Abstract
In addition to their essential role in V(D)J recombination, the RAG proteins function as a transposase capable of inserting the V(D)J recombination intermediate, the signal end DNA fragment, into target DNA. RAG-mediated transposition has been suggested to contribute to genome instability and the development of lymphoid malignancies. Previous studies suggested that the RAG transposase exhibits a target site preference for GC rich sequences and hairpin structures. Here we demonstrate that a transposition hot spot (5′-GCCGCCGGGCC-3′), smaller portions of this hot spot and other GC rich motifs are able to target RAG-mediated transposition. Tracks of GC base pairs have been shown to have an unusually high rate of base pair breathing. Intriguingly, we find that DNA mismatches can efficiently target RAG-mediated transposition and suppress the use of other target sites. Hairpins, however, are not generally preferred targets. Our results indicate that target DNA melting may be a crucial step during RAG-mediated transposition, and that target site selection by the RAG transposase may be intimately linked to mutagenic and metabolic processes that transiently present favorable DNA structures to the transposition machinery.
INTRODUCTION
During lymphocyte development, functional antigen receptor genes are assembled from component V (variable), D (diversity) and J (joining) gene segments through a series of site-specific DNA rearrangement events known as V(D)J recombination (1,2). Conceptually, V(D)J recombination can be divided into two phases: DNA cleavage and DNA repair/joining. DNA cleavage is performed by a protein complex containing the products of two lymphocyte specific genes, RAG1 and RAG2 (3,4). The RAG proteins, in conjunction with a non-specific DNA binding/bending protein HMG1 or HMG2, form a stable DNA–protein complex with a recombination signal sequence (RSS), which is found flanking each gene segment. Subsequently, a second RSS, apparently not yet bound by the RAG proteins (5,6), is recruited to form the synaptic complex within which the RAG proteins catalyze DNA double strand cleavage, separating RSSs from gene segments and yielding signal ends (SEs) containing RSSs, and coding ends containing coding gene segments (1,2). In the second phase of V(D)J recombination, coding ends are processed and joined to form a coding joint and SEs are ligated head-to-head to form a signal joint (7). The transition from DNA cleavage to DNA repair/joining is not well understood. It is thought, however, that after DNA cleavage, SEs and coding ends remain associated with the RAG proteins in a post-cleavage complex, known as the 4-end or cleaved signal complex (8,9). The 4-end complex has been suggested to serve as a critical structure within which DNA ends are protected from degradation prior to DNA joining and oriented to facilitate proper DNA repair/joining (10–13).
In contrast to coding ends, SEs are joined slowly and may persist for hours in developing lymphocytes (14,15), presumably because they remain associated with the RAG proteins in the complex known as the signal end complex (SEC) (8,9,16). In vitro, the RAG proteins in the SEC are able to perform a strand transfer reaction that inserts the signal end fragment into target DNA (17,18). RAG-mediated transposition results in a 3–5 bp target site duplication because the sites of strand transfer on each strand of the target are separated by 3–5 bp. Initial analyses of RAG-mediated transposition suggested that this process is biased toward GC-rich target sequences (17,18). Additionally, altered DNA structures such as the hairpin tips of cruciform DNA have been suggested to be preferred target sites for RAG-mediated transposition (19), although the mechanism by which such DNA structures target RAG-mediated transposition is not known.
RAG-mediated transposition has been proposed to play an essential role in the early evolution of antigen receptor loci, and to cause chromosomal translocations and genome instability in developing lymphocytes (17,18,20). To prevent further modification of antigen receptor loci and to maintain genome stability, it is thought that the transposition activity of the RAG proteins has been selectively suppressed during evolution. Consistent with this notion, RAG-mediated transposition has been shown to be selectively suppressed by high concentrations of Mg++, the C-terminal region of RAG2 and GTP (21–23).
Mobilization of a V(D)J recombination intermediate, presumably by the RAG proteins, has been reported recently in a human peripheral T cell clone (24). Insertion of a TCRα signal end fragment into the X-linked hypoxanthine-guanine phosphoribosyl transferase (HPRT) locus resulted in gene inactivation (24), indicating that RAG-mediated transposition, although rare, can have significant consequences. Target site preference may have a profound influence on the outcome of RAG-mediated transposition. In the present report, we examined the effect of target site sequence and structure on RAG-mediated transposition. Our findings provide evidence for target DNA melting during RAG-mediated transposition and indicate that target site selection of RAG-mediated transposition may not be as random as previously thought.
MATERIALS AND METHODS
Protein purification
The RAG proteins used in the intramolecular transposition reaction were the GST-fusion core RAG1 protein and the his-myc tagged core RAG2 protein. The plasmids expressing the RAG proteins (pEBG-R1C and pEBB-R2C) were co-transfected into 293T cells. The transfected cells were harvested and the RAG proteins were partially purified by a procedure similar to that used in the purification of the GST-core RAG2 fusion protein (25). The RAG1 protein used in the intermolecular transposition assay was the recombinant MBP-core RAG1 fusion protein purified from Escherichia coli BL21 transformed with pCJM233 as described previously (13). The RAG2 protein used was a GST-core RAG2 fusion protein purified from 293T cell transfected with pEBG-2ΔC as described. HMG2 (amino acids 1–185 with a His tag at the N-terminus) was a gift from Isabelle Villey.
DNA and oligonucleotide substrates
The body-labeled in vitro cleavage substrate (+HS 317) was generated by PCR from pC317SB using primers LE1 and Cit4a (18). The –HS 317 substrate was made by a two-step PCR with which the hot spot was replaced with an A-T-rich sequence as shown in Figure 1A. First, two PCRs were carried out using either LE1 or Cit4a in conjunction with the primers bearing the sequence that replaces the hot spot. The gel-purified PCR products were mixed and used as the template for a second PCR using primers LE1 and Cit4a. This two-step PCR mutagenesis effectively replaced the hot spot, as analyzed by restriction enzyme HpaII digest (data not shown). The cold double stranded oligonucleotide substrates were annealed in the 0.5× T4 PNK buffer (New England Biolabs) at a concentration of 1 µM, then diluted to the working concentrations. The quality of the annealed oligonucleotides was examined by a 10% PAGE and visualized by staining with SYBR green. All radioactively labeled oligonucleotide substrates were gel purified. The list of short oligonucleotide targets (31 bp) is as follows (top strand only is shown): +HS target, 5′-cgctcggttgccgccgggcgtactatattga-3′; –HS target, 5′-cgctcggttatactacataatactatattga-3′; target a, 5′-cgctcggttatagccgggcgtactatattga-3′; target b, 5′-cgctcggttgc cgccggtaatactatattga-3′; target c, 5′-cgctcggttatagccggtaatactatattga-3′; target d, 5′-cgctcggttataaccggtaatactatattga-3′; target e, 5′-cgctcggttatagccgataatactatattga-3′; target f, 5′-cgctcggttatagcaggtaatactatattga-3′; target g, 5′-cgctcggttataaccgataatactatattga-3′.
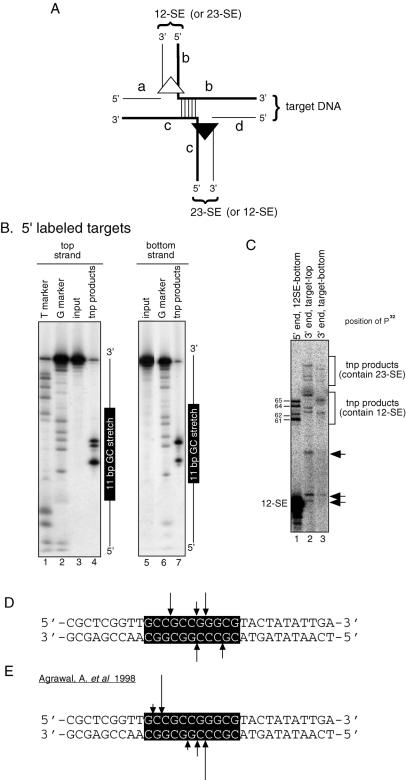
Figure 1.
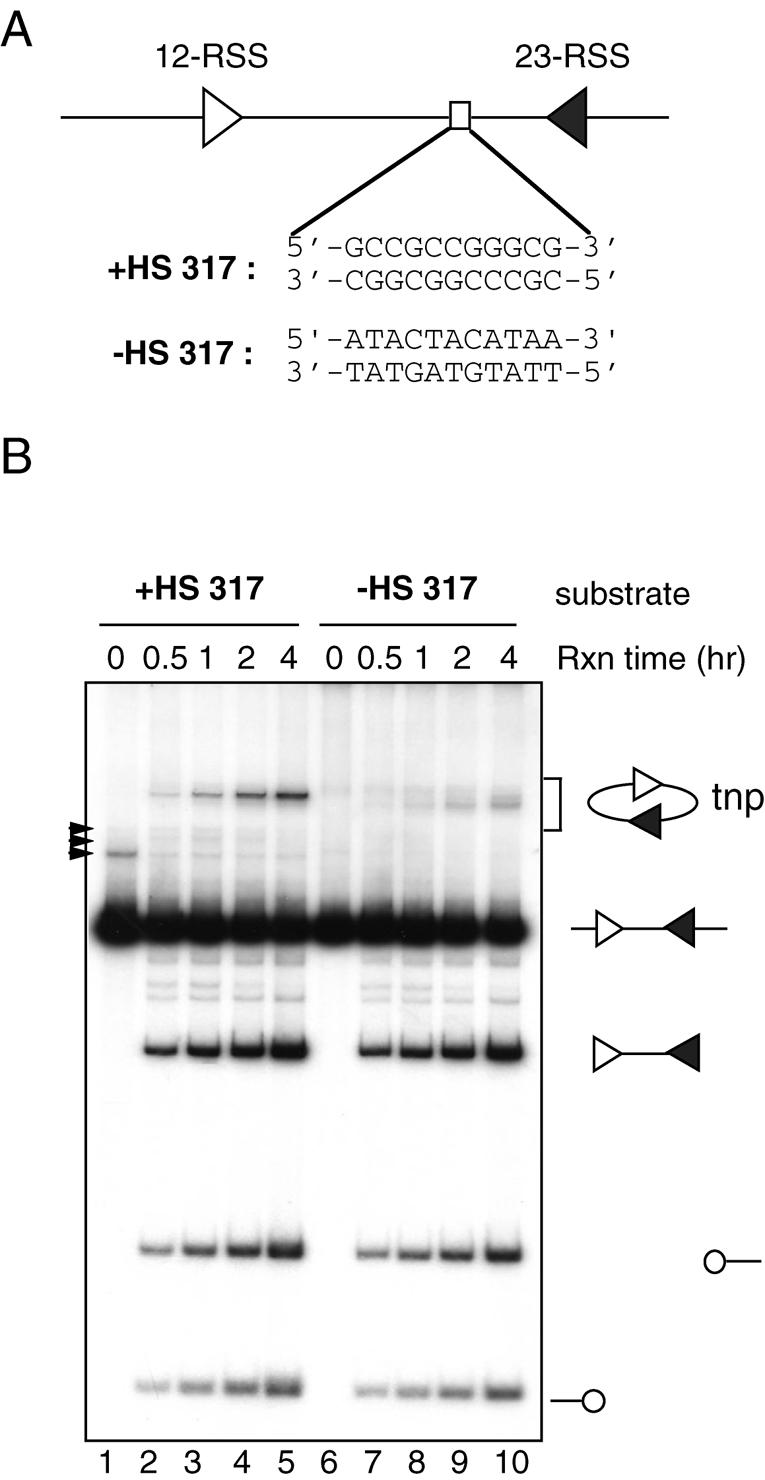
Identification of a hot spot for RAG-mediated intramolecular transposition. (A) Schematic diagram of the in vitro cleavage substrate (+HS 317) depicting the 12-RSS (open triangle), 23-RSS (closed triangle) and putative hot spot for transposition (open rectangle). The sequences of the 11 bp GC stretch as well as the 11 bp AT-rich sequence present in the –HS 317 substrate are shown. (B) Effect of the 11 bp GC stretch on DNA cleavage and transposition was assessed by comparing the reaction products of the DNA substrates with (+HS 317C substrate, lanes 2–5) or without (–HS 317 substrate, lanes 7–10) the 11 bp GC stretch. Structures of the substrates and reaction products are indicated at the right. Open circles indicate hairpin-coding ends and arrowheads indicate non-specific PCR products. tnp, intramolecular transposition products.
Targets containing DNA mismatches were derived from the –HS target and the sequences of DNA mismatches are shown in Figures 5C, and 6A and C. Hairpin targets were made as oligonucleotides in which the 3′ end of the top strand was connected to the 5′ end of the bottom strand with a phosphodiester bond. 100 bp targets (target I, II, III and IV) were derived from the coupled cleavage substrate (+HS 317C). Oligonucleotides were gel purified prior to annealing to form double stranded targets. Changes in sequences are depicted in Figure 7A. The sequence of the target II is the following (top strand only is shown): 5′-gcaggtctccagtaatgacctcagaactccatctggatttgttcagaacgctcggttgccgccgggcgttttttat tggtgagaatcgcagcaacttgtc-3′.
Figure 5.
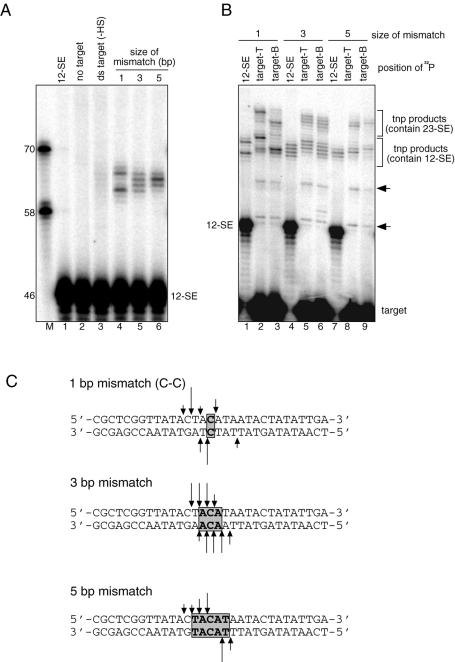
DNA mismatches can stimulate and direct RAG-mediated transposition. (A) Intermolecular transposition reactions were carried out using a double stranded target (lane 3) or the target containing various DNA mismatches as indicated above the lanes [the sequences of these target DNA are shown in (C)]. Sizes of DNA markers and 12-SE are indicated on the left. (B) The origin of transposition products was deduced by comparing those formed using 12-SE labeled at the 5′ end (e.g. lane 1) with those formed using target DNA labeled at the 3′ end of either strand (e.g. lanes 2 and 3). The two groups of transposition products observed in the reaction containing 3′ end-labeled target are indicated on the right. Arrows indicate the transposition products unique to the reactions containing 3′ end labeled target. (C) Sequences of the targets in (A). Gray rectangles indicate the regions of DNA mismatches. Arrows indicate target sites deduced from the same mapping procedure as shown in Figure 3C. The length of the line is roughly proportional to the frequency with which the target site was used.
Figure 6.
All single base pair mismatches stimulate RAG-mediated transposition. (A) Intermolecular transposition reactions were carried out with the –HS targets in which the nucleotides in the central base pair were replaced with different nucleotide combinations, resulting in two targets with perfect base pairing (lanes 3 and 4), and eight targets with single base pair mismatches (lanes 5–12). The definition of the top and bottom strands is based on the –HS substrate. The size of the 12-SE and DNA markers is indicated at the left. (B) Target site mapping of the target containing T-T mismatch (as in Fig. 5B). Arrowheads indicate reaction products unique to reactions containing 3′ end labeled targets, as in Figure 3C. (C) Target sites of the targets containing C-C or T-T mismatch deduced from Figures 5B and 6B, respectively, are indicated as arrows. The length of the line is roughly proportional to the frequency with which the target site was used.
Figure 7.
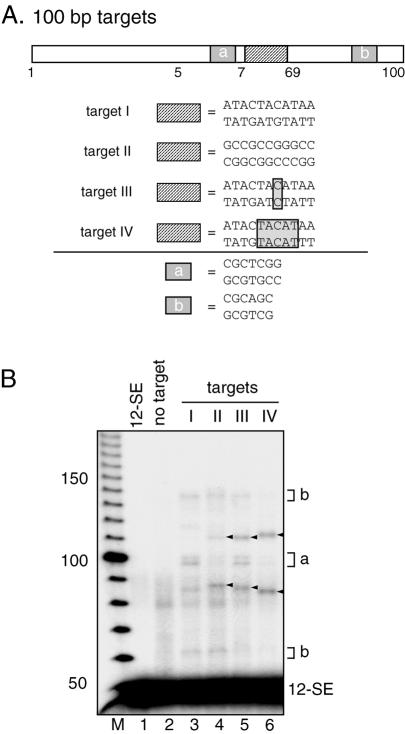
Intermolecular transposition using 100 bp targets. (A) Schematic diagram of 100 bp targets depicting the region of interest where a hot spot or DNA mismatch is located (hatched box), and two GC-rich regions (gray rectangles a and b) which serve as preferred sites of transposition. (B) Intermolecular transposition reactions were carried out in the absence or presence of targets as indicated above the lanes. Arrowheads indicate products that arise due to transposition into the hot spot (lane 3) or mismatches (lanes 4–6). In the absence of the hotspot or mismatches, transposition occurs primarily into regions a and b. M, 10 bp marker.
3′ End labeling of oligonucleotide targets by TdT
3′ End labeling reactions were carried out in a 20 µl reaction containing 4 µl of 5× TdT buffer, 5 pmol of either the top or bottom strand of targets, 8 pmol of 32P cordycepin (5000 Ci/mmol) and 2 µl of TdT (Gibco, 5 U/µl). Reactions were incubated at 37°C for 30 min and terminated by incubating at 70°C for 10 min. Duplex targets were made by annealing the labeled strand with 7 pmol of unlabeled complementary strand. Annealed targets were gel purified to remove single stranded oligonucleotides. The 3′ labeling reaction yielded targets with a 1-nt overhang at the 3′ end of the labeled strand.
Transposition reaction
In vitro couple cleavage/intramolecular transposition reactions were carried out as described (13), except that the RAG proteins used in this study were purified from 293T cells co-expressing both the RAG1 and RAG2 proteins. The amount of the RAG protein used was selected to yield optimal cleavage efficiency. Intermolecular transposition reactions were carried out in a two-step reaction as described (13). Briefly, the signal end complex was formed using equimolar 12- and 23-SE substrates (0.05 pmol) in a 12 µl reaction containing 50 ng of MBP-core RAG1, and 50 ng of GST-core RAG2 in 25 mM MOPS-NaOH (pH 7.0), 75 mM potassium acetate, 100 µg/ml BSA, 4 mM DTT and 5.4 mM CaCl2 at 37°C for 15 min. Subsequently, 3 µl mixtures of 0.2 pmol of labeled target DNA and 25 mM MgCl2 were added to the reaction mixtures and incubated at 37°C for 15–20 min. For the EMSA, the reaction was stopped by placing the reaction on ice and analyzed on a 6% PAGE (80:1). For direct visualization of the ‘b’ strand, the reactions were stopped by adding an equal volume of denaturing dye and incubating the reaction at 100°C for 3–5 min. The reaction products were analyzed on an 8% sequencing gel containing urea.
RESULTS
Identification of a sequence-specific hot spot for RAG-mediated transposition
We previously reported that RAG-mediated transposition is targeted to a particular region of the DNA substrate used in our in vitro intramolecular transposition assay, with 15/41 (37%) of events occurring in a 5 bp portion of the 329 bp substrate (18). One obvious feature of this target region is that it is embedded within an 11 bp GC stretch (5′-GCCGCCGGGCG-3′) (Fig. 1A). Because RAG-mediated transposition is biased toward GC rich target sequences, we considered the possibility that the RAG transposition machinery is targeted to this region by the 11 bp GC stretch. It was also possible, however, that target site selection was biased by positional and topological contraints inherent in the intramolecular transposition assay. To determine if this 11 bp GC stretch is sufficient to constitute a preferred target site for transposition, we first replaced this sequence with an AT rich sequence (5′-ATACTACATAA-3′) and measured the intramolecular transposition efficiency with a standard in vitro cleavage assay involving truncated (core) versions of RAG1 and RAG2, HMG2 and Mg++ (Fig. 1B). Cleavage of the DNA substrate generates a DNA fragment flanked by 2 RSSs (the signal end fragment) and two hairpin-coding ends. After DNA cleavage, the RAG proteins remain stably associated with the signal end fragment and carry out intramolecular transposition in which the 3′ OH groups of the SEs attack the DNA backbone connecting the signal ends. As a result, the linear signal end fragment is converted to a circular DNA containing two 3–5 bp gaps. The intramolecular transposition products can be easily visualized by gel electrophoresis because they migrate above the input substrate in the gel (for example, Fig. 1B, lanes 2–5). As expected, the two DNA substrates were cleaved equally efficiently at all time points examined (Fig. 1B, compare lanes 2–5 with 7–10). Replacement of the 11 bp GC stretch with the AT-rich sequence strongly suppressed the production of the major transposition products observed in the parental substrate (compare lanes 2–5 with lanes 7–10), indicating that the sequence rather than the location of the 11 bp GC stretch is important for targeting the RAG transposition machinery. In addition, the presence of the 11 bp GC stretch appears to enhance overall transposition activity and to suppress the use of other target sites, as suggested by the appearance of other transposition products in the reaction using the DNA substrate lacking the 11 bp GC stretch [note that the use of different target sites gives rise to products with slightly altered mobility in this assay (13,22)]. These results support the idea that the 11 bp GC stretch is a sequence-specific hot spot for RAG-mediated transposition.
The 11 bp GC stretch stimulates RAG-mediated intermolecular transposition into short oligonucleotide targets
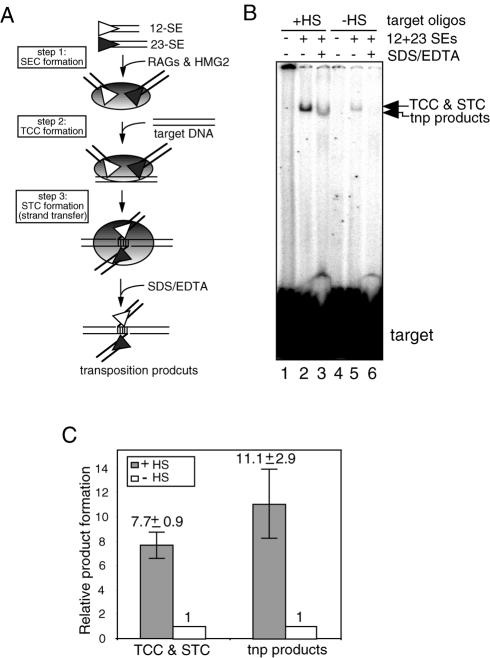
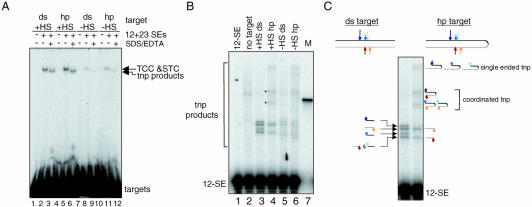
To further characterize this hot spot, we placed the 11 bp GC sequence in a 31 bp target DNA and asked if this sequence acts as a preferred target site for intermolecular transposition. Using oligonucleotide targets allows a rapid assessment of any DNA sequence in the absence of the positional or topological effects inherent in the intramolecular transposition reaction. We first asked if the RAG transposition machinery could preferentially capture the oligonucleotide substrate containing the 11 bp GC stretch using a gel shift assay. Incubation of radioactive target DNA containing the 11 bp GC sequence with the preformed signal end complex resulted in the formation of a slowly migrating band containing two different complexes, the target capture complex (TCC) and strand transfer complex (STC) (Fig. 2B lane 2; a schematic of the transposition reaction is shown in Fig. 2A). The TCC and STC differ in that only in the latter are the target DNA and the SEs covalently linked. To estimate the relative contribution of the TCC and STC to the shifted band, we treated the reaction with SDS/EDTA, which separates the target DNA from the SEs in the TCC and removes the proteins from the transposition products in the STC (illustrated in Fig. 2A). This results in a faster migrating band (the transposition products, lane 3) whose intensity is 30–40% of that of the shifted band in lane 2, indicating that 30–40% of the shifted complex was derived from the STC. The presence of the 11 bp GC stretch stimulates the formation of the TCC and STC by ∼8-fold and the accumulation of the transposition products by 11-fold (compare lane 2 with lane 5 and lane 3 with lane 6, respectively and see Fig. 2C). We conclude that the 11 bp GC stretch is sufficient to stimulate both intramolecular (Fig. 1B) and intermolecular transposition when short oligonucleotide targets are used (Fig. 2B).
Figure 2.
The 11 bp GC stretch stimulates intermolecular transposition into short oligonucleotide targets. (A) Schematic diagram of the DNA-RAG1/2 complexes leading to transposition. Shaded ovals indicate protein complexes. Other symbols as in Figure 1A. (B) TCC and STC formation reactions were carried out to assess the ability of different oligonucleotide substrates to be captured by the RAG transposition machinery. First, the RAG transposition machinery was assembled by incubating the RAG proteins and HMG2 proteins with equimolar amount of the 12-SE and 23-SE in the presence of 5.4 mM CaCl2 at 37°C for 10 min. Formation of the TCC and STC was initiated by adding 5′-end-labeled oligonucleotide target DNA. Deproteinized transposition products were visualized by treating reactions with SDS/EDTA prior to loading on a native 6% (80:1) polyacrylamide gel (lanes 3 and 6). (C) The relative efficiency of TCC and STC formation as well as intermolecular transposition between +HS and –HS targets was quantified and normalized by setting the signal in –HS reactions to 1. The results are the average of four independent experiments.
The 11 bp GC stretch targets RAG-mediated transposition
To determine if RAG-mediated transposition occurs preferentially within the 11 bp GC stretch, we gel purified the shifted band containing the TCC and STC from a large scale reaction at a later time point (60 min) where the majority of the shifted complex was the STC, and analyzed the transposition products on a high resolution sequencing gel (Fig. 3B). As indicated in Figure 3A, only the ‘a’ and ‘d’ strands of the transposition products can be visualized using 5′ end labeled target. Target sites can be estimated by comparing the transposition products to the marker prepared by chemical sequencing reactions. Since the chemical sequencing reactions did not cover every position of the target, and the resulting DNA fragments terminate with a 3′ phosphate group on the preceding nucleotide, whereas the ‘a’ and ‘d’ strands of the transposition products should have 3′ hydroxyl groups, such comparisons could not yield precise mapping. Nonetheless, it was clear that virtually all strand transfer occurred within the 11 bp GC region, with three major bands seen with the top strand labeled substrate and two bands seen with the bottom strand labeled substrate (Fig. 3B, lanes 4 and 7).
Figure 3.
Mapping of transposition target sites. (A) Schematic diagram of the intermolecular transposition product depicting the 12-SE (terminating in an open triangle), the 23-SE (terminating in a closed triangle) and DNA strands (a, b, c and d) relevant to the mapping of target sites. The strands of the target are arbitrarily assigned as the top (a and b) and the bottom (c and d) to better depict the various mapping results; this is not meant to suggest that the 12- or 23-SE inserts preferentially into one strand of the target. (B) Analyses of the ‘a’ and ‘d’ strands of the transposition products. Large scale TCC formation reactions using the +HS target labeled at the 5′ end of either strand were carried out essentially as described in Figure 2A (lane 2). Transposition products from the shifted complex were gel purified and analyzed on a 12% sequencing gel. The T and G markers were generated by chemical sequencing reactions using KnMO4 and DMS, respectively. Locations of the 11 bp GC stretch are indicated at the right. (C) Analyses of the ‘b’ and ‘c’ strands of the transposition products. Intermolecular transposition reactions were carried out using either the 5′ end labeled 12-SE (lane 1), target labeled on the 3′ end of the top strand (lane 2) or bottom strand (lane 3). Reaction products were analyzed on an 8% sequencing gel. Note that 3′-end labeling (lanes 2 and 3) increases the size of the transposition products by 1 nt relative to lane 1. The transposition products containing 12-SE or 23-SE are indicated at the right. Sizes of the transposition products (‘b’ strand) are indicated by the numbers at the left. Arrowheads indicate reaction products unique to reactions containing 3′ end labeled targets. It is not clear how these products are formed. It is possible, however, that they result from attack of the 12-SE or 23-SE at sites close to the 3′ overhang of the labeled targets (a 1-nt overhang is created by the 3′ end labeling reaction). (D) The sequence of the 31-bp target containing the 11 bp GC stretch with target sites indicated by arrows. The length of the line is approximately proportional to the frequency with which the target site was used. (E) Target sites within the 11 bp GC stretch reported in the previous study (18).
To confirm the mapping results, we developed an assay that directly visualizes the DNA strand of the transposition products containing the signal end and the target by 5′ end labeling the bottom strand of the 12-SE (Fig. 3A, ‘b’ strand). Strand transfer of the 12-SE into the target containing the 11 bp GC stretch yielded four major bands (Fig. 3C. lane 1), corresponding to five major transposition products of which three contain the top strand of the target and two contain the bottom strand of the target (Fig. 3C, lanes 2 and 3; Fig. 8C). These four products co-migrated with synthetic oligonucleotide markers designed to correspond to the expected products based on the data of Figure 3B (data not shown). This indicates that the two reactions yield the same products and that the target sites were mapped correctly.
Figure 8.
Hairpin structure is not sufficient to target or stimulate RAG-mediated transposition. (A) TCC and STC formation reactions were carried out to compare the ability of hairpin (hp) and duplex (ds) targets to be captured by the RAG transposition machinery as described in Figure 2A. (B) Target sites were assessed as described in Figure 3C using 5′ end labeled 12-SE. Transposition of the 12-SE into the top strand of the hairpin target results in formation of higher molecular weight transposition products (compare lanes 3 and 4, indicated by asterisks) because in a hairpin structure, the top and bottom strands of the target are covalently linked. They are, however, unlikely to be the result of transposition into the tip of the hairpin since they are present only in the reaction in which the hairpin target contains a hot spot. M (lane 7), oligonucleotide marker corresponding to the transposition product expected for transposition of the 12-SE into the tip of the hairpin containing the 12-SE and entire bottom strand of the +HS target. (C) A schematic diagram depicting various transposition products observed in lanes 3 and 4 (B). Arrows indicate major target sites as mapped in Figure 3.
The ‘b’ and ‘c’ product strands can also be detected by labeling the target DNA at the 3′ end of the top and bottom strands, respectively (Fig. 3A). Such 3′ end labeled targets were also used to map the sites of transposition (Fig. 3C, lanes 2 and 3), yielding results identical to the other mapping assays. In summary, three different approaches have been used to map the sites of transposition into the target containing the 11 bp GC stretch, and all show that transposition occurs at a small number of positions within the GC motif. This strongly suggests that the 11 bp GC stretch is a hot spot for intermolecular transposition. Since mapping of the ‘b’ and ‘c’ strands is more rapid and sensitive (because it does not require gel purification of the transposition products), and yields the same results as mapping of the ‘a’ and ‘d’ strands, we have adopted this mapping method for subsequent experiments.
Curiously, we found that the target sites obtained in the intermolecular transposition assay (Fig. 3D) did not co-localize with those previously obtained in the intramolecular transposition assay (Fig. 3E). The discrepancy is likely due to differences in the sizes of the substrates, topology of the reactions, small differences in the RAG proteins used in these assays and/or differences in the transposition products recovered [some single end events occur in the oligonucleotide assay used here (see below), while only double end events were analyzed by Agrawal et al. (18)]
Characterization of the hot spot
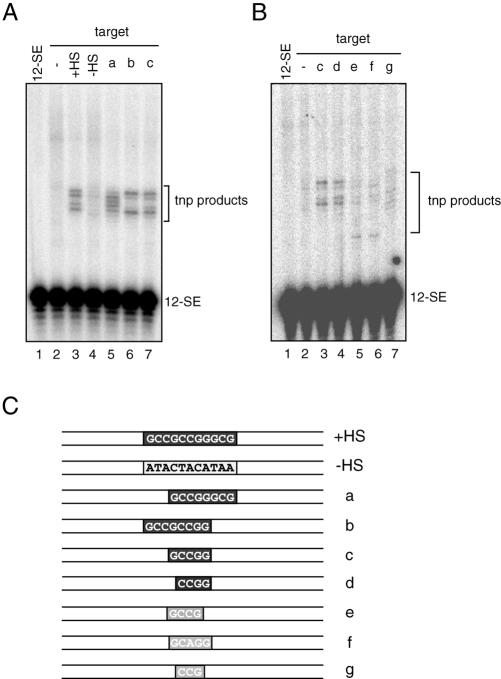
To characterize this hot spot further, we systematically replaced portions of the GC motif with AT-rich sequences (Fig. 4C) and analyzed the ability of these targets to stimulate and direct transposition to the hot spot (Fig. 4A). Removal of the first three GC and/or the last three GC base pairs had only a minor effect on overall transposition efficiency, but substantially altered target site selection (Fig. 4A, compare lane 3 with lanes 5–7, and see below). Since five GC base pairs (5′-GCCGG-3′) still supported efficient transposition, we continued the deletional analyses (Fig. 4B). We found that replacing the first G with A did not substantially change transposition efficiency or target site usage (compare lanes 3 and 4), whereas replacing the last G or the central C with A or T, respectively, reduced the transposition efficiency and altered target site usage (compare lanes 3, 5 and 6, and see below). The evident change in target site usage prompted us to map the sites of strand transfer on these targets (Supplementary Fig. 1, see Supplementary Material available at NAR Online). This confirmed that most transposition occurs within or immediately adjacent to the GC regions, and that changes in the length/sequence of the GC region altered targeting. Taken together, our results indicate that 5′-CCGG-3′ acts as a minimal core sequence sufficient to stimulate and target RAG-mediated transposition in short oligonucleotide targets, and that addition of GC base pairs to the core sequence further enhances targeting efficiency and changes target site selection.
Figure 4.
Characterization of the transposition hot spot. (A) Transposition reactions were carried out using targets bearing the hot spot (lane 3) or various deletions of the hot spot sequence (lanes 5–7) as indicated above each lane and in (C). (B) Further analyses of the 5 bp hot spot in (A). Transposition reactions were carried out as in (A) using the target bearing various mutations of the 5-bp hot spot sequence as indicated above each lane and in (C). (C) Depiction of targets used in (A) and (B) with the area of interest highlighted in boxes. The darkness of the shading of each box indicates the relative efficiency with which a given target stimulates transposition, black being the most efficient target and light gray being the least efficient target. Target sites of transposition into each substrate were mapped and shown in Supplementary Figure 1.
DNA mismatches stimulate and target RAG-mediated transposition
Tracks of four or more GC base pairs have a relatively unstable structure, as indicated by surprisingly high base pair opening rates (26). Thus it is possible that the GC hot spots defined above have a propensity to adopt an unpaired DNA structure that facilitates RAG-mediated transposition. There is considerable precedent for this, since altered DNA structures have been shown to serve as preferred targets for a number of transposases (27–29). To investigate whether the RAG proteins prefer altered target DNA structure(s) for transposition, we introduced various DNA mismatches into the target DNA not containing a hot spot (for target sequence, see Fig. 5C) and measured the efficiency of RAG-mediated transposition. Transposition was strongly stimulated by the presence of 1, 3 or 5 bp mismatches in the target DNA (Fig. 5A, compare lanes 3 with 4–6). Although the various DNA mismatches stimulated transposition to a similar extent, they had distinct effects on target site usage. We thus mapped the sites of transposition into these targets using the procedure described above (data shown in Fig. 5B and results summarized in Fig. 5C). With a 1 bp C-C mismatch, strand transfer occurred predominantly on the same side of the mismatch on both strands (5′ of the mismatch on the top strand and 3′ of the mismatch on the bottom strand; Fig. 5C, first line). As the size of the mismatch increased, target sites remained clustered around the DNA mismatch but now were biased toward the 5′ end of the mismatch on both the top and bottom strands (Fig. 5C). These results strongly suggest that altered DNA structures caused by DNA mismatches can be recognized by the RAG transposition machinery as preferred target sites.
All single base pair mismatches stimulate and target RAG-mediated transposition
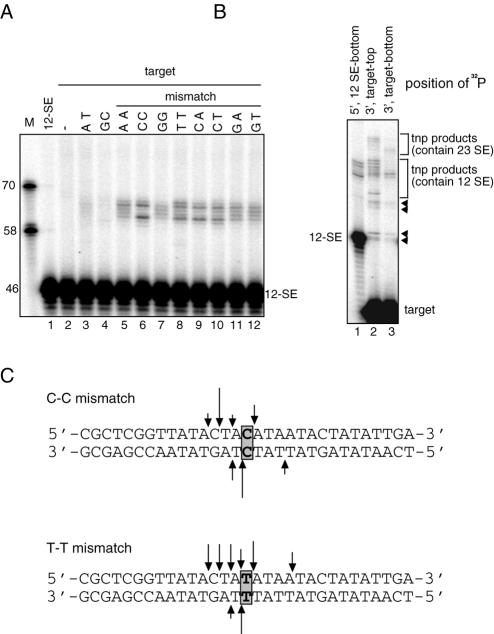
The nucleotides present in the single base pair mismatch may have distinct effects on the DNA structure created by the mismatch, which may affect targeting of the RAG transposition machinery. Using target DNA lacking a GC hot spot, we substituted the central base pair with various nucleotide combinations and assessed the ability of these oligonucleotide substrates to target transposition. All of the combinations resulting in a mismatch stimulated RAG-mediated transposition, whereas the combinations that created perfect double stranded DNA were much less efficient targets (Fig. 6, compare lanes 3 and 4 with lanes 5–12).
To better understand how different DNA mismatches affect target site selection, we also mapped sites of transposition into target DNA bearing a T-T mismatch (Fig. 6B). As was observed with the target containing a C-C mismatch, the target sites were concentrated at one side of the T-T mismatch (Fig. 6C). Target sites were broadly distributed on the top strand but were focused on two major sites on the bottom strand. It is not yet clear what determines target site selection besides the position of the DNA mismatch. These results indicate that the RAG transposition machinery can recognize the altered DNA structures created by various single base DNA mismatches.
Intermolecular transposition reactions using more complex targets
We noticed that RAG-mediated transposition was focused to the central region of the targets even in the absence of a hot spot or DNA mismatch (for example, Figs 4B, lane 2; 6A, lanes 3 and 4). One possibility is that only the central region of the short target (31 bp) is available to the RAG transposase, which would bias results obtained with such targets. Targeting experiments were therefore performed using longer oligonucleotide targets (100 bp) in which the 11 bp GC hot spot or a DNA mismatch (1 or 5 bp) was placed ∼10 bp from the center of the target (Fig. 7A). In the absence of a hot spot or DNA mismatches, RAG-mediated transposition occurred predominantly within two GC-rich regions of the target, labeled a and b (Fig. 7B, lane 3; gray boxes in Fig. 7A). Introduction of a hot spot or DNA mismatch did not stimulate overall transposition efficiency, but their presence substantially altered target site selection (Fig. 7B, compare lane 3 with lanes 4–6). The major new bands present in lanes 4–6 are of the sizes predicted for transposition into the GC hotspot (lane 4) or at the mismatches (lanes 5–6). Furthermore, by moving the 5 bp mismatch to a different site in the 100 bp oligonucleotide, targeting of RAG-mediated transposition to the mismatch was demonstrated to be independent of its location in the target (data not shown). Taken together, these findings indicate that the 11 bp GC hot spot and DNA mismatches are sufficient to target RAG-mediated transposition, and that DNA mismatches are able to stimulate transposition to DNA sequences that are otherwise poor target sequences.
Is a hairpin structure a preferred target for RAG-mediated transposition?
A recent study by Lee et al. found that RAG-mediated transposition could be targeted to the dyad axis of inverted repeats and to hairpins (19). In this study, however, only one synthetic hairpin molecule was analyzed, and hence it was not clear whether the hairpin structure itself, or perhaps some other feature of the particular substrate used, was responsible for the targeting and stimulation of transposition observed. To address this, we created target substrates identical in sequence to those analyzed in Figure 2 (+HS and –HS), but with a hairpin-sealed tip, and assayed for the ability of a hairpin structure to stimulate TCC formation and transposition. In contrast to the findings of Lee et al., we found that a hairpin structure does not significantly stimulate the formation of the TCC or the STC (Fig. 8A, compare lane 2 with 5, and lane 8 with 11), or overall transposition efficiency (compare lane 3 with 6, and lane 9 with 12). This was true whether or not the 11 bp GC hot spot was present in the target. Further analysis of the transposition products, in reactions containing labeled 12-SE donor, suggested that the majority of the target sites were located within the double stranded region of the hairpin targets, and not at the hairpin tip (Fig. 8B). Two major bands present in lane 3 (produced by attack of the 12-SE on the top strand, within the hotspot; see Fig. 3) are absent in lane 4, and are replaced by two new bands whose mobility is that expected for attack of the 12-SE at these same two top strand positions in a hairpin target in which the 23-SE has integrated into the bottom strand (single end insertions in the hotspot on the top strand would yield larger products) (Fig. 8B). Hence, the results suggest that targeting continues to occur within the hotspot despite the presence of the hairpin, and further, that most transposition events (≈70%) are double ended insertions. This is also supported by the absence of similar bands in lane 6, in which the target contains the hairpin but lacks the hotspot (Fig. 8B; lane 7 provides a marker for the product resulting from transposition into the hairpin tip).
These results indicate that, in general, a hairpin structure is not sufficient to stimulate or target RAG-mediated transposition, and that features (presumably of the target) are needed to render hairpins a preferred target site. To search for such features, we analyzed four additional hairpin substrates with different sequences at the tip of the hairpin (see supplementary Fig. 2 for the sequences of hairpin tips analyzed), and found that none of them was able to target or stimulate RAG-mediated transposition (data not shown). We then synthesized the hairpin target used by Lee et al. and found, as they reported (19), that it served as a preferred transposition target (data not shown). Since this hairpin substrate is thus far the only one that exhibits this property, further analysis will be required to identify the additional features that cooperate with the hairpin structure to target and stimulate the reaction. Taken together, our data clearly indicate that hairpin structures are not typically preferred targets of the RAG transposase.
DISCUSSION
Identification of a hot spot for RAG-mediated transposition
In the present study, we have identified a hot spot sequence for RAG-mediated transposition that is constituted of only Gs and Cs (5′-GCCGCCGGGCG-3′). The presence of the hot spot in the target not only focuses transposition to the hot spot but also increases the overall efficiency of both intra- and intermolecular transposition when a short target is used. Formation of TCC/STC is clearly enhanced by the presence of the hotspot, while formation of the strand transfer product appears to be enhanced to an even greater degree (Fig. 2). Taken together, our results suggest that the hot spot stimulates RAG-mediated transposition by two distinct mechanisms: increasing the affinity of the target to the RAG transposition machinery, and facilitating strand transfer.
We would like to propose that melting of the target DNA is required for strand transfer by the RAG transposase, as appears to be the case for other transposases such as Tn10 (29). If this is correct, the mechanism by which the hot spot promotes transposition is likely to be the consequence of high affinity binding of the hot spot containing target to the RAG transposase, since better binding may provide more specific contacts between the target and the RAG proteins, which in turn makes target DNA deformation more energetically favorable. In addition, the hot spot sequence is likely to be more prone to form distorted DNA structures than many other sequences, consistent with the findings that tracts of GC base pairs exhibit high base pair opening rates, indicative of an unstable structure of the double helix (26). It appears that DNA melting is required for many transesterification reactions by the RAG proteins. Like RAG-mediated transposition, several lines of evidence suggest that DNA melting is required for nicking and hairpin formation by the RAG proteins (30,31).
RAG-mediated transposition can be stimulated by and targeted to altered DNA structures, particularly mismatches
Further support for a distorted target DNA structure during RAG-mediated transposition comes from our finding that RAG-mediated transposition is readily stimulated by and directed to the site of a DNA mismatch in a 31 bp target. All possible single base pair mismatches have this effect, indicating that the altered structure rather than the specific sequence of the DNA mismatch is recognized by the RAG transposase. A very similar finding was recently reported for bacteriophage Mu (27), suggesting that target DNA melting during transposition may be a common feature of many transposases. In the case of a single base pair DNA mismatch, it appears that the DNA mismatch cooperates with other information (sequence or GC content) to determine where in the target strand transfer occurs, with biases toward one strand and toward one side of the mismatch observed in some cases.
In a somewhat more complex (100 bp) target, the hot spot or DNA mismatches do not stimulate overall transposition efficiency, but continue to target RAG-mediated transposition, independent of location in the target. The failure of the hot spot or mismatches to stimulate transposition efficiency may be due to the presence of other favorable target sequences. This is suggested by the finding that, in the absence of the hot spot or mismatches, transposition occurs preferentially into GC-rich regions (a and b in Fig. 7). Interestingly, the presence of a 5 bp mismatch suppresses the usage of other target sites and, as a result, RAG-mediated transposition is almost exclusively targeted to the site of DNA mismatch (Fig. 7B, lane 6). Taken together, the results indicate that in a very complex target, such as the mammalian genome, many favorable target sequences will exist, but the presence of mismatches or similar DNA distortions might still significantly influence target site selection.
Our results suggest that results from the gel shift assay for TCC/STC formation (e.g. Fig. 2A) should be interpreted cautiously. All targets containing a single base pair mismatch stimulate strand transfer (Fig. 6A), but only a subset of them (the C-C, C-A and C-T mismatches) stimulate TCC/STC formation using the gel shift assay of Figure 2A (data not shown). While the reasons for this are unclear, it is apparent that not all good transposition targets are detected as such by the gel shift assay.
Hairpins and hybrid joint formation
Lee et al. (19) recently reported that inverted repeats (capable of cruciform formation) and hairpin structures serve to stimulate RAG-mediated transposition. Based on the fact that RAG-mediated hybrid joint formation also involves strand transfer into hairpin (coding) ends, they suggested that hybrid joint formation might in fact represent transposition, with the target (a hairpin end) being a strongly preferred one. Our finding that six different hairpins (with five different sequences at the hairpin tip; see supplementary Fig. 2) failed to serve as good targets for RAG-mediated transposition indicates that hairpins are not generally preferred transposition targets. Based on our results, we would predict that most coding ends would not serve as efficient targets for transposition. Interestingly, RAG-mediated hybrid joint formation by the full-length RAG proteins is extremely inefficient in vivo (32), and is inhibited in vitro by full length RAG2 (22,23). It is likely that virtually all hybrid joint formation in vivo relies on the non-homologous end joining machinery (32), with the RAG proteins serving a non-catalytic, scaffold function (13).
Our observation that mismatches enhance and target RAG-mediated transposition may explain why inverted repeats strongly stimulate transposition and why almost all strand transfer events were observed to occur near their dyad axis (19). Inverted repeats have the potential to form cruciforms, but the cruciforms themselves (consisting of two hairpins) are unlikely to be the preferred target structures. Rather, our data suggest that partially melted intermediates, lying on the pathway between cruciform and standard duplex DNA, are the structures that are targeted preferentially by the RAG-transposase.
Non-random targeting in the genome
Our results suggest that RAG-mediated transposition may not target sequences in the genome as randomly as previously thought. Chromatin is a highly dynamic structure, subject to covalent and conformational modifications by a huge array of different factors and processes. Of particular relevance to RAG-mediated transposition may be transcription, replication, DNA damage/mutation (to create mismatches or other distorted local structures) and DNA repair. Our results indicate that target site selection by the RAG-transposase may be linked to cellular processes that temporarily alter DNA structure [as also suggested by Lee et al. (19)]. We note that HIV integrase, which integrates the HIV genome through a mechanism similar to that of RAG-mediated transposition, exhibits a target site preference for actively transcribed genes (33).
To date, only one unambiguous RAG-mediated transposition event has been identified in vivo (24), suggesting that the transposase activity may be suppressed in vivo and that such events are rare. Our findings that RAG-mediated transposition can be stimulated by and targeted to specific DNA sequences and structures, along with the observation that inverted repeats are preferred targets (19), provide guidelines for the development of more sensitive assays for the detection of RAG-mediated transposition in vivo.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the Schatz laboratory for stimulating discussions. Oligonucleotide synthesis and purification was performed by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. M.C. is a recipient of the Anna Fuller Fund Postdoctoral Fellowship. This work was supported by grant AI32524 to D.G.S. from the National Institutes of Health. D.G.S. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Fugmann S.D., Lee,A.I., Shockett,P.E., Villey,I.J. and Schatz,D.G. (2000) The RAG proteins and V(D)J recombination: complexes, ends and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. (2002) V(D)J recombination: rag proteins, repair factors and regulation. Annu. Rev. Biochem., 71, 101–132. [DOI] [PubMed] [Google Scholar]
- 3.Schatz D.G., Oettinger,M.A. and Baltimore,D. (1989) The V(D)J recombination activating gene (RAG-1). Cell, 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- 4.Oettinger M.A., Schatz,D.G., Gorka,C. and Baltimore,D. (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- 5.Mundy C.L., Patenge,N., Matthews,A.G. and Oettinger,M.A. (2002) Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol., 22, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones J.M. and Gellert,M. (2002) Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J., 21, 4162–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grawunder U. and Harfst,E. (2001) How to make ends meet in V(D)J recombination. Curr. Opin. Immunol., 13, 186–194. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A. and Schatz,D.G. (1997) RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell, 89, 43–53. [DOI] [PubMed] [Google Scholar]
- 9.Hiom K. and Gellert,M. (1998) Assembly of a 12/23 paired signal complex: A critical control point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- 10.Sadofsky M. (2001) The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res., 29, 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leu T.M.J., Eastman,Q.M. and Schatz,D.G. (1997) Coding joint formation in a cell free V(D)J recombination system. Immunity, 7, 303–314. [DOI] [PubMed] [Google Scholar]
- 12.Ramsden D.A., Paull,T.T. and Gellert,M. (1997) Cell-free V(D)J recombination. Nature, 388, 488–491. [DOI] [PubMed] [Google Scholar]
- 13.Tsai C.-L., Drejer,A.N. and Schatz,D.G. (2002) Evidence of a critical architectural function for the RAG proteins in end processing, protection and joining in V(D)J recombination. Genes Dev., 16, 1934–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livàk F. and Schatz,D.G. (1997) Identification of V(D)J recombination coding end intermediates in normal thymocytes. J. Mol. Biol., 267, 1–9. [DOI] [PubMed] [Google Scholar]
- 15.Ramsden D.A. and Gellert,M. (1995) Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev., 9, 2409–2420. [DOI] [PubMed] [Google Scholar]
- 16.Perkins E.J., Nair,A., Cowley,D.O., Van Dyke,T., Chang,Y. and Ramsden,D.A. (2002) Sensing of intermediates in V(D)J recombination by ATM. Genes Dev., 16, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiom K., Melek,M. and Gellert,M. (1998) DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell, 94, 463–470. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal A., Eastman,Q.M. and Schatz,D.G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature, 394, 744–751. [DOI] [PubMed] [Google Scholar]
- 19.Lee G.S., Neiditch,M.B., Sinden,R.R. and Roth,D.B. (2002) Targeted transposition by the V(D)J recombinase. Mol. Cell. Biol., 22, 2068–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth D.B. and Craig,N.L. (1998) V(D)J recombination—a transposase goes to work. Cell, 94, 411–414. [DOI] [PubMed] [Google Scholar]
- 21.Melek M. and Gellert,M. (2000) RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell, 101, 625–633. [DOI] [PubMed] [Google Scholar]
- 22.Tsai C.-L. and Schatz,D.G. (2003) Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J., 22, 1922–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkin S.K., Matthews,A.G. and Oettinger,M.A. (2003) The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J., 22, 1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messier T.L., O’Neill,J.P., Hou,S.M., Nicklas,J.A. and Finette,B.A. (2003) In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J., 22, 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spanopoulou E., Zaitseva,F., Wang,F.-H., Santagata,S., Baltimore,D. and Panayotou,G. (1996) The homeodomain of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell, 87, 263–276. [DOI] [PubMed] [Google Scholar]
- 26.Dornberger U., Leijon,M. and Fritzsche,H. (1999) High base pair opening rates in tracts of GC base pairs. J. Biol. Chem., 274, 6957–6962. [DOI] [PubMed] [Google Scholar]
- 27.Yanagihara K. and Mizuuchi,K. (2002) Mismatch-targeted transposition of Mu: a new strategy to map genetic polymorphism. Proc. Natl Acad. Sci. USA, 99, 11317–11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuduvalli P.N., Rao,J.E. and Craig,N.L. (2001) Target DNA structure plays a critical role in Tn7 transposition. EMBO J., 20, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pribil P.A. and Haniford,D.B. (2000) Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. J. Mol. Biol., 303, 145–159. [DOI] [PubMed] [Google Scholar]
- 30.Ramsden D.A., McBlane,J.F., van Gent,D.C. and Gellert,M. (1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 31.Cuomo C.A., Mundy,C.L. and Oettinger,M.A. (1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol., 16, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekiguchi J., Whitlow,S. and Alt,F. (2001) Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell, 8, 1383–1390. [DOI] [PubMed] [Google Scholar]
- 33.Schroder A.R., Shinn,P., Chen,H., Berry,C., Ecker,J.R. and Bushman,F. (2002) HIV-1 integration in the human genome favors active genes and local hotspots. Cell, 110, 521–529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.