Abstract
Chromosome 1H (approximately 622 Mb) of barley (Hordeum vulgare) was isolated by flow sorting and shotgun sequenced by GSFLX pyrosequencing to 1.3-fold coverage. Fluorescence in situ hybridization and stringent sequence comparison against genetically mapped barley genes revealed 95% purity of the sorted chromosome 1H fraction. Sequence comparison against the reference genomes of rice (Oryza sativa) and sorghum (Sorghum bicolor) and against wheat (Triticum aestivum) and barley expressed sequence tag datasets led to the estimation of 4,600 to 5,800 genes on chromosome 1H, and 38,000 to 48,000 genes in the whole barley genome. Conserved gene content between chromosome 1H and known syntenic regions of rice chromosomes 5 and 10, and of sorghum chromosomes 1 and 9 was detected on a per gene resolution. Informed by the syntenic relationships between the two reference genomes, genic barley sequence reads were integrated and ordered to deduce a virtual gene map of barley chromosome 1H. We demonstrate that synteny-based analysis of low-pass shotgun sequenced flow-sorted Triticeae chromosomes can deliver linearly ordered high-resolution gene inventories of individual chromosomes, which complement extensive Triticeae expressed sequence tag datasets. Thus, integration of genomic, transcriptomic, and synteny-derived information represents a major step toward developing reference sequences of chromosomes and complete genomes of the most important plant tribe for mankind.
Access to the complete genome sequence of an organism provides a direct path to gene identification, understanding gene function, exploring genetic diversity, and correlating this information to phenotypic traits. Application of next generation sequencing (NGS) technology (Shendure and Ji, 2008) for whole genome resequencing may soon become a routine for genome-scale genotyping and haplotype analysis in man. However, such progress is only possible due to the availability of a high-quality reference whole genome sequence—a resource that is still lacking for many of the most important crop species, including the major cereals of the Triticeae tribe.
Barley (Hordeum vulgare) is the number four cereal crop in the world. It is a major resource for animal feed and for the brewing and distilling industry. The genome of barley comprises 5.1 Gbp/1 C (Doležel et al., 1998), is about 12 times the size of the rice (Oryza sativa) genome, and includes over 80% of repetitive DNA (Schulte et al., 2009; Wicker et al., 2009). The size, high repeat content, and costs of conventional Sanger sequencing impede whole genome sequencing in barley. Consequently, only limited knowledge of its genomic sequence has been accumulated so far by dedicated sequencing of barley bacterial artificial chromosome (BAC) contigs in the course of map-based gene isolation (Stein, 2007). Massive data generation and cost efficiency of NGS allows questions on barley genome composition with unprecedented resolution and depth to be addressed. Wicker et al. (2006, 2009) employed pyrosequencing (454/Roche GS20) to survey gene information on selected barley BAC clones (Wicker et al., 2006) and to catalog the composition of the barley genome (Wicker et al., 2009). Moreover, the short-read sequencing by synthesis (Solexa/Illumina GA 1) was used to generate whole genome shotgun sequence information to assist the statistical annotation of DNA motif frequency at whole genome scale for barley (Wicker et al., 2008). Despite the impressive progress, ordering the massive numbers of short reads obtained by NGS to generate genomic scaffolds of the huge Triticeae genomes remains a major challenge.
Instead of sequencing complex cereal genomes containing large fractions of repetitive DNA, smaller genomes of grass species like rice (1 C to approximately 400 Mbp) and Brachypodium distachyon (1 C to approximately 280 Mbp) were suggested as surrogates and models for molecular genomics and positional cloning in cereals with large genomes (Bennetzen and Freeling, 1993; Draper et al., 2001). This strategy is supported by a significant level of colinearity between Poaceae genomes (Moore et al., 1995; Bolot et al., 2009). Moreover, high-quality reference genome sequences for both rice and sorghum (Sorghum bicolor) are available (Sasaki and Sederoff, 2003; Paterson et al., 2009) and provide a platform for large-scale implementation of this approach. Although reference genomes represent very important resources of information for molecular genomics in the Triticeae the potential impact of genome colinearity still is limited and can compromise synteny-based gene isolation, since only 50% of the barley genes remain collinear compared to rice (Gaut, 2002; Stein et al., 2007). This observation has been illustrated during map-based cloning of important genes in wheat (Triticum aestivum; vrn2; Yan et al., 2004) and barley (vrs1; Komatsuda et al., 2007) where orthologs were lacking in rice within otherwise well-preserved colinear genome segments.
An additional option to cope with the complexity of cereal genomes is to isolate individual chromosomes and sequence these individually. The reduced complexity of the sorted chromosome samples facilitates molecular analyses, including the isolation of markers and physical mapping (Doležel et al., 2007). Recently, a physical map of wheat chromosome 3B was constructed based on a BAC library cloned from flow-sorted chromosomes (Paux et al., 2006). A procedure for representative amplification of DNA by multiple displacement amplification (MDA) from sorted barley chromosomes was developed (Simkova et al., 2008). As chromosomal DNA in amounts of a few nanograms can be produced easily, this advance opens new avenues for the wider use of chromosome sorting in Triticeae genomics.
In this study, we demonstrate the potential of high-throughput NGS of flow-sorted chromosomes for genome analysis, sequencing, and the development of a high-resolution gene map. As few as 10,000 copies of chromosome 1H were flow sorted from barley cv Morex and used as a template to assess gene content and genomic composition of this chromosome. Information about sequence conservation and conserved gene content to the rice and sorghum genomes was obtained at unprecedented density and resolution and allowed synteny and homology information to be integrated into a virtual high-density gene map of barley chromosome 1H.
RESULTS
Flow Cytometric Sorting and 454 Sequencing of Barley Chromosomes
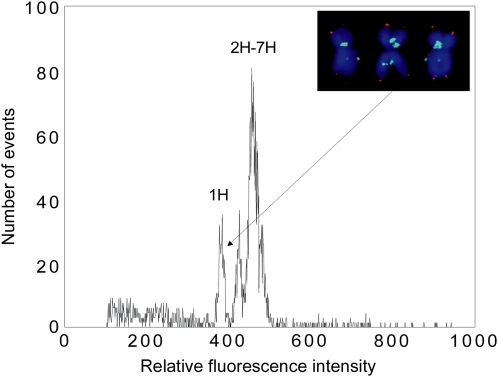
Barley has seven chromosomes that are named 1H through 7H according to their homologous relationship to other Triticeae linkage groups (Linde-Laursen, 1996). Flow-cytometric analysis of chromosome suspensions prepared from Morex resulted in histograms of relative fluorescence intensities (flow karyotypes) with a composite peak representing chromosomes 2H to 7H and a small peak of chromosome 1H (Fig. 1). Chromosome 1H is considerably smaller than chromosomes 2H to 7H and can be easily sorted. The sorted fractions of 1H consisted mainly of chromosome 1H (95.5% ± 0.7%; mean ± sd) as determined by fluorescence in situ hybridization (FISH) on 1,000 sorted chromosomes taken during each sort run (data not shown). The contamination was due to various chromosomes and chromosome fragments. Altogether, five batches of 10,000 chromosomes 1H and five batches of 20,000 chromosomes 1H to 7H were prepared for DNA amplification. The amounts of purified DNA recovered from the sorted chromosomes ranged from 7 to 10 ng and from 10 to 18 ng for chromosome 1H and all chromosomes, respectively. The quantity of DNA obtained after MDA ranged from 3.0 to 5.0 μg DNA in samples with chromosome 1H (whole chromosome amplified 1H = WCA1H), and from 4.5 to 5.6 μg DNA in samples with all chromosomes (1H–7H; whole chromosome amplified all = WCAall).
Figure 1.
Histogram of fluorescence intensity (flow karyotype) obtained after flow-cytometric analysis of 4′,6-diamino-phenylindole stained chromosomes of Morex. The peak of chromosome 1H is well discriminated from the remaining chromosomes forming a composite peak. The insert shows three examples of sorted chromosome 1H after fluorescent labeling of GAA microsatellites (yellow green) and a telomeric repeat (red) using FISH. [See online article for color version of this figure.]
Enrichment of Chromosome 1H Genomic Sequences
Over 3 million sequence reads comprising close to 800 Mb of sequence were obtained from the shotgun sequence of the flow-sorted chromosome 1H (WCA1H; Table I). Considering the 1 C genome size of barley, 5.1 Gb (Doležel et al., 1998), and relative size of chromosome 1H (12.2%; Marthe and Künzel, 1994), the molecular size of 1H can be estimated to be 622 Mb. Assuming a random distribution of sequence reads, every 200 bp a sequence tag is expected. According to the Lander-Waterman model (Lander and Waterman, 1988) at a 1.29-fold sequence coverage, 72.3% of bases from barley chromosome 1H should be represented in the chromosome shotgun sequence dataset.
Table I.
454 sequence read characteristics
Summary of the sequence read characteristics obtained by 454 sequencing of pooled barley chromosomes 1H to 7H (WCAall) and chromosome 1H exclusively (WCA1H).
| Dataset | No. Reads Sequenced Before Masking | Total Basepairs | No. Reads After Masking (%) | Median Read Length | M50 Length | N's | No. Reads with Unique Sequences | Reads with Unique Sequences | No. Percent >90 Masked | Masked RepeatMasker |
|---|---|---|---|---|---|---|---|---|---|---|
| bp | % | % | bp % | |||||||
| WCAall | 381,617 | 99,401,554 | 118,779 (31.1%) | 256 | 259 | 1.96 | 94,889 | 79.9 | 54.3 | 74.4 |
| WCA1H | 3,046,327 | 799,343,261 | 896,421 (29.4%) | 258 | 260 | 2.44 | 813,914 | 90.8 | 56.5 | 77.5 |
We verified the purity in the sorted 1H fractions by comparing the repeat-masked sequence collections from WCA1H to a barley consensus transcript map comprising 2,785 nonredundant EST markers. Chromosome 1H contributed 11.9% (332 markers) of all markers in this map, similar to the relative DNA contribution of chromosome 1H to the entire barley genome (Table II). For the WCA1H sequences, matches were detected to 423 markers of the genome-wide set. A total of 297 out of 332 (89.5%) chromosome 1H located markers were detected whereas only 126 of 2,453 (5.1%) chromosome 2H to 7H markers were hit (cross tab test P value = 0). For sequence data derived from pooled, sorted chromosomes 1H to 7H (WCAall) an even marker detection rate distributed over all chromosomes was observed (Table II). Therefore, based on marker detection rate (89.5%/5.1% = 17.54%) and relative contribution of chromosome 1H to the entire barley genome (87.8%/12.2% = 7.2%), a 126-fold enrichment (17.54% × 7.2%) was observed for WCA1H. This trend was substantiated when using the absolute sequence read counts associated to anchored marker sequences. Of 2,138 individual WCA1H sequence reads anchored to transcript markers, 1,932 (90.4%) were associated with the 297 chromosome 1H markers (Table II; Fig. 2A). Markers located on chromosomes 2H to 7H accumulated less-frequent WCA1H sequence read matches. One-hundred fifteen of all 126 identified 2H to 7H markers (91%) were hit by three or less WCA1H reads (Table II; Fig. 2B).
Table II.
454 read distribution to barley EST-based markers
Sequence reads from pooled chromosomes 1H to 7H (WCAall) and from a chromosome 1H amplified sequence library were compared with 2,785 unique sequence markers anchored on the genetic map of barley. While for WCAall, an even recovery rate over all seven chromosomes was observed, WCA1H is strongly biased toward chromosome 1H. A total of 89.5% of markers recovered and 90.4% of reads are associated with chromosome 1H.
| Chromosome | No. of markers | WCAall
|
WCA1H
|
|||||
|---|---|---|---|---|---|---|---|---|
| No. Reads | No. Marker Detected | Markers Detected | No. Reads | No. Marker Detected | Markers Detected | Anchored Reads | ||
| % | % | |||||||
| 1H | 332 | 37 | 26 | 7.8 | 1,932 | 297 | 89.5 | 90.4 |
| 2H | 468 | 41 | 32 | 6.8 | 38 | 18 | 3.8 | 1.8 |
| 3H | 445 | 45 | 35 | 7.9 | 22 | 13 | 2.9 | 1.0 |
| 4H | 314 | 32 | 30 | 9.6 | 16 | 13 | 4.1 | 0.7 |
| 5H | 492 | 43 | 36 | 7.3 | 57 | 28 | 5.7 | 2.7 |
| 6H | 337 | 19 | 17 | 5.0 | 42 | 29 | 8.6 | 2.0 |
| 7H | 397 | 28 | 22 | 5.5 | 31 | 25 | 6.3 | 1.4 |
| Total | 2,785 | 245 | 198 | 2,138 | 423 | |||
Figure 2.
Detection of gene-based markers by random (WCAall) and chromosome 1H (WCA1H) sequence collections. A, The number of sequence reads of WCAall and WCA1H samples that could be associated to chromosome-anchored sequence markers was plotted. Sequence reads from the WCAall collection were equally distributed over markers anchored to all seven chromosomes while WCA1H reads were highly enriched for chromosome 1H markers. B, The frequency of WCA1H sequence reads obtained for chromosome 1H compared to 2H to 7H gene-based barley markers differed significantly, respectively. The x axis denotes markers anchored on barley chromosomes 1H to 7H, respectively. The y axis plots the number and distribution of WCA1H sequence reads as observed for markers anchored to individual chromosomes (colored lines). The inset depicts values observed for 2H to 7H. [See online article for color version of this figure.]
We calculated the proportion of detected and undetected markers (true/false positives and negatives, respectively) that were identified (true positives: 297; false positives: 126; true negatives: 2,327; false negatives: 35). A recall rate (sensitivity) of 0.895 and specificity of 0.95 was reached. Applying a confusion matrix, the probability for correct classification reached 0.942. These findings were consistent with the estimated purity of enrichment of 95% estimated by microscopic observation of sorted fractions. In summary, cytological as well as molecular evidence based on marker to sequence read association indicated a 95% purity of the barley WCA1H sequence collection. In addition, the sensitivity exceeded the theoretical expectation of 72% derived from the Lander-Waterman model, as 89.5% of the markers located on chromosome 1 were sequence tagged.
Repeat Composition of the Barley Genome and Chromosome 1H
WCA1H and WCAall datasets were compared for content and frequency of individual classes of repeats. Overall similar fractions of 77.5% (WCA1H) and 74.5% (WCAall) were assigned as repetitive elements. For both datasets, the ratio of class I to class II elements was determined to be 11:1 to 12:1 (Table III). The overall frequency of most element types was very similar; however, deviations were detected for class I retroelements contributing a slightly higher percentage to WCA1H (71.1% versus 67.6% in WCAall). In addition, deviations between datasets were found for CACTA-type elements (6% in WCA1H versus 6.4% in WCAall). The relative amount of ribosomal gene sequences was lower in WCA1H (0.04% versus 0.13% in WCAall). This was consistent with the localization of nucleolus organizing regions on barley chromosomes 6H and 7H (Singh and Tsuchiya, 1982), which thus represent regions that should be depleted in WCA1H.
Table III.
Repeat content and composition in WCAall and WCA1H datasets
Sequences from the WCAall as well as the WCA1H collection were analyzed for their repeat content. Similar frequencies of each category were observed in the two collections.
| Type of Repetitive Element | WCAall | WCA1H |
|---|---|---|
| % of genome | % of genome | |
| Class I: retroelement | 67.61 | 71.10 |
| LTR retrotransposon | 66.99 | 70.41 |
| Ty1/copia | 13.41 | 14.44 |
| Ty3/gypsy | 36.44 | 38.56 |
| Unclassified LTR | 17.14 | 17.41 |
| Non-LTR retrotransposon | 0.61 | 0.68 |
| LINE | 0.60 | 0.67 |
| SINE | 0.01 | 0.01 |
| Unclassified retroelement | 0.01 | 0.01 |
| Class II: DNA transposon | 6.44 | 6.00 |
| DNA transposon superfamily | 6.06 | 5.62 |
| CACTA superfamily | 5.59 | 5.19 |
| hAT superfamily | 0.05 | 0.06 |
| Mutator superfamily | 0.24 | 0.22 |
| Tc1/Mariner superfamily | 0.08 | 0.03 |
| PIF/Harbinger | 0.10 | 0.12 |
| Unclassified | 0.01 | 0.01 |
| DNA transposon derivative | 0.24 | 0.26 |
| MITE | 0.24 | 0.26 |
| Helitron | 0.09 | 0.06 |
| Unclassified DNA transposon | 0.05 | 0.06 |
| High copy number gene | 0.13 | 0.04 |
| RNA gene | 0.13 | 0.04 |
| Total | 74.54 | 77.49 |
Estimation of Barley Chromosome 1H Gene Content
To estimate the gene content of chromosome 1H, homology of WCA1H sequence reads to known genes was surveyed by similarity searches against complete reference genomes, namely rice and sorghum, as well as against clustered EST collections from wheat and barley under optimized stringency conditions (Supplemental Fig. S1, A and B). A total of 4,125 and 4,359 homologous rice and sorghum genes were hit, respectively (BLASTX ≥70% identity ≥30 amino acids). From wheat and barley EST collections 5,498 and 4,765 (BLASTN) and 3,923 and 4,154 (BLASTX) ESTs and EST clusters were tagged, respectively (Supplemental Table S1). From the comparisons to the different individual reference datasets a nonredundant gene count was extracted comprising 5,126 genes (TBLASTX; ≥70% and ≥30 amino acids). Given the experimentally observed marker detection rate of approximately 89.5% within the WCA1H dataset, a chromosome 1H content of between 4,600 and 5,800 genes can be estimated. Considering the relative size of chromosome 1H (12%), a total of 38,000 to 48,000 genes can be predicted for the entire barley genome.
Assessment of Conserved Gene Content of Barley Chromosome 1H against Rice and Sorghum
Close syntenic relationships among Poaceae have been known for a long time (Moore et al., 1995). However, the availability of highly enriched chromosome 1H sequence permitted us to infer synteny to rice and sorghum reference genome sequences at the whole chromosome level with a per gene resolution. Using a stringent filter criterion of ≥30 amino acid similarity we analyzed the barley WCA1H sequence reads against the respective rice and sorghum genome assemblies and selected for the best homologs. A similar number and percental range of 4,125 (15.2% of all rice genes) and 4,359 (16% of all sorghum genes) homologous genes were detected, respectively (Supplemental Table S3). Rice chromosomes 5 and 10 as well as sorghum chromosomes 1 and 9 were substantially enriched for putative orthologs and outnumbered the remaining chromosomes of the respective genomes. However, the numbers of putative orthologs provided only a global overview. Therefore, the analysis was refined on the basis of rice and sorghum synteny. Positional information on the respective chromosome was considered and regions containing a high proportion of putative orthologs were depicted (Fig. 3, A and B). Regions with conserved gene content of barley chromosome 1H corresponded to distal regions of both arms of rice chromosome 5 and the distal region of the long arm of rice chromosome 10, respectively. The comparison against sorghum detected such regions for the distal parts of chromosome 9 and the central portion of chromosome 1. A small region of rice chromosome 1 also showed a signal in this analysis. However, subsequent analysis revealed that this region contained a high proportion of protein kinases (26 out of 41 genes) and no apparent synteny to sorghum (data not shown). Generally, genes containing a protein kinase domain are abundant in plant genomes and sequence conservation in the protein kinase domain is usually very high. Therefore, the accumulation of positive matches in this region of rice chromosome 1 indicated rather a false-positive than a true and previously unobserved syntenic region. Due to a lack of detectable syntenic relationship to sorghum and the barley marker scaffold we excluded this region from the subsequent integrative analysis (see below).
Figure 3.
WCA1H sequence reads mapped on the genomes of rice and sorghum. The heatmap is depicting the location of detected rice (A) and sorghum (B) homologous (syntenic) segments. WCA1H sequence reads were anchored on rice and sorghum using BLASTX and the best detectable match. Individual chromosomes were numbered and the size intervals in megabases were given. Regions with conserved gene content to barley chromosome 1H (implied syntenic regions) were obvious and encompassed rice chromosomes 5 and 10 as well as a small region on chromosome 1. For sorghum, similar regions were observed for chromosomes 1 and 9.
Reverse Engineering of an Ordered Gene Map of Barley Chromosome 1H
On the basis of the shotgun read coverage of chromosome 1H, we constructed a virtual gene map of barley chromosome 1H (Fig. 4). Genes from syntenic regions of the rice and sorghum genomes were selected by association with WCA1H sequence reads and were subsequently ordered along the virtual barley chromosome 1H. One hundred and eighty rice and 195 sorghum genes of the syntenic regions could be directly associated to putatively orthologous genetic markers on barley 1H. Their linear order and synteny association provided the framework for integration and deduction of a virtual gene map of barley chromosome 1H. Out of 1,513 and 1,711 genes contained within the 1H syntenic regions of rice and sorghum, WCA1H sequence reads could be assigned to 1,377 (91%) and 1,551 (90.6%) genes, respectively (Supplemental Table S2). Only these rice and sorghum genes were considered for integration into the virtual barley chromosome 1H gene map (Supplemental Table S4). This approach resulted in tentative anchoring of WCA1H derived sequence tags that detected close to 2,000 putatively orthologous genes from rice and sorghum. Best bidirectional hits revealed orthology between rice and sorghum for 1,174 (1,129 with associated marker or read evidence) genes present in the selected syntenic regions from sorghum and rice. In contrast, 277 (18.31%) rice and 452 (26.41%) sorghum genes from these regions were tagged by corresponding sequence matches of WCA1H only but did not exhibit any detectable rice/sorghum orthologous counterpart. Thus, we were able to tentatively allocate 1,858 nonredundant gene loci with associated putative rice/sorghum orthologs on barley chromosome 1H. In addition, 129 map-anchored barley loci without corresponding rice/sorghum ortholog have also been integrated into the 1H gene map. This increased the number of oriented and anchored loci to 1,987, which corresponded to between 34% and 43% of the estimated gene complement of chromosome 1H (Supplemental Tables S2 and S4).
Figure 4.
Schematic representation of marker and synteny guided assembly of an integrated virtual gene map for barley chromosome 1H. Genetically anchored barley markers have been integrated with rice and sorghum genes located in syntenic regions to give an enriched tentative ancestral gene scaffold. WCA1H sequence reads as well as barley EST sequences have been associated with this chromosome matrix and give rise to an ordered integrated gene map of barley chromosome 1H.
The syntenic integration based on information of rice and sorghum provided specifically added value for regions with limited genetic resolution of barley chromosome 1H, i.e. centromeric and subcentromeric regions. Here, sequence identity to collinearly organized homologs (orthologs) of rice and sorghum provided a hypothetical linear order for such barley markers/genes for which linear gene/marker order could not be resolved genetically. Furthermore, the collinear intervals in rice and sorghum that could be framed by cosegregating markers of the barley 1H centromere were carrying as many as 373 genes that were tagged by WCA1H reads. Given that only between 34% to 43% genes are potentially syntenic between barley, rice, and sorghum in this region (see above) it can be postulated that between 850 to 1,100 genes, roughly 20% of all genes of barley 1H, may be located in centromeric and subcentromeric regions exhibiting very low recombination frequency and thus represent genes with limited accessibility based on genetic mapping approaches.
DISCUSSION
A complete genome sequence is a fundamental resource to answer a wide range of basic and applied scientific questions. However, for the Triticeae tribe comprising some of the most important crop species (i.e. wheat, barley), large-scale genomic sequence information is essentially lacking. Whole genome sequencing of barley and wheat is complicated by the huge genome size (1 C to approximately 5.1 Gbp in barley; Doležel et al., 1998; and 1 C to approximately 17 Gbp in wheat; Bennett and Smith, 1976) and the inherent genome complexity caused by a content of 80% to 90% repetitive elements (Smith and Flavell, 1975; Paux et al., 2006). In this study, we combined chromosome sorting and NGS to gain insight at unprecedented density into the gene content of an entire Triticeae chromosome. Integration with high-resolution synteny data from grass model genome sequences of rice and sorghum allowed us to propose a virtually ordered gene inventory of 1,987 anchored genes (39% of sequence-tagged genes) of barley chromosome 1H.
Almost 90% of all genes of chromosome 1H were sequence tagged at only 1.3-fold 454 shotgun sequence coverage. Based on the number of genes detected by 454 sequence reads in the genome reference datasets of rice and sorghum and EST datasets of wheat and barley and a 95% probability of chromosome 1H origin, this translated into a gene content of roughly 5,400 genes for chromosome 1H. Overall 45,000 genes for the entire barley genome can be estimated. This number is very close to a previous estimate based on assembly of 444,652 barley ESTs (28,001 EST contigs + 22,937 EST singles, http://www.harvest-web.org; Close et al., 2008) but it slightly exceeds the annotated gene content of rice (37,544 predicted genes; International Rice Genome Sequencing Project, 2005) and sorghum (34,496 gene models; Paterson et al., 2009). Additional indirect confirmation of our gene content estimate came from end sequencing of approximately 11,000 chromosome-specific BAC clones that suggested a content of 6,000 genes for wheat chromosome 3B (Paux et al., 2006). This wheat chromosome is homologous to barley chromosome 3H (size 755 Mb; Suchankova et al., 2006). Assuming a comparable gene density for both barley chromosomes 1H and 3H, the estimated gene content scales to a content of 6,500 genes for barley chromosome 3H, a similar range of magnitude as estimated for wheat chromosome 3B.
Grass genomes share a significant level of synteny (Moore et al., 1995). Colinearity of Triticeae group 1 chromosomes was recently confirmed to distal regions of both arms of rice chromosome 5 and the distal part of rice chromosome 10 long arm on the basis of several hundred gene-derived markers in barley (Stein et al., 2007) and wheat (Qi et al., 2004), respectively. Here, our study takes this analysis to the level of a complete chromosome view: About 36.2% of all genes detected for chromosome 1H matched to rice and/or sorghum genes located in colinear regions and thus confirmed previously detected synteny. More importantly, the sequence coverage, the high degree of chromosome purity, and corresponding syntenic coverage enabled to imply the extent of syntenic regions with a per gene resolution. No further regions with conserved gene content to the rice and sorghum genomes were observed.
The integration of low-pass shotgun sequencing information of barley chromosome 1H with the colinear gene order of 1,858 nonredundant orthologous rice and sorghum genes allowed us to propose a virtual sequence-based gene order map of an entire Triticeae chromosome. It is noteworthy that syntenic integration also allowed the ordering of genes in regions with limited genetic resolution such as subcentromeric and centromeric regions. Our results indicated that roughly one-fifth of the genes of barley chromosome 1H are possibly located in this region with low recombination frequency. In addition to the currently available sequences of rice and sorghum, genome sequences will soon become available for maize (Zea mays; Pennisi, 2008) and Brachypodium (http://www.brachypodium.org/), of which the latter is evolutionarily considerably closer to barley (Bolot et al., 2009). Such additional information will allow to further refine gene maps derived from low-pass sequencing of flow-sorted chromosomes. Nevertheless, this approach will also meet limitations: Due to translocation of genes in comparison to the synteny scaffolds, an estimated 50% of the detected barley genes cannot be anchored and local rearrangements as well as local duplications like tandemly duplicated genes cannot be resolved. Thus, the presented approach can be seen as a powerful approximation and as a complementary approach to other genetic and physical map-based attempts to develop a complete reference genome sequence of barley and Triticeae in general.
Flow cytometric sorting provides a powerful means to reduce genome complexity since it allows isolation of individual chromosomes (Doležel et al., 2007). In our study we focused on barley chromosome 1H (approximately 622 Mb), which represents about 12% of the barley genome and that can be directly sorted from the remaining six chromosomes (Suchankova et al., 2006). The remaining barley chromosomes 2H to 7H can be sorted separately from wheat-barley ditelosomic addition lines (Suchankova et al., 2006). Such chromosome arms represent between 6% and 9% of the barley genome (301–459 Mbp) and would enable to survey the whole barley genome by NGS low-pass shotgun sequencing at further reduced complexity.
In this study, low-pass shotgun sequencing of flow-sorted chromosomes proved to be efficient to sequence tag the gene content of a whole barley chromosome. Instead of direct sequencing of chromosomal DNA, MDA (Dean et al., 2002) was used to generate microgram quantities of DNA from batches of 10,000 sorted 1H chromosomes. MDA has proven to be useful for highly accurate and representative amplification of human, fungal, and microbial templates (Silander and Saarela, 2008) as well as for flow-sorted barley chromosomes (Simkova et al., 2008). The potential value of this source of DNA for de novo shotgun sequencing and for genome sequence assembly in the Triticeae, however, remains to be determined.
De novo shotgun sequencing has been previously applied to moderately complex plant genomes that exceed the size of individual barley chromosomes and harbor tracks of highly repetitive sequences in the range of several megabases. So far such attempts either relied on Sanger sequencing only or used Sanger and NGS technology in mixed assemblies (Jaillon et al., 2007; Velasco et al., 2007; Paterson et al., 2009). In all cases, however, paired-end sequencing of differently but specifically sized DNA fractions (i.e. genomic plasmid, cosmid, or BAC libraries) was applied to obtain sufficiently sized sequence scaffolds. Since MDA DNA contains a low-amplification bias (Dean et al., 2002; Hosono et al., 2003; Rook et al., 2004) the method might contribute to upcoming strategies for whole chromosome and genome shotgun sequencing and assembly in Triticeae.
CONCLUSION
Low-pass shotgun sequencing of flow-sorted barley chromosome 1H boosted the amount of 1H anchored genes by 6-fold compared to existing map resources. With the integration of syntenic information from other grass genomes unprecedented resolution was achieved. This data will significantly impact cereal genomics: Anchored as well as the unanchored genes determined in this study can be correlated with BAC clone libraries and thus anchored to the emerging physical map of the barley genome (Schulte et al., 2009). In prospect of the rapid improvement of sequencing technology (Shendure and Ji, 2008) and upcoming highly advanced genomic resources for the Triticeae (dense marker frameworks, robust physical maps, reduced DNA sample complexity by chromosome sorting, access to syntenic reference grass genome sequences) the cost-effective generation of sequences for individual chromosome arms and finally the complete barley genome is no longer far out of reach.
MATERIALS AND METHODS
Purification and Amplification of Chromosomal DNA
Intact mitotic chromosomes were isolated by flow cytometric sorting and the purity of the obtained chromosome suspension was determined by FISH essentially as described previously (Suchankova et al., 2006). The DNA of sorted chromosomes was purified and amplified by MDA as described by Šimková et al. (2008).
454 Sequencing
DNA amplified from sorted chromosome 1H (WCA1H) and from sorted chromosomes 1H to 7H (WCAall) was used for 454 shotgun sequencing. Five micrograms of MDA DNA was used to prepare the 454 sequencing library using the GS FLX DNA library preparation kit, following the manufacturer's instructions (Roche Diagnostics). Single-stranded 454 sequencing libraries were quantified by a quantitative PCR assay (Meyer et al., 2008) and processed utilizing a GSFLX standard emPCR kit I and standard LR70 sequencing kit (Roche Diagnostics) according to manufacturer's instructions. For WCA1H, six complete GS FLX sequencer runs (70 × 75 picotiter plates) resulted in 3,046,327 reads with a median read length of 258 bp, yielding 799,343,261 bp of raw sequence data (675,561,265 high-quality bases). Two runs with DNA from pooled chromosomes 1H to 7H (WCAall) using half of a 70 × 75 picotiter plate resulted in overall 381,617 reads (median read length = 259 bp), yielding 99,401,554 bp raw sequence data (90,536,939 high-quality bases). Sequencing details were summarized in Table I. All sequence information generated in this study was submitted to the National Center for Biotechnology Information short read archive under accession number SRP001030.
Sequence Analysis
Analysis of Repetitive DNA and Repeat Masking of Sequences
Initially the content of repetitive DNA per sequence read was identified by analysis with RepeatMasker (http://www.repeatmasker.org) against the MIPS-REdat Poaceae v8.1 repeat library (contains known grass transposons from the Triticeae Repeat Database, http://wheat.pw.usda.gov/ITMI/Repeats, as well as de novo detected LTR retrotransposon sequences from several grass species, e.g. maize [Zea mays]: 12,434, sorghum [Sorghum bicolor]: 7,500, rice [Oryza sativa]: 1,928, Brachypodium distachyon: 466, wheat [Triticum aestivum]: 356, and barley [Hordeum vulgare]: 86 sequences). Subsequently, repetitive regions were masked by vmatch (http://www.vmatch.de) at the following parameters: 55% identity cutoff, 30 bp minimal length, seed length 14, exdrop 5, e value 0.001.
Sequence-Tagged Genes in the WCA1H Sequence Dataset
To estimate the number of barley genes that have been captured in the WCA1H sequence collection, BLAST (Altschul et al., 1990) comparisons were carried out with the repeat-filtered reads against the rice and sorghum proteins/coding sequences as well as against clustered wheat and barley EST collections (HarvEST, http://harvest.ucr.edu/; barley v1.73, assembly 35, wheat v1.16; Rice RAP-DB genome build 4, http://rapdb.dna.affrc.go.jp; sorghum genome annotation v1.4 [http://genome.jgi-psf.org/Sorbi1/Sorbi1.download.ftp.html]; Paterson et al., 2009). The number of tagged genes and the number of gene matching reads were counted after filtering according to the following criteria: (1) the best hit display with a similarity greater than an adjusted species-specific similarity characteristic (see below for definition) and (2) an alignment length ≥30 amino acids (BLASTN 50 bp). A species-adapted similarity cutoff value was calibrated before by performing similarity searches (BLASTX/TBLASTX/BLASTN) of barley EST clusters against rice and sorghum proteins and against wheat ESTs/tentative consensi (similarity cutoff: sorghum 75%, rice 80%, wheat 85%; see Supplemental Fig. S1, A and B).
Identification of Genetic Markers in the WCA1H and WCAall Datasets
The repeat-masked sequence collections from WCA1H and WCAall were compared (BLASTN) against 2,785 nonredundant (of total 2,943) EST-based markers (http://harvest.ucr.edu) under optimized parameters (-r 1 -q -1 -W 9 -G 1 -E 2: -r reward for a nucleotide match, default = 1; -q penalty for a nucleotide mismatch, default = -3; -G cost to open a gap, default = -1; -E cost to extend a gap, default = -1; -W word size, default). Only BLAST matches exceeding a similarity threshold of 98% and an alignment length ≥50 bp were further analyzed.
Comparative Genomics to Rice and Sorghum and Syntenic Integration
The WCA1H dataset was compared (BLASTX) to the reference genomes of rice and sorghum at a filter criterion of ≥30 amino acid similarity. Matched rice and sorghum genes were plotted along their position on the respective chromosomes and the average syntenic content (number of WCA1H matched genes per window size of 10 genes in rice and sorghum, respectively) was computed and visualized in heatmaps.
All rice and sorghum genes contained in syntenic regions in barley that could be delimited by a scaffold of 332 barley chromosome 1H-allocated EST-based markers and that exhibited a match to individual WCA1H 454 sequence reads were selected and integrated, producing a syntenic scaffold. First, putatively orthologous rice and sorghum genes were determined in this set of genes by reciprocal BLASTP searches considering only best matches. Subsequently, genes present either only in rice or sorghum but exhibited matches to WCA1H 454 reads were sorted in between.
All sequence information generated in this study was submitted to the NCBI GenBank short read archive under accession number SRP001030.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence comparisons of barley ESTs against wheat, rice, and sorghum genes.
Supplemental Table S1. Sequence similarities in coding regions between the genomes of rice and sorghum and EST resources from wheat and barley.
Supplemental Table S2. Reconstruction of barley chromosome 1H by using syntenic relationships.
Supplemental Table S3. Comparison of barley chromosome 1H enriched sequences (WCA1H) with chromosomes of rice and sorghum.
Supplemental Table S4. Virtual gene order list of barley chromosome 1H based on syntenic integration.
Supplementary Material
Acknowledgments
We are grateful to Dr. Z. Stehno (Crop Research Institute, Prague, Czech Republic) for providing seeds of barley cv Morex and we kindly acknowledge the excellent technical assistance of D. Werler, I. Heinze, and C. Luge as well as D. Riano-Pachon from www.gabipd.org for support in sequence data submission.
This work was supported by the program Genome Analysis of the Plant Biological System (www.gabi.de) and by grants from the German Ministry of Education and Research (grant no. BMBF FKZ0314000 to N.S., M.P., K.F.X.M., and U.S.). J.D., H.Š., and P.S. were supported by the Czech Republic Ministry of Education, Youth and Sports (grant no. LC06004). N.S., J.D., K.F.X.M., and T.W. participated within the framework of the European Cooperation in Science and Technology program FA0604.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nils Stein (stein@ipk-gatersleben.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB (1976) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci 274: 227–274 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Freeling M (1993) Grasses as a single genetic system: genome composition, collinearity and compatibility. Trends Genet 9: 259–261 [DOI] [PubMed] [Google Scholar]
- Bolot S, Abrouk M, Masood-Quraishi U, Stein N, Messing J, Feuillet C, Salse J (2009) The ‘inner circle’ of the cereal genomes. Curr Opin Plant Biol 12: 119–125 [DOI] [PubMed] [Google Scholar]
- Close TJ, Wanamaker S, Roose ML, Lyon M (2008) HarvEST. In D Edwards, ed, Plant Bioinformatics, Vol 406. Humana Press, New York, pp 161–177
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, et al (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA 99: 5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot (Lond) (Suppl A) 82: 17–26 [Google Scholar]
- Doležel J, Kubaláková M, Paux E, Bartoš J, Feuillet C (2007) Chromosome-based genomics in the cereals. Chromosome Res 15: 51–66 [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge AP (2001) Brachypodium distachyon: a new model system for functional genomics in grasses. Plant Physiol 127: 1539–1555 [PMC free article] [PubMed] [Google Scholar]
- Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154: 15–28 [Google Scholar]
- Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS (2003) Unbiased whole-genome amplification directly from clinical samples. Genome Res 13: 954–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS (1988) Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Linde-Laursen IB (1996) Recommendations for the designation of the barley chromosomes and their arms. Barley Genet Newsl 26: 1–3 [Google Scholar]
- Marthe F, Künzel G (1994) Localization of translocation breakpoints in somatic metaphase chromosomes of barley. Theor Appl Genet 89: 240–248 [DOI] [PubMed] [Google Scholar]
- Meyer M, Briggs AW, Maricic T, Hober B, Hoffner B, Krause J, Weihmann A, Paabo S, Hofreiter M (2008) From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res 36: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution: grasses, line up and form a circle. Curr Biol 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48: 463–474 [DOI] [PubMed] [Google Scholar]
- Pennisi E (2008) Plant sciences: corn genomics pops wide open. Science 319: 1333. [DOI] [PubMed] [Google Scholar]
- Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorák J, Linkiewicz AM, Ratnasiri A, et al (2004) A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook MS, Delach SM, Deyneko G, Worlock A, Wolfe JL (2004) Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am J Pathol 164: 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Sederoff RR (2003) The rice genome and comparative genomics of higher plants. Curr Opin Plant Biol 6: 97–100 [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, et al (2009) The international barley sequencing consortium—at the threshold of efficient access to the barley genome. Plant Physiol 149: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotechnol 26: 1135–1145 [DOI] [PubMed] [Google Scholar]
- Silander K, Saarela J (2008) Whole genome amplification with Phi29 DNA polymerase to enable genetic or genomic analysis of samples of low DNA yield. Methods Mol Biol 439: 1–18 [DOI] [PubMed] [Google Scholar]
- Simkova H, Svensson JT, Condamine P, Hribova E, Suchankova P, Bhat PR, Bartos J, Safar J, Close TJ, Dolezel J (2008) Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genomics 9: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RJ, Tsuchiya T (1982) An improved Giemsa n-banding technique for the identification of barley chromosomes. J Hered 73: 227–229 [Google Scholar]
- Smith DB, Flavell RB (1975) Characterisation of the wheat genome by renaturation kinetics. Chromosoma 50: 223–242 [Google Scholar]
- Stein N (2007) Triticeae genomics: advances in sequence analysis of large genome cereal crops. Chromosome Res 15: 21–31 [DOI] [PubMed] [Google Scholar]
- Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, Kota R, Varshney RK, Perovic D, Grosse I, et al (2007) A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor Appl Genet 114: 823–839 [DOI] [PubMed] [Google Scholar]
- Suchankova P, Kubalakova M, Kovarova P, Bartos J, Cihalikova J, Molnar-Lang M, Endo TR, Dolezel J (2006) Dissection of the nuclear genome of barley by chromosome flow sorting. Theor Appl Genet 113: 651–659 [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, Pruss D, Pindo M, Fitzgerald LM, Vezzulli S, Reid J, et al (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2: e1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Narechania A, Sabot F, Stein J, Vu GT, Graner A, Ware D, Stein N (2008) Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics 9: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N (2006) 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Taudien S, Houben A, Keller B, Graner A, Platzer M, Stein N (2009) A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J (in press) [DOI] [PubMed]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.