Abstract
The conquest of the land by plants required dramatic morphological and metabolic adaptations. Complex developmental programs under tight regulation evolved during this process. Key regulators of plant development are phytohormones, such as cytokinins. Cytokinins are adenine derivatives that affect various processes in plants. The cytokinin signal transduction system, which is mediated via a multistep variant of the bacterial two-component signaling system, is well characterized in the model plant Arabidopsis (Arabidopsis thaliana). To understand the origin and evolutionary pattern of this signaling pathway, we surveyed the genomes of several sequenced key plant species ranging from unicellular algae, moss, and lycophytes, to higher land plants, including Arabidopsis and rice (Oryza sativa), for proteins involved in cytokinin signal transduction. Phylogenetic analysis revealed that the hormone-binding receptor and a class of negative regulators first appeared in land plants. Other components of the signaling pathway were present in all species investigated. Furthermore, we found that the receptors evolved under different evolutionary constraints from the other components of the pathway: The number of receptors remained fairly constant, while the other protein families expanded.
The transition of plants from the aquatic lifestyle to a terrestrial environment is one of the most important events in the evolution of life (Floyd and Bowman, 2007). The colonization of the land by plants and their subsequent diversification led to a series of global environmental changes, which resulted ultimately in the generation of many different habitats and thus were a prerequisite for the establishment of all modern terrestrial ecosystems (Waters, 2003). Based on molecular phylogeny, it has been estimated that the separation between the green algae lineage and the land plant lineage occurred about 725 million years ago (Zimmer et al., 2007). During the transition phase the first land plants faced a number of new challenges, such as a larger temperature range, increased UV radiation, desiccation, and higher gravity (Kenrick and Crane, 1997; Qiu et al., 2006). The plants adapted to the new environmental constraints by large changes in their morphology and their developmental programs, which are also reflected on the physiological and molecular level (Waters, 2003; Bowman et al., 2007). The accurate execution of such developmental programs is assured in part by plant hormones (Bishopp et al., 2006). Plant hormones are thought to have evolved from preexisting elements of the primary metabolism in algae (Kenrick and Crane, 1997). While plants hormones are present in macroalgae, much of their biochemistry, e.g. inactivation by conjugation, has developed only in higher plants (Stirk et al., 2003; Waters, 2003).
Cytokinin is a plant hormone and plays a crucial role in many fundamental processes such as shoot and root development, senescence, chloroplast development and pathogen defense in higher plants, and bud formation in lower plants (Mok and Mok, 2001; von Schwartzenberg et al., 2007). The signaling pathway of this hormone is mediated by a multistep His-to-Asp phospho-relay system, a variant of the bacterial two-component system (TCS; Supplemental Fig. S1). While this type of signaling system is widespread in prokaryotes, it is unique to plants among higher eukaryotes (West and Stock, 2001; Heyl and Schmülling, 2003). Most of the research on the cytokinin signaling pathway has been done using the model plant Arabidopsis (Arabidopsis thaliana; for review, see Bishopp et al., 2006; Heyl et al., 2006; Hwang and Sakakibara, 2006; To and Kieber, 2008). The current model predicts that cytokinin is bound by plasma membrane-localized His kinase receptors via the cyclases/histidine kinases associated sensory extracellular (CHASE) domain (Anantharaman and Aravind, 2001; Mougel and Zhulin, 2001; Heyl et al., 2007). The binding of the ligand leads to the autophosphorylation of the receptors. Subsequently the signal is transferred by phosphorylation of His phosphotransmitter proteins (HPts), which then translocated to the nucleus. Here the HPts activate type-B response regulators (RRs), which belong to the class of Myb-transcription factors. Consequently these transcription factors initiate the transcription of their target genes, one group of which are the type-A RRs. Type-A RR proteins have been shown to be involved in a negative feedback mechanism regulating the activity of the cytokinin signaling pathway (Hwang and Sheen, 2001; To et al., 2004). In Arabidopsis the cytokinin signaling pathway involves more than 30 proteins, leading to a high level of redundancy (Higuchi et al., 2004; Nishimura et al., 2004; To et al., 2004; Mason et al., 2005; Hutchison et al., 2006; Riefler et al., 2006).
In addition to canonical members of the cytokinin signaling pathway there is a group of genes called the type-C RRs (Mizuno, 2004). Originally members of this protein class were regarded as type-A RRs because both protein families share the same domain structure. However, several experiments have shown that in contrast to the type-A RR genes, type-C RR genes cannot be induced by cytokinin (Kiba et al., 2004; Mizuno, 2004; Gattolin et al., 2006). While some experiments hint that type-C RRs might play a role in the cytokinin signaling pathway (Kiba et al., 2004, 2005; Horak et al., 2008), it has yet to be firmly established. Several pseudo RRs have been identified in Arabidopsis, but were shown to play a role in the regulation of the circadian rhythm and not in cytokinin signaling (Mizuno and Nakamichi, 2005).
Following the completion of the rice (Oryza sativa) genome sequencing project, there have also been a few functional genomics approaches for the characterization of the TCS in rice (Ito and Kurata, 2006; Jain et al., 2006; Du et al., 2007). Furthermore, there have been reports on members of the cytokinin signaling pathway in other plants, especially in the cereal maize (Zea mays; Deji et al., 2002; Asakura et al., 2003; Doi et al., 2004; Giulini et al., 2004; Yonekura-Sakakibara et al., 2004; Sugawara et al., 2005).
The wealth of genomic resources provides the unique opportunity to address general questions about how a signaling pathway comes into existence, how proteins get recruited to the pathway, and how the different components including the signaling molecules evolved. The cytokinin signaling pathway offers a unique opportunity to study the evolution of signaling pathways in general for several reasons: (1) the signaling molecule, an N6-substituted adenine derivative, is very ancient, as it is present in very basal organisms such as bacteria (Barciszewski et al., 2007); (2) the different protein domains involved in this signaling pathway are also found in bacteria where they form the archetype of the two-component signaling system (West and Stock, 2001); (3) however, it seems that only in plants those different domains are put together in a way to establish the cytokinin signaling pathway as we can find it in higher plants. Thus the aim of this study was to unravel clues about the origin and the evolution of the signaling pathway of the plant hormone cytokinin by analyzing the genomes of different key plant species.
RESULTS
Selection of Species for Analysis of the Cytokinin Signaling Pathway
For a complete analysis involving all members of a particular pathway of a given species it is essential to have access to the full genome sequence. With this caveat in mind, we focused our analysis on the nuclear genomes of Ostreococcus tauri (Derelle et al., 2006), Chlamydomonas reinhardtii, as model species for unicellular green algae (Merchant et al., 2007), and Volvox carteri (http://genome.jgi-psf.org/Volca1/Volca1.home.html), the multicellular green algae, which serves as a model for the evolution of multicellularity and differentiation (Schmitt, 2001). The latter two algae belong to a sister group of the land plant lineage (Rodriguez-Ezpeleta et al., 2007). Four species of land plants were analyzed in this study. Phylogenetically basal land plants were represented by the moss Physcomitrella patens (Rensing et al., 2008) and the lycophyte Selaginella moellendorfii (http://genome.jgi-psf.org/Selmo1/Selmo1.home.html). Higher plants were represented by the monocot rice (Goff et al., 2002; Yu et al., 2002) and the dicot Populus trichocarpa (Tuskan et al., 2006). Arabidopsis was also included in the analysis, as it is the model plant for cytokinin signaling (for review, see Heyl et al., 2006; Hwang and Sakakibara, 2006; To and Kieber, 2008).
The Complete Set of Proteins of the Cytokinin Signaling Pathway First Appeared in the Basal Land Plant P. patens
The aim of this study was to identify all elements of the cytokinin signaling pathway in the investigated species. Thus the genomes of the selected species were screened using a Hidden Markov model (HMM) search (Eddy, 1998) for the protein domains present in known cytokinin signaling proteins (Supplemental Fig. S1). We first analyzed the selected genomes for proteins that were predicted to have a CHASE domain, as this protein domain has been shown to be responsible for the binding of the cytokinin ligand to a receptor (Heyl et al., 2007). We did not detect any CHASE domain-containing proteins encoded in the genomes of any of the algae species. All other species, including the moss P. patens, which represented the most basal land plant investigated in this study (Qiu et al., 2006), had at least two putative proteins containing this domain (Table I).
Table I.
Number of members of the cytokinin signaling system identified in the different investigated species
The table displays the number of predicted protein-coding sequences for all analyzed genomes and the number of domains identified by HMM search after the filtering step, which were used for the phylogenetic analysis for the different protein families.
| Species | Genes | CHASE | HPt | RR |
|---|---|---|---|---|
| O. tauri | 7,725 | 0 | 1 | 2 |
| C. reinhardtii | 15,143 | 0 | 1 | 4 |
| V. carteri | 15,544 | 0 | 1 | 3 |
| P. patens | 35,938 | 3 | 2 | 14 |
| S. moellendorffii | 22,285 | 2 | 2 | 7 |
| O. sativa | 66,710 | 4 | 2 | 23 |
| P. trichocarpa | 45,555 | 5 | 10 | 32 |
| A. thaliana | 27,855 | 3 | 5 | 22 |
In contrast, all investigated species, including the algal species, contained at least one gene that coded for a phosphotransmitter protein (HPt). The most basal species including the lower land plants, Physcomitrella and Selaginella, showed each two genes coding for this class of proteins. The genomes of the higher land plants showed a major increase in the number of HPts detected. The highest number was found in the poplar (Populus spp.) genome, for which 10 HPt genes were predicted (Table I).
A similar picture emerged for the RR genes. The genomes of the algae species contained only a few RR genes (two to four). In land plants, again starting with the moss P. patens, this gene family started to expand considerably and, similar to the other components investigated in this study, the poplar genome showed the greatest number of predicted proteins (n = 32) containing the RR domain (Table I). A noteworthy exception of the evolutionary pattern detected for the RR proteins was seen in Selaginella. Although this species is considered to be more advanced than Physcomitrella (Qiu et al., 2006), it showed a lower number of RRs.
The Phylogeny of the Cytokinin Receptor Points to Several Duplication Events
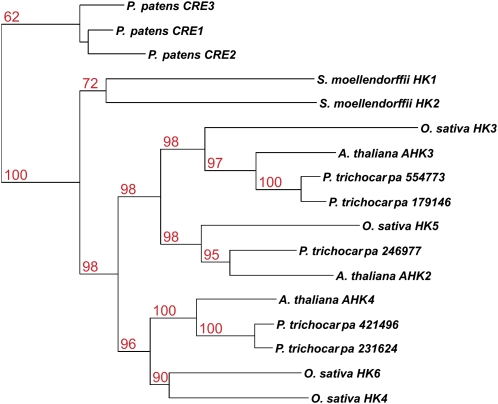
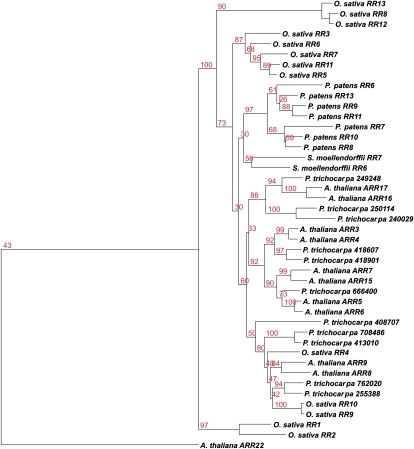
It was shown previously that the cytokinin receptors of maize, rice, and Arabidopsis are more similar to their respective homologs in the other species than to each other (Yonekura-Sakakibara et al., 2004; Ito and Kurata, 2006; Du et al., 2007). This suggested that the expansion of the CHASE domain-containing gene family occurred early in the evolution of this protein family. The inclusion of poplar in the phylogeny of cytokinin receptors further supported this hypothesis. The receptors could be divided into three branches, each containing one of the three Arabidopsis receptors. Within the three branches, duplications in rice and poplar might have led to a further expansion seen in the cytokinin receptor family (Fig. 1). However, the phylogeny of the three cytokinin receptors of Physcomitrella and the two Selaginella receptors was very different from that of the higher plants. Homologs within the two species were more similar to each other than to those of the other species (Fig. 1).
Figure 1.
Phylogenetic tree of CHASE domains in plants. Proteins containing a CHASE domain were identified by a HMM search. A phylogenetic tree of CHASE domains of those proteins was generated using the PHYML program. [See online article for color version of this figure.]
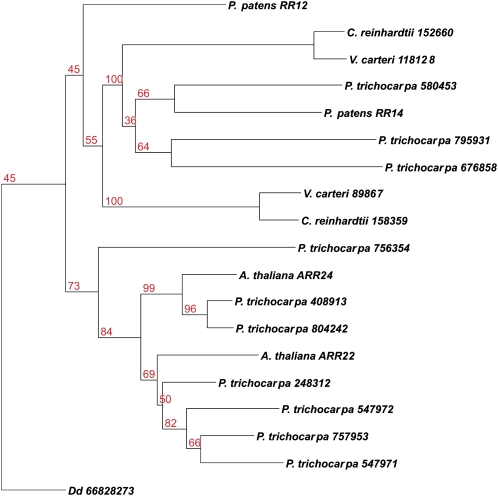
HPt Proteins Diverged Relatively Recently
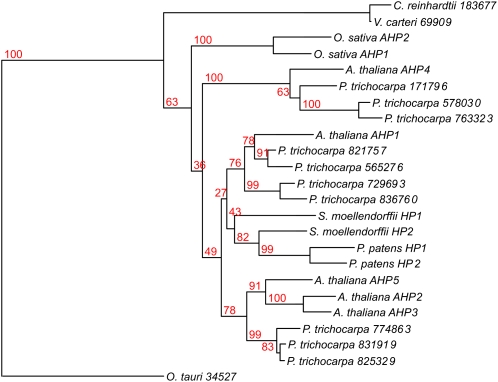
A phylogenetic analysis of the HPt proteins revealed an obvious segregation between the aqueous and land plants. The sequences of the phosphotransmitter proteins of the three algal species were clearly distinct from those of the land plants (Fig. 2). Also within the land plants there was a clear divide between the monocots as represented by the two HPts from the monocot species, rice, and the other plants. These HPts could be further separated into several different clades, two of which were populated by three highly homologous HPts of Arabidopsis and poplar, respectively. Two further branches contained one phosphotransmitter protein from Arabidopsis and three or four from poplar. Surprisingly, another branch that was embedded within the Arabidopsis/Populus clades consisted of the two HPts of Physcomitrella and Selaginella, respectively. As in the cytokinin receptors, the phosphotransmitter proteins of the basal land plants were more similar to each other than to those of any other plant species (Fig. 2). Nevertheless, one has to take into account that the bootstrap values for this subdivision of the clades were rather low in the majority of cases (Fig. 2).
Figure 2.
Phylogenetic tree of HPt domains in plants. Proteins containing a HPt domain were identified by a HMM search. Proteins containing additional domains were eliminated from the analysis. A phylogenetic tree of HPt domains was generated using the PHYML program. [See online article for color version of this figure.]
The Type-C RRs Represent the Most Ancient RR Group
The last group of proteins known to play a role in cytokinin signal transduction is the RR family. The RRs are classically divided into two subgroups based on their domain structure. The type-A RR contain just the RR domain. The type-B RRs contain a Myb domain in addition to the RR domain classifying them as transcription factors (Hwang et al., 2002). Recently a third subgroup was defined as type-C RRs. The type-C RRs have a domain structure similar to the type-A RRs, but their expression is not induced by cytokinin (Kiba et al., 2004; Mizuno, 2004; Horak et al., 2008). Therefore the role of type-C RRs in cytokinin signaling remains unclear.
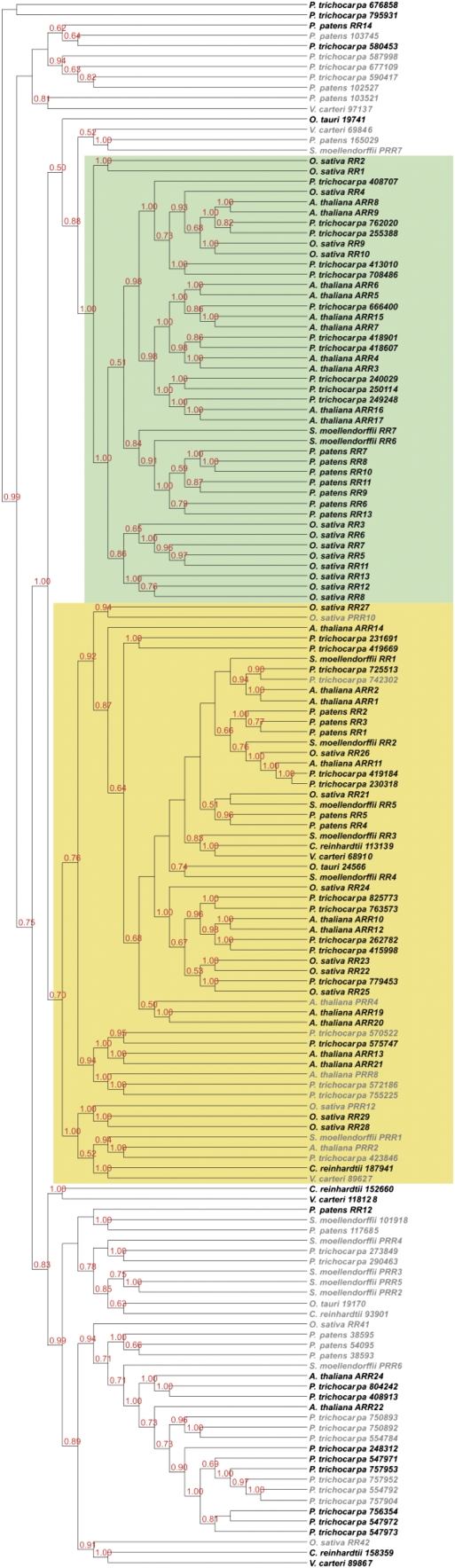
To understand the evolutionary development that led to the diversification of the RRs, we analyzed the RR domain of the all RRs of the different plant species under investigation (Table I). To avoid a distortion of the phylogenetic tree due to a possible omission of the large number of the pseudo RRs, they were also included in this analysis. The type-A and type-B RRs were clearly distinguished as both of them form monophyletic groups (Fig. 3; Table II). Type-B RRs were already found in the unicellular algae used in this study, O. tauri and C. reinhardtii. In contrast, the type-A RRs first appeared in the land plant species, mirroring the evolutionary pattern observed for the CHASE domain-containing His kinase receptors. The third group, the type-C RRs, was highly divergent and contained members from all species investigated with the exception of rice and Selaginella. Rice had previously been reported to contain two type-C RRs: OsRR41 and OsRR42 (Schaller et al., 2007). However, those two rice proteins, as well as a RR from Selaginella (Sm_7704), which also fall into the type-C RR clade, carry mutations of the canonical DDK motif, leading to their annotation as pseudo RRs.
Figure 3.
Phylogenetic tree of all RR domains. Proteins containing a RR domain were identified by a HMM search. Proteins containing additional domains other than a Myb domain were eliminated from the analysis. A phylogenetic tree of RR domains was generated using the MrBayes program. The clade of the type-A RRs is highlighted in green and the clade of the type-B RRs is highlighted in yellow.
Table II.
Number of members of the different RRs classes identified in the different investigated species
The table displays the number RR proteins identified by HMM search and the separation in different groups based on their position in the phylogenetic tree (Fig. 2).
| Species | Total RRs | Type A | Type B | Type C |
|---|---|---|---|---|
| O. tauri | 2 | – | 1 | 1 |
| C. reinhardtii | 4 | – | 2 | 2 |
| V. carteri | 3 | – | 1 | 2 |
| P. patens | 14 | 7 | 5 | 2 |
| S. moellendorffii | 7 | 2 | 5 | – |
| O. sativa | 23 | 14 | 9 | – |
| P. trichocarpa | 32 | 11 | 11 | 10 |
| A. thaliana | 23 | 10 | 11 | 2 |
The Different Types of RRs Seemed to Have Emerged at Different Time Points during Evolution
Members of the type-B RR family were found in all organisms investigated. However the monophyly of type-B RR tree and the low number of pseudo RRs, especially when compared to the type-C RRs, are indicative of a relatively recent divergence (Fig. 3). In Arabidopsis the type-B RRs have been divided into three subgroups based on sequence comparison and expression patterns. Two of those subdivisions are represented by the pairs ARR13/ARR21 and ARR19/ARR20 (Mason et al., 2004; Tajima et al., 2004). Our phylogenetic tree of the type-B RRs produced a similar picture where the type-B RRs of the big subgroup of Arabidopsis clustered together with a high number of type-B RRs from other species. All other type-B RRs were quite divergent to each other, and pairs were only detected for ARR13/ARR21, ARR19/ARR20, and OsRR28/OsRR29, respectively (Fig. 4).
Figure 4.
Phylogenetic tree of type-B RRs. Using the type-B RRs identified (Fig. 3), a tree was generated using the PHYML program. It was based on a full-length sequence alignment of type-B RRs, optimized with the Gblocks program. The type-C RR ARR22 from Arabidopsis was used as outgroup. [See online article for color version of this figure.]
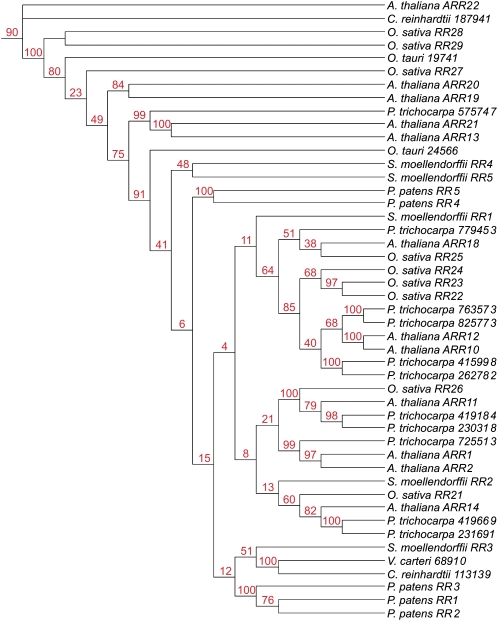
The phylogenetic tree of the type-A RRs could be divided into several branches. Of these clades, one consists of most of the type-A RRs of rice and one is formed by the members of this protein family from Physcomitrella and Selaginella. The type-A RRs from the two dicots Arabidopsis and poplar are interspersed in different clades (Fig. 5).
Figure 5.
Phylogenetic tree of type-A RRs. Using the type-A RRs identified (Fig. 3), a tree was generated using the PHYML program. The tree was based on the RR domain sequences only. The type-C RR ARR22 from Arabidopsis was used as an outgroup. [See online article for color version of this figure.]
Among the RRs, the type-C RRs were the most diverse group. One or two type-C RR was present already in the most basal species used in this study, the green algae. As there was no clear phylogenetic relationship detectable, possibly this group of RRs diverged a long time ago and thus is the most ancient of the RRs (Fig. 6). This hypothesis is further supported by the fact that most pseudo RRs were found among the true RRs of the type C (Fig. 3).
Figure 6.
Phylogenetic tree of type-C RRs. Using the type-C RRs identified (Fig. 3), a tree was generated using the PHYML program. The tree was based on the RR domain sequences only. [See online article for color version of this figure.]
DISCUSSION
The phytohormone cytokinin is involved in many developmental processes in plants and participates in physiological responses to numerous environmental stimuli (Mok et al., 2005). Therefore the evolution of cytokinin signal transduction was likely to be important to plants during the conquest of land. Cytokinin has not only been detected in virtually all plants, ranging from algae to land plants (Stirk et al., 2003), but also in other organisms (Barciszewski et al., 2007). However, the pathway by which the signal is perceived and transmitted has been studied in detail only in Arabidopsis and rice (Heyl et al., 2006; Hwang and Sakakibara, 2006; To and Kieber, 2008). The cytokinin signal transduction offers a unique opportunity to study the establishment and evolution of a signaling pathway and this study shows: (1) P. patens is the most basal plant species containing all the components necessary for cytokinin signaling, and (2) the cytokinin receptor and the other components of this pathway evolve differently.
Common Patterns in the Evolution of Cytokinin Signaling
Our analysis of the components of the cytokinin signaling pathway revealed several interesting aspects of the evolution of this pathway. The most surprising result was that both the cytokinin receptors and the type-A RRs appeared simultaneously in the most basal land plant investigated. In contrast to the other components of the cytokinin signaling pathway, type-A RRs negatively regulate the cytokinin signaling pathway (Hwang and Sheen, 2001; To et al., 2004). While the necessity of the receptor at the beginning of the signaling chain is obvious, our results indicate that a negative regulator is equally important. Possibly, this new signaling pathway could only be established once a negatively acting factor was in place to allow the fine tuning of the signal and to reset the system, so that the original status is reestablished and the pathway is again able to perceive and transduce the signal.
The other components of the cytokinin signal transduction pathway are the HPts and the type-B RRs. The HPts are ubiquitously present in a wide range of organisms from bacteria to eukaryotes (Zhang and Shi, 2005). By contrast the type-B RRs with their signature combination of RR and Myb DNA-binding domains are plant specific (Riano-Pachon et al., 2008). Type-B RRs form a monophyletic group as do the type-A RRs. However, unlike the type-A RRs, type-B RRs are present in all species investigated here. Because type-B RRs were also present in species without either a CHASE domain-containing cytokinin receptor or type-A RRs, type-B RRs might have originally performed some plant-specific function that could have extended or changed to cytokinin signaling.
Cytokinin Receptors Are Under Different Evolutionary Constraints from Other Signaling Components of the Pathway
Most gene families of the cytokinin signaling pathway components are expanded in higher plants. In contrast, the cytokinin receptor gene family was always restricted to two to five members. Even P. patens, the most early diverging species encoding cytokinin receptors in its genome, contains three receptor genes, the same number as Arabidopsis (Table I). This raises the question why their numbers did not increase in the same manner as that of the other components. To gain insight into the forces that drove the evolution of this class of receptor His kinases, it is helpful to consider the evolutionary pattern of the TCS in bacteria. In prokaryotes duplication events can separate the His kinase receptors from their cognate RRs and subsequently both the kinase and the RR evolve at a different pace compared to each other (Alm et al., 2006). A similar mechanism can be envisioned for the His kinase receptors and downstream TCS components, resulting in the altered rate in evolution detected for the cytokinin receptor versus for the other proteins of this signaling pathway. Nevertheless the question remains why there is no increase in the number of receptors, especially as the number of downstream components increases at the same time. An expansion in the number of sequenced genomes of early diverging land plants and algae in the near future might help to address this question.
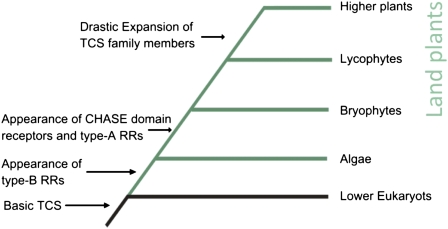
Model for the Evolution of Cytokinin Signal Transduction
Taken together the results of this study allow us to envision a model by which the cytokinin signaling pathway of higher plants came into existence (Fig. 7). The different factors of the TCS, the His kinases, RRs, and His phosphotransfer proteins, were most likely acquired by early eukaryotes via horizontal gene transfer from bacteria (Anantharaman et al., 2007). Because the TCS is shared between plants and fungi but not animals, it is not clear whether the pathway was acquired multiple times or whether it was lost in animals. The latter scenario is supported by the fact that the genomes of several early diverging eukaryotes such as Saccharomyces cerevisiae or Dictyostelium discoideum also contain TCS components (Goffeau, 1996; Eichinger et al., 2005).
Figure 7.
Model of the evolution of the cytokinin signaling pathway.
The origin of a CHASE domain-containing His kinase receptor in this signaling pathway is more puzzling than that of the other components. While the other members of the signaling pathway are found in all plants and in many unicellular eukaryotes, the CHASE domain seemed to have appeared only once the plants conquered the land. To better understand how and when this protein domain was acquired, a PSI-BLAST search against GenBank's nonredundant protein database using the CHASE domain of the cytokinin receptor CRE1/AHK4 of Arabidopsis as a query was conducted. This analysis showed that all CHASE domains of plants are found in one clade clearly distinct from any other groups (Supplemental Fig. S2), indicating a monophyletic origin of the cytokinin receptors of plants. The only known nonplant eukaryote encoding a CHASE domain-containing His kinase receptor is D. discoideum. While this amoeba species apparently uses cytokinin in the regulation of sporulation, this receptor seems not to play a role in cytokinin signaling (Anjard and Loomis, 2008) and thus is more likely part of a different pathway. If the genome of the last common ancestor of amoeba and algae encoded such a receptor, the noncytokinin pathway might not be relevant for algae and therefore the CHASE domain His kinase could have been lost over time in these early plants. However, in response to a new set of abiotic stresses, the landfall of plants led to large-scale morphological and physiological changes. The resulting, more elaborate developmental programs would have required a new or more complex regulation by phytohormones, such as cytokinin. Therefore a CHASE domain-containing His kinase could have been acquired and used in the signaling of this hormone. One possible vector could have been a virus. In this context it is interesting to note that the genome of the virus Ectocarpus siliculosus virus-1, which integrates into the genome of its brown algae host, Ectocarpus siliculosus, encodes a CHASE domain-containing His kinase (PFAM database; Delaroque et al., 2001). Alternatively, the CHASE domain could have been lost only in the algal lineage investigated in this study, and these algae are only a sister lineage to the charaphytes that eventually gave rise to the land plants (Rodriguez-Ezpeleta et al., 2007). Therefore it is conceivable that in those algae, ancestral to land plants, a CHASE domain-containing His kinase was preserved. Sequencing of charaphyte genomes would allow this question to be addressed.
The RRs and the HPts were present in all species investigated and thus might have been acquired early in plant evolution during the endosymbiosis of cyanobacteria (Archibald, 2006). As the type-B RRs are specific to plants and are present in the basal green algae, they might have originally performed a function in the regulation of photosynthesis and later been recruited for the cytokinin signal transduction. In this respect it is noteworthy that cytokinin is involved in chlorophyll retention and chloroplast development (Mok and Mok, 2001). Furthermore it was shown that type-B RRs are only found in photoauxotropic species (Riano-Pachon et al., 2008). The type-A RRs might have evolved from a type-C RR by mutations in their promoters, enabling the binding of cytokinin-activated type-B RRs and other cytokinin-regulated transcription factors, such as the cytokinin response factors (Rashotte et al., 2006), thus turning the type-A RRs into primary response genes for cytokinin. This model of the evolution of the RRs, with the type-C RR being the oldest and type-A RRs being the youngest subgroup, is also supported by the fact that most pseudo RRs have been found in the type-C RRs, while fewer were found in the clade of the type-B RRs, and none were found in the type-A RR branch of the phylogenetic tree (Fig. 3).
In this way, all the components of the cytokinin signaling pathway might have been recruited at different time points during evolution to give rise to the signaling system found in land plants (Fig. 7). A similar scenario highlighting the importance of the conquest of land by plants for the phytohormone signaling was previously postulated not only for cytokinin, but also for auxin and abscisic acid (Rensing et al., 2008). However, one must keep in mind that cytokinin signaling has been investigated in great detail only in Arabidopsis and rudimentarily in rice and poplar, but in none of the other species analyzed. Thus, this study presents a first step toward understanding the evolution of this signaling pathway. Experimental characterization of the cytokinin signaling proteins from other taxa is needed to shed more light on the evolution of this signaling pathway. Furthermore, the availability of more genomic data, especially of key organisms such as Marchantia polymorpha or Mesostigma viride (Qiu et al., 2006) will help to clarify the evolutionary processes that lead ultimately to the cytokinin signaling pathway as it is found in the land plants.
MATERIALS AND METHODS
Data Sources and Preprocessing
Protein sequences of completely sequenced plant genomes were retrieved from the following databases: for Arabidopsis (Arabidopsis thaliana; release version 8) and rice (Oryza sativa; release version 5) protein sequences were downloaded from The Institute for Genomic Research (www.tigr.org). Predicted protein sequences from the genome of Physcomitrella patens (version 1.2), Populus trichocarpa (version 1.1), Chlamydomonas reinhardtii (version 3.1), Ostreococcus tauri (version 2.0), Volvox carteri (version 1.0), and Selaginella moellendorfii (version 1.0) were obtained from the Joint Genome Institute (www.jgi.doe.gov).
Family members of the cytokinin signaling pathway were identified by HMM search (HMMER 2.3.2; Eddy, 1998) using the family-specific domains obtained from the Pfam database (Finn et al., 2006). Cytokinin receptors were searched for the CHASE domain (HMM acc: PF03914.3), HPts for the HPt domain (HMM acc PF01627.13), and RRs for the RR domain (HMM acc PF00072.13). This initial HMM search was followed by a combined automatic and manual filtering step to eliminate falsely identified TCS members, alternative splice variants, and nonfunctional sequences. In the case of the RRs, all sequences containing any other protein domains than the RR or Myb domain, such as the CCT or HisKA domains (that might belong to unrelated pathways such as circadian rhythm; Mizuno, 2004), were removed. Alternative splice variants, which are often a result of the different gene prediction programs used, were identified by the presence of identical domains in the same species, in which case the shorter protein version was eliminated after manual inspection of the sequences. However, we are aware that proteins with splicing variants within the identified domain, such as in the case of OsRR9-2, or unclear annotation, as in the case of Sm_101918, might not be eliminated from the alignment using the above outlined method. As we cannot exclude the possibility that similar problems will also occur in other, freshly annotated genomes and we did not want to introduce a bias into our analysis, those two sequences were also kept in the analysis. The rice sequence Os12g26940 was removed from the dataset because it contains a protein kinase domain in addition to the CHASE, which is not observed in any other genome, and it lacks the HisKA, HATPase_c, and RR domains. Nonfunctional sequences or pseudogenes among the HPts and RRs were identified by scanning the protein sequences for known functional amino acids—the XHQXKGSSXS motif for the HPt proteins and the DDK motif for the RRs according to Hwang et al. (2002). The protein identifiers and domain positions of proteins used in the phylogenetic analysis are shown in Supplemental Table S1.
Phylogenetic Analysis
For each protein family, multiple sequences alignments were generated with Muscle (Edgar, 2004). The phylogenetic relationships were analyzed with two different methods, a maximum likelihood and Bayesian approach, which produced very similar results. The maximum likelihood trees were built with PHYML (Guindon and Gascuel, 2003) using the JTT substitution model (Jones et al., 1992) and four substitution rate categories. Support for each node was gained by nonparametric bootstrap analysis from 100 pseudo replicates. Bayesian analysis was carried out with MrBayes (Huelsenbeck and Ronquist, 2001) under the VT substitution model (Müller and Vingron, 2000), using γ-distributed rates approximated by four rate categories. The analysis was run for at least 200,000 generations. A consensus tree was generated from trees sampled after one-third of the run, when the likelihood estimates had reached stationarity.
For the subtype-specific analysis of RRs, sequences were classified according to the tree of RR domains (Fig. 3). In the case of type-A and type-B RRs, the type-C RR from Arabidopsis, ARR22, was used as an outgroup. A RR from Dictyostelium discoideum (gi:66828273) identified by BLAST search with At_ARR24 (Evalue = 2e-14) was used as an outgroup for type-C RRs. Full-length alignments were optimized with Gblocks because poorly aligned regions, found for example in the domain-flanking regions, often hinder phylogenetic analysis (Talavera and Castresana, 2007). The following settings were used to extract conserved blocks from the alignment with Gblocks: The minimum number of sequences for conserved and flanking positions was set to 50%, the minimum length of a block to two, and the maximum number of contiguous nonconserved positions to 25%. Our method searched for RRs that only contain a RR or the RR and the Myb DNA-binding domain. Sequences lacking a start or stop codon could have contained additional domains in the missing sequence and could have misleadingly appeared in our dataset. Absent start and stop codons were especially abundant among poplar (Populus spp.) sequences and those sequences should be considered with care.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Model of the cytokinin signal transduction pathway.
Supplemental Figure S2. Maximum likelihood tree of the CHASE domains from bacteria and eukaryotes.
Supplemental Table S1. Identifier and domain information of the different proteins used in the phylogenetic analysis.
Supplementary Material
Acknowledgments
Thanks to Thomas Schmülling, Jeffrey Oliver, and the members of the Heyl group for reading and commenting on the manuscript. We especially want to thank Anahid Powell, whose discussions and suggestions greatly helped to improve the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alexander Heyl (heyl@zedat.fu-berlin.de).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alm E, Huang K, Arkin A (2006) The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLOS Comput Biol 2: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L (2001) The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci 26: 579–582 [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L (2007) Comparative genomics of protists: new insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol 61: 453–475 [DOI] [PubMed] [Google Scholar]
- Anjard C, Loomis WF (2008) Cytokinins induce sporulation in Dictyostelium. Development 135: 819–827 [DOI] [PubMed] [Google Scholar]
- Archibald JM (2006) Algal genomics: exploring the imprint of endosymbiosis. Curr Biol 16: 1033–1035 [DOI] [PubMed] [Google Scholar]
- Asakura Y, Hagino T, Ohta Y, Aoki K, Yonekura-Sakakibara K, Deji A, Yamaya T, Sugiyama T, Sakakibara H (2003) Molecular characterization of His-Asp phosphorelay signaling factors in maize leaves: implications of the signal divergence by cytokinin-inducible response regulators in the cytosol and the nuclei. Plant Mol Biol 52: 331–341 [DOI] [PubMed] [Google Scholar]
- Barciszewski J, Massino F, Clark BF (2007) Kinetin-a multiactive molecule. Int J Biol Macromol 40: 182–192 [DOI] [PubMed] [Google Scholar]
- Bishopp A, Mähönen AP, Helariutta Y (2006) Signs of change: hormone receptors that regulate plant development. Development 133: 1857–1869 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K (2007) Green genes—comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Deji A, Sakakibara H, Okumura S, Matsuda T, Ishida Y, Yamada S, Komari T, Kubo T, Yamaya T, Sugiyama T (2002) Accumulation of maize response regulator proteins in mesophyll cells after cytokinin treatment. Biosci Biotechnol Biochem 66: 1853–1858 [DOI] [PubMed] [Google Scholar]
- Delaroque N, Muller DG, Bothe G, Pohl T, Knippers R, Boland W (2001) The complete DNA sequence of the Ectocarpus siliculosus Virus EsV-1 genome. Virology 287: 112–132 [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, et al (2006) Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA 103: 11647–11652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18: 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Jiao F, Chu J, Jin G, Chen M, Wu P (2007) The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics 89: 697–707 [DOI] [PubMed] [Google Scholar]
- Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34: D247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL (2007) The ancestral developmental tool kit of land plants. Int J Plant Sci 168: 1–35 [Google Scholar]
- Gattolin S, Alandete-Saez M, Elliott K, Gonzalez-Carranza Z, Naomab E, Powell C, Roberts JA (2006) Spatial and temporal expression of the response regulators ARR22 and ARR24 in Arabidopsis thaliana. J Exp Bot 57: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D (2004) Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Goffeau A (1996) 1996: a vintage year for yeast and Yeast. Yeast 12: 1603–1605 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Heyl A, Schmülling T (2003) Cytokinin signal perception and transduction. Curr Opin Plant Biol 6: 480–488 [DOI] [PubMed] [Google Scholar]
- Heyl A, Werner T, Schmülling T (2006) Cytokinin metabolism and signal transduction. In P Hedden, S Thomas, eds, Plant Hormone Signaling, Vol 24. Blackwell Publishing, Oxford, pp 93–123
- Heyl A, Wulfetange K, Pils B, Nielsen N, Romanov GA, Schmülling T (2007) Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain. BMC Evol Biol 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J, Grefen C, Berendzen KW, Hahn A, Stierhof YD, Stadelhofer B, Stahl M, Koncz C, Harter K (2008) The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sakakibara H (2006) Cytokinin biosynthesis and perception. Planta 126: 528–538 [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kurata N (2006) Identification and characterization of cytokinin-signalling gene families in rice. Gene 382: 57–65 [DOI] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2006) Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389: 33–39 [Google Scholar]
- Kiba T, Aoki K, Sakakibara H, Mizuno T (2004) Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol 45: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE (2005) Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T (2004) Plant response regulators implicated in signal transduction and circadian rhythm. Curr Opin Plant Biol 7: 499–505 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nakamichi N (2005) Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol 46: 677–685 [DOI] [PubMed] [Google Scholar]
- Mok DW, Mok MC (2001) Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 52: 89–118 [DOI] [PubMed] [Google Scholar]
- Mok MC, Martin RC, Dobrev PI, Vankova R, Ho PS, Yonekura-Sakakibara K, Sakakibara H, Mok DWS (2005) Topolins and hydroxylated thidiazuron derivatives are substrates of cytokinin O-glucosyltransferase with position specificity related to receptor recognition. Plant Physiol 137: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel C, Zhulin IB (2001) CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci 26: 582–584 [DOI] [PubMed] [Google Scholar]
- Müller T, Vingron M (2000) Modeling amino acid replacement. J Comput Biol 7: 761–776 [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al (2006) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA 103: 15511–15516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Riano-Pachon DM, Correa LG, Trejos-Espinosa R, Mueller-Roeber B (2008) Green transcription factors: a Chlamydomonas overview. Genetics 179: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Philippe H, Brinkmann H, Becker B, Melkonian M (2007) Phylogenetic analyses of nuclear, mitochondrial, and plastid multigene data sets support the placement of Mesostigma in the Streptophyta. Mol Biol Evol 24: 723–731 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Doi K, Hwang I, Kieber JJ, Khurana JP, Kurata N, Mizuno T, Pareek A, Shiu SH, Wu P, et al (2007) Nomenclature for two-component signaling elements of rice. Plant Physiol 143: 555–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt R (2001) Volvox carteri: molecular genetics of cell differentiation. Jap J Protozool 34: 7–12 [Google Scholar]
- Stirk WA, Novak O, Strnad M, van Staden J (2003) Cytokinins in macroalgae. Plant Growth Regul 41: 13–24 [Google Scholar]
- Sugawara H, Kawano Y, Hatakeyama T, Yamaya T, Kamiya N, Sakakibara H (2005) Crystal structure of the histidine-containing phosphotransfer protein ZmHP2 from maize. Protein Sci 14: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577 [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Kieber JJ (2008) Cytokinin signaling: two-components and more. Trends Plant Sci 13: 85–92 [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg K, Nunez MF, Blaschke H, Dobrev PI, Novak O, Motyka V, Strnad M (2007) Cytokinins in the bryophyte Physcomitrella patens: analyses of activity, distribution, and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins. Plant Physiol 145: 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER (2003) Molecular adaptation and the origin of land plants. Mol Phylogenet Evol 29: 456–463 [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM (2001) Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26: 369–376 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H (2004) Molecular characterization of cytokinin-responsive histidine kinases in maize: differential ligand preferences and response to cis-zeatin. Plant Physiol 134: 1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
- Zhang W, Shi L (2005) Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology 151: 2159–2173 [DOI] [PubMed] [Google Scholar]
- Zimmer A, Lang D, Richardt S, Frank W, Reski R, Rensing SA (2007) Dating the early evolution of plants: detection and molecular clock analyses of orthologs. Mol Genet Genomics 278: 393–402 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.