Abstract
Fungal species belonging to the genus Trichoderma colonize the rhizosphere of many plants, resulting in beneficial effects such as increased resistance to pathogens and greater yield and productivity. However, the molecular mechanisms that govern the recognition and association between Trichoderma and their hosts are still largely unknown. In this report, we demonstrate that plant-derived sucrose (Suc) is an important resource provided to Trichoderma cells and is also associated with the control of root colonization. We describe the identification and characterization of an intracellular invertase from Trichoderma virens (TvInv) important for the mechanisms that control the symbiotic association and fungal growth in the presence of Suc. Gene expression studies revealed that the hydrolysis of plant-derived Suc in T. virens is necessary for the up-regulation of Sm1, the Trichoderma-secreted elicitor that systemically activates the defense mechanisms in leaves. We determined that as a result of colonization of maize (Zea mays) roots by T. virens, photosynthetic rate increases in leaves and the functional expression of tvinv is crucial for such effect. In agreement, the steady-state levels of mRNA for Rubisco small subunit and the oxygen-evolving enhancer 3-1 were increased in leaves of plants colonized by wild-type T. virens. We conclude that during the symbiosis, the sucrolytic activity in the fungal cells affects the sink activity of roots, directing carbon partitioning toward roots and increasing the rate of photosynthesis in leaves. A discussion of the role of Suc in controlling the fungal proliferation on roots and its pivotal role in the coordination of plant-microbe associations is provided.
Animals and plants are exposed to a complex and dynamic consortia of beneficial (nonpathogenic) bacteria and fungi, such as in the oral cavity, the intestines, and the rhizosphere (Hooper et al., 1998; McFall-Ngai, 1998; Palmer et al., 2001; Hirsch et al., 2003; Dethlefsen et al., 2007). These beneficial interactions usually result in a heightened resistance to prevent subsequent pathogen attacks or establish a commensal association with the host. Therefore, the identification and understanding of the molecular mechanisms that regulate such nonpathogenic associations is warranted.
Among the mutualistic associations in the rhizosphere, the most studied systems are the symbioses between rhizobacteria and leguminous plants and the mycorrhizal associations of fungal species with numerous trees and crops (Hirsch et al., 2003; Sarma et al., 2007; Parniske, 2008). During these interactions, a molecular dialogue between both partners leads to metabolic modifications of host tissues, providing a favorable environment for the symbiont. In the case of mycorrhizal associations, plant-derived carbohydrates are exchanged for essential nutrients supplied by the fungus (Hahn and Mendgen, 2001; Nehls et al., 2001). It has been proposed that enzymes from the plant hydrolyze Suc, and the monosaccharides that are released are transported by the fungal cells (Blee and Anderson, 1998; Nehls et al., 2001). Importantly, the hexose concentration in the apoplast is speculated to control fungal metabolism during the symbiosis, with the monosaccharides acting as nutrients and, at the same time, as signals that regulate gene expression in the fungal cell (Nehls et al., 2001; Nehls, 2008).
Trichoderma species are nonpathogenic, soil-borne (free-living) fungi that also colonize roots of numerous plants (Harman et al., 2004). These fungi are recognized for their important benefits to agriculture, such as their ability to protect crops against diseases and increase crop yield under field conditions (Harman et al., 2004). Trichoderma-treated plants have demonstrated an increase in biomass production (Chang et al., 1986), induced systemic resistance (ISR) to diseases (Yedidia et al., 1999; Djonović et al., 2006, 2007a; Viterbo et al., 2007), increased nutrient uptake, as well as higher efficiency for fertilizer utilization and seed germination rates (Yedidia et al., 2001). The hyphae of Trichoderma grow mainly in the intercellular spaces of roots, where they are restricted to epidermal cell layers and a few cells beyond this level (Yedidia et al., 1999; Harman et al., 2004). This invasion pattern is suggestive of a colonization process strictly controlled by both partners; however, the biochemical pathways involved in the control of such an association are not well understood.
One of the most intriguing outcomes of plant-Trichoderma associations is the systemic control of gene expression in distant leaves. Proteomic and transcriptomic studies have demonstrated that when Trichoderma colonizes plant roots, it induces a systemic change in the expression of plant genes involved in the scavenging of reactive oxygen species, stress responses, isoprenoid oxylipins and ethylene biosynthesis, photosynthesis, photorespiration, and carbohydrate metabolism (Djonović et al., 2006, 2007a; Alfano et al., 2007; Segarra et al., 2007, Shoresh and Harman, 2008). The effect of peptaibols (peptide antibiotics) and the small protein Sm1, both produced by the fungal cells, has been shown to be responsible for the systemic activation of the defense responses in leaves (Djonović et al., 2006; Viterbo et al., 2007). In cucumber (Cucumis sativus) plants, a mitogen-activated protein kinase was also demonstrated to be involved in the signaling events that lead to the induction of resistance (Shoresh et al., 2006). However, the mechanisms that Trichoderma employs to detect the presence of plants in the surroundings to initiate the root invasion remain to be discovered.
By secreting compounds such as organic acids, amino acids, and sugars into the rhizosphere, plants provide carbon-rich resources and also establish a molecular communication with soil microbes that triggers root colonization (Bais et al., 2006). One of the main carbohydrates secreted by plant roots is Suc, which has been detected in high concentrations near root tips (Kraffczyk et al., 1984; Jaeger et al., 1999; Mahmood et al., 2002). However, the function of this exuded disaccharide has been barely explored, with studies mainly focused on its relationship to the uptake of minerals and in the association of plants with bacterial communities (Kraffczyk et al., 1984; Jaeger et al., 1999; Mahmood et al., 2002; Baudoin et al., 2003). On the other hand, it is well established that Suc plays vital roles in the control of plant developmental processes. As the main product of photosynthesis, Suc is produced in source leaves and exported to heterotrophic tissues. Suc controls carbohydrate distribution, acts as an important molecule in carbohydrate-mediated signaling in the plant, and also is degraded by plant cells to yield a carbon source for microbes during plant-microbe associations (Dennis and Blakeley, 2000; Koch, 2004). During the establishment of many plant-pathogen associations, Suc hydrolysis is stimulated in plant cells to enhance the flux of monosaccharides toward the infection site (Biemelt and Sonnewald, 2006). In addition, the presence of invertases (Inv; β-fructofuranosidases belonging to the glycoside hydrolase family GH32) that hydrolyze Suc has been described in some pathogenic fungal species such as Thermomyces lanuginosus, Uromyces fabae, and Aspergillus niger (Chaudhuri et al., 1999; Rubio and Navarro, 2006; Voegele et al., 2006). In the case of U. fabae, the induction of fungal Inv expression was detected upon penetration of the fungus into the leaf with high expression levels in haustoria. In contrast, with the exception of Laccaria bicolor, mycorrhizal fungi lack sucrolytic activities and rely on monosaccharides provided by the plant (Nehls et al., 2001). Most Trichoderma species are able to grow in vitro using Suc as a substrate, suggesting the presence of enzymes capable of hydrolyzing the disaccharide in fungal cells. Moreover, previous studies with Trichoderma virens demonstrated that the expression of tex1 and sm1, involved in the induction of ISR responses, was specifically up-regulated in the presence of Suc or plant roots (Djonović et al., 2006; Viterbo et al., 2007). Based on these observations, we hypothesize plant-derived Suc (similarly to monosaccharides in mycorrhiza) as a nutrient for fungal cells and also as part of the molecular dialogue between Trichoderma and root cells.
In this study, we investigated the presence of sucrolytic activity in T. virens cells and its relevance to colonization of maize (Zea mays) roots by hyphae of T. virens. We describe the identification and functional characterization of tvinv, a gene encoding an intracellular Inv in T. virens. Reverse genetic experiments demonstrated that TvInv is responsible for Suc hydrolysis and the normal growth of T. virens in the presence of Suc. Our data suggest that plant-derived Suc represents not only a carbon source for the fungal cells but also that the extent of root colonization, production of fungal chlamydospores, and systemic induction of photosynthesis in leaves depend upon its metabolism. Although carbohydrate metabolism, mainly monosaccharides, has been shown to be involved in mycorrhizal associations, we are presenting to the best of our knowledge a novel scenario for microbe-host interaction in the rhizosphere. The ability of Trichoderma to transport and metabolize Suc released by plants may be a specific and fine-tuned strategy for root perception and subsequent colonization that makes these fungal species unique among the symbiotic root colonizers.
RESULTS
Identification and Characterization of an Inv from T. virens
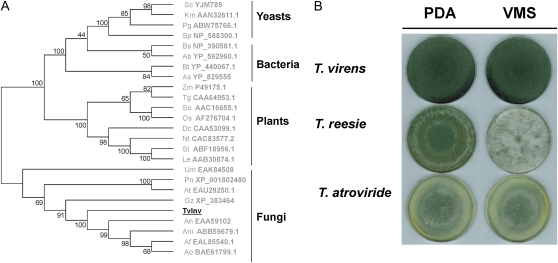
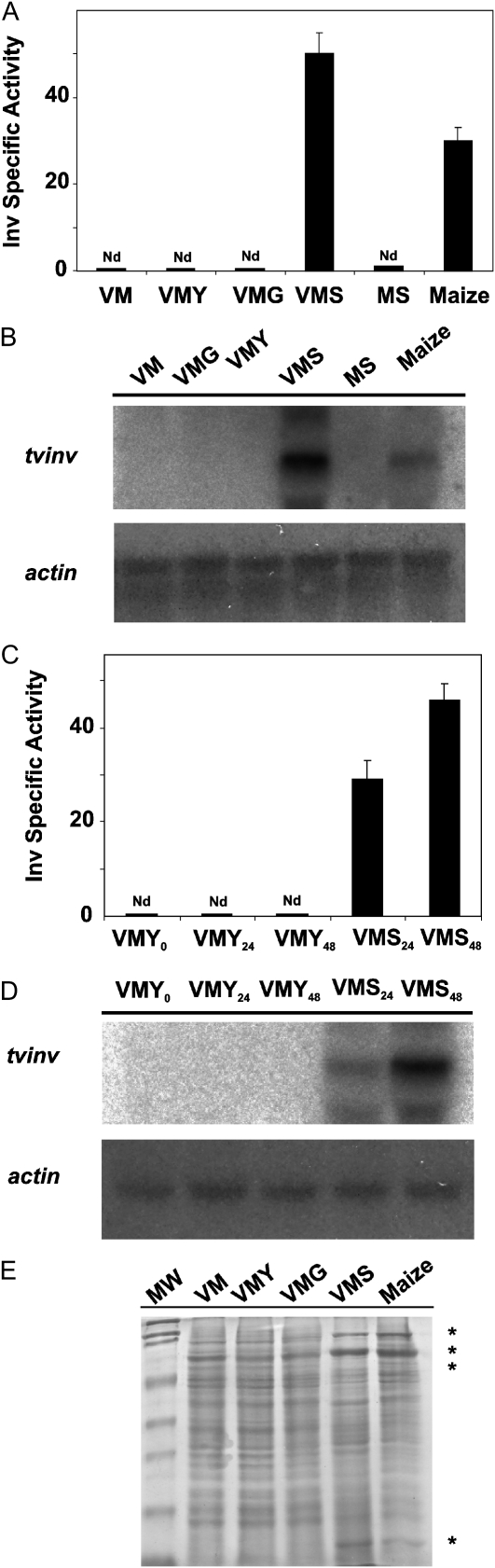
The hypothesis that Suc is part of the molecular communication during Trichoderma-plant associations prompted us to investigate enzymes from T. virens that are involved in Suc metabolism. Using an acid Inv from Saccharomyces cerevisiae (accession no. YJM789) as a query, we identified its homolog (TvInv) in the T. virens Gv29-8 genome (www.genome.jgi-psf.org/Trive1/Trive1.home.html). The genomic sequence contains a predicted 1,902-bp open reading frame coding for a putative 633-amino acid protein (Supplemental Fig. S1). The deduced amino acid sequence is 40% identical to the S. cerevisiae Inv and similar to SucB, the intracellular acid Inv from A. niger (Goosen et al., 2007). TvInv does not contain a transit peptide for secretion or glycosylation sites. Sequence analysis and phylogenetic reconstruction (including homologous sequences from plants, bacteria, and fungi) grouped TvInv with fungal β-fructofuranosidases, levanases, inulinases, and other members of the glycosyl hydrolase family 32 (Fig. 1A). The deduced amino acid sequence of the putative protein displayed the eight conserved motifs associated with catalytic activity in known Inv with acidic optimum pH (Goosen et al., 2007; Supplemental Fig. S1). Among these motifs are four amino acid residues (NDPXG) of the pentapeptide NDPNG, which has been shown to be essential for enzymatic activity in the S. cerevisiae homolog (Reddy and Maley, 1990; Sturm, 1999). Interestingly, we also identified a TvInv homolog sequence in the Trichoderma atroviride genome, but no homolog was detected in the Trichoderma reesei genome (Martinez et al., 2008). A previous report on the carbon utilization profile of T. reesei (Hypocrea jecorina) demonstrated that this fungal species grows very slowly in a medium containing Suc as the sole carbon source (Druzhinina et al., 2006). In good agreement with these observations, T. reesei presented poor hyphal development and sparse sporulation compared with T. virens and T. atroviride when cultured on Vogel's minimal medium supplemented with Suc (VMS; Fig. 1B). To better understand the relevance of TvInv for T. virens growth, we compared gene expression and Inv activity under different cultural conditions. The enzymatic activity was analyzed in protein extracts from fungal mycelia incubated in the presence of different carbon sources or collected from hydroponic systems containing maize seedlings. The enzymatic activity was only detected in fungal cells cultivated in the presence of Suc or in the hydroponic systems (Fig. 2A). Likewise, northern-blot assays demonstrated that tvinv was expressed only in Suc-containing medium or in the presence of plants (Fig. 2B). Additionally, the presence of alternative substrates for growth, such as chitin, fungal cell walls, and starch, failed to induce the expression of tvinv (Supplemental Fig. S2).
Figure 1.
Presence of a putative acid Inv (TvInv) similar to β-fructofuranosidases in the T. virens genome sequence. A, Sequence comparison and phylogenetic reconstruction of β-fructofuranosidases from different organisms, including TvInv. TvInv is presented in boldface and underlined. The tree was constructed with MEGA 4.1 with a bootstrap trial of 1,000. The name of each sequence represents the initials of the organism followed by the GenBank accession number. Ab, Acidobacteria bacterium Ellin345; Af, Aspergillus fumigatus Af293; An, Aspergillus nidulans; Ani, A. niger; Ao, Aspergillus oryzae; As, Arthrobacter sp. FB24; At, Aspergillus terreus; Bs, Bacillus subtilis subsp. subtilis strain 168; Bt, Burkholderia thailandensis E264; Dc, Daucus carota; Gz, Gibberella zeae; Km, Kluyveromyces marxianus; Le, Lycopersicon esculentum (now Solanum lycopersicum); Nt, N. tabacum; Os, Oryza sativa; Pg, Pichia guilliermondii; Pn, Phaeosphaeria nodorum; Sc, S. cerevisiae; So, Saccharum officinarum; Sp, Schizosaccharomyces pombe 972h; St, Solanum tuberosum; Tg, Tulipa gesneriana; Um, Ustilagus maydis 521; Zm, Z. mays. B, In order to compare T. virens, T. atroviride, and T. reesei ability to grow using Suc, spores of each strain were cultured on VMS or PDA medium and photographed after 7 d. [See online article for color version of this figure.]
Figure 2.
TvInv is expressed in the presence of Suc and maize plants. A and B, The levels of enzymatic activity (A) and gene expression (northern blot; B) for TvInv were determined in T. virens hyphae cultured for 4 d in VM or VM supplemented with 1.5% (v/v) glycerol (VMY), Glc (VMG), or Suc (VMS). Also, the expression was tested in fungal tissue that was coinoculated with maize plants in hydroponic systems (Maize) or cultivated in Murashige and Skoog medium (MS) as a control. C and D, The induction of tvinv in the presence of Suc was determined as enzymatic activity (C) or mRNA steady state (northern blot; D). T. virens spores were inoculated in VMY medium, and after 48 h the mycelia was harvested and transferred to fresh VMY or VMS; samples were collected after 0, 24, and 48 h of incubation. All of the experiments were independently repeated twice with similar results. The bars depict mean values ± sd. E, Protein profile in fungal cells incubated in the presence of VM, VMY, VMG, or VMS or when grown with maize plants. Proteins specifically accumulated in cells cultured in VMS or with maize plants are indicated by asterisks. Nd, Not detected.
A time-course experiment demonstrated the presence of TvInv enzymatic activity and mRNA within 24 h of incubation of the fungal hyphae with Suc, increasing after 48 h of culture (Fig. 2, C and D). Inv activity was not detected in culture filtrates from any conditions tested. Specific changes were detected in the intracellular proteomes of T. virens mycelia incubated in the presence of Suc or maize plants in hydroponic systems (Fig. 2E). Compared with the other conditions tested, several polypeptides (indicated by asterisks) differentially accumulated in VMS or in the hydroponic system. These results suggested that the presence of Suc in the medium elicits, in T. virens, very similar responses to those observed in the presence of plant roots, including the up-regulation of tvinv.
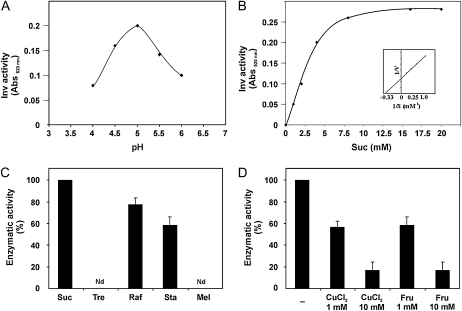
To provide a more complete understanding of TvInv and its biochemical properties, the enzyme was purified from mycelia collected after 48 h of incubation in VMS medium. Protein extracts were prepared, and the enzyme was purified through anion-exchange chromatography followed by gel-filtration chromatography (data not shown). The purified enzyme from T. virens displayed biochemical properties similar to those described for the intracellular Inv from A. niger (Goosen et al., 2007; Fig. 3). The enzyme presented an optimum pH of 5.0 for activity and a Km of 3 ± 0.5 mm for Suc. The purified enzyme hydrolyzed stachyose and raffinose but not trehalose or melizitose, confirming its hydrolytic activity of β-fructofuranosidase. The enzymatic activity was inhibited by Fru and by CuCl2. These results confirmed that T. virens does have an intracellular acid Inv with biochemical properties similar to those of SacB from A. niger and that its expression is only activated in the presence of Suc or plant roots.
Figure 3.
Biochemical characterization of TvInv. A, Optimum pH for activity was assayed in the presence of 50 mm Suc and 100 mm citrate-acetate buffer. This assay was repeated twice with similar results. B, TvInv activity (pH 5) under a gradient of Suc concentration (1–20 mm) was determined, and the Km value for the enzyme was calculated from three different experiments with similar results. The values were calculated from Lineweaver-Burk plots (inset). We present means ± sd of three independent experiments. C, The activity of the purified enzyme was assayed in the presence of 50 mm Suc, trehalose (Tre), raffinose (Raf), stachyose (Sta), and melizitose (Mel) at pH 5. The products of the enzymatic activity were assayed by Somogyi-Nelson methodology. D, The enzymatic activity of TvInv was assayed at pH 5 in the presence of 50 mm Suc with the addition of 0, 1, or 10 mm CuCl2 or Fru. Nd, Not detected.
Loss-of-Function Experiments Confirmed that tvinv Encodes a Functional Acid Inv
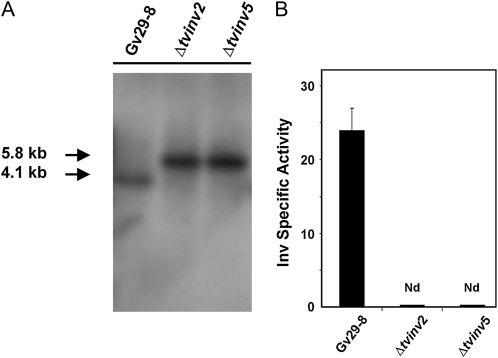
To confirm that tvinv encodes a functional Inv and to perform functional studies, we generated T. virens strains impaired in the expression of tvinv. A disruption cassette was constructed using the double-joint PCR strategy (Supplemental Fig. S3; Kuwayama et al., 2002). The final PCR amplification product was transformed into protoplasts of T. virens Gv29-8, and transformed protoplasts were selected for hygromycin resistance. The stable transformants (18 independent transformants) were evaluated for tvinv disruption by PCR using genomic DNA as a template. Two transformants, designated Δtvinv2 and Δtvinv5, did not contain the wild-type allele (Supplemental Fig. S4A), and no mRNA for tvinv was detected in either of them (Supplemental Fig. S4B). These two null mutants were further analyzed by Southern blotting. The blots were probed with the downstream 1.1-kb DNA fragment used for double-joint PCR (Supplemental Fig. S3, A and B). A 4.1-kb hybridizing fragment was expected for the wild type, a 5.8-kb fragment was expected in the null mutant, and both bands were expected in ectopic integration events (Supplemental Fig. S3A). Both Δtvinv2 and Δtvinv5 presented only the expected hybridization pattern for the null allele (Fig. 4A). As expected, the Inv activity was not detected in protein extracts from the mutant strains cultured in the presence of Suc (Supplemental Fig. S4C) or when inoculated into hydroponic systems containing maize seedlings (Fig. 4B). The absence of Inv activity in protein extracts of the mutant strains confirmed that tvinv encodes the intracellular acid Inv up-regulated in the presence of Suc or maize seedlings.
Figure 4.
Confirmation of tvinv disruption. A, Southern-blot analysis of T. virens wild-type strain (Gv29-8) and tvinv deletion transformants (Δtvinv2 and Δtvinv5). An autoradiograph of a DNA gel blot hybridized with [32P]dCTP-labeled probe is shown in Supplemental Figure S2A. Fifteen micrograms of genomic DNA was digested with HindIII and loaded in each lane. The expected molecular sizes for native and deletion events are indicated on the left. B, TvInv activity in wild-type and mutant cells collected from hydroponic systems. Inv activity was determined in protein extracts from T. virens Gv29-8 and Δtvinv strains coinoculated with maize plants in hydroponic systems. The bar depicts mean values ± sd of two independent experiments. Nd, Not detected.
The Loss of Functional tvinv Suppresses the Ability of T. virens to Grow Using Suc as a Carbon Source
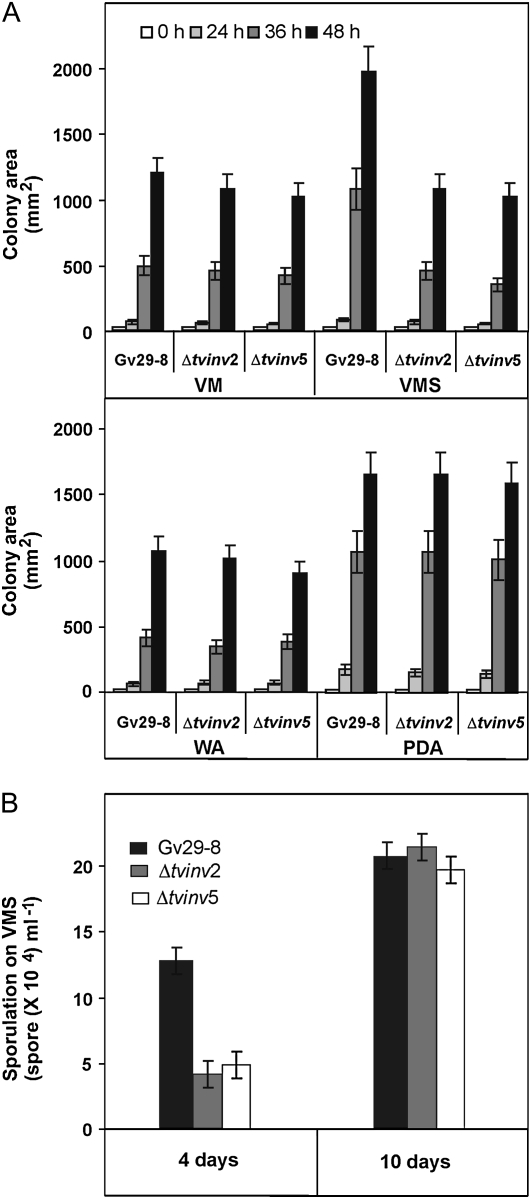
In the rhizosphere, a plethora of natural compounds are delivered by plants and microorganisms. The ability of the soil community to perceive and respond to such stimuli controls its success as a mutualistic ecosystem. Therefore, we sought to determine the role of tvinv in the growth and development of T. virens in the presence of Suc and the extent to which mutant strains were affected in their ability to utilize Suc. Colonies of the null mutant strains, unlike the wild type (Gv29-8), were unable to respond to the addition of Suc (VMS) in the medium. In the presence of Suc, both mutant strains grew at a slower rate, resembling the wild-type strain incubated on minimal medium (Fig. 5A). However, the mutant strains grew at a rate similar to that of Gv29-8 on the other media tested (water-agar [WA], Vogel's minimal medium [VM], or potato dextrose agar [PDA]). When cultured on VMS, initial sporulation in the mutant strains was delayed compared with the wild type (occurring at day 4), but after 10 d in culture, the wild type and mutant strains were similar in their level of sporulation (Fig. 5B). Thus, sporulation initiation was delayed in the mutant strains, reaching similar levels after a prolonged incubation time. No difference in the sporulation process was detected for Gv29-8, Δtvinv2, and Δtvinv5 in WA, VM, or PDA medium.
Figure 5.
Comparison of growth and sporulation among Gv29-8, Δtvinv2, and Δtvinv5. A, The three strains were inoculated in the center of plates containing WA, VM, VMS, or PDA medium. The plates were incubated at 27°C, and growth was monitored at 0, 24, 36, and 48 h. For every strain, three plates with each medium were inoculated in each experiment. The bars depict mean values ± sd of three independent experiments. B, For each strain, a suspension of 103 spores mL−1 was prepared, and 200 μL was spread on each of three VMS plates. After 4 and 10 d, spores were determined from three random plugs (3 mm diameter) per plate. The bars depict mean values ± sd from two independent experiments.
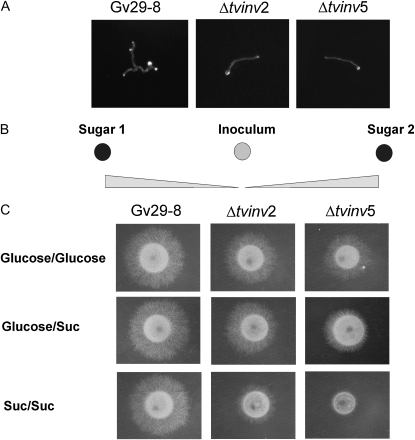
The strains impaired in the expression of tvinv were not affected in their rate of conidial germination with respect to the wild-type strain on WA, VM, VMS, or PDA. However, on VMS medium, a noticeable restriction in the development of the germlings was evident (Fig. 6A). A similar behavior was also observed for Δtvinv chlamydospores, which presented similar germination in WA, VM, VMS, or PDA, but the further development of the germlings was impaired only in VMS (data not shown).
Figure 6.
Germling development and Suc perception by Δtvinv mutants and wild-type strains. A, Germling development of Gv29-8, Δtvinv2, and Δtvinv5 strains after 12 h on VMS medium incubated in moist dark chambers at 27°C. B, Schematic depicting the experimental designs to assess the differential use of sugars by each strain. Spore suspensions for each strain were placed in the center of the plates. Different combinations of two paper discs (sugar 1 and sugar 2) imbibed with 3% (w/v) Suc or 1.5% (w/v) Glc were placed 3 cm away from the center. C, The sugars are expected to diffuse and generate a concentration gradient on both sides of the initial inoculum. Fungal growth was evaluated after 24 h of incubation at 27°C.
Colony morphology and development assays on sugar gradients demonstrated that after conidia germination, the mutant germlings preferably used Glc over Suc to support colony development (Fig. 6, B and C). In this experiment, an aliquot of the spore suspension was placed in the center of a VM plate between two paper discs (sugar 1 and sugar 2) saturated with either Suc or Glc to create a concentration gradient for each sugar after diffusion (Fig. 6B). After incubation for 24 h, the mutant strains preferably grew toward the paper disc saturated with Glc, generating an asymmetric colony (Fig. 6C, row Glc/Suc). The slight growth of the mutants toward the Suc discs is comparable to the slight growth detected in the presence of Suc (Fig. 6C, row Suc/Suc). These results suggested that TvInv is a central component for the utilization of Suc and that Suc may control T. virens development in the rhizosphere.
Disruption of tvinv Disturbs the Symbiotic Association between T. virens and Maize Roots
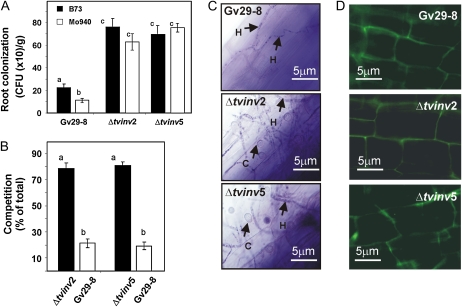
To further analyze the involvement of Suc during plant-Trichoderma associations, we tested the ability of maize plants to exude Suc into the rhizosphere and the ability of strains of T. virens to colonize the roots. By thin-layer chromatography, we detected the presence of Suc in root exudates of 7-d-old maize plants, including the inbred lines B73 and Mo940 as well as the hybrid Silver Queen (Supplemental Fig. S5). To determine the effects that plant-derived Suc may have on the fungal cells, we compared the ability of Gv29-8, Δtvinv2, and Δtvinv5 to colonize roots of B73 (low levels of Suc exudation) and Mo940 (high levels of Suc exudation). The root systems of 2-week-old maize plants grown from seed inoculated with each of the three strains were harvested, weighed, and surface disinfested to eliminate fungal cells attached to the roots (Viterbo et al., 2007). After treatment, the roots were ground and plated on Gliocladium virens specific medium (GVSM; Park et al., 1992). The wild-type strain colonized line B73 at a significantly higher level than Mo949 (Fig. 7A). However, inoculation with either mutant strain resulted in a 4-fold increase in colony-forming units (per gram) with respect to the wild-type strain, and both mutants equally colonized the roots of B73 or Mo940 (Fig. 7A).
Figure 7.
Suc metabolism in T. virens is involved in the coordination of root colonization and fungal development. A, Colonization of maize inbred lines B73 and Mo940 by the wild type (Gv29-8) and mutant strain Δtvinv2 or Δtvinv5. The experiment was performed two times, and colony-forming units (CFUs) were determined in roots of five different plants for each treatment in each experiment. The bars depict mean values ± sd of independent experiments. Bars with different letters differ significantly according to Tukey's HSD test at a significance level of 5%. B, Coinoculation of Gv29-8 and Δtvinv2 or Δtvinv5 on maize roots. The wild-type strain and each of the mutants were equally coinoculated on maize seeds. After 7 d, the root systems were collected and surface sterilized for CFU determination. The ratio of mutant to wild-type colonies was compared for each root system. CFUs were determined in roots of five different plants for each treatment, and the experiment was repeated three times. The bars depict mean values ± sd determined in three independent experiments, and columns with different letters differ significantly according to Tukey's HSD test at a significance level of 5%. C, Bright-field micrographs of root systems of plants grown in hydroponic systems and inoculated with Gv29-8, Δtvinv2, or Δtvinv5. Fungal spores were germinated in VM medium supplemented with 1.5% (v/v) glycerol, and 1 g of fungal hyphae was inoculated into the hydroponic system containing 4-d-old maize plants. Two days after inoculation, roots were collected and photographed. D, Autofluorescence micrographs of maize roots inoculated with Gv29-8, Δtvinv2, or Δtvinv5. Fungal spores were germinated in VM medium supplemented with 1.5% (v/v) glycerol, and 1 g of fungal hyphae was inoculated into the hydroponic system containing 4-d-old maize plants. Two days after inoculation, roots were collected and photographed with a fluorescence microscope.
To understand the effect of plant-derived Suc on root colonization, we examined whether the mutant strains failed to induce plant defense responses that would restrict fungal growth. Maize plants were simultaneously inoculated with the wild type and each of the mutant strains; the number of colony-forming units corresponding to each strain was determined after 2 weeks. By coinoculating with the wild type and a mutant strain, colonization by the wild-type strain was expected to induce any defense response the plant uses to restrict the growth of hyphae. In this case, the colonization extent of the mutant strains would be reduced. The results indicated that the presence of the wild-type strain in the rhizosphere did not have an effect on the colonization extent determined for the mutant strains (Fig. 7B). The difference of root colonization was also observed microscopically after staining the roots for hyphae using a method to detect fungal succinate dehydrogenase activity as described by MacDonald and Lewis (1978). A greater amount of hyphae was detected on roots inoculated with Δtvinv2 or Δtvinv5, and importantly, both mutants formed a greater number of chlamydospores per square millimeter of root than the wild type (4 ± 1.5 and 32.9 ± 4.5 in the wild type and mutants strains, respectively; Fig. 7C).
Similar autofluorescence patterns inside root cells (Fig. 7D) suggest no differential responses of the roots toward the mutant and wild-type strains.
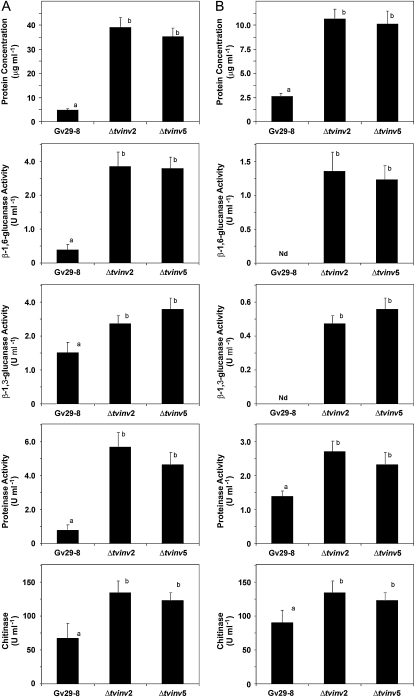
Hydrolytic enzymes play important roles in the root colonization process (Yedidia et al., 1999). In agreement, Δtvinv2 and Δtvinv5 produced higher levels of cell wall-degrading enzymes, which may explain their increased ability to colonize maize roots (Fig. 8). We compared the activity of secreted glucanases, proteinase, and chitinase in both mutants (Δtvinv2 and Δtvinv5) and the wild type when incubated in VMS medium or hydroponic systems with maize plants. In the presence of VMS, both mutants yielded a larger increase (about eight times) in the amount of secreted proteins, β-1,6-glucanase, and proteinase activities in the culture filtrates compared with the wild type (Fig. 8A). This drastic change was also accompanied by a significant increase of the levels of β-1,3-glucanase and chitinase in both mutant strains. A similar effect was also determined when the strains were cultivated in the presence of maize plants. Both Δtvinv2 and Δtvinv5 strains exhibited significant increases in the production of hydrolytic enzymes and secreted proteins compared with Gv29-8 (Fig. 8B). However, such an effect was not observed when the fungal cells were cultured in VM medium supplemented with 1.5% glycerol (data not shown). In good agreement, immunoassays demonstrated higher levels of fungal proteinase polypeptides in protein extracts from maize roots inoculated with Δtvinv2 or Δtvinv5 (Supplemental Fig. S6).
Figure 8.
The activity of hydrolytic enzymes secreted by T. virens is increased in the Δtvinv2 and Δtvinv5 mutant strains. A, Determination of enzymatic activities produced in VMS medium after incubation for 4 d. Spores (106 spores mL−1) from the three strains were germinated on VM supplemented with glycerol for 48 h, and harvested mycelia were inoculated into fresh VMS medium and cultured for an additional 4 d at 25°C. The enzymatic activities were determined in the culture filtrates. The bars depict mean values ± sd of two independent experiments. Bars with different letters differ significantly according to Tukey's HSD test at a significance level of 5%. B, Determination of enzymatic activities produced in hydroponic systems in the presence of maize plants after 4 d of incubation. Spores (106 spores mL−1) from the three strains were germinated on VM medium supplemented with glycerol for 48 h, and mycelia were collected and transferred to the hydroponic systems and then incubated for 4 d at 25°C. The enzymatic activities were determined in the culture filtrates. The bars depict mean values ± sd of two independent experiments. Bars with different letters differ significantly according to Tukey's HSD test at a significance level of 5%.
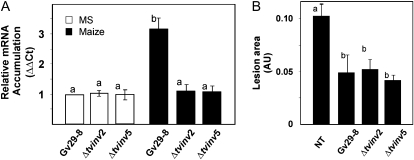
To further investigate the involvement of Suc metabolism in fungus-plant interactions, the expression of sm1 was compared among the mutant and wild-type strains inoculated into hydroponic systems containing maize plants. It was recently described that sm1 is up-regulated in fungal tissues during root colonization. This small protein is part of the molecular dialogue associated with the activation of ISR in maize plants (Djonović et al., 2006, 2007a; Vargas et al., 2008). Reverse transcription followed by real-time PCR experiments showed that the up-regulation of sm1 in the presence of plant roots requires the functional expression of tvinv (Fig. 9A; Supplemental Fig. S7A). However, the mutant strains still were able to induce systemic disease protection in leaves (Fig. 9B; Supplemental Fig. S7B).
Figure 9.
The ability of T. virens to hydrolyze Suc is necessary for the up-regulation of sm1. A, Comparison of the expression of sm1 in Gv29-8, Δtvinv2, or Δtvinv5. Fungal spores were germinated in VM medium supplemented with 1.5% (v/v) glycerol. One gram of fungal hyphae was inoculated into the hydroponic systems containing 4-d-old maize plants or Murashige and Skoog medium (MS) as control. Two days after inoculation, fungal material was collected and sm1 expression was assayed by quantitative real-time RT-PCR. Expression of actin was used as a quantitative internal control. The gene expression data were normalized to the reference control. The data depicted are mean values ± sd of two independent experiments. Bars with different letters differ significantly according to Tukey's HSD test at a significance level of 5%. B, Quantitative analysis of the foliar lesions caused by Colletotrichum graminicola on leaves of maize plants inoculated with Gv29-8, Δtvinv2, or Δtvinv5. The bars depict the average area of lesions (in arbitrary units [AU]) on maize leaves 4 d post inoculation with the pathogen. The data represent mean areas ± sd of five lesions produced on five different plants from two independent experiments. Bars with letters in common did not differ significantly according to Tukey's HSD test at a significance level of 5%. NT, Nontreated.
The Photosynthetic Rate in Leaves of Maize Plants Is Affected by the Ability of T. virens to Use Suc within Roots
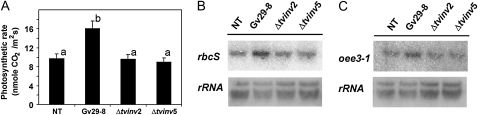
As photosynthesis is the primary energy and carbon supplier supporting the plant, we analyzed the effect on photosynthetic CO2 fixation in leaves when maize roots are colonized by T. virens. Carbon assimilation was measured as an estimate of the rate of CO2 uptake in the distal tip of the third leaf of 14-d-old plants. The results demonstrated that colonization of the maize rhizosphere with T. virens Gv29-8 causes a systemic increase in the uptake of CO2 in leaves (Fig. 10A). Although the mutant strains displayed a greater colonization ability (four times higher) than the wild type (Fig. 7C), neither Δtvinv2 nor Δtvinv5 affected CO2 uptake in leaves compared with nontreated plants (Fig. 10A).
Figure 10.
The expression of tvinv is required for the systemic induction of photosynthesis in maize plants. A, Effect of T. virens strains on photosynthesis. The photosynthetic CO2 uptake was determined in mature leaves of 14-d-old maize plants as an estimation of the photosynthetic rate in maize leaves. The mean ± sd value of two independent experiments is presented. In each experiment, the photosynthetic rate was determined from the third leaf of five different plants and each measurement was repeated twice. Bars with different letters differ significantly according to Tukey's HSD test at a significance level of 5%. B and C, mRNA steady state of rbcS (accession no. X06535) and oee3-1 (accession no. MN_001111878), respectively. Total RNA was extracted from the leaf area where photosynthetic rate was determined. Fifteen micrograms of total RNA was loaded in each treatment and separated on 1% (w/v) agarose gels. The probes were generated using the primers rbcSF/rbcSR and oee31F/oee31R (Supplemental Table S1), and both strands of each probe were sequenced. NT, Nontreated.
To further confirm the systemic effects of T. virens on photosynthesis in maize plants, we analyzed the expression of photosynthesis-related genes in plants inoculated with wild-type T. virens or mutants impaired in the expression of tvinv. Northern-blot assays demonstrated that the accumulation of transcripts of Rubisco small subunit (rbcS) and the oxygen-evolving enhancer 3-1 (oee3-1; from the water-splitting complex) was systemically up-regulated after T. virens Gv29-8 colonized the roots of maize plants (Fig. 10, B and C). The up-regulation of these photosynthetic genes was dependent on Suc degradation in Trichoderma cells, as the up-regulation of these two genes was not detected in plants inoculated with either Δtvinv2 or Δtvinv5. These findings indicate that during the symbiotic association, the degradation of plant-derived Suc in the fungal cells systemically affects gene expression and photosynthetic rate in leaves.
DISCUSSION
Carbohydrates are associated with the most important metabolic processes, such as protein, lipid, and nucleic acid biosynthesis. In animals and plants, sugars have also been described to have important functions as signal molecules regulating gene expression and carbohydrate distribution (Towle, 1995; Kaylor et al., 1997; Pego et al., 2000; Rolland et al., 2001; León and Sheen, 2003; Koch, 2004). In this report, we present evidence that suggests a novel role for Suc metabolism in the coordination of plant interactions with microorganisms in the rhizosphere. The ability of the nonpathogenic fungus T. virens to metabolize Suc is a critical aspect in the control of root colonization of maize plants and for a systemic up-regulation of photosynthesis in leaves. Our findings provide what is to our knowledge a novel scenario for Suc metabolism, photosynthesis, and the control of plant-microbe associations.
In mycorrhizal associations, Suc-derived monosaccharides from the plant support the growth of the fungal cells (Nehls et al., 2001). Mycorrhizal species are unable to directly use Suc and must rely on enzymes from the plant to hydrolyze Suc to deliver monosaccharides for fungal metabolism (Blee and Anderson, 1998; Hahn and Mendgen, 2001). Previous evidence has shown that monosaccharides also act as signals, and their concentration in the apoplast controls the extent of the root invasion (Nehls et al., 2001; Biemelt and Sonnewald, 2006). Both mycorrhiza- and T. virens-plant associations share in common the metabolism of plant-derived carbohydrates; however, different strategies have evolved to obtain these plant resources. Our data suggest that some Trichoderma species have incorporated an intracellular metabolic pathway that facilitates the use of Suc synthesized by the plant. As only oxygenic photosynthetic organisms are able to produce Suc (Salerno and Curatti, 2003), plants are the sole source of Suc in the rhizosphere. The ability of Trichoderma species to use plant-derived Suc in the cytosol may have emerged through coevolution of both organisms, resulting in a commensal relationship.
We demonstrate that the nonpathogenic symbiont T. virens produces TvInv, an intracellular Inv expressed in the presence of Suc or plant roots (Fig. 2). This finding provides novel insights into carbohydrate metabolism and the role of the fungal Inv during the association between T. virens and plants. TvInv is an intracellular Inv with an acidic optimum pH for Suc hydrolysis, and the enzyme displays β-fructofuranosidase activity with biochemical properties similar to those described for an intracellular Inv (SacB) recently characterized from A. niger (Fig. 3; Goosen et al., 2007). BLAST searches in the T. virens genome sequence, using S. cerevisiae Ac-Inv and SacB as queries, retrieved only one homologous sequence (tvinv) displaying the eight motifs conserved in characterized β-fructofuranosidases (Fig. 1A; Supplemental Fig. S1). In addition, two putative membrane-associated Suc transporters, similar to those characterized in plants (Lemoine, 2000), are present in the T. virens genome and complete the pathway for Suc metabolism in Trichoderma cells (W.A. Vargas and C.M. Kenerley, unpublished data). Analysis of strains impaired for tvinv expression showed that TvInv is necessary for the normal growth and development of T. virens in the presence of Suc (Figs. 5 and 6). Moreover, secondary metabolism appears to be also regulated by Suc metabolism in the fungal cells. Thin-layer chromatography experiments demonstrated alteration in the production of secondary metabolites (data not shown) that was also evident by the pigmentation of the culture filtrates (Supplemental Fig. S8). We are pursuing the identification of these metabolites and the metabolic pathways involved in their biosynthesis as possible components of the chemical interaction between T. virens and plants.
Secreted chemical signals from both plants and microbes mediate a complex dialogue that will determine whether an interaction will be malevolent or benign (Bais et al., 2006). Plant roots initiate chemical communication with soil microbes by producing signals that are recognized by the microbes, which in turn produce signals that trigger colonization (Hirsch et al., 2003; Akiyama and Hayashi, 2006; Bais et al., 2006; Bouwmeester et al., 2007; Parniske, 2008; Oldroyd et al., 2009). Only a few chemical signals have been characterized from plants, being mostly restricted to strigolactons and flavonoids exuded by the roots (Hirsch et al., 2003; Akiyama and Hayashi, 2006). Plants can also exude sugars through their roots, but the role of root-exuded sugars is not fully understood (Kraffczyk et al., 1984; Jaeger et al., 1999; Mahmood et al., 2002; Baudoin et al., 2003; Bais et al., 2006). In our studies, we demonstrated that plant-Trichoderma associations are affected by plant-derived Suc, with TvInv activity being crucial for the control of root colonization (Fig. 7). Root colonization by Trichoderma appears to be tightly controlled, with the hyphae restricted to the superficial cell layers of the roots, including the epidermis and some cell layers below (Yedidia et al., 1999). Control of fungal proliferation in mycorrhizal associations has been ascribed to the activation of different responses, including defense mechanisms from the plant cells such as phenyl propanoid biosynthesis, oxidative stress-induced enzymes, and pathogenesis-related genes, as well as the availability of monosaccharides (Gianinazzi-Pearson, 1996; Garcia-Garrido and Ocampo, 2002).
The mechanisms controlling the extent of root colonization by Trichoderma is not fully understood. It was suggested that for penetration of the epidermis and further ingress into the outer cortex, small quantities of cell wall hydrolytic enzymes are produced by the fungus to locally weaken or loosen epidermal cell walls to facilitate the fungal spread into the roots (Yedidia et al., 1999). By ultrastructural studies, Yedidia et al. (1999) showed that the thickening of plant cell walls and callose deposition act as physical barriers that Trichoderma asperellum has to overcome to spread within cucumber roots. In addition, the evidence presented in that report indicated that during root colonization, fungal metabolic status would play an important role in controlling the invasion. T. virens mutant strains impaired in the expression of tvinv, unable to metabolize Suc, presented higher levels of root colonization (Fig. 7). Moreover, we observed that when Gv29-8 and Δtvinv2 or Δtvinv5 were coinoculated on maize roots, the mutant strains still displayed increased colonization ability (Fig. 7B). With the presence of the wild-type strain in the coinoculation experiment, it was expected that any plant defense response to restrict the growth of T. virens (wild type) inside the roots, such as callose deposition or cell wall thickening (Yedidia et al., 1999), would be activated. In this case, root colonization would be reduced for both strains. However, according to the results obtained, the increased root colonization displayed by the Δtvinv mutant strains is independent from such defense responses activated in the roots.
The increased root colonization by Δtvinv2 and Δtvinv5 seems to be controlled by the metabolic status of the fungal cells, with a strong correlation between the production of cell wall-degrading enzymes and the greater ability of the mutant strains to colonize maize roots (Fig. 8). Moreover, the higher levels of fungal proteinase in roots inoculated with Δtvinv2 or Δtvinv5 mutants (Supplemental Fig. S6) suggest that these strains are producing higher levels of hydrolytic enzymes also within the roots. Interestingly, the regulation of the expression of hydrolytic enzymes in different Trichoderma species was reported as being subjected to carbon catabolic repression (Mach et al., 1996; Noronha et al., 2000; Seidl et al., 2008). Carbon catabolic repression of gene expression is a process present in many organisms where Glc and related sugars represses gene expression (Gancedo, 1998). In T. virens, the hydrolysis of Suc provides Glc and Fru, sugars that are involved in the repression of fungal hydrolytic enzymes (Gancedo, 1998; Noronha et al., 2000; Seidl et al., 2008). The control of the expression of enzymes that affect hyphal plasticity (Adams, 2004) ultimately will influence the ability of the fungal cells to spread into roots. When the ability of T. virens to hydrolyze Suc is impaired, catabolic repression mediated by the products of Suc hydrolysis is relieved. The hydrolytic enzymes become derepressed, increasing the level of such enzymatic activities facilitating the fungal dispersion within roots to probe for alternative sources of carbon and energy.
Chlamydospores are produced by many fungi and represent enlarged, thick-walled vegetative cells of varied forms containing condensed cytoplasm (Lin and Heitman, 2005). In some plant pathogenic fungi, chlamydospores have been described as long-term survival structures and important propagules as primary inoculum for plant infection (Abou-Gabal and Fagerland, 1981; Couteaudier and Alabouvette, 1990). The environmental signals for chlamydospore formation in fungi may be species specific and include nutrients, osmolarity, light, temperature, and plant stimulants (Barran et al., 1977; Regulez et al., 1980; Kertesz-Chaloupkova et al., 1998; Kües et al., 1998; Khan et al., 2004). However, the molecular mechanisms of formation and developmental control remain enigmatic (Lin and Heitman, 2005). Species of the genus Trichoderma are also able to form chlamydospores in culture conditions or in the presence of plants (Papavizas, 1985; Chacón et al., 2007), but the environmental conditions that induce this cellular differentiation are unknown. The ability of T. virens to metabolize plant-derived Suc seems to greatly influence chlamydospore differentiation. Even though greater levels of hyphal tissue were present inside roots colonized by the Δtvinv2 and Δtvinv5 mutants, these strains also differentiated a massive number of chlamydospores on the surface of maize roots (Fig. 7C). It is likely that the inability of Δtvinv2 or Δtvinv5 to metabolize root-exuded Suc results in a nutritional stress to the fungal cells growing outside the roots that triggers chlamydospore formation. However, the proliferation of fungal cells inside the roots may be supported by any alternative source of carbon that the fungal cells can withdraw directly from the plant cells. Based on our observations, we speculate that Suc availability during the mutualistic association supplies the nutritional demands of the fungus and regulates the morphological development of T. virens in natural environments.
The beneficial effects of plant-Trichoderma interactions are well described, including protection from plant pathogens and enhancing plant growth and crop yield (Harman et al., 2004). T. virens is known to systematically induce the up-regulation of defense-related genes in plants (Djonović et al., 2006, 2007a; Viterbo et al., 2007; Shoresh and Harman, 2008). Djonović et al. (2006) described the isolation and characterization of a small protein named Sm1, which has the ability to activate ISR in plants. Sm1 was classified as a proteinaceous elicitor that is part of the chemical dialogue and molecular signals that activate plant defenses during the symbiotic association between T. virens and roots. Further experiments demonstrated that the expression of sm1 in the fungal tissue was up-regulated in the presence of plant roots or when cultured in the presence of Suc (Djonović et al., 2006, 2007a; Vargas et al., 2008). In this report, we present further insights into the regulation of sm1 expression as related to the metabolism of plant-derived Suc in Trichoderma cells. Our results suggest that the hydrolysis of Suc is involved in the control of the expression of sm1 in the presence of maize roots (Fig. 9A; Supplemental Fig. S7A). Mechanisms for sugar signaling, involving the phosphorylation of monosaccharides and gene expression regulation, are conserved among different organisms including unicellular eukaryotes such as yeasts (Stulke and Hillen, 1999; Rolland et al., 2001). Moreover, it is known that in plants one of the mechanisms for sugar signaling involves Suc hydrolysis and further phosphorylation of the monosaccharides (Koch, 2004). We speculate that those pathways for sugar signaling might be conserved in T. virens and regulate the intimate association between T. virens and roots. In addition, we tested the ability of Δtvinv2 and Δtvinv5 to activate ISR; even thought both mutant strains were unable to increase the expression of sm1 in the presence of maize roots, the plants were still able to activate ISR in leaves (Fig. 9B; Supplemental Fig. S7B). This phenomenon may be attributed either to basal levels of expression of sm1 or to the presence of other ISR elicitors produced by the fungal cells (Viterbo et al., 2007).
Proteomic experiments demonstrated that in cucumber plants, T. asperellum systemically activates the expression of genes involved in energy and carbohydrate metabolism, including some photosynthetic genes (Segarra et al., 2007; Shoresh and Harman, 2008). In plants, photosynthesis and carbohydrate metabolism establish the basis for the control of plant growth and productivity. Among carbohydrates, Suc is the main photosynthetic product that is transported from source to sink organs (Dennis and Blakeley, 2000). Two of the many functions attributed to Suc in plant cells are as a signal molecule and a controller of carbon partitioning and distribution throughout the plant (Koch, 2004). The expression of photosynthetic genes, such as rbcS, is controlled by Suc and carbohydrate demands from sink tissues, and Ac-Inv activity is known to control the strength of the sink and the sink activity in roots (Urwin and Jenkins, 1997; Sturm, 1999; Pego et al., 2000; Koch, 2004). Increased sink activity in roots represents an enhanced demand for photoassimilates, a higher rate of Suc transport from source leaves, and a release of photosynthesis from the feedback inhibition exerted by sugars (Roitsch, 1999; Pego et al., 2000; Paul and Foyer, 2001). This phenomenon includes the control of the expression of many genes, including the up-regulation of genes involved in photosynthesis (Pego et al., 2000; Paul and Foyer, 2001). In this report, we describe the transcriptional up-regulation of two photosynthetic genes, rbcS and oee31 (Fig. 10, B and C), which is in agreement with a higher photosynthetic rate detected in leaves of maize plants inoculated with T. virens Gv29-8 (Fig. 10A). Recently, the up-regulation of some photosynthesis-related genes was described in plants after treatment with Trichoderma (Segarra et al., 2007; Shoresh and Harman, 2008). We demonstrate that Trichoderma systemically induces the photosynthetic process at different levels, including CO2 fixation and the photochemical production of energy. The up-regulation of oee3-1 (Fig. 10C) suggests that the electron transport in PSII is probably enhanced to support a higher photoassimilation rate in the photosynthetic cells. We found the Trichoderma-mediated up-regulation of photosynthesis is influenced by the expression of tvinv in the fungal cells during the mutualistic association (Fig. 10). We speculate that when Trichoderma colonizes the roots, the up-regulation of tvinv increases the sink activity in roots and consequently the demand for carbon. This increased demand of photoassimilates in roots may release the feedback inhibition of photosynthesis in leaves, stimulate CO2 fixation, and enhance carbon transport toward roots.
Suc degradation in the fungal cells may be considered an extension of plant carbohydrate metabolism that increases the demand for sugars and systemically alters plant metabolism. In mycorrhizal associations, the fungal cells within the root obtain monosaccharides from the plant, affecting carbon metabolism in the whole plant and photosynthesis in leaves (Loewe et al., 2000; Miller et al., 2002; Nehls, 2008). Carbohydrate flow to the symbiotic partner is controlled by the plant through the hydrolysis of Suc. For instance, the importance of plant apoplastic Inv in controlling carbon delivery and root colonization by arbuscular mycorrhiza fungi has been demonstrated in transgenic Nicotiana tabacum plants (Schaarschmidt et al., 2007a, 2007b). Transgenic plants with strongly enhanced Inv activity in leaves resulted in an accumulation of hexoses in source leaves, a decrease of Suc availability for roots, and a reduction of mycorrhization. However, the mechanisms controlling carbon distribution in the plant and arbuscular mycorrhiza colonization are not fully understood. For instance, contrary to the effect observed in plants with a strong increase in Inv activity, mycorrhization was stimulated in plants with a slight increase in leaf Inv activity (Schaarschmidt et al., 2007b). Unexpectedly, arbuscular mycorrhization was not improved in transgenic plants with increased Inv activity in roots that produced high levels of monosaccharides for the fungal cells (Schaarschmidt et al., 2007a). These observations suggest a very tight and complex process that regulates carbohydrate accumulation, distribution, and root colonization by arbuscular mycorrhizae that we do not fully understand. On the other hand, compared with arbuscular mycorrhizae, the metabolic scenario is different for the soil-borne fungus T. virens. In this report, we clearly show the importance of the hydrolysis of plant-derived Suc inside T. virens cells for the symbiotic association with maize roots. In this interaction, T. virens is distinct from mycorrhizal partners, as T. virens has acquired the ability to metabolize Suc without the dependence upon plant Inv. The precise mechanisms associated with Suc metabolism, chemical communication, and plant growth promotion by Trichoderma species are still unclear and will require additional studies to provide a better understanding of such complex processes. Based on the results presented in this and in previously published reports, we introduce Suc as a novel component in the symbiotic association between fungal and root cells as a plant-derived source of carbon and energy with a direct effect on Trichoderma development and growth in the rhizosphere.
MATERIALS AND METHODS
Fungal Strains and Plant Culture
Trichoderma virens Gv29-8 (wild type), T. virens Δtvsp1 (Pozo et al., 2004), Trichoderma atroviride IMI206040, and Trichoderma reesei Q6 were routinely maintained on potato dextrose agar (Difco) unless otherwise indicated. Maize (Zea mays Silver Queen and inbred lines B73 and Mo940) seedlings used in this study were grown in a hydroponic system containing Murashige and Skoog medium (Djonović et al., 2006), in pots containing soil-less mix (Metromix 366; Scotts) or sand, and incubated in a growth chamber at 25°C with a 14-h photoperiod and 60% humidity. Plants grown in pots containing Metromix 366 were inoculated with Trichoderma as reported previously (Djonović et al., 2007a).
Protein Extracts and Enzymatic Assays
Fungal tissue was ground with a mortar and pestle in the presence of liquid nitrogen. The proteins were extracted by treating the ground tissue for 10 min in 50 mm HEPES-NaOH (pH 7.5), 1 mm EDTA, 20% (v/v) glycerol, 20 mm β-mercaptoethanol, and 0.05% (v/v) protease inhibitor cocktail (Sigma). The protein extracts were used for enzymatic assays or enzyme purification. TvInv activity was routinely assayed in reaction mixtures containing 50 mm Suc, 100 mm acetic acid/sodium acetate buffer (pH 5.0), and an aliquot of the protein fraction to be tested. The mixture was incubated at 37°C for different times. The released monosaccharides were quantified with Somogyi-Nelson reagent (Ashwell, 1957).
Purification of TvInv
Crude extracts from fungal tissue grown in liquid Vogel's minimal medium (Vogel, 1956) supplemented with 1.5% (w/v) Suc (VMS) for 48 h were loaded onto High Q support columns (0.5 × 20 cm; Bio-Rad) preequilibrated with 50 mm HEPES-NaOH (pH 6.5), 1 mm EDTA, 5 mm β-mercaptoethanol, and 20% (v/v) glycerol. Proteins were eluted with a 0 to 0.5 m NaCl linear gradient in the equilibration buffer. Fractions containing Inv activity were pooled, concentrated in an Amicon ultrafiltration cell, and further purified by gel filtration through Sephacryl S100 (1.0 × 100 cm; Amersham-Pharmacia) preequilibrated with 50 mm potassium phosphate buffer (pH 6.5), 1 mm EDTA, 10% (v/v) glycerol, 5 mm β-mercaptoethanol, and 150 mm KCl. This column had been previously calibrated using phosphorylase b (97.4 kD), bovine serum albumin (66 kD), carbonic anhydrase (29 kD), and cytochrome c (12 kD) as standards. Since TvInv activity was labile or susceptible to inactivation, fractions from gel-filtration chromatography were collected in tubes containing bovine serum albumin in a final concentration of 5 mg mL−1. The protein was purified approximately 400-fold, and the concentrated fraction was termed the purified enzyme.
Enzyme Characterization
For pH optimum determination, 100 mm citrate-acetate buffer in the pH range 4.0 to 6.0 was added to the reaction medium. Apparent Km determination was performed in the presence of 100 mm acetic acid/sodium acetate buffer (pH 5.0), Suc (1, 2, 4, 8, 16, and 20 mm), and an aliquot of the purified protein. The inhibition of TvInv was assayed in the presence of 1 or 10 mm CuCl2 or Fru plus 5 mm Suc. The substrate specificity was assayed at pH 5.0 in the presence of 50 mm Suc, trehalose, raffinose, stachyose, or melizitose. The products of TvInv activity were quantified using Somogyi-Nelson reagent. In the presence of Fru, the enzymatic activity was determined by Glc oxidase assay as the amount of released Glc (Lloyd and Whelan, 1969).
DNA and RNA Manipulations
Genomic DNA from T. virens was obtained as described previously (Djonović et al., 2006). Total RNA from maize or fungal tissue was prepared using the TRIZOL reagent (Gibco-BRL). RNA quality was analyzed by electrophoresis on agarose gels. Southern- and northern-blot analyses were performed using Hybond-N+ membranes (Amersham Biosciences) according to the manufacturer's suggestions. For northern-blot assays, the probes were PCR-amplified fragments from fungal or maize genomic DNA and radioactively labeled using [32P]dCTP. The fragments amplified corresponded to exons of each gene, and the correct amplification product was confirmed after sequencing. The primers used for the PCR are listed in Supplemental Table S1.
Real-Time PCR Assays
For sm1 expression assays, quantitative real-time reverse transcription (RT)-PCR experiments were conducted as described previously (Vargas et al., 2008) using actin as an internal reference. The experiments were conducted with the QuantiTect SYBR Green RT-PCR kit (Qiagen). The reactions were performed in a 20 μL reaction containing 1× QuantiTect SYBR Green Master Mix, 1× RT QuantiTect Mix, 200 nm primers, and 100 ng of total RNA. The reactions were performed with a 7500 Fast Real-Time PCR System (Applied Biosystems) following the protocols suggested by the manufacturer.
Construction of the tvinv Allele Replacement Cassette by Double-Joint PCR
A DNA fragment consisting of a hygromycin resistance gene (HygB) flanked by DNA regions from the 5′ and 3′ ends of the tvinv gene was amplified by double-joint PCR (Kuwayama et al., 2002). The 5′ (1,354 bp) and 3′ (1,101 bp) fragments of tvinv were amplified using the primer pairs InvUpF/InvUpR and InvDwnF/InvDwnR, respectively (Supplemental Table S1). A 1,430-bp fragment containing the trpC promoter and terminator was amplified from the vector pSCN44 (Fungal Genomic Stock Center) with the primers HygF/HygR (Supplemental Table S1). The three purified fragments were mixed at a 1:3:1 molar ratio and joined by PCR (Kuwayama et al., 2002). The PCR product was used as a template for a final amplification step using the primer pair NestInvF/NestInvR (Supplemental Table S1), which are nested in the 5′ and 3′ fragments of tvinv, respectively. The final PCR product expected was a 3,885-bp DNA fragment with the HygB cassette fused to the 5′ and 3′ regions of tvinv. The two strands of all the DNA fragments generated during the double-joint PCR process were sequenced to assess fidelity.
Fungal Transformation
T. virens protoplasts were prepared and transformed using the lineal DNA fragments according to the method described by Baek and Kenerley (1998). Prototrophic transformants were selected in PDA medium supplemented with hygromycin (100 μg mL−1). Disruption of the tvinv gene in transformants was confirmed by PCR and Southern-blot analysis.
Growth Rate Comparison
Cultures of Gv29-8, Δtvinv2, and Δtvinv5 were compared for colony morphology and radial growth. Spore suspensions (107 spores mL−1) of each strain were prepared, and 3 μL of that suspension was inoculated in the center of WA, VM, VMS, or PDA plates. Plates were visually inspected for production of aerial hyphae, color, and morphology of the colony. The diameter of each colony was recorded at 24, 36, and 48 h of growth at 27°C. Each treatment contained four replicates, and each experiment was repeated three times.
Sugar perception was tested by placing a 3-μL droplet of a spore suspension (107 spores mL−1) in the center of a plate containing VM medium and between two paper discs (sugar 1 and sugar 2). The papers discs were saturated with either 30% (w/v) Suc or 15% (w/v) Glc and placed 3 cm from the inoculation site, establishing a concentration gradient for each sugar after diffusion. The radial symmetry of the colony was evaluated as a measure of the ability of the strains to detect and equally use either Suc or Glc.
Protein Extraction from Roots
Roots of maize seedlings from hydroponic systems were first washed to remove the superficial hyphae of T. virens from the surface. Washed roots were frozen with liquid nitrogen and then ground with mortar and pestle under liquid nitrogen. The total proteins were extracted as described by Hendriks et al. (2003) using 16% (w/v) TCA in diethyl ether and resuspended in Laemmli sample buffer (Laemmli, 1970).
Enzymatic Activity Assays and Protein Quantification in Culture Filtrates
The activity of β-1,3-glucanase, β-1,6-glucanase, endochitinase, and proteinase secreted by the Trichoderma strains used in this study was determined. Enzymatic activities were assayed in the strains when grown in VM or VMS or cultured in Murashige and Skoog medium in the presence or absence (control) of maize seedlings. The activity of β-1,3-glucanase and β-1,6-glucanase was determined in the presence of pustulan and laminarin, respectively, by detecting the reducing sugars released after incubation (Djonović et al., 2007b). Protease and endochitinase activities were determined in the presence of Suc-Ala-Ala-Pro-Phe-pNA (Sigma) and 4-methylumbelliferyl-β-d-N,N',N''-triacetylchitotriose (Sigma), respectively, as recently described (Djonović et al., 2007b). Protein concentration was determined in a microplate assay using the Protein Reagent (Bio-Rad) according to the manufacturer's instructions or by detecting A280 using bovine serum albumin as a standard.
SDS-PAGE of Proteins and Western Blotting
Protein extracts prepared from fungal tissue and from maize roots or leaves were separated on 12% (w/v) polyacrylamide denaturing gels (SDS-PAGE; Laemmli, 1970). The polypeptides were stained with Coomassie Brilliant Blue or blotted onto a nitrocellulose membrane (Hybond C; Amersham) for immunoassays (Renart and Sandoval, 1984). Protein blots were probed with specific antibodies raised against TvSP1 (Pozo et al., 2004).
Infection of Trichoderma-Treated Maize Plants with Colletotrichum graminicola
The inoculation of maize leaves was performed as described previously (Djonović et al., 2007a). Control (nontreated) and treated seeds (coated with chlamydospore preparations of Gv29-8, Δtvinv2, or Δtvinv5) were grown in plastic cylinders containing a soil-less mix (Metromix 366) and incubated in a growth chamber at 25°C. The third leaf of 14-d-old plants was inoculated with 5 μL of a suspension of 6.5 × 104 spores mL−1 C. graminicola. After incubation for 4 d, the leaves were scanned and the infection area was determined and compared among the treatments (Djonović et al., 2007a).
Microscopy and Imaging
Plants in hydroponic systems were inoculated with fungal hyphae, and after 2 d of culture the roots were collected for microscopic visualization and to compare root cell autofluorescence. The fungal hyphae growing on the root surface were stained by the succinate dehydrogenase method as reported by MacDonald and Lewis (1978). This method was originally developed to stain metabolically active arbuscular mycorrhiza inside roots and is based on the reaction catalyzed by succinate dehydrogenase with nitroblue tetrazolium chloride, resulting in the formation of insoluble formazan. Chlamydospores on the root surface were counted on micrographs from five plants per treatment in each of three independent experiments; the results are presented as number of chlamydospores per square millimeter of root ± sd. For autofluorescence, the roots were visualized with an Olympus BX51 fluorescence microscope. Images were recorded using an Olympus DP70 camera and processed with DPController 1.1.165 software.
Bioinformatics and Statistics
Sequence comparisons were performed using deduced amino acid sequences for β-fructofuranosidases retrieved from the databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignments and graphical representations of phylogenetic trees were performed with MEGA (version 4) software (Takamura et al., 2007). The presence of putative subcellular signals or posttranslational sites was calculated using the resources available at ExPASy proteomics tools (http://www.expasy.ch/tools/). Statistical analysis was performed by one-way ANOVA followed by Tukey's honestly significant difference (HSD) test using VassarStat (http://faculty.vassar.edu/lowry/VassarStats.html).
Germination and Conidiation Assays
Spore germination assay was performed as described previously (Baek and Kenerley, 1998). Spore suspensions of each strain were spread on WA-, VM-, VMS-, or PDA-coated slides and incubated for 12 h at 27°C in the dark in moist chambers. Germinated conidia were counted along random transects across the slide. Conidial formation was assayed as described previously (Djonović et al., 2007a). Spore suspensions (103 spores mL−1) of each strain were spread on WA, VM, VMS, or PDA plates, and after 4 or 10 d three plugs were removed from each plate and resuspended in 10 mL of water. Number of spores in the suspension was determined with a hemacytometer.
Root Colonization Assays
The roots of 2-week-old plants inoculated with either the wild type or the mutant strains and grown in Metromix were surface sterilized and ground in the presence of 100 mm phosphate buffer, pH 7, 20 mm MgCl2, and Silwett 77 (root-grinding buffer), and serial dilutions were plated on GVSM medium (Park et al., 1992). Competition between Gv29-8 and either Δtvinv2 or Δtvinv5 for their ability to colonize maize roots was determined by infesting Metromix with equal amounts of T. virens chlamydospore preparation. After 7 d of incubation, roots were processed as above and dilutions were plated on GVSM medium. Colonies that appeared after 3 d were individually transferred to GVSM supplemented with hygromycin. The colonies that were able to grow in the presence of hygromycin corresponded to the mutant strains, and those unable to grow in these conditions corresponded to Gv29-8.
Photosynthetic Uptake of CO2
The uptake of CO2 was determined using the LI-COR LI-6400 portable photosynthesis system. The measurement parameters were set as follows: CO2 flux, 500 μm s−1; sample cell CO2, 360 μmol CO2 mol−1; block temperature, 25°C; photosynthetically active radiation, 600 μmol; area, 6; stomata, 0.50. The measurements (nmol CO2 m−2 s−1) were performed in triplicate on the distal tip of the third leaf of five different plants. The experiment was independently repeated twice.
Collection of Root Exudates and Thin-Layer Chromatography
Maize seeds were surface sterilized using hydrogen peroxide as described previously (Djonović et al., 2007a) and planted in pots containing sterile sand. After 7 d, the plants were carefully removed from the pots and the sand was washed from the roots. Plants with intact root systems were used for exudate collection. Four plants of each line were placed in individual flasks (wrapped with aluminum foil) containing 10 mL of distilled water and incubated under constant light for 4 h with orbital shaking. The liquid suspension was then filter sterilized and lyophilized, and concentrated solids were dissolved in 50 μL of water. The concentrated root exudates were separated by thin-layer chromatography using Silca Gel GF 2000 Uniplate (Analtech) by developing three times with 1-butanol:isopropanol:water (3:12:4). Suc, raffinose, and stachyose were chromatographed in parallel as standards. The positions of the sugars were ascertained with urea-phosphoric acid reagent (Wise et al., 1955).
The sequence data of TvInv have been submitted to the EMBL sequence database under the accession number FM206347.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Deduced amino acid sequence for TvInv.
Supplemental Figure S2. Expression of tvinv in T. virens cells cultured in alternative sources of carbon.
Supplemental Figure S3. Gene-deletion strategy.
Supplemental Figure S4. Screening of T. virens transformants.
Supplemental Figure S5. Detection of Suc in root exudates of maize plants
Supplemental Figure S6. Immunodetection of TvSP1 in protein extracts from maize roots inoculated with T. virens wild type (Gv29-8), Δtvsp1, Δtvinv2, and Δtvinv5.
Supplemental Figure S7. Sm1 expression and ISR induction.
Supplemental Figure S8. Growth of T. virens Gv29-8, Δtvinv2, and Δtvinv5 in VMS medium.
Supplemental Table S1. Primers used in PCR
Supplementary Material
Acknowledgments
We thank Dr. F. Davis for his support in photosynthetic measurements and Dr. S. Djonović and Dr. M. Kolomiets for critical reading of the manuscript.
This work was supported by the U.S. Department of Agriculture National Research Initiative (grant no. 2003–35316–13861 to C.M.K.) and the National Science Foundation (grant no. IOB0445650 to C.M.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Charles M. Kenerley (c-kenerley@tamu.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Abou-Gabal M, Fagerland J (1981) Ultrastructure of the chlamydospore growth phase of Aspergillus parasiticus associated with higher production of aflatoxins. Mykosen 24: 307–311 [DOI] [PubMed] [Google Scholar]
- Adams DJ (2004) Fungal cell wall chitinases and glucanases. Microbiology 150: 2029–2035 [DOI] [PubMed] [Google Scholar]
- Akiyama K, Hayashi H (2006) Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann Bot (Lond) 97: 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano G, Lewis-Ivey ML, Cakir C, Bos JIB, Miller SA, Madden LV, Kamoun S, Hoitink HAJ (2007) Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology 97: 429–437 [DOI] [PubMed] [Google Scholar]
- Ashwell G (1957) Colorimetric analysis of sugars. Methods Enzymol 3: 73–105 [Google Scholar]
- Baek JM, Kenerley CM (1998) The arg2 gene of Trichoderma virens: cloning and development of a homologous transformation system. Fungal Genet Biol 23: 34–44 [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The roles of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 [DOI] [PubMed] [Google Scholar]
- Barran LR, Scheneider EF, Seaman WL (1977) Requirements for the rapid conversion of macroconidia of Fusarium sulphureum to chlamydospores. Can J Microbiol 23: 148–151 [DOI] [PubMed] [Google Scholar]
- Baudoin E, Benizri E, Guckert A (2003) Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol Biochem 35: 1183–1192 [Google Scholar]
- Biemelt S, Sonnewald U (2006) Plant-microbe interactions to probe regulation of plant carbon metabolism. J Plant Physiol 163: 307–318 [DOI] [PubMed] [Google Scholar]
- Blee K, Anderson AJ (1998) Regulation of arbuscule formation by carbon in the plant. Plant J 16: 523–530 [Google Scholar]
- Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G (2007) Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci 12: 1360–1385 [DOI] [PubMed] [Google Scholar]
- Chacón MR, Rodriguez-Galán O, Benítez T, Sousa S, Rey M, Llobell A, Delgado-Jarana J (2007) Microscopic and transcriptomic analyses of early colonization of tomato roots by Trichoderma harzianum. Int Microbiol 10: 19–27 [PubMed] [Google Scholar]
- Chang YC, Baker R, Kleifeld O, Chet I (1986) Increased growth of plants in the presence of the biological control agent Trichoderma harzianum. Plant Dis 70: 145–148 [Google Scholar]
- Chaudhuri A, Bharadwaj G, Maheshwari R (1999) An unusual pattern of invertase activity development in the thermophilic fungus Thermomyces lanuginosus. FEMS Microbiol Lett 177: 39–45 [DOI] [PubMed] [Google Scholar]
- Couteaudier Y, Alabouvette C (1990) Survival and inoculum potential of conidia and chlamydospores of Fusarium oxysporum f. sp. lini in soil. Can J Microbiol 36: 551–556 [DOI] [PubMed] [Google Scholar]
- Dennis DT, Blakeley SD (2000) Carbohydrate metabolism. In BB Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 630–675
- Dethlefsen L, McFall-Ngai M, Relman DA (2007) An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonović S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant Microbe Interact 19: 838–853 [DOI] [PubMed] [Google Scholar]
- Djonović S, Vargas WA, Kolomiets MV, Horndeski M, Wiest A, Kenerley CM (2007. a) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol 145: 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonović S, Vittone G, Mendoza-Herrera A, Kenerley C (2007. b) Enhanced biocontrol activity of Trichoderma virens transformants constitutively coexpressing β-1,3- and β-1,6-glucanase genes. Mol Plant Pathol 8: 469–480 [DOI] [PubMed] [Google Scholar]
- Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP (2006) Global carbon utilization profiles of wild-type, mutant and transformant strains of Hypocrea jecorina. Appl Environ Microbiol 72: 2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM (1998) Yeast carbon catabolite repression. Eur J Biochem 62: 297–313 [DOI] [PubMed] [Google Scholar]
- Garcia-Garrido JM, Ocampo JA (2002) Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. J Exp Bot 53: 1377–1386 [PubMed] [Google Scholar]
- Gianinazzi-Pearson V (1996) Plant cell responses to arbuscular mycorrhizal fungi: getting to the roots of the symbiosis. Plant Cell 8: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen C, Yuan XL, van Munster JM, Ram AF, van der Maarel MJEC, Dijkhuizen L (2007) Molecular and biochemical characterization of a novel intracellular invertase from Aspergillus niger with transfructosylating activity. Eukaryot Cell 6: 674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Mendgen K (2001) Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr Opin Plant Biol 4: 322–327 [DOI] [PubMed] [Google Scholar]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M (2004) Trichoderma species: opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2: 43–56 [DOI] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geingenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Bauer WD, Bird DM, Cullimore J, Tyler B, Yonder JI (2003) Molecular signals and receptors: controlling rhizosphere interactions between plants and other organisms. Ecology 84: 858–868 [Google Scholar]
- Hooper LV, Bry L, Falk PG, Gordon JI (1998) Host-microbial symbiosis in the mammalian intestine: exploring and internal ecosystem. Bioessays 20: 336–343 [DOI] [PubMed] [Google Scholar]
- Jaeger CH III, Lindow SE, Miller W, Clark E, Firestone MK (1999) Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol 65: 2685–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaylor EN, Shih H, Towle HC (1997) Carbohydrate regulation of hepatic gene expression. J Biol Chem 272: 7525–7531 [DOI] [PubMed] [Google Scholar]
- Kertesz-Chaloupkova K, Walser PJ, Garanado JD, Aebi M, Kues U (1998) Blue light overrides repression of asexual sporulation by mating type genes in the basidiomycete Coprinus cinereus. Fungal Genet Biol 23: 95–109 [DOI] [PubMed] [Google Scholar]
- Khan ZU, Ahmad S, Mokaddas E, Chandy R (2004) Tobacco agar, a new medium for differentiating Candida dubliniensis from Candida albicans. J Clin Microbiol 42: 4796–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K (2004) Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7: 235–246 [DOI] [PubMed] [Google Scholar]
- Kraffczyk I, Trolldenier G, Beringer H (1984) Soluble exudates of maize: influence of potassium supply and rhizosphere microorganisms. Soil Biol Biochem 16: 315–322 [Google Scholar]
- Kües U, Granado JD, Hermann R, Boulianne RP, Kertesz-Chaloupkova K, Aebi M (1998) The A mating type and blue light regulate all known differentiation processes in the basidiomycete Coprinus cinereus. Mol Gen Genet 260: 81–91 [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Obara S, Morio K, Katho M, Urushihara H, Tanaka Y (2002) PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res 30: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lemoine R (2000) Sucrose transporters in plants: update on function and structure. Biochim Biophys Acta 1465: 246–262 [DOI] [PubMed] [Google Scholar]
- León P, Sheen J (2003) Sugar and hormone connections. Trends Plant Sci 3: 110–116 [DOI] [PubMed] [Google Scholar]
- Lin X, Heitman J (2005) Chlamydospore formation during hyphal growth in Cryptococcus neoformans. Eukaryot Cell 4: 1746–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JB, Whelan WJ (1969) An improved method for enzymatic determination of glucose in the presence of maltose. Anal Biochem 30: 467–470 [DOI] [PubMed] [Google Scholar]
- Loewe A, Einig W, Shi L, Dizengremel P, Hampp R (2000) Mycorrhiza formation and elevated CO2 both increase the capacity for sucrose synthesis in source leaves of spruce and aspen. New Phytol 145: 565–574 [DOI] [PubMed] [Google Scholar]
- MacDonald RM, Lewis M (1978) The occurrences of some acid phosphatases and dehydrogenases in the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. New Phytol 80: 135–141 [Google Scholar]
- Mach RL, Strauss J, Zeilinger S, Schindler M, Kubicek CP (1996) Carbon catabolite repression of xyn1 (xylanase I-encoding) gene expression in Trichoderma reesei. Mol Microbiol 21: 1273–1281 [DOI] [PubMed] [Google Scholar]
- Mahmood T, Woitke M, Gimmler H, Kaiser WM (2002) Sugar exudation by roots of kallar grass [Leptochloa fusca (L.) Kunth] is strongly affected by the nitrogen source. Planta 214: 887–894 [DOI] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, et al (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26: 553–560 [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ (1998) The development of cooperative associations between animals and bacteria: establishing détente among domains. Am Zool 38: 593–608 [Google Scholar]
- Miller RM, Miller SP, Jastrow JD, Rivetta CB (2002) Mycorrhizal mediated feedbacks influence net carbon gain and nutrient uptake in Andropogon gerardii. New Phytol 155: 149–162 [DOI] [PubMed] [Google Scholar]
- Nehls U (2008) Mastering ectomycorrhizal symbiosis: the impact of carbohydrates. J Exp Bot 59: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Nehls U, Mikolajewski S, Magel E, Hampp R (2001) Carbohydrate metabolism in ectomycorrhizas: gene expression, monosaccharide transport and metabolic control. New Phytol 150: 533–541 [Google Scholar]
- Noronha EF, Kipnis A, Junqueira-Kipnis AP, Ulhoa CJ (2000) Regulation of 36-kDa β-1,3-glucanase synthesis in Trichoderma harzianum. FEMS Microbiol Lett 188: 19–22 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Harrison MJ, Paszkowski U (2009) Reprogramming plant cells for endosymbiosis. Science 324: 753–754 [DOI] [PubMed] [Google Scholar]
- Palmer RJ Jr, Kazmerzak K, Hansen MC, Kolenbrander PE (2001) Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun 69: 5794–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavizas GC (1985) Trichoderma and Gliocladium: biology, ecology, and potential for biocontrol. Annu Rev Phytopathol 23: 23–54 [Google Scholar]
- Park YH, Kenerley CM, Stack JP (1992) Inoculum dynamics of Gliocladium virens associated with roots of cotton seedlings. Microb Ecol 23: 169–170 [DOI] [PubMed] [Google Scholar]
- Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Foyer CH (2001) Sink regulation of photosynthesis. J Exp Bot 52: 1383–1400 [DOI] [PubMed] [Google Scholar]
- Pego JV, Kortstee AJ, Huijser AJ, Huijser C, Smeekens SCM (2000) Photosynthesis, sugars and the regulation of gene expression. J Exp Bot 51: 407–416 [DOI] [PubMed] [Google Scholar]
- Pozo MJ, Baek JM, García JM, Kenerley CM (2004) Functional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet Biol 41: 336–348 [DOI] [PubMed] [Google Scholar]
- Reddy VA, Maley F (1990) Identification of an active site residue in yeast invertase by affinity labeling and site-directed mutagenesis. J Biol Chem 19: 10817–10820 [PubMed] [Google Scholar]
- Regulez P, Ponton J, Dominguez JB, Goni FM, Uruburu F (1980) Lipid composition and the transition from yeast-like to chlamydospores cells of Pullularia pullulans. Can J Microbiol 26: 1428–1437 [DOI] [PubMed] [Google Scholar]
- Renart J, Sandoval IV (1984) Western blots. Methods Enzymol 104: 455–460 [DOI] [PubMed]
- Roitsch T (1999) Source-sink regulation by sugars and stress. Curr Opin Plant Biol 2: 198–206 [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM (2001) Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem Sci 26: 310–317 [DOI] [PubMed] [Google Scholar]
- Rubio MC, Navarro AR (2006) Regulation of invertase synthesis in Aspergillus niger. Enzyme Microb Technol 39: 601–606 [Google Scholar]
- Salerno G, Curatti L (2003) Origin of sucrose metabolism in higher plants: when, how and why? Trends Plant Sci 8: 63–69 [DOI] [PubMed] [Google Scholar]
- Sarma AD, Oehrle NW, Emerich DW (2007) Metabolic intricacies of the symbiotic association between soybean and Bradyrhizobium japonicum: a proteomic outlook. In J Samaj, J Thelen, eds, Plant Proteomics. Springer-Verlag, Berlin, pp 310–325
- Schaarschmidt S, González MC, Roitsch T, Strack D, Sonnewald U, Hause B (2007. a) Regulation of arbuscular mycorrhization by carbon. The symbiotic interaction cannot be improved by increased carbon availability accomplished by root-specifically enhanced invertase activity. Plant Physiol 143: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaarschmidt S, Kopka J, Ludwig-Müller J, Hause B (2007. b) Regulation of arbuscular mycorrhization by apoplastic invertases: enhanced invertase activity in the leaf apoplast affects the symbiotic interaction. Plant J 51: 390–405 [DOI] [PubMed] [Google Scholar]
- Segarra G, Casanova E, Bellido D, Odena MA, Oliveira E, Trillas I (2007) Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics 7: 3943–3952 [DOI] [PubMed] [Google Scholar]
- Seidl V, Gamauf C, Druzhinina IS, Seiboth B, Hartl L, Kubicek CP (2008) The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics 9: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M, Galon A, Chet I (2006) Characterization of a MAPK gene from cucumber required for Trichoderma-conferred plant resistance. Plant Physiol 142: 1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoresh M, Harman GE (2008) The molecular basis of shoot responses of maize seedlings to Trichoderma harzianum T22 inoculation of the root: a proteomic approach. Plant Physiol 147: 2147–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stulke J, Hillen W (1999) Carbon catabolite repression in bacteria. Curr Opin Microbiol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Sturm A (1999) Invertases: primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Towle HC (1995) Metabolic regulation of gene transcription in mammals. J Biol Chem 270: 23235–23238 [DOI] [PubMed] [Google Scholar]
- Urwin NAR, Jenkins GI (1997) A sucrose repression element in the Phaseolus vulgaris rbcS2 gene promoter resembles elements responsible for sugar stimulation of plant and mammalian genes. Plant Mol Biol 35: 929–942 [DOI] [PubMed] [Google Scholar]
- Vargas WA, Djonović S, Sukno SA, Kenerley CM (2008) Dimerization controls the activity of fungal elicitors that trigger systemic induced resistance in plants. J Biol Chem 283: 19804–19815 [DOI] [PubMed] [Google Scholar]
- Viterbo A, Wiest A, Brotman Y, Chet I, Kenerley C (2007) The 18mer peptaibols from Trichoderma virens elicit plant defense responses. Mol Plant Pathol 8: 737–746 [DOI] [PubMed] [Google Scholar]
- Voegele RT, Wirsel S, Möll U, Lechner M, Mendgen K (2006) Cloning and characterization of a novel invertase from the obligate biotroph Uromyces fabae and analysis of expression patterns of host and pathogen invertases in the course of infection. Mol Plant Microbe Interact 19: 625–634 [DOI] [PubMed] [Google Scholar]
- Vogel HJ (1956) A convenient growth medium for Neurospora (medium N). Microbiol Genet Bull 13: 42–43 [Google Scholar]
- Wise CS, Dimler RJ, Davis CE, Rist CE (1955) Determination of easily hydrolysable fructose in dextran preparation. Anal Chem 27: 33–36 [Google Scholar]
- Yedidia I, Benhamou N, Chet I (1999) Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl Environ Microbiol 65: 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedidia I, Srivastva AK, Kapulnik Y, Chet I (2001) Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant Soil 235: 235–242 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.