Abstract
Human CGI-58 (for comparative gene identification-58) and YLR099c, encoding Ict1p in Saccharomyces cerevisiae, have recently been identified as acyl-CoA-dependent lysophosphatidic acid acyltransferases. Sequence database searches for CGI-58 like proteins in Arabidopsis (Arabidopsis thaliana) revealed 24 proteins with At4g24160, a member of the α/β-hydrolase family of proteins being the closest homolog. At4g24160 contains three motifs that are conserved across the plant species: a GXSXG lipase motif, a HX4D acyltransferase motif, and V(X)3HGF, a probable lipid binding motif. Dendrogram analysis of yeast ICT1, CGI-58, and At4g24160 placed these three polypeptides in the same group. Here, we describe and characterize At4g24160 as, to our knowledge, the first soluble lysophosphatidic acid acyltransferase in plants. A lipidomics approach revealed that At4g24160 has additional triacylglycerol lipase and phosphatidylcholine hydrolyzing enzymatic activities. These data establish At4g24160, a protein with a previously unknown function, as an enzyme that might play a pivotal role in maintaining the lipid homeostasis in plants by regulating both phospholipid and neutral lipid levels.
Acylation of glycerol-3-phosphate (G3P) is the first step in the biosynthesis of glycerolipids in plants. Most of the enzymes involved in this pathway were shown to be membrane bound (Somerville and Browse, 1991). However, a soluble G3P acyltransferase has been reported in plants, which acylates G3P to lysophosphatidic acid (LPA) in an acyl-(acyl carrier protein)-dependent manner (Murata and Tasaka, 1997). The role of other soluble enzymes in the glycerolipid biosynthesis pathway is well documented. Cytosolic monoacylglycerol acyltransferase (Tumaney et al., 2001), diacylglycerol acyltransferase (Saha et al., 2006), and LPA phosphatase (Shekar et al., 2002) were shown to be present in the immature seeds of Arachis hypogaea. Recently, cytosolic LPA phosphatase (Reddy et al., 2008) and phosphatidic acid (PA) phosphatase have also been reported in Saccharomyces cerevisiae (Han et al., 2006). In addition, we demonstrated earlier the presence of a soluble LPA acyltransferase (LPAAT) as a part of the cytosolic multienzyme complex for the synthesis of triacylglycerol (TG) in Rhodotorula glutinis (Gangar et al., 2001). In S. cerevisiae, Ict1p catalyzes the acylation of LPA to PA, thereby enhancing phospholipid biosynthesis under cellular stress. A Δict1 deletion strain was shown to be calcofluor white sensitive and exhibited a defective phospholipid biosynthesis, suggesting a role of Ict1p in the maintenance of the cell membranes (Ghosh et al., 2008a).
BLAST analysis of the human genome with the Ict1p sequence resulted in the identification of a gene named CGI-58. Mutations in human CGI-58 are responsible for a rare autosomal recessive genetic disorder known as Chanarin Dorfman syndrome (Zechner et al., 2009). CGI-58 is a member of the α/β-hydrolase family of proteins and has a conserved lipase motif GXNXG, where the Ser is replaced by an Asn. Biochemical characterization of human CGI-58 revealed that it acylates LPA to PA. Heterologous overexpression in yeast showed that expression of CGI-58 enhanced the biosynthesis of total phospholipids, especially PA, phosphatidylethanolamine, and phosphatidylcholine (PC). CGI-58 was found to localize to the lipid bodies isolated from the mice white adipose tissues, but the LPAAT activity in the soluble fraction from adipose tissue was also attributed to CGI-58 (Ghosh et al., 2008b).
So far, a soluble LPAAT from plants has not been identified, although the importance of such enzymes in other experimental systems has been envisaged (Tumaney et al., 2001; Ghosh et al., 2008a). Being aware of the important role of Ict1p and CGI-58 in phospholipid metabolism and in combating stress, we started a systematic search for CGI-58-like proteins in plants. The availability of the complete genome sequence of Arabidopsis (Arabidopsis thaliana) allowed us to perform a comprehensive genome-wide survey of CGI-58 like proteins in Arabidopsis. As will be described in this study, a BLAST analysis of CGI-58 in Arabidopsis revealed At4g24160 as its closest homolog. Biochemical characterization of At4g24160 showed its ability to acylate LPA to PA in an acyl-CoA-dependent manner. The recombinant protein has the capability to hydrolyze TG and PC to a lesser extent. Expression analysis of At4g24160 and its homologs suggests the significance of these genes under various stress conditions. In summary, At4g24160 is a soluble acyltransferase with lipase and phospholipase functions from Arabidopsis belonging to the α/β-hydrolase superfamily of proteins.
RESULTS
Evolutionary Relationship of CGI-58-Like Proteins from Arabidopsis
To search for the homologs of CGI-58 in plants the Arabidopsis genome was screened as described in “Materials and Methods.” After identifying the individual protein sequences, we eliminated the repeated sequences by comparisons with theoretical cDNA and genomic DNA sequences in The Arabidopsis Information Resource database. This strategy led to the identification of 24 proteins that were found to be homologous to CGI-58 (Table I). All 24 of these proteins belong to the α/β-hydrolase or esterase/lipase superfamily.
Table I.
Characteristics and expression analysis of predicted CGI-58-like hydrolases in Arabidopsis
The subcellular localization data were retrieved from the eFP cell browser, and the gene expression data were retrieved from the Genevestigator Web site using the Meta-Analyzer search tool. AGI, Arabidopsis Genome Initiative.
| AGI Code | Molecular Weight | Organelle Expression | Up-Regulated | Down-Regulated | Remarks |
|---|---|---|---|---|---|
| At1g80280 | 71105.2 | Mitochondria | − | ABA, zeatin, salt | α/β-Hydrolase family |
| At1g52750 | 70293.2 | Mitochondria | − | − | α/β-Hydrolase/putative redoxactive |
| At3g24420 | 30022.4 | Cytosol ubiquitous | − | − | α/β-Hydrolase family |
| At3g03990 | 29624.5 | Cell wall/chloroplast | Cold, osmotic | ABA, salt, heat | α/β-Hydrolase family |
| At3g10840 | 50885.0 | Chloroplast | Heat | ABA, salt, hypoxia, wound, osmotic | SARS lipid binding protein |
| At1g13820 | 37958.7 | Chloroplast | − | − | Hydrolase/L- type Ca channel |
| At5g39220 | 36848.6 | Chloroplast/mitochondria/cytosol | ABA, salt, osmotic | Zeatin, genotoxic, drought, cold | α/β-Hydrolase family |
| At4g36530 | 41905.7 | Cytosol/chloroplast | Heat | ABA, zeatin, salt, cold | Hydrolase/ helix-hairpin-helix motif |
| At5g38520 | 40991.9 | Chloroplast | Wound, heat | ABA, zeatin, cold, osmotic | α/β-Hydrolase family |
| At5g19850 | 40104.7 | Chloroplast | Salt, hypoxia, osmotic, heat | − | α/β-Hydrolase family |
| At1g77420 | 42818.9 | Mitochondria | Wound | ABA, zeatin, cold, heat, drought | α/β-Hydrolase family |
| At4g24160 | 46525.4 | Mitochondria | Salt, cold, hypoxia, osmotic | Zeatin, heat | α/β-Hydrolase family |
| At1g17430 | 37851.8 | Mitochondria | ABA, hypoxia | Cold | α/β-Hydrolase family |
| At5g09430 | 35498.0 | Mitochondria | Indole-3-acetic acid, salt | ABA, zeatin, cold, hypoxia, osmotic | α/β-Hydrolase family |
| At2g18360 | 35526.4 | Cell wall/cytosol | Salt, cold, hypoxia, osmotic | ABA, genotoxic, drought | α/β-Hydrolase family |
| At4g36610 | 35621.4 | Chloroplast/cell wall | Cold, hypoxia, osmotic | Salt, wound | Hydrolase associated lipase |
| At5g53050 | 43885.1 | Cytosol/mitochondria | Salt, wound, drought | ABA, cold, heat | Progressive ankylosis protein |
| At4g12830 | 43768.2 | Chloroplast | − | ABA, salt, cold, wound, osmotic | α/β-Hydrolase family |
| At4g25290 | 65166.5 | Mitochondria | − | − | DNA photolyase family |
| At4g24140 | 56210.6 | Plasma membrane | Cold, anoxia, genotoxic | ABA, zeatin, hypoxia | Hydrolase/BDG1-like protein |
| At5g41900 | 53644.8 | Mitochondria | ABA, salt, osmotic, heat | Hypoxia | Hydrolase/BDG1-like protein |
| At1g64670 | 53427.4 | Cytosol ubiquitous | − | ABA, cold, hypoxia | Hydrolase/ BDG1-like protein |
| At5g17720 | 50179.9 | Cytosol/cell wall | Anoxia | Heat, hypoxia | α/β-Hydrolase family |
| At5g17780 | 47685.4 | Nucleus/endoplasmic reticulum/cell wall | Cold, heat | Zeatin, salt, heat | α/β-Hydrolase family |
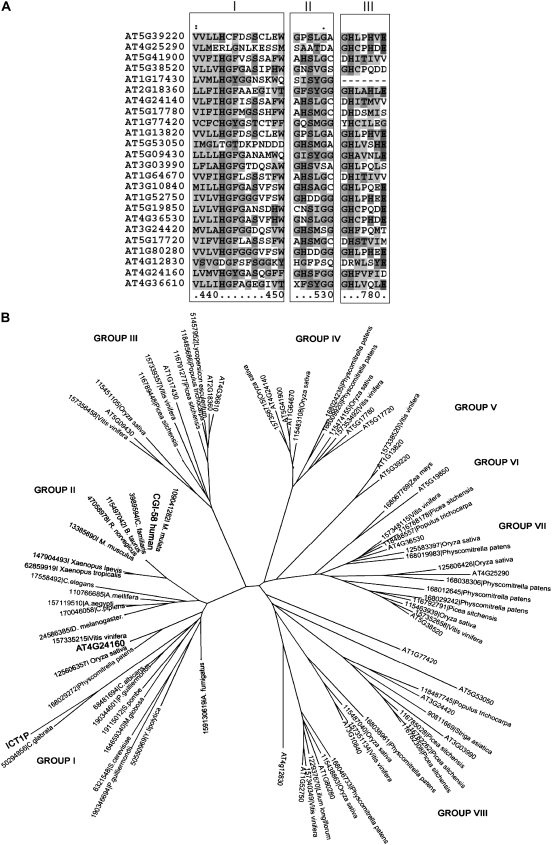
Multiple sequence alignment of all the 24 proteins revealed three conserved motifs (Fig. 1A) and several conserved Ser, Asp, and His residues that may serve as a part of the catalytic triad, the active site in most of the known hydrolases/lipases. Motif I is V(X)3HGF, where X represents hydrophobic residues such as Val, Ile, Phe, Met, and Leu. This is supposedly the lipid binding motif of these polypeptides. The motif II is GXSXG, the signature motif of known lipases, phospholipases, and lysophospholipases. The motif III is H(X)4D/E, previously identified as a signature motif of most acyltransferases. Interestingly, among all the 24 proteins identified, the H(X)4D motif was found only in At4g24160, whereas the other polypeptides contained an H(X)4E motif. In order to gain an insight into CGI-58-like proteins in other plants, a global BLAST search analysis was performed using the plant database at the National Center for Biotechnology Information (NCBI). Multiple sequence alignment revealed the presence of all the three motifs that were seen in Arabidopsis to be conserved in other plant species (data not shown).
Figure 1.
Phylogenetic analysis of CGI-58 and its homologs in yeast, plants, and animals. A, BLAST search was performed using amino acid sequence of CGI-58 followed by ClustalW alignment of the close homologs. B, The evolutionary history was inferred using the neighbor-joining method. The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap tests (500 replicates) is shown next to the branches. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 183 positions in the final data set. Phylogenetic analyses were conducted in MEGA4.
To examine the relationship among CGI-58-like proteins, a topographic dendrogram was constructed (Fig. 1B). ICT1, CGI-58, and At4g24160 were clustered in the same group. However, CGI-58 and its homologs in nonchordates, amphibians, nematode, and higher mammals diverged to form a different subgroup, indicating a final attainment of function in the evolutionary timescale. Interestingly, none of the other 23 CGI-58 homologs from Arabidopsis was present in the same group, suggesting that At4g24160 diverged from other members of this polypeptide family in the evolutionary history to perform specific catalytic functions that may be important in the physiology of the plant. The dendrogram also suggests that the remaining 23 hydrolases have markedly diverged during evolution, attaining specific functions, a feature common among members of the α/β-hydrolase family of proteins. As an example, BODYGUARD 1 domain-containing proteins, At4g24140, At5g41900, and At1g64670, form a distinct group of polypeptides.
Multiple sequence alignment of the proteins present in the groups I and II showed that most of the residues were conserved throughout the family (Fig. 1B). Motifs I, II, and III were distinctly present in all the members of the group, suggesting their probable catalytic role per se. An interesting observation made in the sequence alignment was that the Ser of GXSXG motif is replaced by Asn in all the vertebrates starting from amphibians. The effect of such a conversion needs to be examined. The conversion also suggests the divergence of these polypeptides from the other members of Group I.
Domain Analysis and Subcellular Distribution of CGI-58-Like Proteins in Arabidopsis
Using only the information of the amino acid sequence, it has been difficult to assign a function to proteins of the α/β-hydrolase superfamily, especially because of low sequence similarities among the members investigated. Moreover, members of this superfamily may catalyze a wide variety of reactions acting as lipases, esterases, epoxide hydrolases, acyltransferases, and/or Ser proteases (Nardini and Dijkstra, 1999). To obtain additional information about these proteins, we analyzed the different functional domains and the subcellular localization of CGI-58-like proteins in Arabidopsis. The eFP cell browser was used (Winter et al., 2007; http://www.bar.utoronto.ca/) to retrieve information on subcellular localization, and domain structure was analyzed based on the pfam database (http://www.sanger.ac.uk/Software/Pfam/).
A detailed domain analysis suggested possible biological roles for these hypothetical proteins. At1g52750 possesses a putative redox active protein domain with a very high expression in the mitochondria and a moderate expression in the nucleus. At3g10840 was found to have a SARS lipid binding domain, suggesting a possible association of the protein with long-chain fatty acids (Meier et al., 2006). At1g13820 localized mainly in the chloroplast and harbors a putative l-type calcium channel-like domain. At5g17720, a member of cupin superfamily is probably involved in the plant secondary metabolite production (Dunwell et al., 2000) and localizes to the cell wall, cytosol, mitochondria, and chloroplast. At5g41900 has a moderate expression in the plasma membrane and localizes to the mitochondria. It is related to zona pellucid-like domain-containing proteins, which are known to play structural roles in the plasma membranes (Zazwinska and Affolter, 2004). At4g24140, At1g64670, and At5g41900 were found to contain a domain commonly known as BODYGUARD and may probably be involved in the biosyntheses of cell wall-associated lipids (Kurdyukov et al., 2006). Another member of this group of proteins belongs to the DNA polymerase family of proteins with a putative helix hairpin helix motif, though the subcellular localization suggested a cytosolic association. At4g25290 is related to DNA photolyases, and it is highly expressed in mitochondria followed by the chloroplast. The other proteins in the family do not possess any specialized domain distinction and were grouped under the broad α/β-hydrolase superfamily. Among them, At4g24160 was found to be localized in the mitochondria and the chloroplast. At3g24420 and At5g17780 were found to be intriguing because of their nuclear localization.
Expression Analysis of CGI-58-Like Proteins
The Genevestigator online search tool Meta-Analyzer was used to analyze Arabidopsis Affymetrix microarray data. Relative gene expression was studied in different plant organs, at various growth stages, and under different stress conditions. Many of these genes were found to be up-regulated during biotic stress. The expression profiles of these 24 proteins under different stress conditions are summarized in Table I. At4g24160 was present from the early seedling to the later stages of development but was found to be specifically expressed in the roots. It was up-regulated during Pseudomonas syringae infection and salicylic acid treatment, while abscisic acid (ABA) and zeatin treatment resulted in its down-regulation. The expression pattern of At4g24160 under abiotic stresses like cold, heat, salt, hypoxia, osmotic, genotoxicity, wound, and drought was found to be interesting. At4g24160 is up-regulated under high salt stress with the maximum expression in the roots (eFP database), which is a particularly important observation because salt stress has been shown to be associated with high phospholipid biosynthesis. The other stress conditions that seem to have an effect on the expression of At4g24160 are cold, osmotic stress, and hypoxia.
At4g24160 Is the Closest Homolog of ICT1 and CGI-58
BLAST analysis of Ict1p in the nonredundant database of NCBI revealed plant At4g24160 and human CGI-58 as its closest homologs (Fig. 2). At4g24160, a 418-amino acid protein, is a member of the α/β-hydrolase family of proteins. In addition to the hydrolase domain, At4g24160 possesses an esterase (pfam 00756), hydrolase/acyltransferase (COG 0596), and lysophospholipase domain (COG 2267). A distinct functional motif H(X)4D, a characteristic of most acyltransferases (Heath and Rock, 1998) is also present at the C-terminal region of At4g24160. At the N-terminal region, At4g24160 possesses the highly conserved GXSXG motif found in the known lipases, phospholipases, lysophospholipases, esterases, and Ser proteases.
Figure 2.
At4g24160 is a homolog of ICT1 and CGI-58. BLAST analysis of human CGI-58 with nonredundant protein database available at NCBI was performed. Multiple sequence alignment of proteins homologous to human CGI-58 was carried out using sequences from representative organisms, such as Arabidopsis and yeast. Accession numbers are as follows: NP_057090.2 for Homo sapiens, NP_974605.1 for Arabidopsis, and NP_013200.1 for S. cerevisiae. The conserved H(X)4D and GXNXG motifs are indicated in bold.
Biochemical Characterization of At4g24160
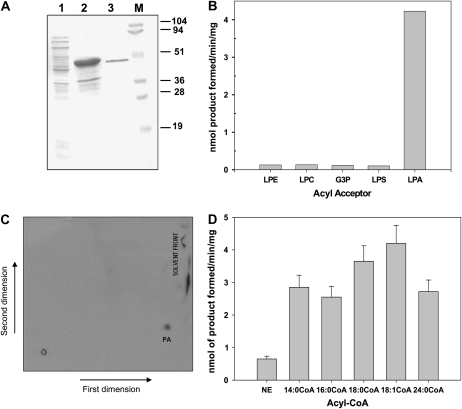
As a prerequisite for biochemical investigations, At4g24160 was cloned in pRSET A and overexpressed in BL21 (DE3) cells. Immunoblot analysis using anti-(His)6 monoclonal antibody was performed to confirm the expression. The recombinant protein was purified by Ni2+-nitrilotriacetic acid (Ni2+-NTA) column chromatography (Fig. 3A).
Figure 3.
Purification of the recombinant At4g24160. A, At4g24160 from E. coli BL21 (DE3) was purified using Ni2+-NTA affinity column chromatography. Proteins were resolved on a 12% SDS-PAGE and stained with Coomassie Brilliant Blue. Lane 1, supernatant of induced lysate; lane 2, pellet of induced lysate; lane 3, the purified At4g24160. M, Molecular mass standard. B, Specificity for cosubstrate (lyso)phospholipids on the acyltransferase activity of purified At4g24160. Values are average of two independent experiments. C, Two-dimensional TLC showing PA as the product formed after the enzymatic acylation of LPA by the purified recombinant At4g24160. First-dimensional solvent system is chloroform:methanol:ammonia (65:35:5, v/v), and the second-dimensional solvent system is chloroform:methanol:acetic acid:water (40:20:5:0.5, v/v). D, Acyl-CoA-dependent formation of PA catalyzed by the purified recombinant At4g24160. Assay was performed with 50 μm [3H]LPA (0.25 μCi/tube), 10 μm acyl-CoA donors, and 5 μg enzyme in a final volume of 100 μL. The reaction was carried out for 20 min. Values are means (±sd) for four independent determinations.
Lysophospholipid Acyltransferase Activity
As one of the most important enzymatic assays, we analyzed lysophospholipid acyltransferase activity of the purified At4g24160 recombinant protein. As can be seen from Figure 3B, At4g24160 was found to be highly specific for LPA as a substrate. Two-dimensional thin-layer chromatography (TLC) analysis confirmed that PA was the product of this enzymatic conversion (Fig. 3C). At4g24160 was found to be capable of using different acyl-CoA donors for the formation of PA; however, the specificity toward oleoyl-CoA was maximum followed by stearoyl-CoA (Fig. 3D). The enzyme showed a protein- and time-dependent incorporation of oleoyl-CoA into PA (data not shown). The LPAAT activity was also analyzed by electron spray ionization-mass spectrometry (ESI-MS; Supplemental Fig. S1, A–C). At this point, it is important to mention that At4g24160 homologs of yeast and human were also shown to exhibit a similar LPAAT activity (Ghosh et al., 2008a, 2008b). ESI-MS analysis also confirmed the LPAAT activity of this recombinant purified enzyme.
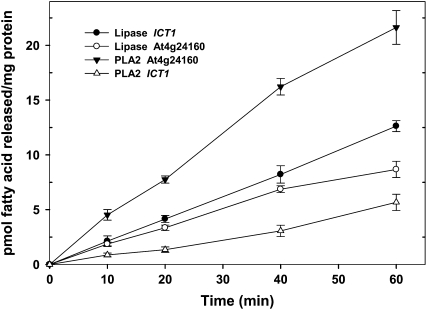
Lipase and Phospholipase Activity
Since the GXSXG motif was found in At4g24160 and in its yeast homolog Ict1p, the purified recombinant proteins of At4g24160 and ICT1 were analyzed for esterase, lipase, and phospholipase A2 activities using para-nitrophenyl stearate, triolein, and PC (dipalmitate), respectively, as substrates. Both the bacterially expressed purified proteins (At4g24160 and Ict1p) were able to hydrolyze triolein and PC in a time-dependent manner. At4g24160 has a higher PLA2 activity (Supplemental Fig. S3, A and B) as compared to the lipase activity (Supplemental Fig. S2, A and B). On the contrary, Ict1p has higher lipase activity (Supplemental Fig. S4, A–C) than the PLA2 activity (Supplemental Fig. S5, A and B). Lipase and phospholipase activities in yeast and plant enzyme were also confirmed by ESI-MS analysis (Fig. 4). Zero minute and no substrate reactions were kept as controls. These analyses revealed that both enzymes hydrolyze triolein to diacylglycerol, monoacylglycerol, and fatty acids and PC in a phospholipase A2-type reaction to lysophosphatidylcholine (LPC) and free fatty acid. On the other hand, both At4g24160 and Ict1p did not show esterase activity (data not shown).
Figure 4.
Time-dependent hydrolysis of TG and PC by the purified recombinant At4g24160. Lipase assay was performed in a reaction buffer containing 50 mm Tris-HCl, pH 7.5, 100 μm sodium taurocholate, and 10 μg enzyme for various time intervals at 30°C in the presence of sonicated suspension of 100 μm [9,10-3H]triolein. Reaction mixtures used for phospholipase A2 assays consisted of 100 μm sonicated vesicles of [2-palmitoyl-9,10-3H]phosphatidylcholine and 10 μg enzyme in total volume of 100 μL. The reaction was carried out at 30°C for various time periods. The activity was assessed as described in “Materials and Methods.” Values are means (±sd) for three independent determinations, and each experiment was done in duplicate.
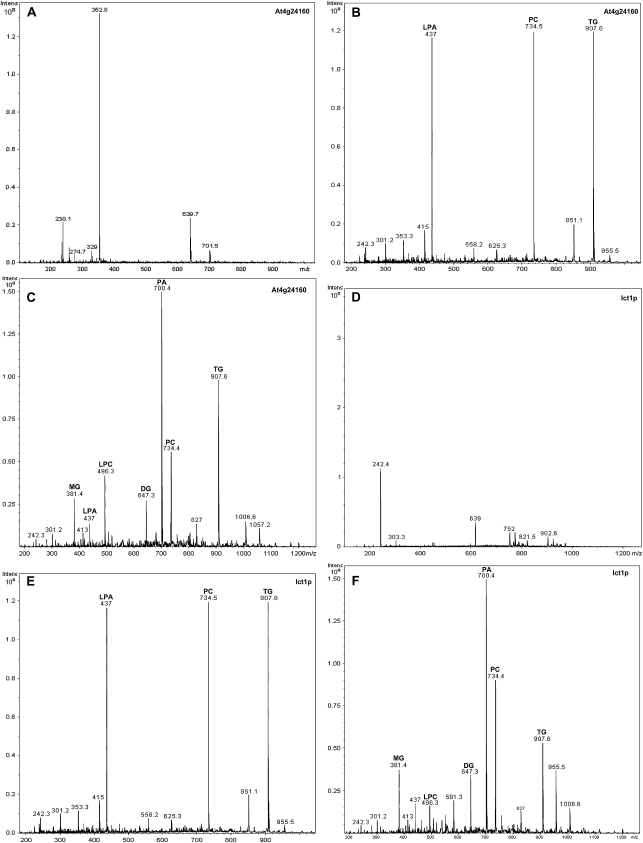
A cocktail assay comprising of all the three substrates, i.e. LPA, TG, and PC, was also performed to determine the substrate preference of both At4g24160 and Ict1p. It was observed that the LPAAT activity for proteins was maximum, and they were capable of hydrolyzing TG and PC to a lesser extent. The PLA2 activity of At4g24160 was higher than its TG lipase activity; however, the case was vice versa for Ict1p. Zero minute and no substrate reactions were kept as controls (Fig. 5, A–F).
Figure 5.
ESI-MS analysis of LPAAT, lipase, and phospholipase activities (cocktail assay). A, Control reaction (without substrate) was performed with assay buffer and 10 μg At4g24160 enzyme in a final reaction volume of 100 μL. The reaction was carried out for 40 min at 30°C. The lipids were extracted with butanol, dried, and analyzed by ESI-MS. The ESI analysis was done in a positive mode. The control reaction clearly reveals that the products formed are not contaminants from the enzyme source. B, Control reaction (without enzyme) was performed with assay buffer, 1 mm LPA (1-oleoyl), 10 μm oleoyl-CoA, 1 mm PC (dipalmitate), and 1 mm triolein in a final reaction volume of 100 μL. The reaction was carried out for 40 min at 30°C. The lipids were extracted with butanol, dried, and analyzed by ESI-MS. The ESI analysis was done in a positive mode. The neutral lipid molecules were sodiated. The control reaction clearly reveals that the products formed are not contaminants from the substrate. C, A cocktail assay was performed with assay buffer and 10 μg purified recombinant At4g24160 enzyme and three substrates, i.e. 1 mm LPA (1-oleoyl), 10 μm oleoyl-CoA, 1 mm PC (dipalmitate), and 1 mm triolein, in a final reaction volume of 100 μL. The reaction was carried out for 40 min at 30°C. The ESI analysis was done in positive mode and the neutral lipids were sodiated. The added LPA was acylated to PA in the presence of oleoyl-CoA. TG was hydrolyzed to diacylglycerol (DG) and monoacylglycerol (MG), whereas LPC was formed from the hydrolysis of PC. The LPAAT activity of the purified At4g2160 was the most pronounced followed by its PLA2 activity. The TG lipase activity was found to be the least. D, Control reaction (without substrate) was performed with assay buffer and 10 μg Ict1p enzyme in a final reaction volume of 100 μL. The reaction was carried out for 40 min at 30°C. The lipids were extracted with butanol, dried, and analyzed by ESI-MS. The ESI analysis was done in a positive mode. This control clearly reveals that the products formed are not contaminants from the enzyme source. E, Control reaction (without enzyme) was performed with assay buffer, 10 μm oleoyl-CoA, 1 mm LPA (1-oleoyl), 1 mm PC (dipalmitate), and 1 mm triolein in a final reaction volume of 100 μL. The reaction was carried out for 40 min at 30°C. The lipids were extracted with butanol, dried, and analyzed by ESI-MS. The ESI analysis was done in a positive mode. The neutral lipid molecules were sodiated. The control reaction clearly reveals that the products formed are not contaminants from the substrate. F, A cocktail assay was performed with assay buffer and 10 μg Ict1p enzyme and three substrates, i.e. 10 μm oleoyl-CoA, I mm LPA (1-oleoyl), 1 mm PC (dipalmitate), and 1 mm triolein, in a final reaction volume of 100 μL. The reaction was carried out for 40 min at 30°C. The ESI analysis was done in positive mode, and the neutral lipids were sodiated. The LPA provided was acylated to PA in the presence of oleoyl-CoA. TG was hydrolyzed to diacylglycerol and monoacylglycerol, whereas as LPC was formed from the hydrolysis of PC. The LPAAT activity of the purified Ict1p was the most pronounced followed by its TG lipase activity. The PLA2 activity was found to be the least.
Lysophospholipase Activity
Since At4g24160 was found to possess a putative lysophospholipase domain, we analyzed the LPA lysophospholipase activity of the purified enzyme using [3H]LPA; however, no lysophospholipase activity was detected. The same experiment was also done by ESI-MS using unlabeled LPA (1-oleoyl), and the entire substrate provided was recovered at the end of the reaction without any product formation, clearly indicating the absence of lysophospholipase activity (Supplemental Fig. S6, A and B).
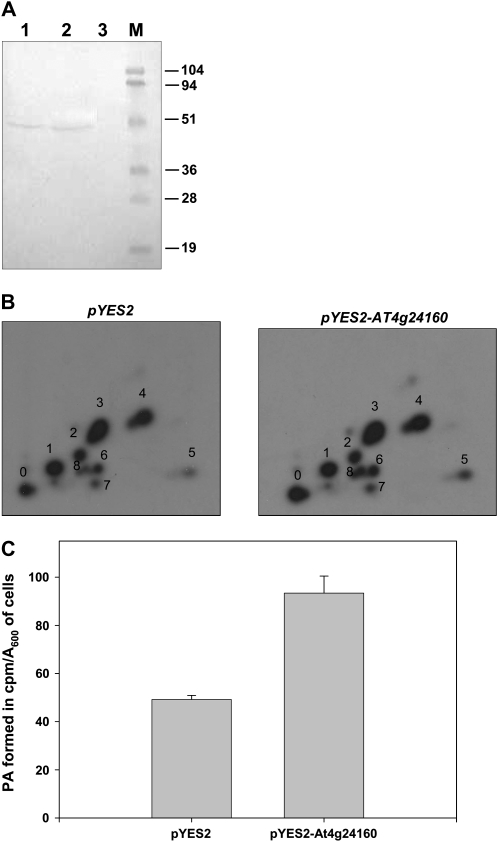
At4g24160 Overexpression Alters Phospholipids in S. cerevisiae
To study the effect of At4g24160 and Ict1p overexpression on the levels of cellular phospholipids and neutral lipids, S. cerevisiae was transformed with pYES2-At4g24160 and pPS189-ICT1. Immunoblotting with anti-Ict1p antibodies confirmed the expression of the At4g24160 protein (Fig. 6A). Overexpression of At4g24160 in yeast led to an approximately 2-fold increase in PA as analyzed by [32P]labeling of phospholipids in vivo (Fig. 6, B and C). In addition, total phospholipids were quantified as described by Broekhuyse (1968). An increase of 1.5-fold in total phospholipid was observed when At4g24160 was overexpressed in S. cerevisiae. The phospholipids in the wild type were found to be 23.8 mg PL/(A600 = 5) of cells, whereas on At4g24160 overexpression the total phospholipids were estimated to be 36.8 mg PL/(A600 = 5) of cells. Besides the increase in total phospholipids, At4g24160 overexpression showed an approximately 47% increase in PA. The overexpression of ICT1 increased the total cellular phospholipid levels by 1.9-fold and a decrease of nearly 3-fold in the PA formation was observed in ICT1 deleted strain (Ghosh et al., 2008a). Thus, the overexpression of At4g24160 and Ict1p resulted in an increase in PA formation.
Figure 6.
Heterologous expression of At4g24160 in S. cerevisiae enhances the phosphatidic acid level and TG hydrolysis. A, pRSETA-At4g24160 (lane 1) transformed E. coli BL21 (DE3) as a control to check that the protein expressed (At4g24160) in yeast is the same as that in bacteria and is not an isoform expressed under protein overexpression or stress conditions. pYES2-At4g24160 (lane 2) transformed yeast and pYES2 (lane 3) vector were grown overnight in the presence of Gal, and cells (A600 = 5) were lysed using glass beads. The proteins were separated by 12% SDS-PAGE, and immunoblotting was performed using anti-Ict1p antibodies at a dilution of 1:1,000 (v/v). M, Molecular mass standard. B, Yeast cells overexpressing At4g24160 and the vector control were grown for 24 h in Gal media in the presence of 200 μCi of [32P]orthophosphate. Lipids were extracted from cells (A600=25) and resolved on two-dimensional silica-TLC using chloroform:methanol:ammonia (65:35:5, v/v) as first-dimensional and chloroform:methanol:acetone:acetic acid:water (50:10:20:15:5, v/v) as second-dimensional solvent systems. Lipids are indicated as 1, phosphatidylinositol; 2, phosphatidylserine; 3, PC; 4, phosphatidylethanolamine; 5, PA; and 6 to 8, unknown. 0 indicates the origin. C, The amount of [32P]orthophosphate incorporated into PA is represented as the cpm per A600 of cells per 24 h of labeling.
Moreover, yeast cells overexpressing At4g24160 and Ict1p were labeled with [14C]acetate in an induction medium to address the question of their effect on neutral lipid metabolism, which suggested that At4g24160 and Ict1p may also be involved in TG turnover in yeast cells (data not shown).
DISCUSSION
LPAAT is a crucial enzyme controlling the metabolic flow of LPA into the pool of PA, which plays a key role in many physiological aspects, such as cell signaling, cell polarity, and apoptotic signaling cascades in higher eukaryotes (Park et al., 2004). It was also shown that PA specifically regulates the genes involved in modulating cell shape and organization. PA has also been found to target proteins to the membranes, thereby modulating their catalytic activity (Fang et al., 2001; Anthony et al., 2004; Zhang et al., 2004; Huang et al., 2006). A recent study also demonstrated that PA is involved in the regulation of stomatal movements by influencing the ABA level. It binds to the ABA-interacting protein phosphatase to signal ABA-promoted stomatal closure, whereas phospholipase D and PA interact with G protein to mediate ABA inhibition of stomatal opening. This signaling is essential for maintaining the hydration status of plants (Mishra et al., 2006).
In Arabidopsis, five AtLPAAT1 to AtLPAAT5 genes encoding LPAATs have been reported based on their amino acid sequence similarity (Kim and Huang, 2004). Arabidopsis has one plastidial LPAAT, AtLPAAT1, and the deletion of this gene is known to be embryonic lethal (Kunst et al., 1988). AtLPAAT2 encodes another LPAAT, which is ubiquitous and endoplasmic reticulum located. A related gene, AtLPAAT3, is highly expressed in pollen presumably for a rapid membrane turnover in the pollen tube (Kim et al., 2005). All AtLPAATs identified so far are membrane bound through transmembrane domains. In contrast, several soluble acyltransferases have been identified from various plant sources (Murata and Tasaka, 1997; Tumaney et al., 2001; Saha et al., 2006). This study provides an additional example of a soluble acyltransferase in plants whose enzymatic properties deserve our attention as biochemists and cell biologists.
Based on the BLAST analysis, 24 proteins were identified in Arabidopsis that are homologs of CGI-58 and Ict1p (Fig. 1A). Among these polypeptides At4g24160 is the closest homolog to the two templates, and in the dendrogram analysis it is grouped (Fig. 1B) in close vicinity to CGI-58 and ICT1. All the three proteins possess no transmembrane domains. Most notably, none of the known AtLPAATs fulfill the latter criterion. Earlier studies have shown that Ict1p in S. cerevisiae is highly expressed during organic solvent exposure (Miura et al., 2000; Matsui et al., 2006) and acylates LPA to PA, thereby bringing about an overall increase in the biosyntheses of membrane phospholipids (Ghosh et al., 2008a). A Δict1 mutant was found to have reduced phospholipid biosynthesis and showed sensitivity to calcofluor, suggesting a defect in membrane formation (Miura et al., 2000). Our recent analysis also revealed that the overexpression of Ict1p not only increased the level of phospholipid but also facilitated the degradation of TG, resulting in an enhanced supply of fatty acyl-CoA for PA synthesis and membrane biogenesis during solvent stress. Similarly, the human homolog of Ict1p, CGI-58, was found to be capable of acylating LPA to PA in an acyl-CoA-dependent manner. Mutations in CGI-58 are responsible for a rare hereditary disorder known as Chanarin Dorfman syndrome. Fibroblasts from such patients showed an abnormal biosynthesis of phospholipids (Ghosh et al., 2008b). Ict1p and human CGI-58 both encode a soluble acyl-CoA-dependent LPAAT responsible for enhanced phospholipid synthesis. Here, we demonstrate that the purified recombinant At4g24160 is capable of acylating LPA to PA (Fig. 3B), and it is, to our knowledge, the first soluble LPAAT to be reported in plants.
Interestingly, At4g24160 was also found to possess lipase and phospholipase activities similar to Ict1p (Figs. 4 and 5). Another single polypeptide exhibiting esterase, lipase, and phospholipase activity was reported earlier in the plant system. Thermally stable alkaline lipase from rice bran was shown to hydrolyze para-nitrophenyl palmitate, TG, and phospholipids (Bhardwaj et al., 2001). In addition, adipocyte triglyceride lipase, a single polypeptide, was shown to have phospholipase A2, TG hydrolase, and acylglycerol transacylase activities (Jenkins et al., 2004; Zechner et al., 2009). Here, we report for the first time, to our knowledge, a soluble protein in the plant system, At4g24160, which exhibits acyl-CoA-dependent LPAAT, TG lipase, and phospholipase activities with the LPAAT activity being most profound. The TG lipase and PLA2-specific activities of At4g24160 and Ict1p were found to be low in comparison to the previously demonstrated Arabidopsis lipases. The low activities could be attributed to the heterologous expression of the enzyme in Escherichia coli. The TG lipase-specific activity of sugar-dependent 1 was found to be 40 μmol/mg protein/min and was determined by using the protein purified from yeast (Eastmond, 2006). The baculovirus-expressed AtLip1 (At2g15230) showed a TG hydrolysis with the specific activity of 45 μmol/mg protein/min. However, the E. coli-expressed protein did not show TG hydrolysis (El-Kouhen et al., 2005). On the other hand, our data clearly establish a detectable TG lipase and PLA2 activity for the E. coli-expressed recombinant protein. In the ESI-MS cocktail assay where the recombinant enzyme was presented with a mixture of substrates at the end of reaction period, the products were analyzed by MS (Fig. 5) that revealed the presence of all the three enzyme activities.
In plants, PA plays a pivotal role as a multifunctional stress signal. PA is generated either by the action of phospholipase D or by the sequential action of phospholipase C and diacylglycerol kinase (van Leeuwen et al., 2004; Wang, 2004; Testerink and Munnik, 2005). At4g24160 can generate PA by acylating LPA to PA, or the diacylglycerol produced by the hydrolysis of TG can be channeled toward PA synthesis. Hence, At4g24160 might be involved in generating an independent cytosolic PA pool. The PLA2 activity of this enzyme might keep a check on membrane biogenesis in the presence of excess PA formed. The gene expression analysis also revealed that At4g24160 was highly expressed during salt stress, emphasizing the role of this gene in plant stress signaling. In conclusion, data presented here provide important insights into the probable physiological role of At4g24160 in maintaining lipid homeostasis, thereby regulating the cellular physiology of plants.
MATERIALS AND METHODS
Materials
At4g24160 clone was obtained from the Arabidopsis Biological Resource Center. [1-14C]Oleoyl-CoA (54 mCi/mmol), [14C]acetate (51 mCi/mmol), [9,10-3H]triolein (53 Ci/mmol), [2-palmitoyl-9,10-3H]phosphatidylcholine (92.3 Ci/mmol), and [9,10-3H]LPA (47 Ci/mmol) were purchased from Perkin-Elmer Biosciences. [32P]Orthophosphate (5000 Ci/mmol) was obtained from the Board of Radiation and Isotope Technology, Bhabha Atomic Research Centre (Mumbai, India). Silica gel 60F254 TLC plates were from Merck. Oligonucleotide primers, TG, diacylglycerol, phospholipids, lysophospholipids, and solvents were purchased from Sigma-Aldrich. Acyl-CoA donors were obtained from Avanti Polar Lipids Polyclonal antibodies were raised against Ni2+-NTA affinity-purified recombinant Ict1p as described (Ghosh et al., 2008a).
Bioinformatics Analysis
Sequence Retrieval, Alignment, and Comparison
cDNA, EST, and protein sequences were identified by searching public databases available at NCBI (http://www.ncbi.nlm.nih.gov) and The Arabidopsis Information Resource (http://www.arabidopsis.org/) with the BLAST algorithms (Altschul et al., 1990, 1997). Sequences were aligned using the ClustalX program (Thompson et al., 1994). The nonredundant protein sequence database was searched using default parameters, and the sequences with E value of 10−5 and score of 80 were retrieved and subjected to sequence alignment.
Phylogenetic Tree Construction
The obtained multiple sequence alignment was subjected to bootstrap resampling. These bootstrap replicate alignments were then used to construct phylogenetic trees by the neighbor-joining method (Saitou and Nei, 1987). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4 software (Tamura et al., 2007). The results were analyzed using the bootstrap method (1,000 replicates) to provide confidence level for tree topology (Felsenstein, 1996). Consensus trees summarizing the topologies found among the bootstrap replicate trees are presented. The tree topology was generated by neighbor joining, with bootstrap support at critical nodes indicated as percentage.
Examination of Conserved Protein Domains
Conserved protein domains were examined using the conserved domain database at NCBI (http://www.ncbi.nih.gov/Structure/cdd/cdd.shtml) and pfam database (http://pfam.sanger.ac.uk/; Bateman et al., 2000).
Gene Expression Analysis
The Genevestigator online search tool Meta-Analyzer (http://www.genevestigator.ethz.ch) was used to retrieve microarray expression data.
Cloning and Expression of At4g24160
pUNI vector containing At4g24160 open reading frame was used as a template for amplification of the gene. Forward primer (5′-ATGGATCCATGAACTTGAGCCGTTTTGCTT-3′) and reverse primer (5′-ATGAATTCCTAAACCAATCGTAGACCATCTAGG-3′) were used. PCR (1 min denaturation at 94°C, 1 min annealing at 55°C, and 1 min elongation at 72°C) was performed using Pfu polymerase for 30 cycles with 10 pmol concentration of each primer. The purified PCR product and pRSET A vector (N-terminal His tag) were digested with BamHI and XhoI and ligated directionally. The construct was transformed into Escherichia coli BL21 (DE3) cells and induced with 1 mm isopropylthio-β-galactoside for 4 h at 37°C. The cell pellet was resuspended in lysis buffer containing 50 mm Tris-HCl (pH 8.0) and 300 mm NaCl. Cells were disrupted by sonication. The inclusion bodies were separated and solubilized in lysis buffer containing 6 m urea and 25 mm imidazole. The 10,000g supernatant of solubilized inclusion bodies was allowed to bind to the Ni2+-NTA matrix. The column was washed with lysis buffer containing 25 mm imidazole. The bound protein was eluted with 250 mm imidazole in lysis buffer. Fractions (1 mL each) were collected and analyzed on 12% SDS-PAGE followed by Coomassie Brilliant Blue staining.
For overexpression of At4g24160 in Saccharomyces cerevisiae, full-length At4g24160 cDNA was subcloned from pRSET A into pYES2 vector at the BamHI-EcoRI site and transformed into yeast cells by lithium chloride method (Schiestl and Gietz, 1989). Transformants were selected on synthetic minimal medium devoid of uracil (SM-U) containing 2% (w/v) Glc and were grown to the late log phase. The cells were harvested by centrifugation and inoculated at an A600 of 0.4 in SM-U medium containing 2% (w/v) Gal and grown for 24 h. To confirm the expression, cells (A600 = 5) were resuspended in 50 mm Tris-HCl (pH 7.5) and 2% (w/v) SDS and then disintegrated using glass beads. The proteins were separated by 12% SDS-PAGE and transferred onto a nitrocellulose membrane. The overexpression of At4g24160 was confirmed using anti-Ict1p antibodies at a dilution of 1:1,000 (v/v).
LPAAT Assay
The reaction mixture contained 10 μm [1-14C]oleoyl-CoA (110,000 dpm/assay), 1 to 5 μg enzyme, and 50 μm LPA (1-oleoyl) in assay buffer with a total volume of 100 μL. The reaction was carried out at 30°C for 10 min and terminated by extracting the lipids (Ghosh et al., 2008a, 2008b). Lipids were also analyzed by two-dimensional TLC using chloroform:methanol:ammonia (65:35:5, v/v) as the first-dimensional solvent system and chloroform:methanol:acetic acid:water (40:20:5:0.5, v/v) as the second-dimensional solvent system. The TLC plates were subjected to autoradiography, and the PA spots were scraped and counted with toluene-based scintillation cocktail. Control incubations were carried out for zero time and in the absence of enzyme. The control value was subtracted from the actual assay value, and the specific activity was calculated after the correction. For ESI-MS analysis, unlabeled substrates 10 μm oleoyl-CoA and 1 mm LPA (1-oleoyl) were used. The reactions were stopped by addition of 100 μL of butanol. The samples were dried and subjected to ESI-MS analysis. The mass by charge peaks obtained were analyzed using the lipid metabolites and pathways strategy (http://www.lipidmaps.org).
Lipase Assays
The purified recombinant At4g24160 and Ict1p (10 μg protein) were incubated in a reaction buffer containing 50 mm Tris-HCl, pH 8.0, for 40 min at 30°C in the presence of a sonicated suspension of 1 mm triolein Reactions were stopped by addition of 100 μL of butanol. The butanol layer containing the lipids was washed with 50 mm sodium acetate. The samples were dried and subjected to ESI-MS analysis. The mass by charge peaks obtained were analyzed using the lipid metabolites and pathways strategy.
Lipase assay was also performed in a reaction buffer containing 50 mm Tris-HCl, pH 7.5, and 100 μm sodium taurocholate for 40 min at 30°C in the presence of sonicated suspension of 100 μm [9,10-3H]triolein (0.25 μCi/tube). Reactions were stopped by addition of 100 μL of butanol, and the samples were resolved on a silica-TLC plate along with oleic acid standard using chloroform:methanol:ammonia (65:35:5, v/v). The region corresponding to the oleic acid standard was scraped and quantified by liquid scintillation counting. The control value was subtracted from the actual assay value, and the specific activity was calculated after the correction.
For esterase assay, para-nitrophenyl stearate (2.5 mm) was used as the substrate, and the hydrolytic product para-nitrophenol was monitored at 410 nm. The absorbance was measured against the reference cell to which buffer had been added instead of dialyzed enzyme solution.
Phospholipase Assays
The reaction mixture contained 1 mm sonicated vesicles of dipalmitate and 10 μg enzyme in a total volume of 100 μL assay buffer (0.05 m Tris-HCl, pH 7.5, and 2 mm dithiothreitol). The reaction was carried out at 30°C for 40 min and terminated by extracting the lipids with butanol. The lipids were dried and analyzed by ESI-MS. The mass-to-charge ratio (m/z) peaks of the substrate and the product were identified using lipid maps.
Radiometric assay consisted of 100 μm sonicated vesicles of [2-palmitoyl-9,10-3H]phosphatidylcholine (1 μCi/assay) and 10 μg enzyme in total volume of 100 μL assay buffer (0.05 m Tris-HCl, pH 7.5, and 2 mm dithiothreitol). The reaction was carried out at 30°C for 40 min and terminated by extracting the lipids by the method of Bligh and Dyer (1959). Lipids were analyzed by silica-TLC (Bhardwaj et al., 2001). The region of the plate corresponding to free fatty acid standard was scraped and counted with toluene-based scintillation cocktail.
Cocktail Assays
The purified recombinant At4g24160 and Ict1p (10 μg protein) were incubated in a reaction buffer containing 50 mm Tris-HCl, pH 8.0, for 40 min at 30°C in the presence of a sonicated suspension of 10 μm oleoyl-CoA, 1 mm LPA (1-oleoyl) , 1 mm triolein, and 1 mm PC (dipalmitate). The reaction was terminated by extracting the lipids with butanol. A sodium acetate wash was given to the butanol layer. The lipids were dried and analyzed by ESI-MS. The m/z peaks of the substrate and the product were identified using lipid maps.
ESI-MS Analysis
The butanol soluble fraction was dried and suspended in HPLC-grade methanol and subjected to ESI-MS analysis (Bruker Esquire 3000 plus electrospray ion trap instrument).The sample was applied directly into the ESI source through a polytetrafluoroethylene line at the rate of 4 μL/min. The ESI-MS settings used were as follows: turbo electrospray inonization source was maintained at 260°C, and the data were collected in the positive ion mode. The experiments were repeated three times, and a similar spectrum was obtained in all the cases. The m/z peaks obtained were analyzed by lipid maps (http://www.lipidmaps.org).
[32P]Orthophosphate Incorporation into Phospholipids
pYES2-At4g24160 and pYES2 transformants were grown to the late log phase in 5 mL SM-U containing 2% (w/v) Glc and then transferred to 50 mL of the same media, such that the absorbance was 0.1. The cells were grown till the absorbance reached 3. A600 = 0.4 cells were inoculated in a fresh medium containing 2% (w/v) Gal and 200 μCi [32P]orthophosphate and grown for 24 h. Cells (A600 = 25) were harvested by centrifugation, and lipids were extracted and analyzed by two-dimensional TLC. The solvents for the first dimension were chloroform:methanol:ammonia (65:35:5, v/v), and solvents for the second dimension were chloroform:methanol:acetic acid:water (40:20:5:0.5, v/v; Ghosh et al., 2008a).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. LPAAT activity analysis of At4g24160 by ESI-MS.
Supplemental Figure S2. TG lipase assay for recombinant purified At4g24160 by ESI-MS.
Supplemental Figure S3. Phospholipase A2 assay for recombinant purified At4g24160 ESI-MS.
Supplemental Figure S4. TG lipase assay for recombinant purified Ict1p by ESI-MS.
Supplemental Figure S5. Phospholipase A2 assay for recombinant purified Ict1p by ESI-MS.
Supplemental Figure S6. Lysophospholipase activity of At4g24160 by ESI-MS.
Supplementary Material
This research was supported by a grant from the Department of Biotechnology, Government of India, New Delhi (a program supporting nonconventional yeast to R.R.), and the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (Projects 18857 and W901–B05 to G.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ram Rajasekharan (lipid@biochem.iisc.ernet.in).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bogre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL (2000) The Pfam protein families database. Nucleic Acids Res 28: 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K, Raju A, Rajasekharan R (2001) Identification, purification, and characterization of a thermally stable lipase from rice bran. A new member of the (phospho) lipase family. Plant Physiol 127: 1728–1738 [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Broekhuyse RM (1968) Phospholipids in tissues of the eye. Isolation, characterization and quantitative analysis by two dimensional thin-layer chromatography of diacyl and vinylether phospholipids. Biochim Biophys Acta 152: 307–315 [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Khuri S, Gane P (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 64: 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kouhen K, Blangy S, Ortiz E, Gardies AM, Ferte N, Arondel V (2005) Identification and characterization of a triacylglycerol lipase in Arabidopsis homologous to mammalian acid lipases. FEBS Lett 579: 6067–6073 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266: 418–426 [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J (2001) Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942–1945 [DOI] [PubMed] [Google Scholar]
- Gangar A, Karande AA, Rajasekharan R (2001) Isolation and localization of a cytosolic 10 S triacylglycerol biosynthetic multienzyme complex from oleaginous yeast. J Biol Chem 276: 10290–10298 [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Ramakrishnan G, Chandramohan C, Rajasekharan R (2008. b) CGI-58, the causative gene for Chanarin-Dorfman syndrome, mediates acylation of lysophosphatidic acid. J Biol Chem 283: 24525–24533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Ramakrishnan G, Rajasekharan R (2008. a) YLR099C (ICT1) encodes a xoluble acyl-CoA-dependent lysophosphatidic acid acyltransferase responsible for enhanced phospholipid synthesis on organic solvent stress in Saccharomyces cerevisiae. J Biol Chem 283: 9768–9775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 281: 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RJ, Rock CO (1998) A conserved histidine is essential for glycerolipid acyltransferase catalysis. J Bacteriol 180: 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Gao L, Blanchoin L, Staiger CJ (2006) Heterdimeric capping protein from arbidopsis is regulated by phosphatidic acid. Mol Biol Cell 4: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW (2004) Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J 7Biol Chem 279: 48968–48975 [DOI] [PubMed] [Google Scholar]
- Kim HU, Huang AH (2004) Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol 134: 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Li Y, Huang AHC (2005) Ubiquitous and endoplasmic reticulum–located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17: 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst LJ, Browse J, Somerville C (1988) Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc Natl Acad Sci USA 85: 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov WS, Faust A, Nawrath C, Baer S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Metraux JP, et al (2006) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Hirayama T, Kuroda K, Shirahige K, Ashikari T, Ueda M (2006) Screening for candidate genes involved in tolerance to organic solvents in yeast. Appl Microbiol Biotechnol 71: 75–79 [DOI] [PubMed] [Google Scholar]
- Meier C, Aricescu AR, Assenberg R, Robin T, Gilbert RJC, Grimes JM, Stuar DI (2006) The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure 14: 1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra G, Zhang W, Deng F, Zhao J, Wang X (2006) A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266 [DOI] [PubMed] [Google Scholar]
- Miura S, Zou W, Ueda M, Tanaka A (2000) Screening of genes involved in isooctane tolerance in Saccharomyces cerevisiae by using mRNA differential display. Appl Environ Microbiol 66: 4883–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N, Tasaka Y (1997) Glycerol-3-phosphate acyltransferase in plants. Biochim Biophys Acta 1348: 10–16 [DOI] [PubMed] [Google Scholar]
- Nardini M, Dijkstra BW (1999) Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol 9: 732–737 [DOI] [PubMed] [Google Scholar]
- Park J, Gu Z, Lee Y, Yang Z, Lee Y (2004) Phosphatidic acid induces leaf cell death in Arabidopsis by activating the rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol 134: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Singh AK, Rajasekharan R (2008) The Saccharomyces cerevisiae PHM8 gene encodes a soluble magnesium-dependent lysophosphatidic acid phosphatase. J Biol Chem 283: 8846–8854 [DOI] [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds: molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Shekar S, Tumaney AW, Rao TJ, Rajasekharan R (2002) Isolation of lysophosphatidic acid phosphatase from developing peanut cotyledons. Plant Physiol 128: 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J (1991) Plant lipids: metabolism, mutants, and membranes. Science 252: 80–87 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Nucleic Acids Res 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Testerink C, Munnik T (2005) Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci 10: 368–375 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaney AW, Sekhar S, Rajasekharan R (2001) Identification, purification, and characterization of monoacylglycerol acyltransferase from developing peanut cotyledons. J Biol Chem 276: 10847–10852 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Okrész L, Bögre L, Munnik T (2004) Learning the lipid language of plant signaling. Trends Plant Sci 9: 378–384 [DOI] [PubMed] [Google Scholar]
- Wang X (2004) Lipid signaling. Curr Opin Plant Biol 7: 329–336 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase D alpha1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zazwinska A, Affolter M (2004) A family of genes encoding zona pellucida (ZP) domain proteins is expressed in various epithelial tissues in Drosophila embryogenesis. Gene Expr Patterns 4: 413–421 [DOI] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A (2009) Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res 50: 3–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.