, the expression of NPY was decreased in an in vitro shear stress model. Taken together, our observation demonstrates that the expression of NPY in the bone cells is responsive to mechanical stress. This shows some similarity to the regulation of NPY by different stressor elements in the brain [Thorsell et al., 1998]. The potential mechanism and the signaling cascades within the osteoblast lineage require further studies.
NPY signaling is known to occur via the Y receptors, with the Y1 and Y2 being implicated in the regulation of bone formation. In this study, we evaluated whether NPY acting through its Y1 receptor could influence osteoblastic differentiation. Our findings indicate that osteoblasts cultured in the presence of NPY exhibited decreased expression of markers of differentiation, OC, bone sailoprotein, and DMP1 and a decrease in the extent of mineralization. This finding mirrors an earlier observation in which osteoblast cultures from mice lacking Y1 exhibited increased differentiation potential and high bone mass, while in another study NPY infusion in mice resulted in decreased osteoid width [Baldock et al., 2007; Allison et al., 2008]. Taken together these observations suggest a potential role for the local NPY-Y1 signaling in osteoblast biology.
The NPY-Y1 receptors are G-protein-coupled receptors and have been shown to inhibit the activity of adenylate cyclase and consequently decreases the cAMP levels, we therefore hypothesized that the mechanism underlying NPY regulation of osteoblast maturation lies in its ability to decrease the intracellular cAMP levels in these cells [Amano et al., 2007]. To test this hypothesis, we cultured mouse calvarial osteoblasts with or without forskolin in the presence of NPY or NPY-Y1 receptor inhibitor BIB03304. NPY significantly reduced forskolin-induced cAMP in cultured osteoblasts. The inhibitory effect of NPY on cAMP levels in cultured bone cells appears to be mediated through the Y1 receptors, although this does not exclude the possibility of other undiscovered pathways.
To summarize our study define that NPY is expressed by osteoblast lineage cells and underlines the potential for a local signaling pathway that can regulate the osteoblast lineage activity.
Acknowledgments
This work was supported by grants from the National Institutes of Health, NIAMS (R03-AR053275) and institutional support to I.K. through NIH grant UDE016495A.
References
- Aarden EM, Burger EH, Nijweide PJ. Function of osteocytes in bone. J Cell Biochem. 1994;55:287–299. doi: 10.1002/jcb.240550304. [DOI] [PubMed] [Google Scholar]
- Allen JM, Adrian TE, Tatemoto K, Polak JM, Hughes J, Bloom SR. Two novel related peptides, neuropeptide Y (NPY) and peptide YY (PYY) inhibit the contraction of the electrically stimulated mouse vas deferens. Neuropeptides. 1982;3:71–77. doi: 10.1016/0143-4179(82)90001-4. [DOI] [PubMed] [Google Scholar]
- Allison S, Baldock P, Enriquez R, Lin E, During M, Gardiner E, Eisman J, Sainsbury A, Herzog H. Critical interplay between neuropeptide Y and sex steroid pathways in bone and adipose tissue homeostasis. J Bone Miner Res. 2008 doi: 10.1359/jbmr.081013. [DOI] [PubMed] [Google Scholar]
- Amano S, Arai M, Goto S, Togari A. Inhibitory effect of NPY on isoprenaline-induced osteoclastogenesis in mouse bone marrow cells. Biochim Biophys Acta. 2007;1770:966–973. doi: 10.1016/j.bbagen.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Baldock PA, Sainsbury A, Couzens M, Enriquez RF, Thomas GP, Gardiner EM, Herzog H. Hypothalamic Y2 receptors regulate bone formation. J Clin Invest. 2002;109:915–921. doi: 10.1172/JCI14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldock PA, Sainsbury A, Allison S, Lin EJ, Couzens M, Boey D, Enriquez R, During M, Herzog H, Gardiner EM. Hypothalamic control of bone formation: Distinct actions of leptin and y2 receptor pathways. J Bone Miner Res. 2005;20:1851–1857. doi: 10.1359/JBMR.050523. [DOI] [PubMed] [Google Scholar]
- Baldock PA, Allison SJ, Lundberg P, Lee NJ, Slack K, Lin EJ, Enriquez RF, McDonald MM, Zhang L, During MJ, Little DG, Eisman JA, Gardiner EM, Yulyaningsih E, Lin S, Sainsbury A, Herzog H. Novel role of Y1 receptors in the coordinated regulation of bone and energy homeostasis. J Biol Chem. 2007;282:19092–19102. doi: 10.1074/jbc.M700644200. [DOI] [PubMed] [Google Scholar]
- Celeste AJ, Rosen V, Buecker JL, Kriz R, Wang EA, Wozney JM. Isolation of the human gene for bone gla protein utilizing mouse and rat cDNA clones. EMBO J. 1986;5:1885–1890. doi: 10.1002/j.1460-2075.1986.tb04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese C, Rowe D, Kream B. Construction of DNA sequences complementary to rat alpha 1 and alpha 2 collagen mRNA and their use in studying the regulation of type I collagen synthesis by 1,25-dihydroxyvitamin D. Biochemistry. 1984;23:6210–6216. doi: 10.1021/bi00320a049. [DOI] [PubMed] [Google Scholar]
- Harris SE, Gluhak-Heinrich J, Harris MA, Yang W, Bonewald LF, Riha D, Rowe PS, Robling AG, Turner CH, Feng JQ, McKee MD, Nicollela D. DMP1 and MEPE expression are elevated in osteocytes after mechanical loading in vivo: Theoretical role in controlling mineral quality in the perilacunar matrix. J Musculoskelet Neuronal Interact. 2007;7:313–315. [PMC free article] [PubMed] [Google Scholar]
- Hill EL, Elde R. Distribution of CGRP-, VIP-, D beta H-, SP-, and NPY-immunoreactive nerves in the periosteum of the rat. Cell Tissue Res. 1991;264:469–480. doi: 10.1007/BF00319037. [DOI] [PubMed] [Google Scholar]
- Holler J, Zakrzewicz A, Kaufmann A, Wilhelm J, Fuchs-Moll G, Dietrich H, Padberg W, Kuncova J, Kummer W, Grau V. Neuropeptide Y is expressed by rat mononuclear blood leukocytes and strongly down-regulated during inflammation. J Immunol. 2008;181:6906–6912. doi: 10.4049/jimmunol.181.10.6906. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- Kalajzic I, Braut A, Guo D, Jiang X, Kronenberg MS, Mina M, Harris MA, Harris SE, Rowe DW. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Kalajzic Z, Li H, Wang LP, Jiang X, Lamothe K, Adams DJ, Aguila HL, Rowe DW, Kalajzic I. Use of an alpha-smooth muscle actin GFP reporter to identify an osteoprogenitor population. Bone. 2008;43:501–510. doi: 10.1016/j.bone.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King PJ, Williams G, Doods H, Widdowson PS. Effect of a selective neuropeptide Y Y(2) receptor antagonist, BIIE0246 on neuropeptide Y release. Eur J Pharmacol. 2000;396:R1–R3. doi: 10.1016/s0014-2999(00)00230-2. [DOI] [PubMed] [Google Scholar]
- Kuo LE, Kitlinska JB, Tilan JU, Li L, Baker SB, Johnson MD, Lee EW, Burnett MS, Fricke ST, Kvetnansky R, Herzog H, Zukowska Z. Neuropeptide Y acts directly in the periphery on fat tissue and mediates stress-induced obesity and metabolic syndrome. Nat Med. 2007;13:803–811. doi: 10.1038/nm1611. [DOI] [PubMed] [Google Scholar]
- la Fleur SE, Ji H, Manalo SL, Friedman MI, Dallman MF. The hepatic vagus mediates fat-induced inhibition of diabetic hyperphagia. Diabetes. 2003;52:2321–2330. doi: 10.2337/diabetes.52.9.2321. [DOI] [PubMed] [Google Scholar]
- Lemos VS, Bucher B, Cortes SF, Takeda K. Inhibition of [Ca2+]i transients in rat adrenal chromaffin cells by neuropeptide Y: Role for a cGMP-dependent protein kinase-activated K+ conductance. Eur J Neurosci. 1997;9:1144–1152. doi: 10.1111/j.1460-9568.1997.tb01468.x. [DOI] [PubMed] [Google Scholar]
- Lundberg P, Allison SJ, Lee NJ, Baldock PA, Brouard N, Rost S, Enriquez RF, Sainsbury A, Lamghari M, Simmons P, Eisman JA, Gardiner EM, Herzog H. Greater bone formation of Y2 knockout mice is associated with increased osteoprogenitor numbers and altered Y1 receptor expression. J Biol Chem. 2007;282:19082–19091. doi: 10.1074/jbc.M609629200. [DOI] [PubMed] [Google Scholar]
- Parker RM, Herzog H. Regional distribution of Y-receptor subtype mRNAs in rat brain. Eur J Neurosci. 1999;11:1431–1448. doi: 10.1046/j.1460-9568.1999.00553.x. [DOI] [PubMed] [Google Scholar]
- Raimondi L, Banchelli G, Matucci R, Stillitano F, Pirisino R. The direct stimulation of Gi proteins by neuropeptide Y (NPY) in the rat left ventricle. Biochem Pharmacol. 2002;63:2063–2068. doi: 10.1016/s0006-2952(02)00986-3. [DOI] [PubMed] [Google Scholar]
- Sahu A, Kalra PS, Crowley WR, Kalra SP. Functional heterogeneity in neuropeptide-Y-producing cells in the rat brain as revealed by testosterone action. Endocrinology. 1990;127:2307–2312. doi: 10.1210/endo-127-5-2307. [DOI] [PubMed] [Google Scholar]
- Sainsbury A, Schwarzer C, Couzens M, Herzog H. Y2 receptor deletion attenuates the type 2 diabetic syndrome of ob/ob mice. Diabetes. 2002;51:3420–3427. doi: 10.2337/diabetes.51.12.3420. [DOI] [PubMed] [Google Scholar]
- Sar M, Sahu A, Crowley WR, Kalra SP. Localization of neuropeptide-Y immunoreactivity in estradiol-concentrating cells in the hypothalamus. Endocrinology. 1990;127:2752–2756. doi: 10.1210/endo-127-6-2752. [DOI] [PubMed] [Google Scholar]
- Tatemoto K. Neuropeptide Y: Complete amino acid sequence of the brain peptide. Proc Natl Acad Sci USA. 1982;79:5485–5489. doi: 10.1073/pnas.79.18.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y—A novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature. 1982;296:659–660. doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- Thorsell A, Svensson P, Wiklund L, Sommer W, Ekman R, Heilig M. Suppressed neuropeptide Y (NPY) mRNA in rat amygdala following restraint stress. Regul Pept. 1998:75–76. 247–254. doi: 10.1016/s0167-0115(98)00075-5. [DOI] [PubMed] [Google Scholar]
- Wadhwa S, Godwin SL, Peterson DR, Epstein MA, Raisz LG, Pilbeam CC. Fluid flow induction of cyclo-oxygenase 2 gene expression in osteo-blasts is dependent on an extracellular signal-regulated kinase signaling pathway. J Bone Miner Res. 2002;17:266–274. doi: 10.1359/jbmr.2002.17.2.266. [DOI] [PubMed] [Google Scholar]
- Weinbaum S, Cowin SC, Zeng Y. A model for the excitation of osteocytes by mechanical loading-induced bone fluid shear stresses. J Biomech. 1994;27:339–360. doi: 10.1016/0021-9290(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Wilding JP, Gilbey SG, Lambert PD, Ghatei MA, Bloom SR. Increases in neuropeptide Y content and gene expression in the hypothalamus of rats treated with dexamethasone are prevented by insulin. Neuroendocrinology. 1993;57:581–587. doi: 10.1159/000126410. [DOI] [PubMed] [Google Scholar]
- Wong GL, Cohn DV. Target cells in bone for parathormone and calcitonin are different: Enrichment for each cell type by sequential digestion of mouse calvaria and selective adhesion to polymeric surfaces. Proc Natl Acad Sci USA. 1975;72:3167–3171. doi: 10.1073/pnas.72.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lu Y, Kalajzic I, Guo D, Harris MA, Gluhak-Heinrich J, Kotha S, Bonewald LF, Feng JQ, Rowe DW, Turner CH, Robling AG, Harris SE. Dentin matrix protein 1 gene cis-regulation: Use in osteocytes to characterize local responses to mechanical loading in vitro and in vivo. J Biol Chem. 2005;280:20680–20690. doi: 10.1074/jbc.M500104200. [DOI] [PubMed] [Google Scholar]
- Yang K, Guan H, Arany E, Hill DJ, Cao X. Neuropeptide Y is produced in visceral adipose tissue and promotes proliferation of adipocyte precursor cells via the Y1 receptor. FASEB J. 2008;22:2452–2464. doi: 10.1096/fj.07-100735. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, Sinha R, Virkkunen M, Mash DC, Lipsky RH, Hu XZ, Hodgkinson CA, Xu K, Buzas B, Yuan Q, Shen PH, Ferrell RE, Manuck SB, Brown SM, Hauger RL, Stohler CS, Zubieta JK, Goldman D. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452:997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]


