Abstract
Glycolysis is a central metabolic pathway that, in plants, occurs in both the cytosol and the plastids. The glycolytic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate with concomitant reduction of NAD+ to NADH. Both cytosolic (GAPCs) and plastidial (GAPCps) GAPDH activities have been described. However, the in vivo functions of the plastidial isoforms remain unresolved. In this work, we have identified two Arabidopsis (Arabidopsis thaliana) chloroplast/plastid-localized GAPDH isoforms (GAPCp1 and GAPCp2). gapcp double mutants display a drastic phenotype of arrested root development, dwarfism, and sterility. In spite of their low gene expression level as compared with other GAPDHs, GAPCp down-regulation leads to altered gene expression and to drastic changes in the sugar and amino acid balance of the plant. We demonstrate that GAPCps are important for the synthesis of serine in roots. Serine supplementation to the growth medium rescues root developmental arrest and restores normal levels of carbohydrates and sugar biosynthetic activities in gapcp double mutants. We provide evidence that the phosphorylated pathway of Ser biosynthesis plays an important role in supplying serine to roots. Overall, these studies provide insights into the in vivo functions of the GAPCps in plants. Our results emphasize the importance of the plastidial glycolytic pathway, and specifically of GAPCps, in plant primary metabolism.
Glycolysis is a central metabolic pathway that is present, at least in part, in all living organisms. Its fundamental role is to oxidize hexoses to generate ATP, reducing power and pyruvate, and to produce precursors for anabolism (Plaxton, 1996). In recent years, additional nonglycolytic functions such as regulation of transcription or apoptosis have also been attributed to glycolytic enzymes (Kim and Dang, 2005). In plants, glycolysis occurs in both the cytosol and the plastids (Plaxton, 1996; Fig. 1). It is thought that each glycolytic pathway is involved in the generation of specific products of the primary metabolism, although the contribution and the degree of integration of both pathways are not well known. The plastidic and cytosolic glycolytic pathways interact through highly selective transporters present in the inner plastid membrane (Weber et al., 2005), which may suggest that key glycolytic intermediates are fully equilibrated in both compartments. In addition, the functions of the glycolytic pathway in plastids of heterotrophic tissues (amyloplasts and leucoplasts) and in the chloroplasts might be very different. Because of this, it is difficult to define the contribution of each glycolytic enzyme and pathway to the primary metabolism of plant cells using only biochemical approaches. Direct molecular or genetic evidence corroborating the in vivo contribution of glycolytic enzymes to specific primary metabolite production is generally lacking.
Figure 1.
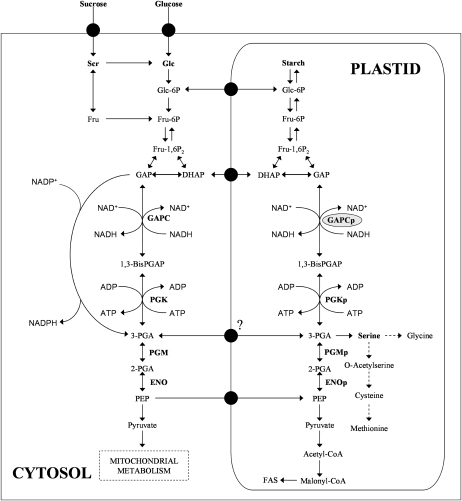
Schematic representation of glycolysis in a plant cell. Emphasis is given to the plastidic and cytosolic glycolytic reactions catalyzed by GAPDH (GAPC, cytosolic isoform; GAPCp, plastidial isoform) and phosphoglycerate kinase (PGK, cytosolic isoform; PGKp, plastidial isoform). 1-3BisPGAP, 1,3-Bisphosphoglycerate; DHAP, dihydroxyacetone phosphate; ENO, enolase; FAS, fatty acid synthesis; 2-PGA, 2-phosphoglycerate; PGM, phosphoglycerate mutase. Broken lines indicate several enzymatic reactions. Adapted from Schwender et al. (2003).
The glycolytic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reversibly converts the glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate by coupling with the reduction of NAD+ to NADH (Fig. 1). In addition to the cytosolic NAD+-specific GAPDH (GAPC), another NAD+-dependent GAPDH (GAPCp) biochemical activity has been described in the plastids of land plants (Meyer-Gauen et al., 1994; Backhausen et al., 1998). GAPCp arose through gene duplication from cytosolic GAPC in early chloroplast evolution (Petersen et al., 2003). Recently, the regulation and functional characterization in vivo of the cytosolic plant GAPC has been reported (Hajirezaei et al., 2006; Holtgrefe et al., 2008; Rius et al., 2008). However, little is known about the physiological function and relevance of the plastidic GAPCps. Petersen et al. (2003) suggested that GAPCp plays a specific role in glycolytic energy production in nongreen plastids and that it is absent in the chloroplasts of angiosperms. Backhausen et al. (1998) postulated that GAPCp would be essential for starch metabolism during the dark period in green and nongreen plastids. GAPCp, along with the phosphoglycerate kinase, was shown to be involved in the production of ATP needed for starch metabolism (Backhausen et al., 1998).
An important function of chloroplast glycolysis in the dark and in nongreen plastids is to participate in starch breakdown for energy production but also to generate primary metabolites for anabolic pathways such as fatty acid and amino acid synthesis (Plaxton, 1996; Ho and Saito, 2001; Andre et al., 2007; Baud et al., 2007). The strategic location of GAPCps in the glycolytic pathway could make them important players in one or all of the processes described above. Theoretically, the plastidic glycolytic enzymatic reactions between the triose phosphate pool (glyceraldehyde-3-phosphate and dihydroxyacetone phosphate) and phosphoenolpyruvate (PEP) are reversible (Buchanan et al., 2000). In addition, translocators of triose phosphate, 3-phosphoglycerate (3-PGA), and PEP that could equilibrate the pools of these metabolites between the cytosol and the plastid have been described (Flügge, 1999; Fischer and Weber, 2002; Knappe et al., 2003). All of these facts make it difficult to predict the consequences of knocking out the plastidic GAPCps for plant metabolism and development.
Here, we investigate the role of the Arabidopsis (Arabidopsis thaliana) GAPCps by a gain- and-loss-of-function approach. We have obtained gapcp double mutants that show a drastic phenotype of root developmental arrest. The mutants have impaired sugar and amino acid accumulation patterns as compared with wild-type plants. We present genetic evidence that GAPCp has an important role in plant development by affecting the Ser supply to roots. An important function for GAPCp in providing substrates for the phosphorylated pathway of Ser biosynthesis in nonphotosynthetic organs is proposed. The relationship between carbon and nitrogen metabolism is also discussed.
RESULTS
Identification and Expression Analysis of Plastid-Localized GAPDHs
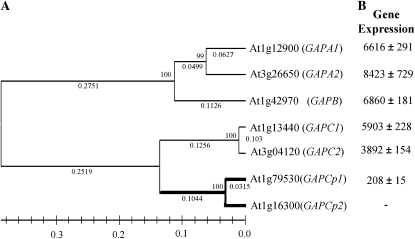
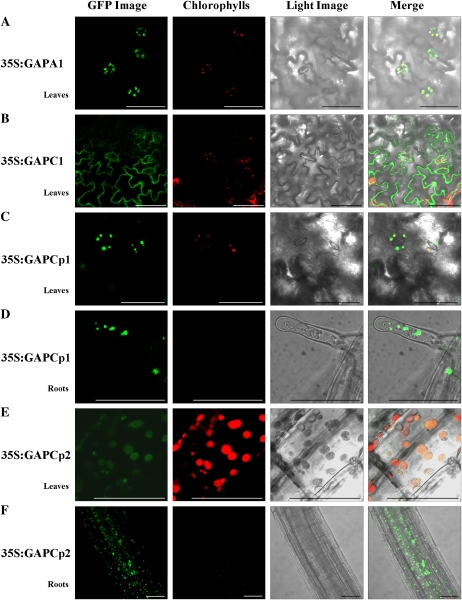
In The Arabidopsis Information Resource database (http://www.arabidopsis.org), we found a family of seven genes encoding putative phosphorylating GAPDHs (Fig. 2A). Four of them (GAPC1, GAPC2, GAPCp1, and GAPCp2) encode for glycolytic enzymes, and three of them (GAPA1, GAPA2, and GAPB) participate in the photosynthetic reductive carbon cycle. According to the ChloroP prediction server (http://www.cbs.dtu.dk/services/ChloroP/), two of the glycolytic isoforms, GAPCp1 (At1g79530) and GAPCp2 (At1g16300), showing 93% amino acid identity, have a putative N-terminal plastid/chloroplast localization signal. To investigate the subcellular localization of GAPCp1 and GAPCp2, we stably expressed both C- and N-terminal GFP fusion protein constructs in Arabidopsis. Both constructs complemented the gapcp double mutant (see below), although the C-terminal GAPCp-GFP fusion showed more efficient complementation compared with the N-terminal GFP-GAPCp protein fusion. In leaves, GAPCp1 and GAPCp2 could be localized in both chloroplasts and nongreen plastids (Fig. 3, C–F). In roots, GAPCp1 and GAPCp2 showed a plastidial localization (Fig. 3, D and F).
Figure 2.
Phylogenetic tree of the Arabidopsis phosphorylating GAPDH proteins, and gene expression levels. A, The phylogenetic tree was constructed from an alignment of deduced amino acid sequences as described in “Materials and Methods.” The bootstrap value from 100,000 replicates is given at each node. The scale shows the length of the maximum possible bootstrap value (100). Branch length is given under each segment according to the algorithm specified in “Materials and Methods.” B, Gene expression levels of the Arabidopsis GAPDH isoforms based on microarray experiments performed on 15-d-old seedlings.
Figure 3.
Subcellular localization of GAPDH isoforms by stable expression of GFP-GAPDH fusion proteins in Arabidopsis. A and B, Localization of chloroplastic and cytosolic isoforms GAPA1 and GAPC1. C and D, Chloroplastic/plastidic localization of GAPCp1 in leaves (C) and roots (D). E and F, Chloroplastic/plastidic localization of GAPCp2 in leaves (E) and roots (F). GFP fluorescence, chlorophyll fluorescence, light image, and merged image are presented from left to right. Bars = 50 μm.
According to microarray databases (https://www.genevestigator.ethz.ch/gv/index.jsp), GAPCp gene expression changes throughout Arabidopsis development, with the highest GAPCp expression occurring in 14- to 18-d-old plants. Therefore, we performed microarray experiments on 15-d-old plants using Affymetrix microarrays. Because a single probe for GAPCp1 and GAPCp2 is present in these microarrays, it is conceivable that the expression data obtained are the sum of the expression of both genes. The gene expression levels of these two isoforms were between 2% and 5%, as compared with other GAPDH isoforms (Fig. 2B).
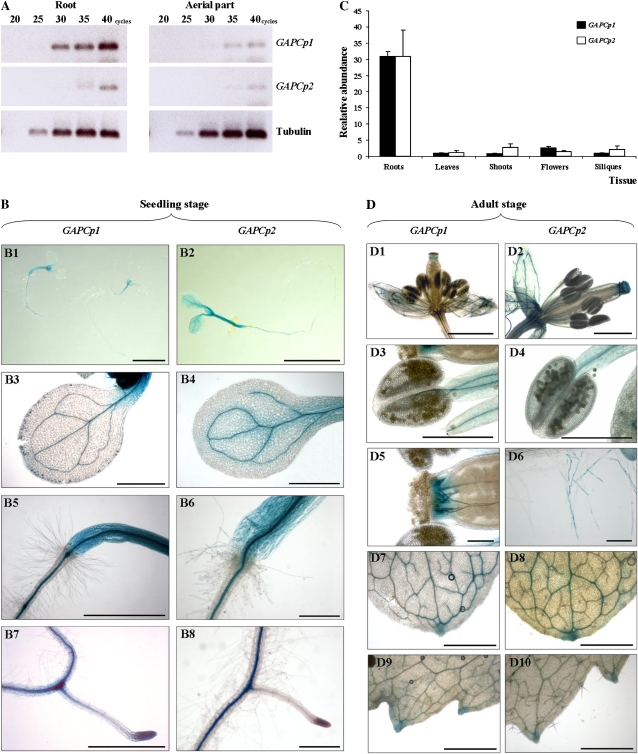
Expression patterns of GAPCp1 and GAPCp2 were assessed by reverse transcription (RT)-PCR and by analysis of promoter-GUS fusions. At the seedling stage, GAPCp1 and GAPCp2 are mainly expressed in roots as compared with the aerial part (Fig. 4A). The GUS expression analyses showed that GAPCp1 is mainly expressed in shoots and root vasculature and also in leaf veins (Fig. 4B). GAPCp2 follows a similar expression pattern to that of GAPCp1 (Fig. 4B). At the adult stage, GAPCp1 and GAPCp2 transcripts were detected in all organs studied: roots, shoots, leaves, flowers, and siliques (Fig. 4C). GAPCp1 and GAPCp2 were most abundantly expressed in roots, but expression was also clearly observed in shoots and flowers. Again, GUS activity was mainly associated with the vascular tissue (Fig. 4D).
Figure 4.
Expression analysis of GAPCp1 and GAPCp2 genes. A, RT-PCR analysis of GAPCp1 and GAPCp2 in the aerial part and roots of 17-d-old seedlings. B, Expression of GUS under the control of GAPCp1 and GAPCp2 promoters in seedlings (B1 and B2), cotyledons (B3 and B4), shoots (B5 and B6), and roots (B7 and B8) of 6- to 10-d-old plants. C, Relative gene expression of GAPCp1 and GAPCp2 in adult plants. Values are means ± sd (n = 3). D, Expression of GUS under the control of GAPCp1 and GAPCp2 promoters in flowers (D1 and D2), anthers (D3 and D4), pistils (D5), roots (D6), and leaves (D7–D10) of adult plants. Bars = 5 mm (B1 and B2), 1 mm (B3–B8, D1, D2, and D7–D10), and 0.25 mm (D3–D6).
Phenotypic Characterization of gapcp Mutants
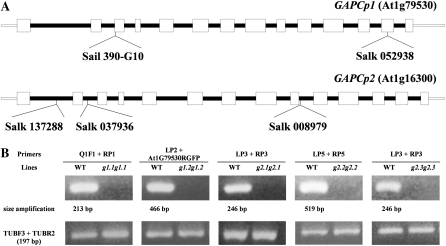
In order to shed light on the in vivo function of GAPCps, a gain- and-loss-of-function approach was followed. Multiple independent T-DNA insertion mutant lines affecting GAPCp1 and GAPCp2 were obtained. The presence and genomic locations of the T-DNA insertions were verified by PCR with genomic DNA and sequencing of PCR products for five T-DNA insertion mutant lines (SAIL_390_G10 and SALK_052938 for GAPCp1 and SALK_137288, SALK_008979, and SALK_037936 for GAPCp2). These mutant alleles were named gapcp1.1, gapcp1.2, gapcp2.1, gapcp2.2, and gapcp2.3, respectively. The exact location of each T-DNA insertion is presented in Figure 5A. RT-PCR analysis showed that transcript levels in all five mutant alleles were undetectable (Fig. 5B). None of the single mutants of either GAPCp1 or GAPCp2 presented a clear visual phenotype (Fig. 6A). Five different double gapcp1 gapcp2 mutant allele combinations were generated (gapcp1.1 gapcp2.1, gapcp1.1 gapcp2.2, gapcp1.1 gapcp2.3, gapcp1.2 gapcp2.1, and gapcp1.2 gapcp2.2).
Figure 5.
Characterization of gapcp1 and gapcp2 mutants. A, Genomic organization of gapcp T-DNA mutant lines. Open boxes represent exons, and solid lines represent introns. The T-DNA insertion point in each gapcp mutant is shown along with the position of primers used for genotyping. B, Detection of the GAPCp transcript in the gapcp1 and gapcp2 mutants by RT-PCR analysis. Total RNA was extracted from wild-type (WT) or mutant roots, and RT-PCR was performed with the specific primers for each T-DNA insertion located at both sides of the T-DNA insertion. Forty PCR cycles were done. For simplicity, g stands for gapcp.
Figure 6.
Phenotypic characterization of gapcp mutants and complemented lines. A, Phenotypic changes in the gapcp mutant. Plants were grown for 18 d on one-fifth-strength MS plates. Double mutants (g1.1g1.1 g2.1g2.1, g1.1g1.1 g2.2g2.2, and g1.2g1.2 g2.1g2.1) showed arrested root development. Single mutants (g1.1g1.1 G2G2), heterozygous lines (g1.1g1.1 G2g2.1), and gapcp double mutant complemented with a genomic GAPCp1 construct (g1.1g1.1 g2.1g2.1 PG1:G1) had normal root development as compared with control plants (WT). At bottom are data of the final root length, root growth rate, and aerial part fresh weight (FW) of the wild type, double mutant, and complemented line. Measurements (mean ± sd; n ≥ 30) were made in plants grown with or without 1% (w/v) Suc. B and C, Size of the root epidermal cells is reduced in the gapcp double mutant. Electron microscopy of wild-type (B) and gapcp double mutant (C) roots is shown. Measurements are means ± sd (n = 3). D and E, Bright-field inverted microscopy images showing that the size of the epidermal cells in leaves is not reduced in the gapcp double mutant (E) as compared with the wild type (D). Measurements are means ± sd (n = 3). F, gapcp double mutants (g1.1g1.1 g2.1g2.1, g1.1g1.1 g2.2g2.2, and g1.1g1.1 g2.3g2.3) are dwarf in the adult stage. Heterozygous plants (g1.1g1.1 G2g2.1) are not different from wild-type plants. G, Complementation of gapcp double mutants with a genomic GAPCp1 construct (g1.1g1.1 g2.1g2.1 PG1:G1) rescues the wild-type phenotype. For simplicity, g stands for gapcp and G stands for GAPCp. Bars = 1 cm (A), 50 μm (B–E), and 10 cm (F and G).
Although seedlings appear to be wild type at germination, all gapcp double mutants could be phenotypically identified after 10 to 12 d of growth on plates based on a severe arrest of root development (Fig. 6A). The root phenotype observed in the mutants is in agreement with the pattern of promoter activity observed (Fig. 4, A, B5–B8, and C). Eighteen days after germination, the root length of the gapcp double mutant was approximately 8-fold shorter and the root growth rate was about 11-fold lower than those of the wild type (Fig. 6A). Formation of lateral roots was not affected in gapcp double mutants, but the size of the root epidermal cells was about 50% smaller than in wild-type plants (Fig. 6, B and C). The phenotype of arrested root development was associated with a reduction of the growth of the aerial part, as determined by fresh weight (Fig. 6A). However, the size of the leaf epidermal cells was not significantly modified (Fig. 6, D and E). This result would indicate that the smaller size of gapcp double mutant plants is attributable primarily to reduced root cell expansion.
Adult gapcp double mutant plants grown in the greenhouse have pleiotropic growth defects resulting in severe dwarf phenotypes, with short root systems and sterility (Fig. 6F). The five double mutant lines presented similar phenotypes, although with different degrees of severity (Fig. 6F). To further corroborate the genetic linkage of GAPCps with the observed phenotypes, we examined the progeny of four different genotypes. Heterozygous gapcp mutants were self-crossed, and the segregation genotype was confirmed by PCR. In all cases, the dwarf phenotype of adult plants segregated with the double mutant genotype (Supplemental Table S1). Plants with wild-type phenotypes carried at least one wild-type allele at the GAPCp1 or GAPCp2 locus, with a ratio of about 1:2 (wild type:heterozygous), as expected for the segregation of a unique locus. Heterozygous GAPCp plants were visually indistinguishable from wild-type plants, indicating that double GAPCp mutation is required in order to achieve the observed phenotype. These results indicate that the gapcp phenotypes are genetically linked to both the GAPCp1 and GAPCp2 loci and that the two genes are dominant. Finally, we were able to complement all of the gapcp double mutant phenotypes with a genomic construct of the GAPCp1 locus (Fig. 6, A and G).
Since other glycolytic mutants show clear sugar phenotypes, we investigated whether gapcp double mutant growth and development were affected by sugars. Two different experiments were carried out: germination assays and growth measurements. The response of gapcp double mutants to exogenous sugars is normal in germination. gapcp double mutant seeds do not show delayed germination, do not need added sugar in order to establish on agar plates, and can directly develop in soil (defined as developing true leaves, shoots, and flowers). Also, gapcp double mutants do not show a phenotype in response to the addition of high sugar levels during germination, as observed for other mutants in the plastidic glycolytic pathway (Supplemental Fig. S1). Finally, the presence of exogenous Suc (1%, w/v) improves the general growth of gapcp double mutants, as observed for wild-type plants, but does not rescue the root growth arrest (Fig. 6A).
Because all five mutant lines generated show similar phenotypes, further experiments were mainly performed with one or two lines.
Total GAPDH Activity Is Not Modified, But Plastidic Activity Is Reduced in gapcp Double Mutants
Despite the strong phenotype, total GAPDH activity in gapcp double mutant seedlings was not affected as compared with that of wild-type plants (Table I). Both NAD+- and NADP+-dependent activities were measured in crude extracts from roots and aerial parts obtained at the end of the dark period and in the middle of the light period. No significant difference in activity between the wild type and double mutants was observed in any of the conditions tested (Table I). These results indicate that the activity of GAPCp1 and GAPCp2 is low and may be masked by the bulk activity of other GAPDH isoforms present in the extract. In order to observe differences in GAPDH activity, it was necessary to assay chloroplast/plastid-enriched preparations. In this case, we found a reduction in the NAD+-dependent GAPDH activity of about 25% in plastid-enriched fractions from gapcp double mutants as compared with controls (Table I). Since we did not use pure plastid preparations, the remaining GAPDH activity in our fractions could be due to contamination from other GAPDH sources.
Table I.
GAPDH activity
Specific GAPDH activity [μmol NAD(P) min−1 mg−1 protein] in the wild type and gapcp1.1gapcp1.1 gapcp2.1gapcp2.1 (g1.1g1.1 g2.1g2.1) double mutants was assayed in root and aerial part crude extracts during the light period and at the end of the dark period. Activity was also assayed in chloroplast- and plastid-enriched fractions. Values represent means ± sd (n ≥ 3). * Significant at P < 0.05.
| Organ | Fraction | Collection Time | Substrate | Wild Type | g1.1g1.1 g2.1g2.1 |
|---|---|---|---|---|---|
| Aerial part | Crude extract | Light | NAD+ | 0.58 ± 0.02 | 0.55 ± 0.03 |
| Dark | NAD+ | 0.53 ± 0.02 | 0.61 ± 0.06 | ||
| Light | NADP+ | 0.370 ± 0.001 | 0.42 ± 0.03 | ||
| Chloroplast fraction | Light | NAD+ | 0.055 ± 0.026 | 0.042 ± 0.018 | |
| Light | NADP+ | 0.089 ± 0.011 | 0.124 ± 0.021 | ||
| Plastid fraction | Light | NAD+ | 0.17 ± 0.02 | 0.13 ± 0.01* | |
| Light | NADP+ | 0.14 ± 0.02 | 0.12 ± 0.02 | ||
| Roots | Crude extract | Light | NAD+ | 0.50 ± 0.04 | 0.50 ± 0.03 |
| Dark | NAD+ | 0.72 ± 0.01 | 0.74 ± 0.03 | ||
| Light | NADP+ | 0.009 ± 0.002 | 0.014 ± 0.002 | ||
| Plastid fraction | Light | NAD+ | 0.28 ± 0.04 | 0.21 ± 0.02* | |
| Light | NADP+ | 0.023 ± 0.004 | 0.018 ± 0.005 |
Carbohydrate and Lipid Profiling of gapcp Double Mutants and GAPCp-Overexpressing Plants
Disruption of GAPCp could lead to impairment of lipid, carbohydrate, and amino acid metabolism. In order to investigate the importance of the contribution of GAPCps to these metabolic pathways, a biochemical characterization of both gapcp double mutants and GAPCp-overexpressing plants was carried out.
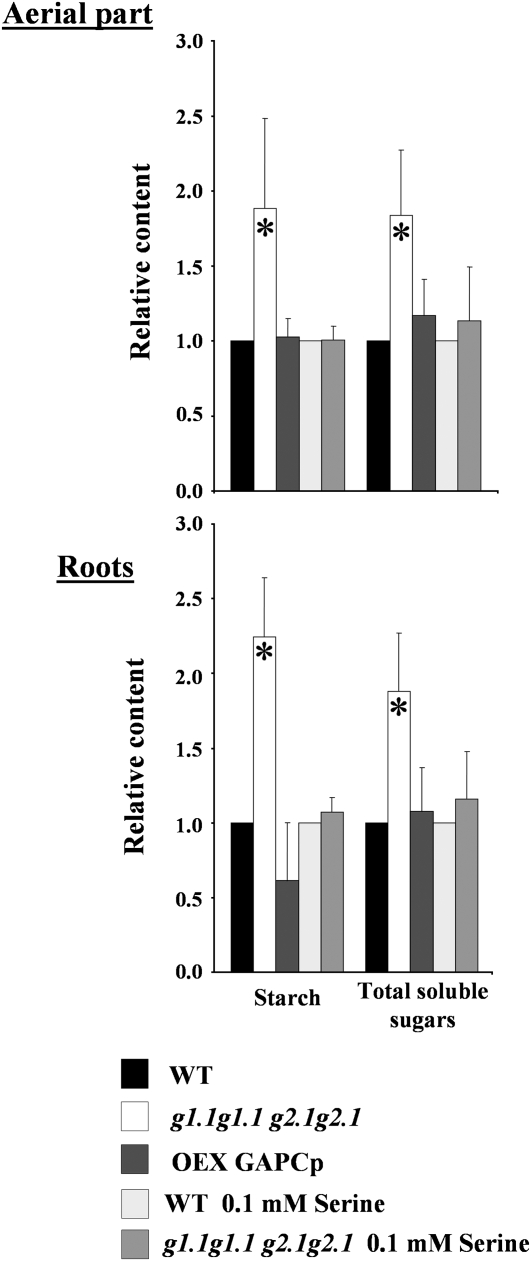
The starch and total soluble sugar contents were increased by more than 80% in the aerial part and roots of the gapcp double mutants as compared with controls (Fig. 7). In addition, the intracellular levels of the starch precursor molecule, ADP-Glc, in the aerial part of gapcp double mutants were 26% higher than those of wild-type plants (9.80 ± 0.49 versus 7.76 ± 0.56 nmol g−1 dry weight, respectively). Overexpressing GAPCp did not significantly change the carbohydrate levels in the aerial part, although a trend toward a reduction in the starch content in roots was observed as compared with control plants (Fig. 7).
Figure 7.
Carbohydrate profiling of gapcp double mutants and GAPCp-overexpressing plants. Starch and total soluble sugars in the aerial part and roots of 18- to 21-d-old wild-type (WT), gapcp double mutant (g1.1g1.1 g2.1g2.1), and 35S-GAPCp-overexpressing (Oex GAPCp) plants. Seeds were sown on plates with one-fifth-strength MS medium for 8 to 10 d. After phenotypic selection of gapcp double mutants as described in “Materials and Methods,” seedlings were transferred to one-fifth-strength MS medium with or without 0.1 mm Ser for an additional 10 d. Values (mg g−1 fresh weight) were normalized to the mean response of the wild type (starch: 1.75 ± 0.33 in wild-type aerial part, 0.19 ± 0.05 in wild-type roots, 2.03 ± 0.4 in wild-type aerial part with Ser, 0.17 ± 0.05 in wild-type roots with Ser; soluble sugars: 0.31 ± 0.10 in wild-type aerial part, 0.87 ± 0.15 in wild-type roots, 0.41 ± 0.06 in wild-type aerial part with Ser, 1.10 ± 0.15 in wild-type roots with Ser). For simplicity, g stands for gapcp. * Significant at P < 0.05.
An analysis of a range of enzymes closely associated with starch and Suc metabolism revealed that the aerial part of gapcp double mutants has relatively high activities of enzymes involved in the synthesis of Suc, such as Suc phosphate synthase (EC 2.3.1.14); in the synthesis of starch, such as total starch synthase (EC 2.4.1.21) and starch phosphorylase (EC 2.4.1.1); and in the synthesis of ADP-Glc, such as ADP-Glc pyrophosphorylase (EC 2.7.7.27) and Suc synthase (EC 2.4.1.13; Table II). Invertase (EC 3.2.1.26; involved in Suc breakdown) and total amylolytic (involved in starch breakdown) activities were normal in the gapcp double mutants (data not shown).
Table II.
Activity of enzymes associated with starch and Suc metabolism
Enzyme activities in extracts from the aerial part of the wild type and gapcp1.1gapcp1.1 gapcp2.1gapcp2.1 (g1.1g1.1 g2.1g2.1) double mutants cultured in the presence or absence of 0.1 mm Ser. Activities are given in units g−1 dry weight. Values represent means ± sd (n = 3). * Significant at P < 0.05.
| Enzyme | Wild Type | g1.1g1.1 g2.1g2.1 | Wild Type with 0.1 mm Ser | g1.1g1.1 g2.1g2.1 with 0.1 mm Ser |
|---|---|---|---|---|
| Suc synthase | 1,529 ± 195 | 2,002 ± 36* | 1,569 ± 134 | 1,614 ± 78 |
| ADP-Glc pyrophosphorylase | 1,437 ± 54 | 1,701 ± 68* | 1,518 ± 50 | 1,450 ± 97 |
| Suc phosphate synthase | 1,028 ± 221 | 1,486 ± 39* | 1,191 ± 76 | 1,116 ± 111 |
| Starch synthase | 86.5 ± 5.3 | 116.0 ± 9.3* | 95.8 ± 5.8 | 82.7 ± 8.5 |
| Starch phosphorylase | 276.1 ± 19.5 | 390.6 ± 27.6* | 358.9 ± 45.5* | 151.6 ± 36.9* |
Total fatty acid content was not changed in seeds of either heterozygous (gapcp1.1gapcp1.1 GAPCp2gapcp2.1) or 35S:GAPCp-overexpressing plants as compared with wild-type controls, although small differences were found in the fatty acid composition (Supplemental Table S2).
gapcp Double Mutants Are Ser Deficient
gapcp double mutants had a reduced carbon-to-nitrogen ratio in roots (9.7 in the wild type versus 8.7 in gapcp double mutant), indicating that its amino acid metabolism could be affected. In accordance with this reduced carbon-to-nitrogen ratio, total free amino acids in gapcp double mutant roots were increased by more than 50% as compared with control plants (Table III). Ala and the nitrogen transport amino acids Gln and Asn were mainly responsible for the increase in the total amount of free amino acids in the gapcp double mutants (Supplemental Table S3). Ser was the only amino acid whose content was significantly reduced, by 17% (Table III). As a result of this, the relative abundance of Ser in gapcp double mutants was reduced by 44% as compared with wild-type roots. We also observed a reduction in the relative abundance of other amino acids, such as the Ser derivative Gly, but to a lesser extent (21% reduction; Supplemental Table S3).
Table III.
Ser, total amino acids, and protein content of 3-week old wild-type and gapcp double mutant (g1.1g1.1 g2.1g2.1) plants sampled in the middle of the light period, after 24 h in the dark (24HD), or after 5 d in the dark (5DD)
Data in parentheses are as follows: a percentage of the absolute values normalized to the mean value calculated for the wild type grown in the light; b percentage of relative abundance normalized to the mean value calculated for the wild type. Plants were grown in one-fifth-strength MS medium as described in “Materials and Methods.” 5DD plants were supplemented with 1% (w/v) Suc in the growing medium. Data are means ± sd (n ≥ 3). Values that are significantly different from the wild type are shown in boldface.
| Parameter | Light-Grown Plants
|
24HD
|
5DD
|
|||
|---|---|---|---|---|---|---|
| Wild Type | g1.1g1.1 g2.1g2.1 | Wild Type | g1.1g1.1 g2.1g2.1 | Wild Type | g1.1g1.1 g2.1g2.1 | |
| Aerial part | ||||||
| Ser (mg g−1 dry weight) | 1.74 ± 0.01 | 1.90 ± 0.01 (110)a | 0.92 ± 0.14 (53)a | 0.82 ± 0.12 (47)a | 0 ± 0 (0)a | 0.01 ± 0.01 (1)a |
| Ser (% relative abundance) | 8.00 ± 0.04 | 8.62 ± 0.02 (108)b | 2.17 ± 0.15 | 1.70 ± 0.17 (78)b | 0 ± 0 | 0.01 ± 0.01 (0)b |
| Total amino acids (mg g−1 dry weight) | 21.72 ± 0.03 | 22.06 ± 0.11 (102)a | 42.22 ± 5.43 (194)a | 48.46 ± 4.2 (223)a | 114.17 ± 2.79 (526)a | 109.81 ± 6.52 (506)a |
| Protein content (mg g−1 dry weight) | 88 ± 9 | 72 ± 8 | 73.6 ± 8.2 | 58.4 ± 7.3 | 19.6 ± 2.4 | 6.3 ± 3.0 |
| Roots | ||||||
| Ser (mg g−1 dry weight) | 1.02 ± 0.07 | 0.85 ± 0.08 (83)a | 1.73 ± 0.45 (170)a | 1.04 ± 0.22 (102)a | 0.28 ± 0.02 (28)a | 0.28 ± 0.01 (28)a |
| Ser (% relative abundance) | 7.58 ± 0.59 | 4.20 ± 0.88 (56)b | 12.70 ± 0.23 | 5.66 ± 0.64 (45)b | 2.31 ± 0.20 | 1.06 ± 0.00(46)b |
| Total amino acids (mg g−1 dry weight) | 13.50 ± 1.79 | 20.56 ± 2.45 (152)a | 13.65 ± 3.72 (101)a | 18.88 ± 2.97 (140)a | 12.25 ± 0.41 (91)a | 26.79 ± 1.19 (198)a |
| Protein content (mg g−1 dry weight) | 27.9 ± 4.5 | 21.5 ± 2.5 | 15 ± 2 | 11.4 ± 2.5 | 13.2 ± 1.5 | 5.8 ± 0.8 |
In the aerial part of gapcp double mutants, the total amino acid content was not increased and the amount of Ser was not reduced as compared with controls (Table III). The most drastic change observed in the aerial part of gapcp double mutants was the relative increase (4-fold increase) of Met (Supplemental Table S3).
In 35S:GAPCp-overexpressing plants, there was a reduction in the total amount of free amino acids in both the aerial part and roots as compared with controls (Supplemental Table S3). Drastic changes in the relative abundance of amino acids were not found in these 35S:GAPCp-overexpressing plants (Supplemental Table S3).
Ser is mainly synthesized through two pathways: the light-dependent glycolate pathway and the phosphorylated pathway. In order to separate the contribution of these two pathways to Ser biosynthesis, we determined the amino acid composition in plants grown for 24 h (24HD) or 5 d (5DD) in the dark. In the aerial part, the total amino acid content was increased in a similar way in both the wild type and gapcp double mutants as the duration of the dark period lengthened (2-fold increase in 24HD and 5-fold increase in 5DD). The amounts of most of the amino acids increased, but those of Ser and Gly were drastically reduced (Table III; Supplemental Table S3). Specifically, the content of Ser decreased by 50% in 24HD and was close to zero in 5DD in both controls and gapcp double mutants (Table III). A slight but significant reduction in the relative abundance of Ser was found in 24HD gapcp double mutants as compared with controls.
In roots, the increase in the total free amino acid content was only observed in the gapcp double mutants and was not as drastic as that found in the aerial part (only 2-fold increase in 5DD; Table III). As in the light-grown plants, Ala, Gln, and Asn were mainly responsible for the increase in the total amount of free amino acids in the gapcp double mutant roots (Supplemental Table S3). The evolution of the Ser levels in roots grown in darkness was different from that of the aerial part. The Ser contents were even higher in roots from 24HD wild-type plants and gapcp double mutants as compared with light-grown wild-type plants (Table III). However, the differences in the Ser content between wild-type and gapcp double mutant plants were increased as compared with light-grown plants (significant reductions of 40% of Ser content and 55% of relative abundance in gapcp double mutants as compared with controls). No depletion of Ser was observed in 5DD roots (Table III). Both wild-type and gapcp double mutants still had 28% of the Ser content present in the controls grown in the light (Table III). In 5DD plants, the relative abundance of Ser in the gapcp double mutant was still 54% lower than in the wild type.
Contrary to the amino acid content, there was a trend toward a decrease in the protein content of gapcp double mutants as compared with controls, but this decrease was only significant in 5DD plants (Table III).
Ser Supplementation Rescues Root Development and Restores Normal Levels of Carbohydrates and Sugar Biosynthetic Activities in gapcp Double Mutants
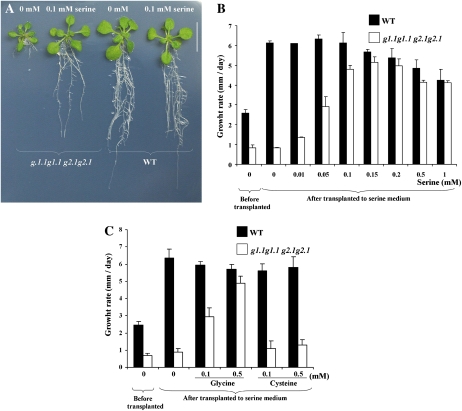
gapcp double mutant seedlings grown on medium supplemented with Ser no longer displayed arrested root development (Fig. 8A). The recovery of root growth was concentration dependent (Fig. 8B). Most importantly, adding Ser to the growth medium restored normal starch, total soluble sugars (Fig. 7), and ADP-Glc (7.58 ± 0.65 nmol g−1 dry weight) contents in the gapcp double mutants as compared with controls. The activity of enzymes involved in Suc and starch biosynthesis in the gapcp double mutants was also reduced to control levels or even lower by Ser supplementation (Table II). Gly supplementation was able to partly complement the root phenotype, but the addition of Cys had no effect (Fig. 8C).
Figure 8.
Ser rescues the arrested root development in gapcp double mutants. A to C, Seeds from wild-type (WT) and gapcp double mutant (g1.1g1.1 g2.1g2.1) plants were germinated in one-fifth-strength MS medium for 8 to 10 d and then transplanted for an additional 10 d to a medium with or without 0.1 mm Ser (A). The same experiment described in A was conducted with different concentrations of Ser (B) and Gly or Cys (C). Data are means ± sd (n ≥ 30). For simplicity, g stands for gapcp.
gapcp Double Mutants Have Altered Gene Expression
To assess the effect of GAPCp mutation on gene expression levels, a microarray analysis comparing gapcp double mutant and wild-type seedlings was performed. Full details of the microarray analysis are given in Supplemental Table S4, and a summary of the results is provided in Figure 9.
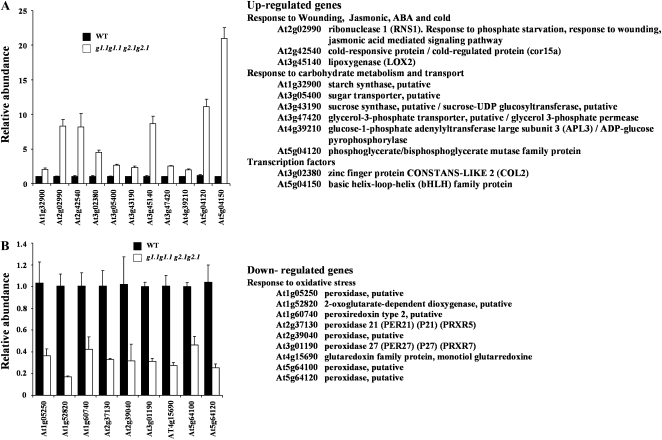
Figure 9.
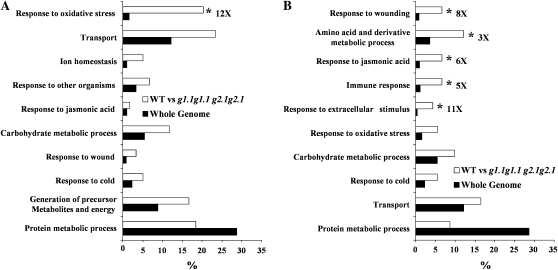
Functional categorization of the genes differentially expressed in the gapcp double mutant. Down-regulated (A) and up-regulated (B) transcripts (2-fold increase) in gapcp double mutant as compared with the wild-type (WT) control were sorted by their putative functional categories and compared with the whole genome according to the FatiGO tool. For simplicity, g stands for gapcp. * Significantly enriched function at adjusted P < 0.05.
In the gapcp double mutant, 274 genes were deregulated (more than 2-fold difference relative to the wild type; P < 0.05) as compared with wild-type plants. Among this population, 106 genes were down-regulated and 168 genes were up-regulated.
The gene expression of all other GAPDH isoforms was not significantly modified in the gapcp double mutant. None of the genes encoding enzymes located immediately upstream or downstream of GAPDH (aldolase and phosphoglycerate kinase) or other enzymes farther away from GAPDH in the glycolytic pathway were modified in the gapcp double mutant.
In order to identify processes related to or affected by GAPCp, an analysis of genes according to Genevestigator (https://www.genevestigator.ethz.ch/gv/index.jsp) and the FatiGO tool in Babelomics (http://babelomics.bioinfo.cipf.es/EntryPoint?loadForm=fatigo) was performed. According to the Genevestigator analysis, most of the down-regulated genes in the gapcp double mutant were mainly expressed in roots, which is in accordance with the observed root phenotype. Figure 9A shows a summary of functional enrichment analysis according to the FatiGO tool. Among the genes down-regulated in the gapcp double mutant, there is a significant enrichment of genes involved in oxidative stress response (12-fold increase; adjusted P value of 2.9 e−8). There is no clear anatomical pattern of up-regulated genes in the gapcp double mutant, as was observed for down-regulated genes. The most enriched categories among the up-regulated genes in the gapcp double mutant are presented in Figure 9B. There is a significant enrichment in genes involved in stress responses such as wounding (8-fold increase; adjusted P value of 0.014), response to jasmonic acid (6-fold increase; adjusted P value of 0.02), and immune response (5-fold increase; adjusted P value of 0.03). There is also a significant enrichment in genes of amino acid derivatives and metabolic processes (3.3-fold increase; adjusted P value of 0.02) and response to extracellular stimulus (11-fold increase; adjusted P value of 0.02). Deregulation of several genes was reconfirmed by quantitative real-time PCR (QRT-PCR; Fig. 10). These include some genes from significantly enriched categories such as peroxidases of unknown function, genes responding to wounding, jasmonic acid, and cold (lipoxygenase [LOX2], ribonuclease [RNS1], and cold-responsive protein [cor15a]), and genes involved in carbohydrate metabolism (Glc-1-P adenyltransferase APL3, putative starch synthase, and putative Suc synthase). The QRT-PCR measurements globally confirmed the changes in transcript level in the gapcp double mutant observed in the microarrays.
Figure 10.
Quantification of the changes in transcript levels using real-time PCR. A, A selection of up-regulated genes in gapcp double mutants. B, A selection of down-regulated genes in gapcp double mutants. At right, the putative functions of the selected genes are presented. WT, Wild type.
DISCUSSION
Arabidopsis Has Two Functionally Redundant GAPCps
We have shown that the Arabidopsis At1g79530 (GAPCp1) and At1g16300 (GAPCp2) genes encode plastidial isoforms of GAPDH that could correspond to the isoforms biochemically described in other plant species (Backhausen et al., 1998; Meyer-Gauen et al., 1998). The lack of visual phenotypes in single gapcp mutants or even in heterozygous plants, the complete complementation of the mutant phenotypes with either GAPCp1 or GAPCp2, along with their similar expression pattern indicate that both GAPCp genes are functionally redundant in Arabidopsis.
Our results suggesting that the activity of GAPCps is low as compared with the cytosolic isoforms are in agreement with those in the literature (Eastmond and Rawsthorne, 2000). Those authors found NAD+-dependent GAPDH activity in embryo plastids to be about 10% of that in the cytosol. This is also in agreement with the very low gene expression level of the GAPCp isoforms in Arabidopsis. Nevertheless, the activity of the plastidic isoforms could also be masked by an activation of cytosolic GAPDHs in the gapcp double mutants as a compensatory effect. In this respect, a trend to an increase in the NAD+-dependent GAPDH activity in the dark was observed in crude extracts from both aerial parts and root fractions of gapcp double mutants.
gapcp Double Mutants Are Severely Affected in Growth and Development
gapcp double mutant seedlings are visually indistinguishable from wild-type plants until several days (8–10) after germination. However, dramatic phenotypes such as arrested root development, dwarfism, and sterility are observed in seedlings and mature plants. These observations indicate that GAPCp is mainly required at a specific plant developmental stage. Recently, a reverse genetics approach was used to investigate the biological role of the cytosolic GAPDH GAPC1 (Rius et al., 2008). In the gapc1 mutant, drastic changes in the glycolytic and Krebs cycle intermediates and a reduction of total GAPDH activity by more than 50% were found. However, phenotypes of this mutant are much milder than those observed in the gapcp double mutants presented in this study. Specifically, the gapc1 mutant shows normal leaves and roots, and only fruits display alterations in size and weight, but plants are fertile. On the other hand, GAPC inhibition by an antisense approach indicated that GAPC plays a minor role in the regulation of morphology and metabolism in potato (Solanum tuberosum) plants (Hajirezaei et al., 2006). According to current knowledge, there is a near equilibrium of the key glycolytic metabolite pools between cytosol and plastid (Schwender et al., 2003). It could be assumed that knocking out a glycolytic reaction in one compartment may not lead to severe phenotypic changes if this reaction can be performed in the other glycolytic pathway. Our results emphasize the importance of the glycolytic plastidial pathway, and specifically of the “minor GAPDH isoforms” the GAPCps, in plant metabolism. They also indicate that compartmentalization is very important in plant glycolysis and that probably not all of the glycolytic metabolites are in equilibrium between the cytosol and plastids.
GAPCp Activity Is Critical for Ser Biosynthesis in Roots
The root growth arrest caused by GAPCp loss of function can be rescued by Ser supplementation to the growth medium. These results provide in vivo evidence that Ser is crucial for root development. Ser biosynthesis in plants proceeds mainly by two pathways: the glycolate pathway, which takes place in the peroxisomes and is associated with photorespiration; and the so-called phosphorylated pathway, which takes place in the plastids and uses 3-PGA as a precursor (Ho et al., 1998, 1999a, 1999b; Ho and Saito, 2001). The sequential reactions of GAPDH and phosphoglycerate kinase provide the 3-PGA that is converted to Ser through the phosphorylated pathway (Ho and Saito, 2001). Since we demonstrated that GAPCp is localized to the plastids, its suppression can only restrict substrate supplies to the phosphorylated pathway.
It has been suggested that the phosphorylated pathway plays an important role in supplying Ser to roots or other nonphotosynthetic tissues (Ho and Saito, 2001). Also, the phosphorylated pathway could be important in photosynthetic organs, especially in the dark when the pathway of carbon flux from glycolate to Ser ceases to function (Kleczkowski and Givan, 1988; Ho and Saito, 2001). However, no genetic evidence has been provided to date about the significance of this pathway in plants.
In order to separate the contribution of the glycolate (light-dependent) and the phosphorylated pathways to Ser biosynthesis, we measured the Ser content in plants after a growth period in the dark. It is well documented that the glycolate pathway is very important in the biosynthesis of Ser in the photosynthetic organs of C3 plants (Somerville and Somerville, 1983; Kleczkowski and Givan, 1988), but the in vivo contribution of alternative Ser biosynthetic pathways in these organs is still not well understood. In this respect, the activity of the phosphoglycerate dehydrogenase, the first committed enzyme of the phosphorylated pathway, is enhanced in the dark in both roots and leaves in Arabidopsis (Waditee et al., 2007). We found a significant decrease in the relative abundance of Ser in the aerial part of gapcp double mutants grown for 24 h in the darkness as compared with controls. This difference was not observed in light-grown plants. These results imply that the phosphorylated pathway may contribute to Ser biosynthesis in leaves in dark conditions. It could also be possible that there are no substrate limitations for the phosphorylated pathway in plastids/chloroplasts in leaves under light conditions, since 3-PGA could be provided from other sources such as the Calvin cycle or photorespiration, but some limitations may occur in the dark. Nevertheless, we found that Ser is the only amino acid whose content is completely depleted in the aerial part of both wild-type and gapcp double mutant plants grown for 5 d in the dark. These results suggest that the quantitative contribution of the phosphorylated pathway to Ser biosynthesis in the aerial part of Arabidopsis is minor. There have been many discussions about the role of the “wasteful” process of photorespiration in plants. Recently, mutants of Arabidopsis Gly decarboxylase have been found to be lethal under nonphotorespiratory conditions, providing evidence for the nonreplaceable function of this enzyme in vital metabolic processes other than photorespiration (Engel et al., 2007). According to our data, an obvious function of the Gly decarboxylase-Ser hydroxymethyltransferase system that may not have been given sufficient consideration would be to provide Ser to photosynthetic organs.
Our results show that the phosphorylated pathway plays a more relevant role in the roots than in the aerial part under both dark and light conditions. First, in all assayed conditions, the Ser abundance in roots was lower in the gapcp double mutant as compared with the wild type, which corroborates that lack of GAPCp activity restricts substrate supplies to the phosphorylated pathway in this organ. Second, in both wild-type and gapcp double mutant roots, the Ser content did not drop in samples collected after 24 h in the darkness and was not depleted after 5 d. This means that the quantitative contribution of the phosphorylated pathway to Ser biosynthesis in roots is far more important than in the aerial part.
The differences in Ser abundance between the gapcp double mutant and wild-type plants were enhanced when samples were collected from plants grown in the dark. The plant usually manages to provide relatively constant levels of all amino acids (Nikiforova et al., 2006). In order to compensate for amino acid deficiency in roots, we assume that part of the Ser synthesized through the glycolate pathway during the day is transported via phloem to the roots. However, our results indicate that transport of Ser from aerial parts is not enough to compensate for the Ser deficiency in the roots of gapcp double mutants even in the light.
Thus, our results provide in vivo evidence that the phosphorylated Ser pathway is crucial in supplying Ser to nonphotosynthetic tissues such as roots. In situ hybridization analysis of plastidic phosphoserine aminotransferase, an enzyme participating in the Ser phosphorylated pathway, revealed that this gene is preferentially expressed in the meristem tissue of root tips (Ho et al., 1998). These results are consistent with the arrest of primary root growth found in the gapcp double mutants, suggesting that GAPCp provides the substrate to plastids for the Ser biosynthesis needed in the root meristem. 3-PGA could be supplied by other sources to the plastids. PEP imported from the cytosol could provide 3-PGA to the plastids through the reactions catalyzed by phosphoglycerate mutase and enolase, two reactions that are theoretically reversible. In addition, at least two transport systems are thought to translocate 3-PGA between cytosol and plastids, the specific triose phosphate translocator and the Glc phosphate translocator (Flügge, 1999; Fischer and Weber, 2002; Knappe et al., 2003). Since our results suggest a deficiency of 3-PGA in the gapcp double mutant plastids, this means that either the low activity of phosphate translocators or the low flow of PEP to 3-PGA is limiting the substrates required for Ser biosynthesis necessary for root growth, at least during some developmental stages.
No direct (in vivo) evidence of the function of GAPCp has been reported previously. We propose that the main function of GAPCp and phosphoglycerate kinase is to supply the 3-PGA for Ser biosynthesis and that these activities are seemingly a limiting step in this pathway. Other essential functions attributed to GAPCp and phosphoglycerate kinase, such as the production of ATP and NADH, may not be crucial for several reasons: (1) ATP can be produced endogenously in the plastid by other enzymes such as pyruvate kinase or transported from the cytosol (Reiser et al., 2004); (2) NADH can be provided by the pyruvate dehydrogenase reaction in plastids and also by photosynthesis and import of reducing equivalents generated in the mitochondria or cytosol (Schwender et al., 2003); and (3) ATP and NADH levels would not be limiting in gapcp double mutant plastids, since the exogenous supply of the end product, Ser, overrides all of the phenotypes studied.
Little is known about whether some amino acids play any specific role in plant development. This is partly due to the difficulty of separating their metabolic and regulatory functions. Using a nonlethal deficiency mutant, a crucial role for His homeostasis in root meristem maintenance has been demonstrated (Mo et al., 2006). Also, it has been shown that Trp deficiency produces retardation of aerial organ development by affecting cell expansion in Arabidopsis (Jing et al., 2009). By reducing the content of Ser in gapcp double mutant roots, a drastic arrest of its development was observed. Although this root growth phenotype in gapcp double mutants could be attributed to an indirect effect of Ser deficiency, for instance as a consequence of protein synthesis inhibition, a more direct effect of Ser or its derivatives on root development cannot be ruled out. In this respect, the homeostasis of Ser and/or derivative molecules may also be important to maintain root development.
Ser Deficiency in Roots of gapcp Double Mutants Stimulates the Biosynthesis of Starch and Soluble Sugars in the Aerial Part of the Plant
The increased starch and soluble sugar content in the gapcp double mutants would suggest that this mutant is unable to metabolize starch. However, Ser supply to the roots reestablishes normal growth and carbohydrate levels, corroborating that GAPCp is not crucial for starch metabolism but for Ser biosynthesis. Starch is synthesized and catabolized only in plastids, but carbon obtained from starch degradation enters the glycolytic pathway in the cytosol as hexoses (Taiz and Zeiger, 2006). So the blockage in the plastidial glycolytic pathway in the gapcp double mutants could be short circuited through the cytosolic pathway, where the highly active cytosolic GAPDHs could metabolize the triose phosphates. Metabolites could then reenter the plastidial pathway as pyruvate, 3-PGA, or PEP.
Starch and glycogen are the main storage carbohydrates in plants and bacteria, respectively. The metabolism of these reserve polysaccharides is highly regulated and connected to amino acid metabolism in response to the physiological needs of the cell. In the bacterium Escherichia coli, amino acid starvation elicits the “stringent response,” a pleiotropic physiological change that down-regulates nucleic acid and protein synthesis and up-regulates the expression of genes involved in glycogen biosynthesis. E. coli mutants impaired in the synthesis of amino acids such as Ser are known to display a glycogen-excess phenotype as a result of the stringent response, which disappears when Ser is added to the culture medium (Eydallin et al., 2007). In plants, very little is known of how stringent conditions regulate starch metabolism, but (1) the accumulation of high levels of starch and ADP-Glc in the aerial part of the Ser-deficient gapcp double mutants, and (2) the accumulation of normal levels of these metabolites in the mutant when Ser is included in the culture medium clearly point to the occurrence of GAPCp-dependent mechanisms connecting carbon and nitrogen metabolism.
Several starch and Suc biosynthetic enzymes are up-regulated in the aerial part of the Ser-deficient gapcp double mutants and recover normal levels of activity when mutants are cultured in the presence of exogenously added Ser. Taken together, these data strongly indicate that, essentially similar to the case of the glycogen-excess E. coli mutants impaired in Ser biosynthesis, the starch-excess phenotype of the gapcp double mutants is ascribed to an activation of the starch biosynthetic machinery by still undefined molecular mechanisms. Thus, our results suggest that GAPCp activity is important to control carbon metabolic fluxes toward amino acid and starch biosynthesis and that Ser deficiency stimulates sugar biosynthesis. These results also suggest that amino acid homeostasis seemingly represents a main signal directing starch accumulation in the aerial part of the plant.
gapcp Double Mutants Have Altered Gene Expression
Several genes involved in carbon transport and metabolism were up-regulated in the gapcp double mutant, supporting the hypothesis that carbohydrate metabolism is deregulated upon GAPCp inactivation. Although expression of genes that encode for Ser biosynthetic enzymes was not induced, the gapcp double mutant was enriched in transcripts of genes involved in amino acid derivatives and metabolic processes.
Interestingly, mutation of GAPCp mainly affected the expression of genes involved in oxidative stress. A possible explanation of the alteration of genes of oxidative stress in gapcp double mutants is that GAPCp could be participating in reactive oxygen species (ROS) or redox signaling pathways, as has been described in animals and yeast (Morigasaki et al., 2008; Rodríguez-Pascual et al., 2008). In this way, the plant cytosolic GAPDH has been described as a potential target of hydrogen peroxide, and it has also been implicated in the suppression of the ROS (Hancock et al., 2005; Baek et al., 2008). Null mutants of cytosolic GAPDH showed increased ROS accumulation (Rius et al., 2008). The two major sites of ROS production in plant cells are the chloroplast and the mitochondria (Millar et al., 2001; Moller, 2001). Several retrograde signals have been reported to trigger signaling from chloroplasts, including redox and ROS signals (Woodson and Chory, 2008). GAPCp, as a plastid-localized NADH producer/consumer enzyme, could be a redox sensor participating in these retrograde signaling pathways.
MATERIALS AND METHODS
Bioinformatics
GAPDH genes were initially identified in The Arabidopsis Information Resource (http://www.arabidopsis.org/). Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 3.1 (Kumar et al., 2004). Predicted full-length protein sequences were aligned using ClustalW2. The neighbor-joining method with 100,000 bootstrap replications and the Poisson's correction model were used to elaborate the cladogram. The percentage of identity between different GAPDHs was obtained by aligning pair sequences using bl2seq at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Putative chloroplast/plastid localization sequences were identified using the ChloroP prediction server (Emanuelsson et al., 1999). Functional characterization of microarray data was done using Genevestigator (Grennan, 2006) and the FatiGO tool in Babelomics (Al-Shahrour et al., 2006). Significance of enriched functional groups is given according to the adjusted P value from Fisher's exact test after correcting for multiple testing using the false discovery rate procedure of Benjamini and Hochberg (1995).
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds (ecotype Columbia 0) were supplied by the European Arabidopsis Stock Center (Scholl et al., 2000) and by the Arabidopsis Biological Resource Center (ABRC; http://www.biosci.ohio-state.edu/∼plantbio/Facilities/abrc/index.html). Unless stated otherwise, seeds were sterilized and sown on 0.8% (w/v) agar plates containing one-fifth-strength Murashige and Skoog (MS) medium with Gamborg vitamins buffered with 0.9 g L−1 MES (adjusted to pH 5.7 with Tris). After a 4-d treatment at 4°C, seeds were placed in a growth chamber (Sanyo; MLR-351H) at 22°C with a 16-h-day/8-h-night photoperiod at 100 μmol m−2 s−1. Plates were supplemented with different concentrations of sugars and amino acids as indicated in the table and figure legends. For selection of GAPCp-overexpressing plants, half-strength MS plates supplemented with 0.5% (w/v) Suc and appropriate selection markers were used. Some plantlets and seeds were also grown in greenhouse conditions in pots filled with a 1:1 (v/v) mixture of vermiculite and fertilized peat (KEKILA 50/50) irrigated with demineralized water as needed.
Because gapcp double mutants are sterile, seeds from heterozygous plants were used for germination experiments and for lipid quantification (gapcp1GAPCp1 gapcp2gapcp2 or gapcp1gapcp1 GAPCp2gapcp2). For the rest of the physiological and molecular studies, double homozygous gapcp plants were identified in segregating populations by the root phenotype after 8 to 10 d and then subcultured on medium containing different concentrations of sugars or amino acids for an additional 8 to 12 d, as indicated in the table and figure legends.
Primers
All primers used in this work are listed in Supplemental Table S5.
T-DNA Mutant Isolation and Characterization
gapcp1.1 (SAIL_390_G10) and gapcp1.2 (SALK_052938) alleles of the At1g79530 gene and gapcp2.1 (SALK_137288), gapcp2.2 (SALK_008979), and gapcp2.3 (SALK_037936) alleles of the At1g16300 gene were identified in the SIGnAL Collection database at the Salk Institute (Alonso et al., 2003). Mutants were identified by PCR genotyping using gene-specific primers and left border primers of the T-DNA insertion (LB1_SAIL for SAIL lines and LBa1 for Salk lines). The gene-specific primers for the different alleles were as follows: for gapcp1.1, LP1 and RP1; for gapcp1.2, LP2 and RP2; for gapcp2.1, LP3 and RP3; for gapcp2.2, LP5 and RP5; and for gapcp2.3, LP4 and RP4. T-DNA insertions were confirmed by sequencing the fragments amplified by the T-DNA internal borders and gene-specific primers. gapcp double mutants (gapcp1.1 gapcp2.1, gapcp1.1 gapcp2.2, gapcp1.1 gapcp2.3, gapcp1.2 gapcp2.1, and gapcp1.2 gapcp2.2) were generated by crossing single gapcp mutants and identified by PCR genotyping with the same primers described for single mutants. Since GAPCp1 and GAPCp2 are located on the same chromosome, double mutants were obtained by crossing single mutants and searching for the recombinant plants in the segregating population.
Cloning and Plant Transformation
Standard methods were used to make the gene constructs (Sambrook and Russell, 2001). The cDNA corresponding to gene GAPCp1 (At1g79530; supplied by the ABRC; C00197) was PCR amplified using primers At1g79530FXhoI and C00197RBamHI in order to introduce XhoI and BamHI sites. The amplified cDNA fragment was placed under the control of the 35S promoter by cloning it into a modified pGreen II plant transformation vector (Hellens et al., 1993) named p336 kindly provided by Dr. Jeff Harper (University of Nevada). A cDNA from gene GAPCp2 (At1g16300) was obtained by RT (first-strand cDNA synthesis kit for RT-PCR; Roche) using total RNA as template (RNeasy plant mini kit; Qiagen). This cDNA was PCR amplified using primers At1g16300FXhoI and AT1g16300RBamHI in order to introduce XhoI and BamHI sites. The amplified fragment was cloned in the p336 plant transformation plasmid described above.
For gene promoter-reporter fusions, 1.5- and 1.6-kb fragments were PCR amplified from bacterial artificial chromosomes T8K14 and F309 (supplied by the ABRC). These fragments correspond to regions −1,521 to +18 and −1,601 to −1 relative to GAPCp1 and GAPCp2 translation start codons, respectively. Primers used for the amplifications were PromAt1g79530FHindIII and PromAt1g79530RSpeI for the GAPCp1 promoter and PromAt1g16300FXbaI and PromAt1g16300RNcoI for the GAPCp2 promoter. Plasmid pCAMBIA1303 (CAMBIA) was used to fuse the promoter fragments to the GUS gene using the sites indicated in the respective primer names.
For genomic complementation, a 5.5-kb fragment including 1,520 nucleotides upstream of the ATG was PCR amplified from bacterial artificial chromosome T8K14 using primers At11g795390FBamHI and At1g79530RSpeI. This fragment was cloned into a plant transformation plasmid derived from pFP101 (http://www.isv.cnrs-gif.fr/jg/alligator/) kindly provided by Prof. Lola Peñarrubia (Universitat de Valencia) using BamHI and SpeI sites.
For GAPCp-GFP, GAPC-GFP, and GAPA-GFP fusions, GAPDH cDNAs were PCR amplified with the following primers: At1g79530FGFP and At1g79530RGFP for GAPCp1, At1g16300FGFP and At1g16300RGFP for GAPCp2, At1g13440FGFP and At1g13440RGFP for GAPC1, and At1g12900FGFP and At1g12900RGFP for GAPA1. PCR products were cloned in the pCR8/GW/TOPO plasmid (Invitrogen). These cDNAs were subcloned in the plasmids pMDC43 and pMDC83 (Curtis and Grossniklaus, 2003) using the Gateway technology with Clonase II (Invitrogen). All PCR-derived constructs were verified by DNA sequencing.
GAPCp-overexpressing plants were obtained by transforming wild-type plants using the floral dipping method (Clough and Bent, 1998) with Agrobacterium tumefaciens carrying pSOUP and the p336-35S-GAPCps cDNAs. Phenotypic characterization of overexpressing plants was performed as for T-DNA mutants. For stable expression of GAPC1-GFP and GAPA1-GFP, wild-type plants were transformed as described above. For genomic complementation and stable expression of GAPCp-GFP fusions, double gacp1 gapcp2 mutants were transformed. Because gapcp1 gapcp2 double mutants were unable to produce seeds, we transformed the progeny of heterozygous plants (gapcp1.1gapcp1.1 GAPCp2gapcp2.1 or GAPCp1gapcp1.1 gapcp2.1gapcp2.1) with the different constructs. Transformants were selected by antibiotic selection, and gapcp double mutants were verified by PCR genotyping as described above. Additional primers At1g79530FBProm and At1g79530R3Ex were used to differentiate genomic transgene insertion from the endogenous At1g79530. LP390_G10.2 was also used for genotyping double mutants in transgenic GFP-GAPCp plants. Single insertion homozygous T3 lines were selected for characterization.
Enzyme Assays and Plastid Isolation
For GUS activity assays, plant organs were incubated in GUS buffer (100 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.1% [v/v] Triton X-100, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 2 mm 5-bromo-4-chloro-3-indonyl-β-d-glucuronic acid; Duchefa) overnight at 37°C. The plant material was cleared either in 70% (v/v) ethanol or Hoyer's solution before microscopic observation. At least six independent transgenic lines showed identical GUS-staining patterns and only differed in the expression level of GUS.
For GAPDH activity, whole extracts and plastid-enriched fractions were obtained. Frozen tissues from material collected at the end of the dark period and at the middle of the light period were ground in liquid nitrogen and resuspended in extraction buffer (50 mm HEPES-KOH, pH 7.5, 2 mm MgCl2, 5 mm EDTA, 5 mm β-mercaptoethanol, and 1:100 [v/v] protease inhibitor; Sigma P9599). The supernatant was obtained after centrifugation at 15,000g for 20 min at 4°C. For plastid-enriched fraction isolation, 2 to 4 g of fresh material was homogenized at 4°C in a buffer containing 50 mm HEPES-KOH, pH 7.5, 200 mm sorbitol, 100 mm KCl, 5 mm EDTA, 5 mm β-mercaptoethanol, and 1:100 (v/v) protease inhibitor cocktail. The homogenate was filtered through several layers of museline and centrifuged at 150g for 5 min. The supernatant was further centrifuged at 2,200g for 30 s and at 2,200g for 10 min. The resulting pellet was washed several times in homogenization buffer and resuspended in the same buffer without sorbitol. The protein content was quantified using the Bio-Rad protein assay kit. GAPDH activity of the extracts was assayed according to Hara et al. (2005) with minor modifications. The assay was performed in a final volume of 1 mL containing 10 mm Tris-HCl, pH 8.5, 20 mm sodium arsenate, 2 mm NAD(P)+, 0.5 mm dithiothreitol, 2 mm dl-glyceraldehyde-3-phosphate, and 20 μg of protein. Reactions were initiated by addition of glyceraldehyde-3-phosphate and were linear for at least 4 min. The rate of reduction of NAD+ at 25°C was measured at 340 nm in triplicate with several extracts. The NADP+-dependent activity was measured as above in a reaction buffer containing 50 mm Tris-HCl, pH 8, 5 mm MgCl2, 0.5 mm dithiothreitol, 2 mm NADP+, and 2 mm dl-glyceraldehyde-3-phosphate. In order to differentiate between the phosphorylating and nonphosphorylating NADP+ GAPDHs, enzyme activities were assayed with and without 20 mm sodium arsenate. Data presented are the arsenate-dependent activities.
Measurements of ADP-Glc pyrophosphorylase, starch phosphorylase, Suc synthase, total starch synthase, and Suc phosphate synthase activities were assayed as described by Baroja-Fernández et al. (2004). We define 1 unit of enzyme activity as the amount of enzyme that catalyzes the production of 1 μmol of product per minute.
Microscopy
Leaf epidermal peels were directly observed using a bright-field inverted microscope (Leica DM IRB). Roots were prepared using the critical-point drying method before observation with a field emission scanning electronic microscope (Hitachi S4100-FEG). Observation of GFP fluorescence was carried out with a confocal microscope (Leica TCS-SP).
Microarrays
Total RNAs from three pools of 15-d-old seedlings (gapcp double mutants and wild type) vertically grown on one-fifth-strength MS plates at the same time as described above were extracted using the RNeasy plant mini kit (Qiagen). RNA integrity was determined using RNA 6000 Nano Labchips in an Agilent 2100 Bioanalyzer following the manufacturer's protocol. The purified RNA (8 μg) was used to generate first-strand cDNA in a RT reaction (One-Cycle Target Labeling and Control Reagents; Affymetrix). After second-strand synthesis, the double-stranded cDNAs were used to generate copy RNA (cRNA) via an in vitro transcription reaction. The cRNA was labeled with biotin, and 20 μg of the labeled cRNA was fragmented. The size distribution of the cRNAs and fragmented cRNAs was assessed using an Eppendorf Biophotometer and electrophoresis. Fragmented cRNA (15 μg) was added to 300 μL of hybridization solution, and 200 μL of this mixture was used for hybridization on Arabidopsis ATH1 Genome Arrays for 16 h at 45°C. The standard wash and double-stain protocols (EukGE-WS2v5-450) were applied using an Affymetrix GeneChip Fluidics Station 450. The arrays (three replicates of each line) were scanned on an Affymetrix GeneChip scanner 3000. Fluorescence images were normalized with the software GCOS from Affymetrix. Data were analyzed and compared using the dChip software (Li, 2008; http://www.hsph.harvard.edu/∼cli/complab/dchip/) using threshold criteria of 2-fold and P < 0.05.
RT-PCR
Total RNA was extracted from seedlings and adult plants as for microarray experiments. RNA was treated with RNase-free DNase (Promega). RNA (0.5 μg) was reverse transcribed using polyT primers and the first-strand cDNA synthesis kit for RT-PCR (Roche) according to the manufacturer's instructions. QRT-PCR was performed using a 5700 sequence detector system (Applied Biosystems) with the Power SYBR Green PCR MasterMix (Applied Biosystems) according to the manufacturer's protocol. Each reaction was performed in triplicate with 1 μL of the first-strand cDNA in a total volume of 25 μL. Data are means of three biological samples. The specificity of the PCR amplification was confirmed with a heat dissociation curve (from 60°C to 95°C). Efficiency of the PCR was calculated, and different internal standards were selected (Czechowski et al., 2005) depending of the efficiency of the primers. Relative mRNA abundance was calculated using the comparative threshold cycle method according to Pfaffl (2001). For semiquantitative RT-PCR, cDNAs were PCR amplified in a final volume of 100 μL per reaction. Samples were collected after 20, 25, 30, 35, and 40 cycles. Primers used for PCRs are listed in Supplemental Table S5.
Metabolite Determination
Starch and total soluble sugars were determined with the ENZYTEC starch kit (ATOM). ADP-Glc was determined by HPLC as described by Muñoz et al. (2005) using a Waters Associated Alliance 2695 HPLC system connected to a Whatman Partisil-10SAX (4.6 mm × 250 mm) WCS analytical column and a Waters 996 Photodiode Array Detector. A computer with Waters Millenium32 Chromatograph Manager Software was connected to the HPLC system. The A254 was extracted from the photodiode array for quantifying ADP-Glc. Nitrogen, carbon, and hydrogen contents were determined with a microanalyzer (LECO CHNS-932; Leco Corporation).
Amino acid content was determined by HPLC after derivatization with diethyl ethoxymethylenemalonate (Alaiz et al., 1992). Seed oil quantification by fatty acid methyl ester analysis was done according to Garcés and Mancha (1993) using a Hewlett-Packard 58900 with a split-splitless injector at 250°C, a Supelco SP-2380 column (60-m × 0.25-mm diameter × 0.2-μm film), and a flame ionization detector at 250°C. The oven temperature was 165°C for 20 min, followed by a ramp of 2°C min−1 to 200°C.
Arabidopsis Genome Initiative locus identifiers of Arabidopsis genes used in this article are as follows: GAPCp1 (At1g79530), GAPCp2 (At1g16300), GAPC1 (At1g13440), and GAPA1 (At1g12900).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Germination response of gapcp mutants to sugars and osmoticum.
Supplemental Table S1. Dwarf phenotype and gapcp mutant genotypes are linked.
Supplemental Table S2. Fatty acid composition of mature seeds from wild-type plants, 35S:GAPCp-overexpressing plants (Oex GAPCp), and heterozygous mutant plants (g1.1g1.1 G2g2.1).
Supplemental Table S3. Amino acid composition and protein content of 3-week-old wild-type (WT), gapcp double mutant (g1.1g1.1 g2.1g2.1), and 35S:GAPCp-overexpressing (Oex GAPCp) plants sampled in the middle of the light period after 24 h in the dark (24HD) or after 5 d in the dark (5DD)
Supplemental Table S4. Microarray data.
Supplemental Table S5. Primers used in this work.
Supplementary Material
Acknowledgments
We thank Julian Schroeder (University of California San Diego) for supporting initial research on this project. We thank the Servei Central de Suport a la Investigació Experimental of the Universitat de Valencia for technical assistance, Manuel Alaiz and Arturo Cert (Instituto de la Grasa, Consejo Superior de Investigaciones Científicas) for amino acid and lipid determination, and Prof. Lynne Yenush (Universidad Politécnica de Valencia) for critical reading of the manuscript. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants.
This work was supported by the European Union (Sixth Framework programme, grant no. MOIF–CT–2004–50927), by the Spanish Government (grant no. BFU2006–01621/BFI), by the Valencian Government (grant nos. PROMETEO/2009/075 and ACOMP/2009/328), by a Formación de Profesorado Universitario research fellowship from the Spanish Government to B.C.-M., and by the National Institutes of Health and the National Science Foundation (grant nos. GM060396 and MCB0417118, respectively, to Julian Schroeder at University of California San Diego).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Roc Ros (roc.ros@uv.es).
The online version of this article contains Web-only data.
References
- Alaiz M, Navarro JL, Giron J, Vioque E (1992) Amino acid analysis by high-performance liquid chromatography after derivatization with diethyl ethoxymethylenemalonate. J Chromatogr A 591: 181–186 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Tárraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J (2006) BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res 34: W472–W476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhausen JE, Vetter S, Baalmann E, Kitzmann C, Scheibe R (1998) NAD-dependent malate dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta 205: 359–366 [Google Scholar]
- Baek D, Jin Y, Jeong JC, Lee HJ, Moon H, Lee J, Shin D, Kang CH, Kim DH, Nam J, et al (2008) Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry 69: 333–338 [DOI] [PubMed] [Google Scholar]
- Baroja-Fernández E, Muñoz FJ, Zandueta-Criado A, Morán-Zorzano MT, Viale AM, Alonso-Casajus N, Pozueta-Romero J (2004) Most of ADP·glucose linked to starch biosynthesis occurs outside the chloroplast in source leaves. Proc Natl Acad Sci USA 101: 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S (2000) Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol 122: 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu U, Morgenthal K, Weckwerth W, Pärnik T, Keerberg O, Bauwe H (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eydallin G, Viale AM, Morán-Zorzano MT, Muñoz FJ, Montero M, Baroja-Fernández E, Pozueta-Romero J (2007) Genome-wide screening of genes affecting glycogen metabolism in Escherichia coli K-12. FEBS Lett 581: 2947–2953 [DOI] [PubMed] [Google Scholar]
- Fischer K, Weber A (2002) Transport of carbon in non-green plastids. Trends Plant Sci 7: 345–351 [DOI] [PubMed] [Google Scholar]
- Flügge UI (1999) Phosphate translocators in plastids. Annu Rev Plant Physiol Plant Mol Biol 50: 27–45 [DOI] [PubMed] [Google Scholar]
- Garcés R, Mancha M (1993) One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal Biochem 211: 139–143 [DOI] [PubMed] [Google Scholar]
- Grennan AK (2006) Genevestigator: facilitating Web-based gene-expression analysis. Plant Physiol 141: 1164–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirezaei MR, Biemelt S, Peisker M, Lytovchenko A, Fernie AR, Sonnewald U (2006) The influence of cytosolic phosphorylating glyceraldehyde 3-phosphate dehydrogenase (GAPC) on potato tuber metabolism. J Exp Bot 57: 2363–2377 [DOI] [PubMed] [Google Scholar]
- Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ (2005) Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol Biochem 43: 828–835 [DOI] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7: 665–674 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Ellis TH, Lee D, Turner L (1993) Repeated sequences as genetic markers in pooled tissue samples. Plant Mol Biol 22: 153–157 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito K (1999. a) Plastidic pathway of serine biosynthesis: molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. J Biol Chem 274: 11007–11012 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Saito K (1999. b) Regulation of serine biosynthesis in Arabidopsis: crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem 274: 397–402 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Yamazaki M, Saito K (1998) Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20: 243–259 [DOI] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant 133: 211–228 [DOI] [PubMed] [Google Scholar]
- Jing Y, Cui D, Bao F, Hu Z, Qin Z, Hu Y (2009) Tryptophan deficiency affects organ growth by retarding cell expansion in Arabidopsis. Plant J 57: 511–521 [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV (2005) Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30: 142–150 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Givan CV (1988) Serine formation in leaves by mechanisms other than the glycolate pathway. J Plant Physiol 132: 641–652 [Google Scholar]
- Knappe S, Flügge UI, Fischer K (2003) Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol 131: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Li C (2008) Automating dChip: toward reproducible sharing of microarray data analysis. BMC Bioinformatics 9: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Gauen G, Herbrand H, Pahnke J, Cerff R, Martin W (1998) Gene structure, expression in Escherichia coli and biochemical properties of the NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Pinus sylvestris chloroplasts. Gene 209: 167–174 [DOI] [PubMed] [Google Scholar]
- Meyer-Gauen G, Schnarrenberger C, Cerff R, Martin W (1994) Molecular characterization of a novel, nuclear-encoded, NAD(+)-dependent glyceraldehyde-3-phosphate dehydrogenase in plastids of the gymnosperm Pinus sylvestris L. Plant Mol Biol 26: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Millar H, Considine MJ, Day DA, Whelan J (2001) Unraveling the role of mitochondria during oxidative stress in plants. IUBMB Life 51: 201–205 [DOI] [PubMed] [Google Scholar]
- Mo X, Zhu Q, Li X, Li J, Zeng Q, Rong H, Zhang H, Wu P (2006) The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol 141: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Morigasaki S, Shimada K, Ikner A, Yanagida M, Shiozaki K (2008) Glycolytic enzyme GAPDH promotes peroxide stress signaling through multistep phosphorelay to a MAPK cascade. Mol Cell 30: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz FJ, Baroja-Fernández E, Morán-Zorzano MT, Viale AM, Etxeberria E, Alonso-Casajús N, Pozueta-Romero J (2005) Sucrose synthase controls both intracellular ADP glucose levels and transitory starch biosynthesis in source leaves. Plant Cell Physiol 46: 1366–1376 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Bielecka M, Gakière B, Krueger S, Rinder J, Kempa S, Morcuende R, Scheible WR, Hesse H, Hoefgen R (2006) Effect of sulfur availability on the integrity of amino acid biosynthesis in plants. Amino Acids 30: 173–183 [DOI] [PubMed] [Google Scholar]
- Petersen J, Brinkmann H, Cerff R (2003) Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J Mol Evol 57: 16–26 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE (2004) Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol 136: 3524–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol 148: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Pascual F, Redondo-Horcajo M, Magán-Marchal N, Lagares D, Martínez-Ruiz A, Kleinert H, Lamas S (2008) Glyceraldehyde-3-phosphate dehydrogenase regulates endothelin-1 expression by a novel, redox-sensitive mechanism involving mRNA stability. Mol Cell Biol 28: 7139–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J, Ohlrogge JB, Shachar-Hill Y (2003) A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J Biol Chem 278: 29442–29453 [DOI] [PubMed] [Google Scholar]
- Somerville SC, Somerville CR (1983) Effect of oxygen and carbon dioxide on photorespiratory flux determined from glycine accumulation in a mutant of Arabidopsis thaliana. J Exp Bot 34: 415–424 [Google Scholar]
- Taiz L, Zeiger E (2006) Plant Physiology, Ed 4. Sinauer Associates, Sunderland, MA
- Waditee R, Bhuiyan NH, Hirata E, Hibino T, Tanaka Y, Shikata M, Takabe T (2007) Metabolic engineering for betaine accumulation in microbes and plants. J Biol Chem 282: 34185–34193 [DOI] [PubMed] [Google Scholar]
- Weber AP, Schwacke R, Flügge UI (2005) Solute transporters of the plastid envelope membrane. Annu Rev Plant Biol 56: 133–164 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.