Abstract
Brassinosteroids (BRs) are involved in many developmental processes and regulate many subsets of downstream genes throughout the plant kingdom. However, little is known about the BR signal transduction and response network in monocots. To identify novel BR-related genes in rice (Oryza sativa), we monitored the transcriptomic response of the brassinosteroid deficient1 (brd1) mutant, with a defective BR biosynthetic gene, to brassinolide treatment. Here, we describe a novel BR-induced rice gene BRASSINOSTEROID UPREGULATED1 (BU1), encoding a helix-loop-helix protein. Rice plants overexpressing BU1 (BU1:OX) showed enhanced bending of the lamina joint, increased grain size, and resistance to brassinazole, an inhibitor of BR biosynthesis. In contrast to BU1:OX, RNAi plants designed to repress both BU1 and its homologs displayed erect leaves. In addition, compared to the wild type, the induction of BU1 by exogenous brassinolide did not require de novo protein synthesis and it was weaker in a BR receptor mutant OsbriI (Oryza sativa brassinosteroid insensitive1, d61) and a rice G protein alpha subunit (RGA1) mutant d1. These results indicate that BU1 protein is a positive regulator of BR response: it controls bending of the lamina joint in rice and it is a novel primary response gene that participates in two BR signaling pathways through OsBRI1 and RGA1. Furthermore, expression analyses showed that BU1 is expressed in several organs including lamina joint, phloem, and epithelial cells in embryos. These results indicate that BU1 may participate in some other unknown processes modulated by BR in rice.
Brassinosteroids (BRs) are essential growth regulators, involved in many physiological processes, e.g. cell expansion and division, vascular bundle differentiation, skotomorphogenesis, flowering, senescence, abiotic, and biotic stresses (Szekeres et al., 1996; Clouse and Sasse, 1998; Nakashita et al., 2003; Yu et al., 2008). The main components of BR signaling have been mostly identified in Arabidopsis (Arabidopsis thaliana; Belkhadir and Chory, 2006). BRs are perceived by a Leu-rich repeat receptor kinase known as BRASSINOSTEROID INSENSITIVE1 (BRI1; Li and Chory, 1997; Kinoshita et al., 2005). BRI1 heteromerizes with the BRI1 ASSOCIATED KINASE1, and this protein complex phosphorylates BRASSINOSTEROID SIGNALING KINASEs (BSKs; Nam and Li, 2002; Tang et al., 2008). BSKs inhibit the kinase activity of BRASSINOSTEROID INSENSITIVE2 (BIN2), which encodes a GLYCOGEN SYNTHASE KINASE3-like kinase. In the absence of BR, BIN2 phosphorylates two transcription factors, BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-EMS-SUPRESSOR1 (BES1), and abolishes their functions and BR response (He et al., 2002; Yin et al., 2002). On the other hand, BRs induce dephosphorylation of BZR1 and BES1, which are transcriptional regulators of target genes in the nucleus (He et al., 2002; Yin et al., 2002).
BZR1 and BES1 have been identified by genetic screens based on skotomorphogenesis (Wang et al., 2002; Yin et al., 2002). In Arabidopsis, it has been reported that hypocotyls of BR-deficient and -insensitive mutants, such as constitutive photomorphogenic dwarf and bri1, are shortened and deetiolated (Szekeres et al., 1996; Li and Chory, 1997). In contrast, hypocotyls of constitutive BR-response mutants are not shortened and deetiolated by a BR deficiency that was artificially induced by brassinazole (BRZ; Asami et al., 2000). The bzr1-D mutant was screened as a BRZ-resistance gain-of-function mutant (Wang et al., 2002). A suppressor of a bri1 mutant, bes1-D, also showed phenotypes similar to bzr1-1D (Yin et al., 2002).
We have previously isolated the brassinosteroid deficient1 (brd1) mutant that had a defective BR biosynthetic gene, BRD1 (OsBR6ox/OsDWARF) in rice (Oryza sativa; Mori et al., 2002). Analyses of brd1 mutants clarified many BR effects in rice. The brd1 mutant showed severe dwarfism, sterility, small seeds, abnormal vascular bundles, shortened internodes, and inhibition of elongation of the coleoptile and mesocotyl in the dark (Hong et al., 2002; Mori et al., 2002). The lamina joint, which is a border region between the leaf blade and sheath, is an especially sensitive organ to BR and it is severely bent after exposure to active BRs (Wada et al., 1981). Loss-of-function mutants of BR biosynthesis, d2 and d11, have erect leaves because of BR deficiency (Hong et al., 2003; Tanabe et al., 2005). In addition, BR-deficient rice mutants have much shorter grains than the wild type (Mori et al., 2002; Hong et al., 2003; Tanabe et al., 2005). On the other hand, hyperproduction of BR by overexpression of DWARF4, which encodes a BR biosynthetic gene, increased seed size and yield in Arabidopsis (Choe et al., 2001). In rice, overexpression of DWARF4 increased grain filling, but there was no mention of its effect on grain size (Wu et al., 2008).
Some components of BR signaling have been also identified in rice. OsBRI1 and OsBZR1 have been respectively identified as counterparts of BRI1 and BZR1 in Arabidopsis and their loss-of-function mutants display BR-deficient mutant-like phenotypes such as erect leaves (Yamamuro et al., 2000; Bai et al., 2007). OsBZR1 directly binds to the promoter of DWARF AND LOW TILLERING (DLT) gene, whose product is needed for the full BR response in rice (Tong et al., 2009). Recently, rice heterotrimeric G protein alpha 1 (RGA1), known to be involved in gibberellic acid signaling and disease resistance (Ueguchi-Tanaka et al., 2000; Suharsono et al., 2002), was also implicated as a BR signaling component (Wang et al., 2006; Oki et al., 2009). Loss-of-function mutant of RGA1, d1, displayed less sensitivity to brassinolide (BL) in lamina joints and primary roots. Since expression patterns of OsBZR1-regulated genes were not altered by BL in d1 mutants, RGA1 may be involved in a BR signaling pathway distinct from that of OsBZR1 (Oki et al., 2009).
Some transcriptional regulators identified in BR signaling encode basic/helix-loop-helix (bHLH) proteins in Arabidopsis. BES1 interacts with another transcription factor BES1-INTERACTING MYC-like1 (BIM1), which encodes a MYC-like bHLH protein, and both proteins together bind to a promoter region of the target gene to regulate expression (Yin et al., 2005). In addition, three redundant bHLH proteins, BR ENHANCED EXPRESSION1 (BEE1) to BEE3, are products of early BR response genes and they are required for the full BR response (Friedrichsen et al., 2002). Although some bHLH proteins play a critical role in BR signaling in Arabidopsis as described above, little is known in rice. Here, we report the identification and characterization of a novel rice gene BRASSINOSTEROID UPREGULATED1 (BU1), which encodes a bHLH protein but presumably lacks a basic region, by using rice plants overexpressing and repressing BU1.
RESULTS
Isolation of BR-Induced Genes
To identify novel BR-regulated genes, RNA were obtained from young shoots of 50-d-old brd1 mutants, with or without 40 nm BL treatment for 24 h and analyzed using a 22K rice microarray. Ninety six of 21,938 genes were up-regulated by BL application, as defined by a more than 2-fold difference in the fold-change values that represent ratios of hybridization signals between BL-treated and control brd1 plants. Several Arabidopsis proteins reportedly involved in BR signaling, such as BEE1 to BEE3 and BIM1 to BIM3, contain bHLH motifs. Therefore, we searched the bHLH motif among the 96 up-regulated genes and identified three genes. In this article, we report one of the three genes with the bHLH motif that we have designated as BU1 (Os06g0226500), whose induction by exogenous BL has been confirmed by real-time PCR analysis both in brd1 and wild-type plants (Supplemental Fig. S1; Fig. 4A). We will provide details of our microarray results and other up-regulated genes in future publications.
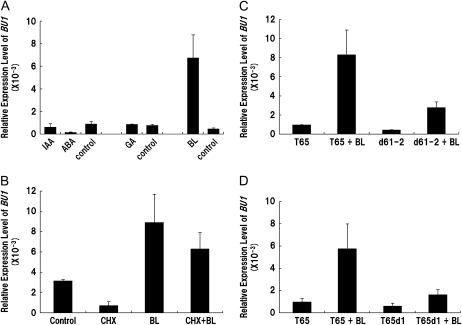
Figure 4.
Expression pattern of BU1 analyzed by real-time PCR. Total RNA used in these experiments were isolated from shoots of 2-week-old seedlings after a variety of chemical treatments for 24 h. Data are the average of three or four independent experiments and normalized by 18S rRNA. The error bars indicate sd. A, Response of BU1 to various plant hormones. IAA 10 μm, ABA 100 μm, GA 10 μm, BL 1 μm. B, Effect of CHX on BU1 induction. Wild-type (Nipponbare) plants were treated with mock, CHX, BL, and both CHX and BL. Two-week-old seedlings were pretreated with or without 100 μm CHX for 3 h, before 1 μm BL or mock treatment for 24 h. C and D, Expression of BU1 in response to BL treatment in either d61-2 and d1 or the wild type (T65). Plants were treated with 1 μm BL for 24 h.
In Silico Analysis of BU1 Protein
Transcription factors with the bHLH motif regulate a large array of target genes and play a pivotal role in cellular signaling in living organisms (Ledent and Vervoort, 2001; Toledo-Ortiz et al., 2003; Li et al., 2006). A bHLH protein has two functional domains, the basic region and the HLH region. The former is required for binding to DNA to regulate the expression of genes, while the latter is required for interaction with other bHLH proteins for hetero- or homodimerization (Murre et al., 1989). In rice, more than 160 bHLH genes have been identified (Li et al., 2006). Although the BU1 protein is not described by Li et al. (2006), it has an HLH motif and is included in the bHLH protein family (Fig. 1A). Since BU1 lacks the basic region needed for binding to DNA, it is classified into group D of bHLH proteins, the non-DNA-binding protein family (Li et al., 2006). In rice, there are three proteins that have high identity with BU1. However, Os02g0747900 may not be a functional protein, since Os02g0747900 lacks one helix motif of the HLH domain (Fig. 1A). BU1 and these proteins have a high similarity with PACLOBUTRAZOL RESISTANCE1 (PRE1) of Arabidopsis, an HLH protein that also lacks the basic region. Since PRE1 functions in GA signaling (Lee et al., 2006), we expect that BU1 may also play an important role in plant hormone signaling, especially BR. Therefore, we used phylogenetic analysis to determine the similarity of the HLH domain of BU1 to those of BEE1 and BIM1, which are bHLH proteins related to BR signaling in Arabidopsis (Fig. 1B). Since the neighbor-joining tree indicates that the similarity between BU1 and BEE1 or BIM1 is very low, BU1 and its homologs may be a novel protein family involved in BR signaling. In contrast to BU1, because the induction of the three homologs by exogenous BL was not detected in the wild-type shoot (data not shown), we preferentially analyzed BU1 in this article.
Figure 1.
Protein sequence analysis of BU1. A and B, Alignment (A) and phylogenic tree (B) of HLH domains for BU1 (Os06g0226500), its three homologous proteins, PRE1, and two bHLH proteins associated with BR signaling, BEE1 and BIM1. The top five protein sequences are full length. HLH domains are indicated below the sequences. Black and gray backgrounds indicate identical and similar amino acids, respectively. The bar corresponds to 0.1 amino acid substitutions per site.
BR Action Phenocopies Rice Plants Overexpressing BU1
To characterize BU1, we made transgenic rice plants overexpressing BU1 (BU1:OX) under the control of a maize (Zea mays) Ubiquitin promoter. Many T0 plants showed enhanced bending of the lamina joint, but most of them were sterile. As a result, we obtained seeds from only two overexpressed lines, BU1:OX-3 and 4 (Fig. 2A). The expression level of BU1 is higher in OX-3 and much higher in OX-4 than in wild-type plants (Fig. 2B). BU1:OX showed increased bending of the lamina joint toward the abaxial side depending on the expression levels of BU1 (Fig. 2C) and had larger grains than the wild type (Fig. 2, D and E). Since it is known that bending of the lamina joint and grain size are regulated by BR (Yamamuro et al., 2000; Mori et al., 2002; Hong et al., 2003), these results suggest that BU1 may be involved in BR signaling or biosynthesis. In addition, spikelet numbers were drastically reduced in OX-4, although they were similar in the wild type and OX-3 (data not shown). This observation may be the result of poor growth due to high transcript levels of BU1 in OX-4.
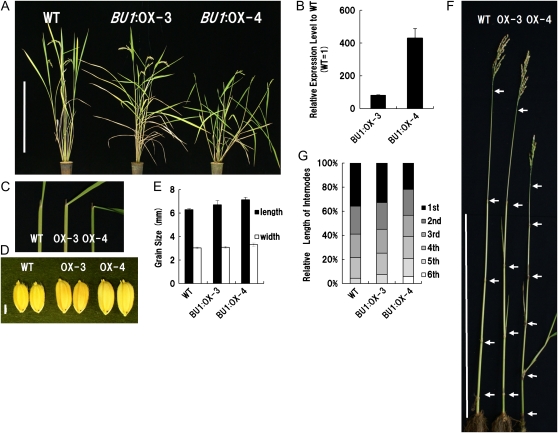
Figure 2.
Phenotypes of rice plants overexpressing BU1 (BU1:OX). A, Gross morphology of the wild type (WT, left), BU1:OX-3 (center), and BU1:OX-4 (right) at the heading stage. Bar = 50 cm. B, Relative expression level of BU1 in BU1:OX-3 and -4 to the wild type, analyzed by real-time PCR and normalized by RUBQ2. The error bars indicate sd. C, Lamina joint of second leaf from flag leaf in the wild type and BU1:OX-3 and -4. BU1:OX showed increased bending of the lamina joint. D and E, Grain morphology and size of the wild type and BU1:OX-3 and -4. BU1:OX-3 and -4 (center, right) have larger grains than that of the wild type (left). Bar = 2 mm. The error bars indicate sd (n = 10). F and G, Photograph (F) and length as a percentage (G) of internodes in the wild type and BU1:OX-3 and -4 (n = 8). Uppermost internode of BU1:OX-3 and -4 was shorter, but its two lower internodes were longer than the wild type. Bar = 50 cm.
BU1:OX also has some characteristic abnormalities in the pattern of internode elongation (Fig. 2, F and G), progression of tillers in nodes of aerial part (Fig. 2F), and lignification and development of crown roots in these nodes (Supplemental Fig. S2). The first internode was severely shortened, but the fifth and sixth internodes were slightly elongated. Panicles did not emerge perfectly from the flag leaf sheath as a result of the shortened first internode in OX-4.
BU1 Is Involved in BR Signaling, Not BR Biosynthesis
In Arabidopsis, a BR signal component, BZR1, was isolated by genetic screens using elongation of hypocotyls with or without BRZ (Wang et al., 2002). Similar to hypocotyls in Arabidopsis, elongation of the mesocotyls and second internodes in rice grown in complete darkness was severely dependent on BR (Yamamuro et al., 2000). To ascertain whether BU1 functions in BR signaling or biosynthesis, we used the inhibition of elongation of the mesocotyl and second internode in dark with or without BRZ, similar to the test performed in Arabidopsis hypocotyls. Mesocotyls and second internodes in the wild type were elongated in half-strength Murashige and Skoog medium without BRZ, but shortened in half-strength Murashige and Skoog medium containing BRZ (Fig. 3, A and B). On the other hand, in BU1:OX-4, the length of the second internodes was not affected by BRZ. Mesocotyls of OX-4 were greatly elongated compared to the wild type and the elongation was not much affected by BRZ. These results indicate that BU1:OX-4 is less sensitive to BRZ than the wild type.
Figure 3.
Involvement of BU1 in BR signaling. A and B, Effect of BRZ. Etiolated seedlings of the wild type (WT) and BU1:OX-4 were grown in complete darkness with or without 10 μm BRZ2001 for 2 weeks prior to measurement of the length of the second lower internodes and mesocotyls. Arrows indicate nodes. Arrowheads indicate mesocotyls, organs between lowest nodes and seeds. Bar = 5 cm. The error bars indicate sd (n = >9). C, Gross morphology of d61, a BR-insensitive mutant that has an inactive BR receptor OsBRI1 (left), and d61 overexpressing BU1 (right). The erect leaf phenotype of d61 was complemented by overexpression of BU1. Bar = 50 cm. D, Culm of d61 (left) and d61 overexpressing BU1 (right). Arrows indicate nodes. Bar = 10 cm.
Bending of the lamina joint is very sensitive to BR. BU1:OX shows enhanced bending of the lamina joint as described above. In contrast, the loss-of-function mutant of OsBRI1, d61, has erect leaves and it is less sensitive to BL. We generated d61 plants overexpressing BU1 and observed the resulting phenotypes, especially their lamina joints. BU1 overexpressor in the d61 background showed increased bending of the lamina joint (Fig. 3C) and a pattern of internode elongation similar to BU1:OX-4 (Fig. 3D), indicating that overexpression of BU1 complemented the BR-deficient phenotype of d61.
In addition, the wild type and OX-4 did not differ markedly in their level of expression of BRD1, a gene that encodes a P450 that catalyzes the conversion of 6-deoxocastasterone to castasterone (CS; Supplemental Fig. S3A). We also measured endogenous BR contents in BU1:OX-4 and the wild type. There were no obvious differences between OX-4 and the wild type in the amounts of both CS (presumably the most active endogenous BR in rice) and 6-deoxocastasterone, the main precursor of CS (Supplemental Fig. S3B). These results indicate that the biosynthesis of BRs is not activated in BU1:OX. Therefore, BU1 is a signaling component of BR in rice.
BU1 Is an Early Response Gene by BL through Dual Pathways, OsBRI1 and RGA1
To test the response to other plant hormones, we performed real-time PCR analysis using RNA samples of seedlings subjected to a variety of hormones and chemical treatments. Previous research showed that genes induced by auxin overlapped with those induced by BR, and both hormones synergistically modulate plant architecture (Yin et al., 2002; Nemhauser et al., 2004). However, expression of BU1 was not induced by indole-3-acetic acid (IAA) or by GA3. On the other hand, abscisic acid (ABA), a known antagonist to BR response, repressed expression of BU1 (Fig. 4A). The mechanism whereby ABA represses the primary signaling outputs of BR in Arabidopsis (Zhang et al., 2009) may be conserved in rice as well.
BU1 was greatly up-regulated by exogenous BL in wild-type plants. In addition, BU1:OX showed many BR-related phenotypes. These results suggest that BU1 may act as an early response gene in BR signaling. We tested the response of BU1 expression to exogenous BL treatment with cycloheximide (CHX), an inhibitor of de novo protein synthesis. A previous report in Arabidopsis indicated that induction of primary response genes by BR signaling, e.g. BEE1 to BEE3, did not need de novo protein synthesis (Friedrichsen et al., 2002). Induction of BU1 by exogenous BL was not affected by CHX (Fig. 4B). This result indicates that induction of BU1 does not need de novo protein synthesis and BU1 is a primary response gene to BR signaling.
To determine whether the induction of BU1 by endogenous BRs is through OsBRI1, a transmembrane BR receptor, we compared the response of BU1 to exogenous BL between the d61-2 (OsbriI) mutant and its wild type, japonica cv Taichung 65 (T65; Fig. 4C). In T65, induction of BU1 by exogenous BL was 8.7 times more than the control, and in the d61-2 mutant, 6.6 times greater than the control. The expression level of BU1 in d61-2 after BL treatment was about one-half compared to that in T65. These results indicate that BU1 is less sensitive to exogenous BL in d61-2 compared to T65 and it is induced through OsBRI1 by BRs. On the other hand, the sensitivity of BU1 to exogenous BL still remained in d61-2. This result suggests that the induction of BU1 by BRs may be also through another pathway distinct from the one associated with OsBRI1. Recent research showed that RGA1, encoding the rice heterotrimeric G protein alpha subunit, was also involved in BR signaling in rice (Wang et al., 2006; Oki et al., 2009). Therefore, we compared the response of BU1 to exogenous BL between T65 and the d1 mutant, a loss-of-function mutant of RGA1 in the T65 background (Fig. 4D). In the wild type (T65), induction of BU1 by BL was 5.9-fold to control but only 2.7-fold in the d1 mutant. The expression level of BU1 in d1 after BL treatment was about one-third compared to that in T65. These results indicate that BU1 is less sensitive to exogenous BL in d1 and induced through both OsBRI1 and RGA1 pathway by BRs.
Expression Analysis of BU1
We compared the expression level of BU1 in various organs by real-time PCR analysis (Fig. 5A). The expression level of BU1 is high in the lamina joint in vegetative organs. Aside from the lamina joint, BU1 is highly expressed in the panicle especially at heading stage. To analyze the expression pattern of BU1 in detail, we inserted a 2-kb segment of the promoter region of BU1 to drive the GUS gene, then transformed rice plants with this gene cassette. We performed the histochemical GUS assay using these transgenic plants (Fig. 5B). GUS staining was observed in various organs. We examined the lamina joint region of the second and eighth leaf (Fig. 5B, 1 and 2), leaf blade and sheath near the lamina joint in 9-d-old seedlings (Fig. 5B, 3). The veins in the leaf blade and sheath were well stained (Fig. 5B, 4 and 5). Using a light microscope, we observed staining in phloem in transverse sections of leaf blades (Fig. 5B, 6). In the reproductive stage, staining was observed in the ovule and filaments before anthesis (Fig. 5B, 7). In particular, staining was observed in the vascular bundles of the ovule, lemma, and palea (Fig. 5B, 8–10). In the seed, epithelial cells were stained in the embryo 3 d after imbibition (Fig. 5B, 11 and 12). The expression of BU1 in the lamina joint, verified by two independent experiments (Fig. 5, A and B, 2), strongly suggests that BU1 is involved in bending of the lamina joint in wild-type plants. In addition, the results of GUS staining suggest that expression of BU1 that was detected by real-time PCR in the leaf blade and panicle may be localized in the vascular bundles (phloem) in these organs.
Figure 5.
Expression pattern of BU1. A and C, Expression analysis of BU1 in various organs and seeds by real-time PCR. Data are the average of three or four independent experiments and normalized by 18S rRNA (A) and RUBQ2 (C). A and C, Total RNA was isolated from various organs (A) and embryo and endosperm of 10 dehusked seeds (C). YP, Young panicle; DAH, days after heading. The error bars indicate sd. B, Histochemical GUS staining of rice plants harboring BU1∷GUS. 1 and 2, Close-up view of second lamina joint region of 9-d-old seedling and eighth lamina joint region. 3, Nine-day-old seedling. Leaf blade was stained. 4 and 5, Leaf blade (4) and sheath (5) of 9-d-old seedling. 6, Transverse section of leaf blade. 7, Flower before anthesis. 8, Close-up view of stigma and ovule. 9, Transverse section of ovule. 10, Palea and lemma of young panicle. Stained blue line is a vascular bundle. 11 and 12, Embryo (11) and cross section of embryo (12) 3 d after imbibition. Epithelial cells were stained.
Expression of BU1 is higher in the embryo than in the endosperm; however, BU1 expression is not influenced by imbibition (Fig. 5C). Although elongation of the second internode and mesocotyl is very sensitive to BR and these organs were elongated in BU1:OX compared to the wild type, in the presence of BRZ in dark (Fig. 3, A and B), GUS staining was not observed in 10-d-old seedlings grown in the dark (data not shown). This result suggests that BU1 may not be involved in BR response in the absence of light in wild-type plants.
To determine the intracellular localization of BU1 protein, we constructed a plasmid DNA for fusion protein in which BU1 was fused to the C terminus of an enhanced GFP (eGFP) that we subsequently introduced into rice coleoptiles via particle gun bombardment. Dispersed fluorescent signal of eGFP-BU1 fusion protein throughout the coleoptile cells was observed (Supplemental Fig. S4). Transcription factors are basically localized in the nucleus. Therefore, BR signal transduction by BU1 may be mediated by a different mechanism.
Rice Plants Suppressing Both BU1 and Its Homologs Show Erect Leaves
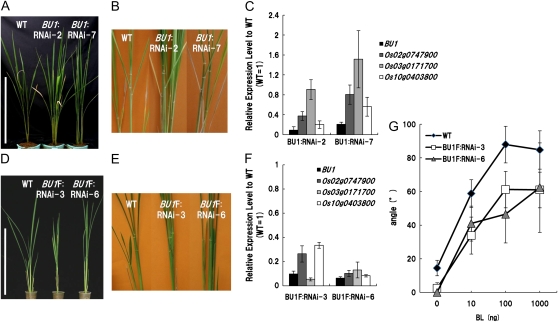
We generated transgenic plants suppressing BU1 using the 3′-untranslated region (340 bp) as a trigger region (BU1:RNAi), and investigated the effect of silencing by morphological evaluation. The angle of the lamina joint and other morphological characters in rice suppressing BU1 did not markedly change as compared to wild-type plants (Fig. 6, A–C). As described in Figure 1A, there are three proteins that have a high similarity in amino acid sequence to BU1. We supposed that these three proteins may have functions that overlapped with those of BU1, therefore we generated transgenic plants suppressing both BU1 and these homologs using the whole open reading frame region (270 bp) of BU1 as a trigger region (BU1F:RNAi). BU1F:RNAi plants showed erect leaves in contrast to BU1:OX (Fig. 6, D–F). In addition, we compared the sensitivity between BU1F:RNAi and the wild type to BL by using the lamina joint bending assay. In BU1F:RNAi, the angles of the lamina joint were narrower than those of the wild type at all concentrations of BL (Fig. 6G), indicating that the sensitivity of BU1F:RNAi to BL is lower than that of the wild type. Moreover, the expression levels of BU1 homologs were reduced in the lamina joint of BRZ-treated wild-type seedlings as BU1 (Supplemental Fig. S5). These results indicate that BU1 controls bending of the lamina joint and the three homologs have redundant function with BU1 in the lamina joint. However, we did not observe changes of phenotype in the vascular bundles of BU1F:RNAi (data not shown).
Figure 6.
Phenotype of rice plants suppressing BU1. A, Gross morphology of the wild type (WT) and BU1:RNAi-2 and -7 lines, suppressing BU1 at vegetative phase. Bar = 50 cm. B, Close-up view of the lamina joint region of the wild type and BU1:RNAi-2 and -7. C, Relative expression level of BU1 and its homologs (Os02g0747900, Os03g0171700, Os10g0403800) in BU1:RNAi-2 and -7 to the wild type, analyzed by real-time PCR and normalized by RUBQ2. The error bars indicate sd. D, Gross morphology of the wild type and BU1F:RNAi-3 and -6 lines, suppressing BU1 and its three homologs at vegetative phase. Bar = 50 cm. E, Close-up view of the lamina joint region of the wild type and BU1F:RNAi-3 and -6. F, Relative expression level of BU1 and its homologs in BU1F:RNAi-3 and -6 to the wild type, analyzed by real-time PCR and normalized by RUBQ2. The error bars indicate sd. G, BL response at the lamina joint of the wild type, BU1F:RNAi-3, and BU1F:RNAi-6. The error bars indicate sd (n > 3).
DISCUSSION
Although some BR signaling components have been isolated in Arabidopsis, little is known about BR signaling components in rice, other than OsBZR1 and DLT functioning in the OsBRI1 pathway and RGA1. In this study, we demonstrate that BU1, a small HLH protein, is a novel BR signaling component. BU1:OX shows many characteristic phenotypes associated with BR effects, e.g. enhanced bending of the lamina joint, large grain, and elongation in dark-grown seedlings. In addition, BU1:OX is resistant to BRZ and overexpression of BU1 in d61, a BR receptor mutant, complements the BR-insensitive erect leaf phenotype of d61. These results indicate that BU1 is involved in BR signaling, but not biosynthesis. Real-time PCR analysis showed that BU1 is a primary response gene of BR signaling and up-regulated through both OsBRI1 and RGA1 pathways by BRs. The expression pattern of BU1 also provides information about the native function of BU1. From analysis of phenotypes of BU1F:RNAi plants and the lamina joint bending assay, it is clear that BU1 controls the bending of the lamina joint.
BU1:OX shows abnormal phenotypes that includes previously known and also possibly new BR effects. The length of the first internode of culm is shortened but that of the fifth internode is elongated, and the sixth internode protrudes in BU1:OX (Fig. 2, F and G). d61 has a relatively longer first internode and shorter lower internodes (Fig. 3D) in comparison to BU1:OX. This result suggests that alternation of length of internodes in BU1:OX may be a characteristic BR effect on growth and development of culm. Progression of tillers in lower aerial nodes (Fig. 2F) may also be a possible BR effect, since rice plants overexpressing DWARF4, a BR biosynthetic gene, have more tillers whereas dlt mutants have less tillers (Wu et al., 2008; Tong et al., 2009). Severe lignification and abnormal development of crown roots in nodes was observed in BU1:OX. It is known that epidermal cell death in nodes correlates with root emergence in rice and it is mediated by ethylene (Steffens and Sauter, 2009). Since ethylene biosynthesis is activated by BR (Arteca et al., 1983; Yi et al., 1999), this observation may be a BR effect mediated via ethylene biosynthesis. Moreover, most of BU1:OX plants displayed severe sterility. Plants suppressing OsSPY, a negative regulator of GA signaling and BR biosynthesis, rarely developed fertile flowers (Shimada et al., 2006). Overexpression of genes related to plant hormones often causes severe sterility.
The expression pattern of BU1 revealed by the histochemical GUS assay suggests that BU1 may be involved not only in bending of the lamina joint but also in vascular differentiation in the leaf blade, leaf sheath, and floral organs. The characterization of the loss-of-function mutant of BRI1 and its homologs, BRL1 and BRL3, in Arabidopsis and OsBRI1 in rice showed that BR plays an important role in vascular differentiation, enhances the differentiation of the xylem, and inhibits the differentiation of the phloem (Caño-Delgado et al., 2004; Nakamura et al., 2006). However, we could not detect morphological changes in vascular bundles of BU1:OX and BU1F:RNAi. According to the results of the GUS assay, BU1 is mainly expressed in the phloem, a vital organ for long-distance transport of photosynthetic products, nutrients, ions, proteins, and hormones. It is possible that BU1 may regulate gene expression related to transport or unknown function in the phloem. The epithelium is also an important organ for the transport of nutrients, ions, and sugars from the endosperm, and GAs from the embryo. However, the expression level of BU1 was scarcely affected by imbibition. This result suggests that BU1 in the epithelium may not be involved in expression of growth-related genes but homeostatic-related genes. Therefore, genes regulated by BU1 in the epithelium may be different from those in the phloem.
In this study, we showed that BU1 is a novel primary response gene to BR signaling through both OsBRI1 and RGA1. Previous reports describe that OsBZR1 functions as a transcriptional repressor for down-regulating DLT and BR biosynthesis genes such as AtBZR1 (He et al., 2005; Bai et al., 2007; Tong et al., 2009). On the other hand, real-time PCR clearly showed that BU1 is a primary response gene up-regulated by exogenous BL, suggesting that BU1 may be up-regulated by a novel transcription factor (maybe an activator) distinct from OsBZR1.
In addition, we compared the expression of known BR-marker genes between the wild type and two independent BU1:OX lines. However, BR-marker genes, OsXTR1, OsXTR3, OsBLE2, and OsBLE3 (Uozu et al., 2000; Yang et al., 2003, 2006), were not up-regulated in BU1:OX (data not shown). Therefore, downstream genes of BU1 may be different from known BR-related genes regulated by the OsBZR1 pathway in rice.
BU1 protein is categorized as a putative non-DNA-binding bHLH protein, since it lacks a basic region (Fig. 1A). Therefore, downstream genes of BU1 may be regulated by more complex mechanisms, different from the general mechanism of transcriptional regulation of target genes by a transcription factor. Molecular mechanism of non-DNA-binding HLH proteins is understood well in human Inhibitor of DNA binding (Id) proteins. Id proteins heterodimerize with other DNA-binding bHLH protein partners via HLH motif and abolish their functions as transcription factors, binding to the cis-element of their target genes (Benezra et al., 1990; Sun et al., 1991). Consequently, expression of their target genes by partners of Id proteins is abolished by Id proteins. Likewise, BU1 may also interact or form a complex with putative BR-negative regulators (maybe bHLH proteins), and inhibit their functions as transcription factors. Taking into account these studies and perspectives, we propose a model of BR signaling in rice (Fig. 7).
Figure 7.
BR signaling model in rice. Induction of BU1 by BL is partially inhibited in OsBRI1 mutant (d61-2) and RGA1 mutant (d1). In addition, BU1 is the primary response (up-regulated) gene to BR signaling. Therefore, BU1 may be a novel mediator of a distinct pathway from OsBZR1, because OsBZR1 functions as the transcription repressor. Moreover, regulation of downstream genes by BU1 may be through a complex mechanism like the regulation of human Id protein.
BU1 has high similarity with PRE1, encoding a HLH protein involved in GA signaling in Arabidopsis (Lee et al., 2006). In addition, elongation of internodes in darkness is affected not only by BRs but also by GAs, suggesting that BU1 may also be involved in GA signaling. To test whether BU1 is also a GA signal component, we performed two kinds of experiments, the second leaf sheath elongation assay and the α-amylase induction assay. First, we compared the effect of paclobutrazol, an inhibitor of GA biosynthesis, against second leaf sheath elongation in BU1:OX and its wild type. BU1:OX showed shortened leaf sheaths and displayed sensitivity to paclobutrazol similar to the wild type (Supplemental Fig. S6, A and B). Second, we examined α-amylase induction by GA in BU1:OX-4 and BU1F:RNAi-6 seeds by using the starch plates. For example, slender rice, a constitutive GA response mutant, produces and secretes amylase from embryoless half seeds without GA addition and a plaque zone around half seeds is observed on agar plates after staining by iodine (Ikeda et al., 2001). Production of amylase from embryoless half seeds of OX-4 and RNAi was observed only on GA applied agar plates (clear zones), but not on control plates. There was no significant difference in sensitivity to GA relative to the wild type (Supplemental Fig. S6C). These results indicate that BU1 is not directly involved in GA signaling. Our study suggests that many non-DNA-binding bHLH proteins may play an important role not only in GA signaling but also in some plant hormone signaling similar to the function of human Id proteins, although little is known about functions of HLH proteins in plants.
BRs play an important role in plant morphogenesis. In this study, we show that BU1 controls bending of the lamina joint via BR signaling. BR-defective and -insensitive mutants, which have erect leaves, have higher biomass and grain yield than wild-type plants at high planting density, since they may increase light capture for photosynthesis by their erect leaves (Morinaka et al., 2006; Sakamoto et al., 2006). Therefore, control of bending of the lamina joint via regulating expression of BU1 may lead to increased biomass and grain yield. In addition, BU1:OX displays a valuable phenotype, large grains. Further studies of downstream effects of BU1 may provide important insights for engineering the plant's architecture and increasing grain yield.
MATERIALS AND METHODS
Plant Material and Growth Condition
All transgenic and wild-type plants were generated from rice (Oryza sativa japonica ‘Nipponbare’), except for the d61-2 mutant, d1 mutant, and their wild type, which were obtained from T65. All transgenic plants were grown in isolated green houses at 28°C.
For mRNA analysis of plant hormones including brassinolide or CHX treatment, dehusked seeds of rice plants were surface sterilized and sown on one-half-strength Murashige and Skoog medium containing 3% (w/v) Suc and 0.4% (w/v) Gelrite (Wako Pure Chemicals). Seedlings were transplanted into Kimura's B solution medium about 10 d after sowing (Sato et al., 1996). After 4 d, seedlings were incubated with 1 μm BL (Daiichi Fine Chemical), 10 μm IAA (Wako Pure Chemicals), 100 μm ABA (Wako Pure Chemicals), 10 μm GA3 (Sigma Aldrich), or an equal volume of the pertinent solvent (dimethyl sulfoxide [DMSO], NaOH, ethanol) for 24 h. For CHX treatment, plants were pretreated with 100 μm CHX (Nacalai Tesque) for 3 h before BL treatment. Plants were grown in a growth chamber at 25°C under long-day conditions (14 h light [60–70 μmol m−2 s−1]/10 h dark).
For etiolation analysis, transgenic plants and the wild type were germinated and grown on half-strength Murashige and Skoog medium supplemented with 10 μm BRZ2001 (Sekimata et al., 2001) or its equal volume of solvent (DMSO) in a dark chamber at 25°C. The length of the mesocotyl and second internode were measured after 2 weeks.
For the second leaf sheath elongation assay, seeds of transgenic plants and the wild type were sterilized, sown on distilled water for 2 d at 28°C, and germinated. Germinated seeds were sown on half-strength Murashige and Skoog medium supplemented with 2 μm paclobutrazol (Wako Pure Chemicals) or its equal volume of solvent (DMSO), and grown at 25°C under long-day conditions. After a week, the lengths of the second leaf sheath were measured. The method described by Sekimata et al. (2001) was used as reference in this experiment.
Oligo DNA Microarray Analysis
We used a rice 22K oligo DNA microarray kit (G2554A; Agilent Technology) containing 21,938 oligonucleotides based on the sequence data of the rice full-length cDNA project (Kikuchi et al., 2003; Yazaki et al., 2004). Total RNA was extracted using Isogen (Wako Pure Chemicals) and further purified with an RNeasy plant kit (Qiagen) according to the manufacturer's instructions. RNA amplification, labeling, hybridization, scanning, and image analysis were performed according to the manufacturer's instructions (Agilent Technology). The microarray results were filtered to select candidate clones with P-value log ratios of less than 0.01. A 2.0-fold expression cutoff was applied, and the cases in which dye-swapped replications passed this cutoff were scored as differential expression.
Vector Construction and Rice Transformation
To generate overexpression lines, the full-length cDNA of BU1 (AK071601), provided by the Rice Genome Resource Center in the National Institute of Agrobiological Sciences, was cloned into the SfiI site of the pRiceFox vector (Nakamura et al., 2007) between the maize (Zea mays) Ubiquitin promoter and the nopaline synthase terminator. To generate RNAi lines, the PCR product that was designed to trigger RNAi was first subcloned into the pENTR entry vector (Invitrogen). PCR products were amplified using the following primer pairs: BU1:RNAi 5′-CACCGCTCGTGCCCTGTAGCTA-3′ and 5′-GGACGACTCTACTGCATCAAGGA-3′; BU1F:RNAi 5′-CACCGACGATGTCGAGCCGGAG-3′ and 5′-GCGGAGGCTGCGGATGATCTC-3′. Using the Gateway system, the fragment was cloned into the pANDA destination vector through a LR clonase reaction (Miki and Shimamoto, 2004; Miki et al., 2005). The rice plants were transformed according to the Agrobacterium-mediated method of Toki et al. (2006).
Quantitative Real-Time Reverse Transcription-PCR Analysis
After RNA extraction, first-strand cDNAs were synthesized from equal amounts of total RNA (1 μg/reaction) with a PrimeScript II first-strand cDNA synthesis kit (TaKaRa Bio) in a total volume of 20 μL, as described by the manufacturer. Synthesized cDNAs were used for real-time PCR. Real-time PCR was performed with Thermal Cycler Dice TP800 system (Takara Bio) using SYBR Premix Ex Taq II (Takara Bio). The following primer pairs were used for real-time PCR: BU1 5′-GTAGCCAGCTTGATCTCATCTC-3′ and 5′-GGGACGACTCTACTGCATCA-3′; RUBQ2 5′-GGTCATCCCGAGCCTCTGTT-3′ and 5′-GCAAATGAGCAAATTGAGCA-3′; 18S rRNA 5′-GGAAGGAGAAGTCGTAACAAGG-3′ and 5′-CAGGGTCACGACAATGATCC-3′; Os02g0747900 5′-TAGCACTAGGGGCCTAAGCA-3′ and 5′-AAGGGAGAGGGAAAAACCAA-3′; Os03g0171700 5′-GGTATGTACTGTACCAGGGCTAA-3′ and 5′-ATCTCTGAGCTTATCAAAGCAGCT-3′; Os10g0403800 5′-GGTCAAACTCGAAGCCCTTT-3′ and 5′-TCCATTCGATTGTCCGTAGA-3′.
Histochemical GUS Activity Assay
For analysis of BU1 expression, a 2.0-kb promoter region was amplified by PCR with primer pairs 5′-GGGGTACCCTCGACAATTTGACATGCCGTGCAT-3′ and 5′-GGTCTAGAGAAGCAGAAAGGGAGAGGGGGCA-3′, and cloned into the pSMAHdN632L-M2GUS vector between the KpnI and XbaI sites in front of the GUS gene. T1 plants were selected with 50 μg mL−1 hygromycin and used for GUS assay. GUS staining was performed according to the method of Kosugi et al. (1991). Transverse section and organs were observed under a LEITZ DMR light microscope (Leica) and a VHX-500 digital microscope (Keyence).
Lamina Joint Bending Assay
Detailed method is described in Fujioka et al. (1998). Seeds of transgenic plants and the wild type were dehusked, sterilized, and grown on half-strength Murashige and Skoog medium at 25°C under long-day conditions. After a week (when third leaf began to emerge), 1 μL of ethanol:DMSO (9:1, v/v) solution containing 0, 10, 100, and 1,000 ng BL was spotted on the lamina joint of the second leaf of seedlings. After 3 d, the angles of the lamina joint were measured.
Quantification of Endogenous BRs
Mature shoots (BU1:OX, 50 g fresh weight; vector control, 50 g fresh weight) grown in soil in an isolated greenhouse for 2 months were used for the experiment. The detailed method is described in Mori et al. (2002).
Subcellular Localization of BU1 Protein
The BU1 cDNA fragment was cloned into a multicloning site in frame at the C terminus of EGFP in the pSAT6-EGFP-C1 vector (Tzfira et al., 2005). This construct was introduced into coleoptiles of 5-d-old rice seedlings by particle bombardment method (Bio-Rad Biolistic PDS-1000/He particle delivery system, Bio-Rad; Heiser, 1992; Sanford et al., 1993). After incubation at 28°C for 24 h, GFP fluorescence was observed using a fluorescent microscope.
α-Amylase Induction Assay
Five embryoless half seeds per plate were dehusked and sterilized. These half seeds were placed on starch plates (0.2% [w/v] starch and 2% [w/v] agar) with or without 2 μm GA3. These plates were incubated for 5 d at 28°C in darkness. For detection of secreted amylase, iodine vapor was applied to plates (Ueguchi-Tanaka et al., 2000). Purple zones indicated unhydrolyzed starch, while clear zones (plaques, not turned to purple) indicated starch hydrolyzed by the amylase present in the half seeds.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_001063738 (BU1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Result of real-time PCR analyses of BU1.
Supplemental Figure S2. Close-up view of third node of culm in the wild type, BU1:OX-3, and BU1:OX-4.
Supplemental Figure S3. Role of BU1 in BR biosynthesis.
Supplemental Figure S4. Subcellular localization of eGFP-BU1 fusion protein.
Supplemental Figure S5. Expression level of BU1 and its homologs analyzed by real-time PCR.
Supplemental Figure S6. BU1 is not involved in GA signaling.
Supplementary Material
Acknowledgments
We thank Dr. Hidemi Kitano (Bioscience and Biotechnology center, Nagoya University) for providing the d61 mutant. We thank Dr. Motoyuki Ashikari (Bioscience and Biotechnology Center, Nagoya University) for providing T65d1 mutant. We thank Dr. Hiroaki Ichikawa (National Institute of Agrobiological Sciences, Japan), Dr. Ko Shimamoto (Nara Institute of Science and Technology, Japan), and Dr. Tzvi Tzfira (State University of New York) for kindly providing pRiceFox and pSMAHdN632L-M2GUS, pANDA, and pSAT6-EGFP, respectively. We thank Kyomi Shibata (Teikyo University, Japan) for quantification of BRs. We thank Akiko Hashimoto, Yumiko Yoshida, and Keiko Takeuchi (Institute of Society for Techno-Innovation of Agriculture, Forestry and Fisheries, Japan) for microarray assistance. We thank Haruko Onodera, Kazuko Ono, Satoru Maeda, Lois Ishizaki, Tomiko Senba, and Chiyoko Umeda (National Institute of Agrobiological Sciences, Japan) for advice and support in rice transformation and for overall technical assistance.
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences (to M.M. and T.A.), and by a Grant-in Aid from the Ministry of Education, Culture, Science and Technology of Japan (S0801019 to T.Y.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masaki Mori (morimasa@affrc.go.jp).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Arteca RN, Tsai DS, Schlagnhaufer C, Mandava NB (1983) The effects of brassinosteroid on auxin-induced ethylene production by etiolated mung bean segments. Physiol Plant 59: 539–544 [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J (2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulator of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Noguchi T, Takatsuto S, Yoshida S (1998) Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49: 1841–1848 [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser W (1992) Optimization of Biolistic Transformation Using the Helium-Driven PDS-1000/He System. Bio-Rad Bulletin 1688. Bio-Rad, Hercules, CA
- Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32: 495–508 [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y (1991) Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA-GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Res 19: 1571–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent V, Vervoort M (2001) The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res 11: 754–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, Soh MS (2006) Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, et al (2006) Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, et al (2002) Isolation and characterization of a rice dwarf mutant with defect in brassinosteroid biosynthesis. Plant Physiol 130: 1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M (2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al (1989) Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58: 537–544 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Fujioka S, Sunohara H, Kamiya N, Hong Z, Inukai Y, Miura K, Takatsuto S, Yoshida S, Ueguchi-Tanaka M, et al (2006) The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol 140: 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Hakata M, Amano K, Miyao A, Toki N, Kajikawa M, Pang J, Higashi N, Ando S, Toki S, et al (2007) A genome-wide gain-of function analysis of rice genes using the FOX-hunting system. Plant Mol Biol 65: 357–371 [DOI] [PubMed] [Google Scholar]
- Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33: 887–898 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki K, Inaba N, Kitagawa K, Fujioka S, Kitano H, Fujisawa Y, Kato H, Iwasaki Y (2009) Function of the alpha subunit of rice heteromeric G protein brassinosteroid signaling. Plant Cell Physiol 50: 161–172 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Sanford JC, Smith FD, Russell JA (1993) Optimizing the biolistic process for different biological applications. Methods Enzymol 217: 483–509 [DOI] [PubMed] [Google Scholar]
- Sato H, Imiya Y, Ida S, Ichii M (1996) Characterization of molybdenum cofactor mutant of rice, Oryza sativa L. Plant Sci 119: 39–47 [Google Scholar]
- Sekimata K, Kimura T, Kaneko I, Nakano T, Yoneyama K, Takeuchi Y, Yoshida S, Asami T (2001) A specific brassinosteroid biosynthesis inhibitor, Brz2001: evaluation of its effects on Arabidopsis, cress, tobacco, and rice. Planta 213: 716–721 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Steffens B, Sauter M (2009) Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 21: 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K (2002) The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 99: 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D (1991) Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 11: 5603–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, et al (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17: 776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) Dwarf and low-tillering, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58: 803–816 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97: 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozu S, Tanaka-Ueguchi M, Kitano H, Hattori K, Matsuoka M (2000) Characterization of XET-related genes of rice. Plant Physiol 122: 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Marumo S, Ikekawa N, Morisaki M, Mori K (1981) Brassinolide and homobrassinolide promotion of lamina inclination of rice seedling. Plant Cell Physiol 22: 323–325 [Google Scholar]
- Wang L, Xu YY, Ma QB, Li D, Xu ZH, Chong K (2006) Heterotrimeric G protein alpha subunit is involved in rice brassinosteroid response. Cell Res 16: 916–922 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20: 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Matsuoka M, Iwasaki Y, Komatsu S (2003) A novel brassinolide-enhanced gene identified by cDNA microarray is involved in the growth of rice. Plant Mol Biol 52: 843–854 [DOI] [PubMed] [Google Scholar]
- Yang G, Nakamura H, Ichikawa H, Kitano H, Komatsu S (2006) OsBLE3, a brassinolide-enhanced gene, is involved in the growth of rice. Phytochemistry 67: 1442–1454 [DOI] [PubMed] [Google Scholar]
- Yazaki J, Kojima K, Suzuki K, Kishimoto N, Kikuchi S (2004) The rice PIPELINE: a unification tool for plant functional genomics. Nucleic Acids Res 32: D383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi HC, Joo SJ, Nam KH, Lee JS, Kang BG, Kim WT (1999) Auxin and brassinosteroid differentially regulate the expression of the three members of the 1-aminocyclopropane-1-carboxylate synthase family in mung bean (Vigna radiata L.). Plant Mol Biol 41: 443–454 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Li L, Guo M, Chory J, Yin Y (2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X (2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA 106: 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.