Abstract
The phytohormone auxin plays a critical role for plant growth by regulating the expression of a set of genes. One large auxin-responsive gene family of this type is the small auxin-up RNA (SAUR) genes, although their function is largely unknown. The expression of the rice (Oryza sativa) SAUR39 gene showed rapid induction by transient change in different environmental factors, including auxin, nitrogen, salinity, cytokinin, and anoxia. Transgenic rice plants overexpressing the SAUR39 gene resulted in lower shoot and root growth, altered shoot morphology, smaller vascular tissue, and lower yield compared with wild-type plants. The SAUR39 gene was expressed at higher levels in older leaves, unlike auxin biosynthesis, which occurs largely in the meristematic region. The transgenic plants had a lower auxin level and a reduced polar auxin transport as well as the down-regulation of some putative auxin biosynthesis and transporter genes. Biochemical analysis also revealed that transgenic plants had lower chlorophyll content, higher levels of anthocyanin, abscisic acid, sugar, and starch, and faster leaf senescence compared with wild-type plants at the vegetative stage. Most of these phenomena have been shown to be negatively correlated with auxin level and transport. Transcript profiling revealed that metabolic perturbations in overexpresser plants were largely due to transcriptional changes of genes involved in photosynthesis, senescence, chlorophyll production, anthocyanin accumulation, sugar synthesis, and transport. The lower growth and yield of overexpresser plants was largely recovered by exogenous auxin application. Taken together, the results suggest that SAUR39 acts as a negative regulator for auxin synthesis and transport.
An optimum level of auxin is required for several aspects of plant growth and developmental processes, including cell division, differentiation, and elongation, vascular development, root and shoot architecture, organ patterning, and tropism (for review, see Woodward and Bartel, 2005; Teale et al., 2006). Auxin exerts these effects by altering the expression of numerous genes (for review, see Abel and Theologis, 1996; Hagen and Guilfoyle, 2002). The early auxin-responsive genes, which are rapidly and transiently induced in response to auxin, have been grouped into three major families: auxin/indoleacetic acid (Aux/IAA), Gretchenhagen-3 (GH3), and small auxin-up RNA (SAUR; for review, see Hagen and Guilfoyle, 2002). The transcription of many of the Aux/IAA genes is transiently induced by auxin (Abel et al., 1995; for review, see Abel and Theologis, 1996). Aux/IAA proteins have been shown to function as negative regulators of auxin response factor proteins (Ulmasov et al., 1997). Some of the GH3 genes have also been shown to be induced by auxin, and these genes encode enzymes that conjugate free IAA with amino acids (Staswick et al., 2005).
The transcripts of the SAUR genes also accumulate within minutes after application of auxin (Jain et al., 2006c). However, the functions of the members of this gene family are largely unknown (for review, see Woodward and Bartel, 2005; Jain et al., 2006c). SAUR genes have been identified in different plants such as soybean (Glycine max; McClure et al., 1989), tobacco (Nicotiana tabacum; Roux et al., 1998), and maize (Zea mays; Knauss et al., 2003), with a total of 78 (http://www.arabidopsis.org/) and 58 SAUR genes identified in Arabidopsis (Arabidopsis thaliana; for review, see Hagen and Guilfoyle, 2002) and rice (Oryza sativa; Jain et al., 2006c), respectively. Some of the SAUR genes are mainly expressed in the elongation tissues of maize and soybean (McClure and Guilfoyle, 1989; Gee et al., 1991; Knauss et al., 2003), suggesting their role in auxin-mediated cell elongation. In rice, some SAUR genes were found to be differentially expressed in various tissues (Jain et al., 2006c). Recently, an AtSAUR32 gene has been shown to be involved in apical hook development in Arabidopsis (Park et al., 2007). The SAUR transcripts and proteins degrade rapidly after induction (Knauss et al., 2003), with the instability of the SAUR mRNA being conferred by the presence of conserved downstream destabilizing elements in their 3′ untranslated region (Gil and Green, 1996; Jain et al., 2006c). Some of the SAUR proteins have been shown to bind with Ca+-binding/calmodulin proteins in maize and Arabidopsis (Yang and Poovaiah, 2000; Reddy et al., 2002). However, whether the SAUR genes are simply a downstream target of the auxin response factor transcription factors or actually involved in auxin signaling is not yet known.
Here, we performed a detailed characterization of a SAUR gene using overexpresser (OX) plants and show that this SAUR39 gene acts as a negative regulator of auxin biosynthesis and transport in rice. Overexpression of the SAUR39 gene in rice resulted in reduced free IAA levels and lower auxin transport. The OX plants had reduced shoot and root growth, altered shoot morphology, and lower yield than wild-type plants, which can be partially compensated for by the addition of exogenous auxin.
RESULTS
Identification and Molecular Characteristics of a SAUR Gene
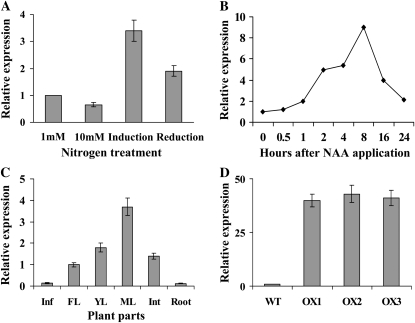
We conducted a whole genome transcript profile analysis in rice grown under different nitrogen (N) conditions. Among the N-responsive genes, Os09g0545300 was up-regulated by transient change of N, that is by N induction (switching plants from low to high N) and by N reduction (switching plants from high to low N) 2 h before harvesting, and real-time PCR analysis confirmed that the Os09g0545300 gene was greater than 3-fold up-regulated by N induction and about 2-fold up-regulated by N reduction (Fig. 1A). Gene Expression Omnibus profiles (http://www.ncbi.nlm.nih.gov/) and Genevestigator (https://www.genevestigator.ethz.ch/gv/index.jsp) database analysis reveals that this gene is also up-regulated in rice by salinity stress (Walia et al., 2005), application of cytokinin (Hirose et al., 2007), and anoxia (Lasanthi-Kudahettige et al., 2007), indicating that its expression is induced by transient changes in many different environmental stimuli.
Figure 1.
Identification, auxin response, and expression pattern of the SAUR39 gene. A, Expression of the SAUR39 gene in shoots of wild-type rice plants grown at 1 or 10 mm N for 4 weeks. Two hours before harvest, plants were switched from 1 to 10 mm N (induction) and from 10 to 1 mm N (reduction). B, Expression of SAUR39 after application of NAA in shoots. The wild-type rice plants were grown for 4 weeks, and shoots were harvested at the indicated time points after application of 4 μm NAA. C, Expression of SAUR39 in different tissues of wild-type plants. Inf, Inflorescence; FL, flag leaf; YL, young leaf; ML, mature leaf; Int, internode. D, Expression of SAUR39 in shoots of wild-type (WT) and OX lines, measured by real-time PCR.
The Os09g0545300 gene belongs to the SAUR gene family in rice. A total of 58 SAUR genes have been identified in rice, and this gene is designated as SAUR39 (Jain et al., 2006c). The SAUR39 gene has an auxin-responsive element in its 5′ untranslated region, a destabilizing element in its 3′ untranslated region, and no introns (Supplemental Fig. S1), which are characteristics of SAUR family members (Jain et al., 2006c). This gene encodes a protein of 171 amino acids (Supplemental Fig. S1).
Several of the SAUR family genes are known to be rapidly induced after exogenous auxin application in different plants (McClure et al., 1989; Abel and Theologis, 1996; for review, see Woodward and Bartel, 2005; Jain et al., 2006c; Teale et al., 2006), with the exception of the AtSAUR32 gene in Arabidopsis (Park et al., 2007). To confirm whether the SAUR39 gene is responsive to exogenous auxin application, 4-week-old rice wild-type plants were supplied with 4 μm naphthaleneacetic acid (NAA) and harvested at the indicated time points (Fig. 1B). The transcript level in shoots started to increase from 30 min after NAA application, kept accumulating up to 8 h, and declined thereafter. The expression of SAUR39 in rice wild-type plants was highest in the mature leaves followed by young leaves and the flag leaf, with very low transcript levels in inflorescence and roots (Fig. 1C). Its expression kept increasing in leaves until physiological maturity and was still quite high in senescing leaves (Supplemental Fig. S2), showing that mature leaves have higher transcript levels than younger leaves.
Overexpression of SAUR39 Inhibits Growth and Yield of Rice Plants
To understand the molecular function of the SAUR39 gene in rice, OX plants were generated. The full-length cDNA of SAUR39 was amplified and cloned into a binary vector, and transgenic rice plants were generated through Agrobacterium tumefaciens-mediated transformation using a constitutively expressed ubiquitin promoter (Christensen and Quail, 1996). Ten independent T1 transgenic lines were generated, and half of them had a single insertion site for the SAUR39 gene, as genotyping results showed a 3:1 segregation ratio in T2 plants (data not shown). Transgene expression level was measured in three of these five lines by real-time PCR, and transgenic lines had 40- to 43-fold higher expression of SAUR39 in leaves as compared with wild-type plants (Fig. 1D). These three lines were further tested in their T2 and T3 generations, and all of the lines showed similar differential growth, biochemical, and expression results compared with the wild type; the data of two transgenic lines are presented hereafter.
The transgenic plants overexpressing SAUR39 were smaller than wild-type plants (Fig. 2). The OX plants had significantly fewer leaves and tillers, with shorter plant height and lower yield compared with the wild type (Tables I and II). The primary root length in OX plants was smaller, and they had significantly fewer lateral roots (Table III). These plants displayed 33% and 35% reduction in shoot and root dry biomass, respectively, as compared with wild-type plants (Table I). In rice plants, senescence in the older leaves starts even when the plants are still at the vegetative stage and extends upward as the plant grows (Lee et al., 2001). The start of senescence of older leaves was much earlier in OX plants when the plants were 3 weeks old, whereas wild-type plants had no symptoms of senescence (Fig. 2A); 1 week later, the number of senescing leaves was significantly higher in OX plants than in wild-type plants (Table I). Interestingly, when the vegetative stage in plants was complete, the angle of leaves in OX plants started to increase (data not shown). In 10-week-old OX plants, both younger and older leaves turned more horizontal, whereas wild-type plants still had a smaller leaf angle (Fig. 2B). The flowering and maturity in both OX and wild-type plants occurred at similar times (data not shown). At maturity, shoot dry weight, number of spikes and spikelets, and seed yield per plant were significantly lower in OX plants than in wild-type plants (Table II).
Figure 2.
Growth of wild-type (WT) and SAUR39-OX rice plants. A, Three-week-old plants. Arrows indicate start of senescence in OX plants. B, Ten-week-old plants. Arrows indicate wider angle of leaves in OX plants.
Table I.
Growth parameters of 4-week-old wild-type and SAUR39-OX rice plants
The data are means ± se (n = 10 plants). Asterisks indicate values of wild-type plants that are significantly different from values of OX lines within that treatment (P < 0.05, Fisher's protected lsd test).
| Treatment and Plant | Leaf Number | Senescing Leaves | Total Tillers | Shoot Length | Shoot Dry Biomass | Root Dry Biomass |
|---|---|---|---|---|---|---|
| cm | g | g | ||||
| Nutrient solution without NAA | ||||||
| Wild type | 18.3 ± 1.4* | 1.9 ± 0.22* | 3.5 ± 0.22* | 23.1 ± 2.1* | 2.4 ± 0.21* | 0.48 ± 0.05* |
| OX1 | 14.2 ± 1.5 | 3.1 ± 0.32 | 2.8 ± 0.23 | 19.1 ± 1.5 | 1.6 ± 0.19 | 0.31 ± 0.04 |
| OX2 | 14.5 ± 1.7 | 3.3 ± 0.35 | 2.7 ± 0.27 | 19.3 ± 1.7 | 1.7 ± 0.18 | 0.33 ± 0.04 |
| Nutrient solution with 0.04 μm NAA | ||||||
| Wild type | 18.7 ± 2.0 | 1.8 ± 0.17* | 3.6 ± 0.29 | 24.0 ± 2.2 | 2.5 ± 0.24 | 0.50 ± 0.05 |
| OX1 | 17.1 ± 1.9 | 2.2 ± 0.19 | 3.3 ± 0.30 | 22.2 ± 1.9 | 2.1 ± 0.21 | 0.41 ± 0.05 |
| OX2 | 17.3 ± 1.8 | 2.1 ± 0.18 | 3.2 ± 0.33 | 22.2 ± 2.0 | 2.2 ± 0.19 | 0.42 ± 0.05 |
Table II.
Yield and yield attributes in wild-type and SAUR39-OX rice plants
The data are means ± se (n = 10 plants). Asterisks indicate values of wild-type plants that are significantly different from values of OX lines within that treatment (P < 0.05, Fisher's protected lsd test).
| Treatment and Plant | Total Tillers | Shoot Dry Weight | Spikes | Spikelets | Seed Yield |
|---|---|---|---|---|---|
| g | g | ||||
| Nutrient solution without NAA | |||||
| Wild type | 6.6 ± 0.3* | 8.9 ± 0.6* | 4.1 ± 0.3* | 396 ± 26* | 7.7 ± 0.5* |
| OX1 | 5.6 ± 0.3 | 7.5 ± 0.6 | 3.4 ± 0.3 | 295 ± 21 | 5.5 ± 0.4 |
| OX2 | 5.5 ± 0.4 | 7.7 ± 0.5 | 3.5 ± 0.3 | 304 ± 27 | 5.6 ± 0.4 |
| Nutrient solution with 0.04 μm NAA | |||||
| Wild type | 6.8 ± 0.3 | 9.1 ± 0.6 | 4.2 ± 0.3 | 407 ± 25 | 7.8 ± 0.6 |
| OX1 | 6.1 ± 0.4 | 8.2 ± 0.6 | 3.9 ± 0.4 | 361 ± 24 | 6.9 ± 0.6 |
| OX2 | 6.2 ± 0.4 | 8.3 ± 0.6 | 3.9 ± 0.4 | 368 ± 26 | 7.0 ± 0.5 |
Table III.
Effects of auxin (NAA) and auxin transport inhibitor (NPA) on root growth in rice plants
Rice seedlings were grown hydroponically, NAA or NPA was mixed in nutrient solution and applied to 3-d-old seedlings, and observations were taken 3 d later. The data are means ± sd (n = 8–10 plants). Asterisks indicate values of wild-type plants that are significantly different from values of OX lines within that treatment (P < 0.05, Fisher's protected lsd test).
| Treatment and Plant | Primary Root Length | Lateral Roots per Plant Primary Root |
|---|---|---|
| cm | ||
| Control | ||
| Wild type | 15.6* ± 1.2 | 102* ± 12 |
| OX1 | 12.9 ± 1.0 | 78 ± 8 |
| OX2 | 13.1 ± 1.0 | 80 ± 9 |
| 0.2 μm NAA | ||
| Wild type | 12.0 ± 1.0 | 98 ± 10 |
| OX1 | 11.1 ± 1.0 | 94 ± 9 |
| OX2 | 11.5 ± 1.1 | 96 ± 11 |
| 5 μm NPA | ||
| Wild type | 14.8* ± 1.1 | 78* ± 8 |
| OX1 | 12.1 ± 0.9 | 60 ± 7 |
| OX2 | 11.7 ± 1.0 | 58 ± 6 |
SAUR39-OX Plants Had Lower Free IAA and Less Polar Auxin Transport
SAUR39-OX plants were smaller with less root volume (Fig. 2; Tables I and III). Since auxin is known to play a role in plant growth and lateral root formation (Woodward and Bartel, 2005; Teale et al., 2006; Chhun et al., 2007), free IAA level was measured in wild-type and OX plants. Figure 3A shows that free IAA in shoots of 4-week-old OX plants was 20% lower than in wild-type plants. To determine whether auxin transport was also affected in OX plants, 4-cm stem segments from the base of 4-week-old plants were basipetally incubated for 3 h in [3H]IAA, and stem segments were further divided into four parts. All of the stem segments in the OX plants had significantly lower [3H]IAA (Table IV), suggesting that these plants had lower auxin transport than wild-type plants. The synthetic auxin transport inhibitor N-1-naphthylphthalamic acid (NPA), at a concentration of 100 μm, similarly reduced [3H]IAA movement in transgenic and wild-type plants (Table IV).
Figure 3.
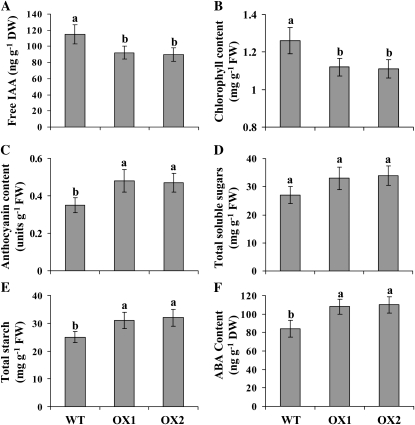
Biochemical analysis in wild-type (WT) and SAUR39-OX rice plants. Free IAA (A), chlorophyll (B), anthocyanin (C), total soluble sugars (D), total starch (E), and ABA (F) contents in shoots of 4-week-old wild-type and OX rice plants. Data are means ± sd (n = 3–5). Bars with different letters indicate significant differences at P < 0.05 (Fisher's protected lsd test). DW, Dry weight; FW, fresh weight.
Table IV.
Polar auxin transport in stems of rice plants
Four-centimeter stem segments of 4-week-old wild-type and OX plants were incubated in [3H]IAA solution for 3 h, and segments were further divided into four sections of 1 cm each. Section 1 was submerged in [3H]IAA solution, and section 4 indicates the farthest section. The data are means ± sd (n = 6 plants). All values of wild-type plants are significantly different from those of OX lines (P < 0.05, Fisher's protected lsd test).
| Treatment and Plant | [3H]IAA in Stem Section
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| cpm | ||||
| Basipetal (without NPA) | ||||
| Wild type | 291,451 ± 9,472 | 33,645 ± 5,142 | 5,268 ± 426 | 1,271 ± 45 |
| OX1 | 286,412 ± 9,112 | 20,348 ± 3,751 | 3,457 ± 415 | 745 ± 75 |
| OX2 | 289,231 ± 9,282 | 20,521 ± 3,695 | 3,507 ± 395 | 761 ± 57 |
| Basipetal (with 100 μm NPA) | ||||
| Wild type | 290,122 ± 9,108 | 2,933 ± 458 | 590 ± 68 | 280 ± 30 |
| OX1 | 281,332 ± 10,102 | 2,065 ± 216 | 414 ± 43 | 198 ± 21 |
| OX2 | 287,385 ± 9,514 | 2,202 ± 254 | 408 ± 52 | 207 ± 24 |
SAUR39-OX Plants Had Altered Chlorophyll, Anthocyanin, Sugar, and Abscisic Acid Contents
Different biochemical assays were performed on 4-week-old plants in order to identify the traits associated with the observed smaller plant growth and yield. The leaves of OX plants were paler than those of wild-type plants (Fig. 2A), and chlorophyll analysis confirmed that leaves of OX plants had significantly lower chlorophyll content than leaves of wild-type plants (Fig. 3B). The anthocyanin content was 37% higher in OX leaves compared with the wild type (Fig. 3C). Sugar and starch content increases when plants grow older and leaves start to senesce (Pourtau et al., 2006; Wingler et al., 2006). The OX plants had slightly higher total soluble sugars (Fig. 3D) and significantly higher total starch content than wild-type plants (Fig. 3E). Abscisic acid (ABA) is an important plant growth hormone; its level increases in plants under abiotic stress (Seo and Koshiba, 2002), and the ABA level was significantly higher in OX plants (Fig. 3F).
SAUR39-OX Plants Had Smaller Vascular Tissue
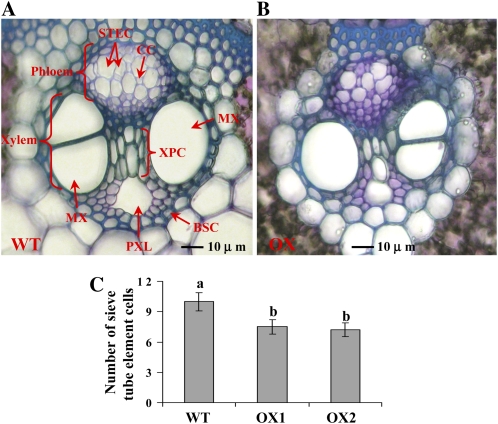
Since the polar auxin transport was lower (Table IV), along with smaller roots and inhibited shoot growth in OX plants (Tables I and III; Fig. 2), stem cross-sections were stained with toluidine blue to dissect the vascular tissue system of OX plants. Figure 4, A and B, shows that vascular tissue in OX shoot was smaller than in the wild type, with both phloem and xylem tissues being smaller. Auxin is actively transported from shoot toward root through the phloem (Teale et al., 2006). In the phloem, sieve tube element cells are important for translocation of photosynthates and auxin. The number of sieve tube element cells was significantly lower in OX shoots (Fig. 4C), suggesting that long-distance transport of auxin might be less in OX plants.
Figure 4.
Vascular tissue in rice stems. A and B, Stem cross-sections of wild-type (WT; A) and OX (B) plants stained with toluidine blue. C, Number of sieve tube element cells. Data are means ± se; bars with different letters indicate significant differences at P < 0.05 (Fisher's protected lsd test). The photograph and data are representative of sections taken from at least three different plants and eight to 10 sections in each plant from 4-week-old rice plants. BSC, Bundle sheath cells; CC, companion cells; MX, metaxylem; PXL, protoxylem lacuna; STEC, sieve tube element cells; XPC, xylem parenchyma cells.
SAUR39 Protein Is Localized in the Cytoplasm
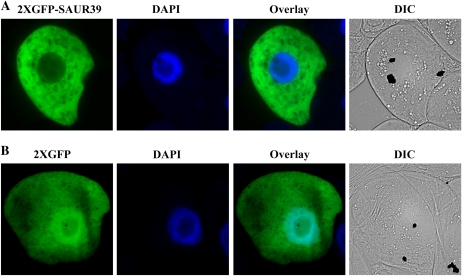
To gain further insight into the function of the SAUR39 protein at the subcellular level, initially a 35S-yellow fluorescent protein (YFP)-SAUR39 construct was developed using the Gateway-compatible vector pEarleyGate 104 (Earley et al., 2006). The 35S-YFP-SAUR39 and its control (35S-YFP) plasmids were biolistically bombarded separately into tobacco BY2 cells. Both YFP-SAUR39 (Supplemental Fig. S3A) and YFP (Supplemental Fig. S3B) proteins were observed to be in the nucleus and cytoplasm, based on fluorescent images acquired through epifluorescence microscopy. The YFP-SAUR39 protein (approximately 46.1 kD) and the control YFP protein (approximately 26.9 kD) had molecular mass values below the limit to avoid passive diffusion of a protein between nucleus and cytoplasm, given that the exclusion limit for protein diffusion is approximately 50 kD (Grebenok et al., 1997). To avoid this artifact, the 35S-GFP-GFP-SAUR39 (2XGFP-SAUR39) construct with higher molecular mass protein was used for subcellular localization. The 35S-2XGFP-SAUR39 and its control 35S-2XGFP constructs were prepared using the PRTL2 GFP-MCS vector (Shockey et al., 2006), their plasmids were biolistically bombarded into the tobacco BY2 cells, and fluorescent images were acquired via epifluorescence microscopy. The 2XGFP-SAUR39 protein has a molecular mass of approximately 73 kD, which is well above the exclusion limit, and was localized only in the cytoplasm (Fig. 5A). In contrast, the control 2XGFP protein (approximately 53.8 kD, which is quite close to the exclusion limit) was localized in both the nucleus and cytoplasm (Fig. 5B). It should be noted that GFP is known to give a green fluorescence signal in both the nucleus and cytoplasm (Grebenok et al., 1997). These results confirm that the SAUR39 protein was exclusively localized in the cytoplasm (Fig. 5A). Furthermore, analyses by the online prediction programs TargetP 1.1 (Emanuelsson et al., 2000) and WoLF PSORT (Horton et al., 2007) also predict the SAUR39 protein to be localized in the cytoplasm, with SAUR39 not having any nucleus- or organelle-specific localization signals.
Figure 5.
Subcellular localization of transiently expressed 2XGFP-SAUR39 (A) and 2XGFP (B) proteins. The plasmids were transformed biolistically into tobacco BY2 cells, and images were observed by epifluorescence microscopy. From left to right are the subcellular localization of 2XGFP-SAUR39 or 2XGFP fusion protein, nuclei stained with 4′,6-diamidino-2-phenylindole (DAPI), overlay of the two images, and differential interference contrast (DIC) images.
Transcript Profiling Reveals That Differential Expression of Genes Correlates with Altered Auxin Synthesis and Transport as Well as Chlorophyll, Anthocyanin, and Sugar Contents in SAUR39-OX Plants
To investigate which sets of genes were affected in OX plants, RNA was extracted in shoots of 4-week-old OX and wild-type plants and hybridized to a rice Affymetrix GeneChip whole genome array. To identify differentially expressed genes in OX plants compared with wild-type plants, a 2.0-fold change threshold and P < 0.05 were set as criteria to analyze the profiling data. A total of 1,505 genes were identified as statistically significant, with 1,094 genes up-regulated by 2-fold or greater (Supplemental Table S1) and 411 genes down-regulated by 2-fold or less (Supplemental Table S2).
The expression of SAUR39 was approximately 40-fold higher in OX plants in the microarray analysis (Supplemental Table S3), which is similar to what was observed in the real-time PCR analysis (Fig. 1D). In addition, expression of one more SAUR gene and five genes from the GH3 and Aux/IAA auxin-responsive gene family was also differentially expressed (Supplemental Table S3). The free IAA level was lower in OX plants (Fig. 3A). However, none of the auxin biosynthesis genes were observed to be differentially expressed in the transcript profiling data. These genes could be screened out based on the statistical criteria used or not yet annotated. Therefore, a search was performed of the rice genome for the homologous genes to those from Arabidopsis involved in auxin biosynthesis, and their expression levels were determined by real-time PCR. Relative expression of some of the genes involved in auxin biosynthesis (TRP1, TRP2, TRP5, and YUCCA6) was slightly lower in shoots of the OX plants by 0.6-, 0.71-, 0.63-, and 0.72-fold, respectively, as compared with wild-type plants (Supplemental Table S3). The transport of auxin was lower in OX plants (Table IV), and in the transcript profiling data, an auxin efflux carrier component 6 (PIN6) gene was repressed over 3-fold (Supplemental Table S3). Also, the transcript level of a gene encoding a cAMP-dependent protein kinase/protein kinase G/protein kinase C (AGC)-like kinase was down-regulated, with the AGC kinases having been proposed to regulate polar auxin transport (for review, see Galvan-Ampudia and Offringa, 2007). In addition, the relative expression of an auxin influx carrier gene, AUX1-like, was lower in the OX plants (Supplemental Table S3).
The expression of 23 genes that have putative functions in photosynthesis and chlorophyll production was down-regulated in OX plants, with the exception of a putative red chlorophyll catabolite reductase, which has been shown to be involved in chlorophyll breakdown (Rodoni et al., 1997) and was up-regulated. The transcription of five genes with putative roles in senescence was higher in OX plants (Supplemental Table S3), which correlates with an early progression of senescence in the OX plants (Fig. 2). The transcript levels of 12 genes involved in anthocyanin synthesis, such as PAL, CHS, F3H, and AGT, were increased 2.3- to 10.5-fold in OX plants (Supplemental Table S3), which supports the marked increase in anthocyanin content in the OX plants (Fig. 3C). The up-regulation of several genes involved in sugar synthesis and sugar transport in OX plants (Supplemental Table S3) correlates with higher total sugar and starch in OX plants (Fig. 3, D and E).
The SAUR proteins have been shown to bind with Ca+-binding/calmodulin proteins (Yang and Poovaiah, 2000; Reddy et al., 2002). Consistent with these reports, transcript levels of 12 Ca+-binding/calmodulin genes and five calcium-transporting ATPase genes were higher in the OX plants. The OX plants showed stunted growth and lower yield, which is characteristic of plants grown under biotic and/or abiotic stress. Thus, it is not surprising that 27 genes related to disease, pathogenesis, and oxidative, drought, salinity, and heat stresses were up-regulated in OX plants. Some genes involved in hormone synthesis and the response to ethylene, gibberellin, ABA, cytokinin, and salicylic acid were also differentially expressed in OX plants (Supplemental Table S3). The expression levels of several genes governing plant architecture were analyzed by real-time PCR. The D3 (Ishikawa et al., 2005), HTD1 (Zou et al., 2006), D10 (Arite et al., 2007), and FC1 (Takeda et al., 2003) genes that have been known to negatively regulate lateral branching and/or knockout mutant lines of these genes had higher lateral branching in rice. Relative expression of D3 (MAX2/ORE9), HTD1 (MAX3), D10 (MAX4/RMS1/DAD1), and FC1 (OsTB1) was up-regulated in shoots of OX plants by 1.8-, 1.9-, 1.7-, and 3.1-fold, respectively, as compared with the wild type (Supplemental Table S3). Overexpression of FON1 and EUI genes has been shown to lead to smaller floral meristems (Suzaki et al., 2004) and dwarf plants (Zhu et al., 2006) in rice, and the transcript levels of these genes were 1.8- and 2.7-fold higher in OX plants, respectively (Supplemental Table S3).
The validity of the expression differences detected in the microarray was examined by real-time PCR by analyzing the relative expression of 11 genes that were either up- or down-regulated (Supplemental Table S4). The real-time PCR and microarray results were consistent with each other, although the exact magnitude of difference was slightly different for some genes.
Exogenous Application of Auxin Rescued the SAUR39-OX Phenotype
Auxin synthesis and transport were less in OX plants (Fig. 3A; Table IV), resulting in lower root growth and thereby inhibiting shoot growth (Tables I and III; Fig. 2). To test whether exogenous auxin application could rescue the OX phenotype, different concentrations and forms of auxin (NAA, IAA, and 2,4-dichlorophenoxyacetic acid) were applied along with the nutrient solution to seedlings grown hydroponically. All of the different types of auxin gave similar results in terms of rescue of the OX phenotype (data not shown), with the results of only the NAA treatment shown here. The NAA application reduced the root length (Table III), which is characteristic of exogenous auxin effects on roots (for review, see Woodward and Bartel, 2005). Importantly, the number of lateral roots per plant primary root, which was significantly lower in OX plants compared with wild-type plants under control treatment, was statistically similar in both OX and wild-type plants when 0.2 μm NAA was applied (Table III). In contrast, application of the auxin transport inhibitor NPA at a concentration of 5 μm led to a significantly reduced number of lateral roots per plant primary root in both OX and wild-type plants compared with the control (Table III). In addition, the number of lateral roots per plant primary root was lower in OX plants than in wild-type plants with NPA.
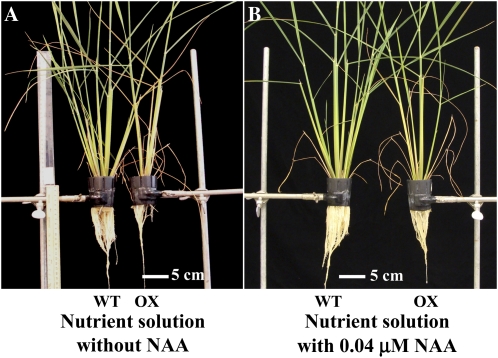
To confirm whether the beneficial effect of exogenous NAA application on seedlings could be reflected in plants until maturity, they were grown with low concentration of NAA (0.04 μm). Figure 6 and data in Tables I and II show that plant growth and yield in terms of the number of leaves and tillers, shoot length, shoot and root biomass, number of spikes and spikelets, and seed yield increased with 0.04 μm NAA in OX plants. Under these conditions, the OX plants were similar in growth characteristics to wild-type plants. Their growth and yield parameters were slightly lower, yet this was not statistically significant.
Figure 6.
Rice wild-type (WT) and SAUR39-OX plants. A, Plants grown in regular nutrient solution without NAA. B, Plants grown in nutrient solution with 0.04 μm NAA. Photographs were taken when plants were 7 weeks old.
DISCUSSION
The SAUR gene family comprises genes that are responsive to auxin (for review, see Abel and Theologis, 1996; Woodward and Bartel, 2005; Jain et al., 2006c). Apart from SAUR, there are two more auxin-responsive gene families called Aux/IAA and GH3. In rice, 58 SAUR, 31 Aux/IAA, and 12 GH3 genes have been identified (Jain et al., 2006a, 2006b, 2006c), while in Arabidopsis, there are 78 SAUR (http://www.arabidopsis.org/), 28 Aux/IAA, and 20 GH3 genes (Hagen and Guilfoyle, 2002). The molecular function of some of the Aux/IAA and GH3 genes have been reported in Arabidopsis and other plants (Abel et al., 1995; Ulmasov et al., 1997; Gray et al., 2001; Zenser et al., 2001; Staswick et al., 2005). However, the physiological function of any SAUR gene remains unknown. Previously, attempts have been made to obtain altered growth phenotypes in different SAUR gene loss-of-function mutant plants (Jain et al., 2006c; Park et al., 2007). However, functional redundancy and compensatory functions of conserved members of the SAUR gene family might be expected to prevent the presentation of phenotypic differences in loss-of-function mutants (Gil and Green, 1996; Park et al., 2007).
The SAUR39 gene was identified in rice microarray studies as responding to transient changes in N conditions (Fig. 1A). This gene also responds to salinity stress (Walia et al., 2005), application of cytokinin (Hirose et al., 2007), anoxia (Lasanthi-Kudahettige et al., 2007), and exogenous auxin (Fig. 1B). The SAUR39 gene is the only member of this gene family that responds to transient change in multiple external inputs. Therefore, a detailed physiological, biochemical, and molecular functional analysis was performed on SAUR39-OX rice plants. The OX plants were smaller than wild-type plants, showing the negative effect of overexpression of the SAUR39 gene on normal growth. The shoots of the OX plants had less free IAA and decreased auxin transport compared with the wild type, which can explain why the OX plants had fewer lateral roots and smaller root biomass, thereby reducing shoot growth.
The reduced growth rate corresponded with a number of other phenotypic and biochemical changes. The OX plants had lower chlorophyll content (Fig. 3B), which correlates with repression of several genes involved in photosynthesis and chlorophyll synthesis (Supplemental Table S3). The progression of senescence in older leaves of these plants was faster than in wild-type plants (Fig. 2; Table I). The early loss of photosynthetic capacity in senescing leaves and lower chlorophyll content contributed to lower grain yield in OX plants. The early progression of senescence in OX plants was accompanied by an increase in sugar and starch content (Fig. 3, D and E). It has been shown that sugar and starch levels increase during senescence (Pourtau et al., 2006; Wingler et al., 2006) and also when plants are grown under different abiotic stress conditions (Abbasi et al., 2007). The higher sugar and starch in OX plants was accompanied by up-regulation of some genes related to sugar synthesis and transport. The up-regulation of sugar-responsive genes and increased accumulation of sugar and starch in OX plants might be ascribed in part to the lower IAA levels in these plants. Ohto et al. (2006) reported an inverse relationship between auxin and sugar levels in Arabidopsis, with exogenous IAA application leading to a repression of sugar-responsive genes.
Increased sugar levels in plants induce anthocyanin accumulation, which is also considered to be a marker of abiotic stress in plants (Mita et al., 1997; Chalker-Scott, 1999; Ohto et al., 2006). Anthocyanin accumulation was higher in OX plants, along with an increase in transcript level for several genes involved in anthocyanin synthesis. ABA accumulates in plants with increasing abiotic stress (Seo and Koshiba, 2002), and the ABA content was higher in the OX plants (Fig. 3F). The higher anthocyanin and ABA levels and smaller size of the OX plants indicate that these plants were growing like wild-type plants do under stress. Indeed, several genes related to pathogenesis and oxidative, drought, salinity, and heat stress were up-regulated in the OX plants.
Flavonoids are known to repress auxin transport in Arabidopsis (Brown et al., 2001; Lazar and Goodman, 2006; Peer and Murphy, 2007). Similar results were obtained in our study, with shoots of OX plants having more anthocyanin and less polar auxin transport than wild-type plants. Since auxin is required for root growth and lateral root development, lower transport of auxin toward roots in OX plants could possibly be the reason for the lower number of lateral roots in these plants. The smaller vascular tissue system of OX plants (Fig. 4) is consistent with the idea that these plants have lower translocation of photosynthates and long-distance transport of auxin. Vascular development in plants requires auxin signaling, and Sachs (1981) has hypothesized that auxin flux is required for differentiation of vascular strands.
Auxin biosynthesis occurs mainly in the shoot apical meristem, with its highest level in the youngest plant parts (for review, see Woodward and Bartel, 2005). In contrast, the transcript level of SAUR39 was low in young leaves and higher in older leaves (Fig. 1C). The analysis of free IAA (Fig. 3A) and auxin transport (Table IV) suggests that constitutive overexpression of SAUR39 resulted in less production of auxin and reduced auxin transport. This was confirmed by the recovery of a more normal phenotype in OX plants supplied with exogenous auxin (Fig. 6). Lateral root number, shoot and root biomass, and grain yield were significantly increased in OX plants with an exogenous supply of NAA (Tables I–III). The effect of lower free IAA levels in OX plants was most pronounced during the reproductive stage, in that the shoot architecture was changed along with a wider angle for the OX leaves compared with wild-type plants (Fig. 2B). This is not surprising, since auxin has a role in organ patterning (for review, see Woodward and Bartel, 2005; Teale et al., 2006).
In conclusion, our results suggest that SAUR39 might be involved in auxin signaling, and its constitutive up-regulation negatively regulated auxin biosynthesis and transport. Lower auxin levels would be predicted to increase the expression of sugar-responsive genes, thereby repressing photosynthetic genes. The plants were stressed in this situation and accumulated more anthocyanin, which further reduced auxin transport, with all of these factors leading to reduced root and shoot growth. The expression of SAUR39 in wild-type plants is substantially increased under a wide array of environmental stress conditions. It is not unreasonable to assume that this would eventually lead to a similar down-regulation of auxin synthesis and transport and negative growth effects under stress, as observed here in OX plants under normal growth conditions. Hence, in wild-type rice plants, destabilization and down-regulation of the SAUR39 transcript, after a transient induction by a change in external input, is required for optimum auxin synthesis, thereby allowing for normal growth. Dissecting the molecular mechanism of how the SAUR39 gene regulates auxin synthesis and transport would be of considerable interest in delineating how auxin modulates plant growth under ideal and environmental stress conditions.
MATERIALS AND METHODS
Plant Growth Conditions
Rice (Oryza sativa japonica ‘Donjin’) plants were grown in a 1:4 mixture of peat moss and vermiculite (SunGro Horticulture Canada). Seeds were soaked overnight in water to get even and faster germination. The nutrient solution was added once per week until harvest and contained 5 mm NH4NO3, 4 mm MgSO4, 5 mm KCl, 5 mm CaCl2, 1.5 mm KH2PO4, 0.1 mm Fe-EDTA, 0.5 mm MES (pH 6.0), 9 μm MnSO4, 0.7 μm ZnSO4, 0.3 μm CuSO4, 46 μm NaB4O7, and 0.2 μm Na2MoO4. Plants were grown in a growth room with 12 h of light (approximately 500 μmol m−2 s−1) at 29°C, 12 h of dark at 23°C, and 65% relative humidity. To analyze the effect of exogenous auxin on root growth, seedlings were grown hydroponically in 2-L buckets; auxin or auxin transport inhibitor was applied to 3-d-old seedlings, and observations for root elongation and number of lateral roots were taken 3 d later. Different forms of auxin (IAA, NAA, and 2,4-dichlorophenoxyacetic acid) were tested, and similar results were obtained with all three forms. However, NAA treatment gave more consistent and reproducible results. NPA was applied as an auxin transport inhibitor. To analyze the effect of exogenous auxin application on plant growth and yield, plants were grown hydroponically with 18 plants in a 35-L plastic container. NAA (0.04 μm) was added with the nutrient solution, and the solution was replaced every week. Growth conditions and the nutrient solutions were as described above.
Microarray Analysis
Total RNA was isolated using Qiagen RNeasy columns. Microarray hybridization was performed according to Zhu et al. (2003). Five micrograms of total RNA from each sample was used to synthesize double-stranded cDNAs. Labeled complementary RNA, synthesized from the cDNA, was hybridized to the rice Affymetrix GeneChip whole genome array (catalog no. 900601; Affymetrix). This array contains 50,188 probe sets representing 50,119 known and predicted rice genes. Among them, 39,201 showed high homology to the nontransposable element-related protein-coding sequences. On average, each gene contains approximately 11 perfect match probes, selected from the 3′ end of the coding region. The hybridization signal of the arrays was acquired by the GeneChip scanner 3000 and quantified by MAS 5.0 (Affymetrix). The probe set measurement was summarized as a weighted average value of all probes in a set, subtracting the bottom 5% of average intensity of the entire array using a custom algorithm. The overall intensity of all probe sets of each array was further scaled to a target intensity of 100 to enable direct comparison. The data analysis was conducted using GeneSpring 7.3 (Agilent). The data were normalized with a default setting of the program followed by gene filtering, which required that each gene must have either a “P” or “M” flag in the three replicate samples. The genes with 2-fold change were identified, and ANOVA was used to screen for significantly differentially expressed genes (Welch t test P value cutoff at 0.05). For microarray analysis, two independent biological experiments were conducted, and within each experiment, treatments were replicated three times.
Transgenic Rice Plants
The construct to overexpress SAUR39 was made using a ubiquitin promoter (Christensen and Quail, 1996). Agrobacterium tumefaciens-mediated transformation was performed, and the T1 transgenic seeds were harvested. Phosphomannose isomerase tests were used for genotyping to detect the selectable marker phosphomannose isomerase (Negrotto et al., 2000). Almost half of the independent T1 lines had a single insertion of the SAUR39 gene, as genotyping results showed a 3:1 segregation ratio in T2 plants.
Expression Analysis by Quantitative Real-Time PCR
Total RNA was isolated from plant tissues using TRIZOL reagent (Invitrogen). To eliminate any residual genomic DNA, total RNA was treated with RQ1 RNase-free DNase (Promega). The first-strand cDNA was synthesized from total RNA using the Reverse Transcription System kit (Promega). Primer Express 2.0 software (Applied Biosystems) was used to design the primers. Primer sequences for each gene are given in Supplemental Table S5. Real-time PCR was performed according to Kant et al. (2006). Relative quantification values for each target gene were calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001) using ACTIN2 as an internal reference gene for comparing data from different PCR runs or cDNA samples. To ensure the validity of the 2−ΔΔCT method, 2-fold serial dilutions of cDNA from control plants were used to create standard curves, and the amplification efficiencies of the target and reference genes were approximately equal (Livak and Schmittgen, 2001).
Biochemical Assays
Frozen shoot tissue from 4-week-old plants was used for the following biochemical assays. Endogenous free IAA and ABA were analyzed according to the method of Chiwocha et al. (2005) by HPLC-electrospray-tandem mass spectrometry using deuterated internal standards. Chlorophyll was assayed according to Arnon (1949). Relative anthocyanin content was analyzed based on Neff and Chory (1998). Soluble sugars were extracted with 80% ethanol, supernatants were pooled, and total soluble sugars were assayed according to Dubois et al. (1956). Starch was extracted according to Delatte et al. (2005) and quantified using a commercial total starch assay kit (Megazyme; http://www.megazyme.com).
Polar Auxin Transport Assay
Polar auxin transport was measured by modification of the procedure described by Lazar and Goodman (2006). Four-centimeter-long stem segments taken from similar positions in OX and wild-type plants were placed in glass test tubes with the apical end submerged in 50 μL of buffer (5 mm MES, 1% [w/v] Suc, 800 nm IAA, and 600 nm [3H]IAA, pH 5.5) in the presence or absence of 100 μm NPA. Segments were incubated for 3 h at room temperature in the dark and dissected into 1-cm-long sections. The amounts of radioactivity in the sections were measured in a liquid scintillation counter after 2 d of incubation at room temperature.
Vascular Tissue Staining
Sections were taken from similar positions near the middle of shoots in 4-week-old OX and wild-type plants, stained with 0.05% toluidine blue for 2 to 3 min, rinsed with water, and mounted on slides. The sections were viewed immediately on a Leica DM LS2 microscope.
Subcellular Localization of SAUR39
To determine the subcellular localization of SAUR39 protein, the coding sequence of its gene was amplified from rice cDNA by PCR and cloned into the Gateway-compatible vector pEarleyGate 104 (Earley et al., 2006) having a YFP reporter gene. The YFP-SAUR39 protein had a molecular mass of approximately 46.1 kD, which is below the limit to avoid passive diffusion of a protein between the nucleus and cytoplasm (Grebenok et al., 1997). Therefore, 35S-GFP-GFP-SAUR39 (2XGFP-SAUR39) and its control 35S-2XGFP constructs were also prepared. For this, the GFP coding sequence was amplified from PRTL2 GFP-MCS (Shockey et al., 2006) by PCR. Both the GFP and SAUR39 PCR products were then ligated to PRTL2 GFP-MCS. All of these constructs (YFP-SAUR39, YFP, 2XGFP-SAUR39, and 2XGFP) were biolistically bombarded separately into tobacco (Nicotiana tabacum) BY2 cells, and fluorescent images were acquired 6 h post bombardment through epifluorescence microscopy. The nuclei of BY2 cells were stained with 4′,6-diamidino-2-phenylindole.
Statistics
The results shown are representative of three independent experiments, and within each experiment treatments were replicated three times, unless otherwise stated. Data were statistically analyzed by Fisher's protected lsd test using SAS statistical software (SAS Institute).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK241264.1 and Os09g0545300 (SAUR39).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence analysis of the SAUR39 gene.
Supplemental Figure S2. Expression pattern of the SAUR39 gene at different growth stages and plant parts in wild-type rice plants.
Supplemental Figure S3. Fluorescent images of transiently expressed YFP-SAUR39 protein.
Supplemental Table S1. Up-regulated genes in shoots of OX plants compared with wild-type plants in microarray analysis.
Supplemental Table S2. Down-regulated genes in shoots of OX plants compared with wild-type plants in microarray analysis.
Supplemental Table S3. Selected genes differentially regulated in shoots of OX plants compared with wild-type plants in microarray analysis.
Supplemental Table S4. Comparison of relative transcript abundance of some genes measured by real-time PCR versus microarray analysis.
Supplemental Table S5. Primers used for real-time PCR analysis of gene expression.
Supplementary Material
Acknowledgments
We thank Dr. Satinder Gidda (University of Guelph) for help in subcellular localization; Lewis Melville and Dr. Larry Peterson (University of Guelph) for their guidance in vascular tissue staining; and Eddie Bondo (Syngenta Biotechnology) for assistance with microarray experiments.
This work was supported by the Natural Sciences and Engineering Research Council of Canada and the Ontario Research and Development Challenge Fund.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Steven J. Rothstein (rothstei@uoguelph.ca).
The online version of this article contains Web-only data.
References
- Abbasi AR, Hajirezaei M, Hofius D, Sonnewald U, Voll LM (2007) Specific roles of alpha- and gamma-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiol 143: 1720–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 [DOI] [PubMed] [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol 126: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott L (1999) Environmental significance of anthocyanins in plant stress responses. Photochem Photobiol 70: 1–9 [Google Scholar]
- Chhun T, Uno Y, Taketa S, Azuma T, Ichii M, Okamoto T, Tsurumi S (2007) Saturated humidity accelerates lateral root development in rice (Oryza sativa L.) seedlings by increasing phloem-based auxin transport. J Exp Bot 58: 1695–1704 [DOI] [PubMed] [Google Scholar]
- Chiwocha SD, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross AR, Kermode AR (2005) The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J 42: 35–48 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5: 213–218 [DOI] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles K, Hamilton J, Rebers P, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia CS, Offringa R (2007) Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci 12: 541–547 [DOI] [PubMed] [Google Scholar]
- Gee MA, Hagen G, Guilfoyle TJ (1991) Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs. Plant Cell 3: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Green PJ (1996) Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J 15: 1678–1686 [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Grebenok RJ, Pierson E, Lambert GM, Gong FC, Afonso CL, Haldeman-Cahill R, Carrington JC, Galbraith DW (1997) Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J 11: 573–586 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hirose N, Makita N, Kojima M, Kamada-Nobusada T, Sakakibara H (2007) Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol 48: 523–539 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, Onishi K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46: 79–86 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006. a) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6: 47–59 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Tyagi AK, Khurana JP (2006. b) The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct Integr Genomics 6: 36–46 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006. c) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88: 360–371 [DOI] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29: 1220–1234 [DOI] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L (2003) The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278: 23936–23943 [DOI] [PubMed] [Google Scholar]
- Lasanthi-Kudahettige R, Magneschi L, Loreti E, Gonzali S, Licausi F, Novi G, Beretta O, Vitulli F, Alpi A, Perata P (2007) Transcript profiling of the anoxic rice coleoptile. Plant Physiol 144: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM (2006) MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proc Natl Acad Sci USA 103: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RH, Wang CH, Huang LT, Chen SC (2001) Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J Exp Bot 52: 1117–1121 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1989) Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 243: 91–93 [DOI] [PubMed] [Google Scholar]
- McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ (1989) Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita S, Murano N, Akaike M, Nakamura K (1997) Mutants of Arabidopsis thaliana with pleiotropic effects on the expression of the gene for beta-amylase and on the accumulation of anthocyanin that are inducible by sugars. Plant J 11: 841–851 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrotto D, Jolley M, Beer S, Wenck A, Hansen G (2000) The use of phosphomannose-isomerase as a selectable marker to recover transgenic maize plants (Zea mays L.) via Agrobacterium transformation. Plant Cell Rep 19: 798–803 [DOI] [PubMed] [Google Scholar]
- Ohto MA, Hayashi S, Sawa S, Hashimoto-Ohta A, Nakamura K (2006) Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant Cell Physiol 47: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Park J-E, Kim Y-S, Yoon H-K, Park C-M (2007) Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci 172: 150–157 [Google Scholar]
- Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563 [DOI] [PubMed] [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A (2006) Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224: 556–568 [DOI] [PubMed] [Google Scholar]
- Reddy VS, Ali GS, Reddy AS (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Rodoni S, Vicentini F, Schellenberg M, Matile P, Hortensteiner S (1997) Partial purification and characterization of red chlorophyll catabolite reductase, a stroma protein involved in chlorophyll breakdown. Plant Physiol 115: 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Bilang J, Theunissen BH, Perrot-Rechenmann C (1998) Identification of new early auxin markers in tobacco by mRNA differential display. Plant Mol Biol 37: 385–389 [DOI] [PubMed] [Google Scholar]
- Sachs T (1981) The control of the patterned differentiation of vascular tissues. Adv Bot Res 9: 151–262 [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY (2004) The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131: 5649–5657 [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33: 513–520 [DOI] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, et al (2005) Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 139: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57: 391–399 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2000) Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275: 3137–3143 [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J (2001) Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci USA 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen W, Provart N, Chang HS, Guimil S, Su W, Estes B, Zou G, Wang X (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J 1: 59–70 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, et al (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W, Zhao X, Pan X, Xie Q, Zhu L (2006) The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48: 687–698 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.