Abstract
Although allelic diversity of genes has been reported to play important roles in different physiological processes, information on allelic diversity of defense-responsive genes in host-pathogen interactions is limited. Here, we report that a pair of allelic genes, OsWRKY45-1 and OsWRKY45-2, which encode proteins with a 10-amino acid difference, play opposite roles in rice (Oryza sativa) resistance against bacterial pathogens. Bacterial blight caused by Xanthomonas oryzae pv oryzae (Xoo), bacterial streak caused by Xanthomonas oryzae pv oryzicola (Xoc), and fungal blast caused by Magnaporthe grisea are devastating diseases of rice worldwide. OsWRKY45-1-overexpressing plants showed increased susceptibility and OsWRKY45-1-knockout plants showed enhanced resistance to Xoo and Xoc. In contrast, OsWRKY45-2-overexpressing plants showed enhanced resistance and OsWRKY45-2-suppressing plants showed increased susceptibility to Xoo and Xoc. Interestingly, both OsWRKY45-1- and OsWRKY45-2-overexpressing plants showed enhanced resistance to M. grisea. OsWRKY45-1-regulated Xoo resistance was accompanied by increased accumulation of salicylic acid and jasmonic acid and induced expression of a subset of defense-responsive genes, while OsWRKY45-2-regulated Xoo resistance was accompanied by increased accumulation of jasmonic acid but not salicylic acid and induced expression of another subset of defense-responsive genes. These results suggest that both OsWRKY45-1 and OsWRKY45-2 are positive regulators in rice resistance against M. grisea, but the former is a negative regulator and the latter is a positive regulator in rice resistance against Xoo and Xoc. The opposite roles of the two allelic genes in rice-Xoo interaction appear to be due to their mediation of different defense signaling pathways.
Plant pathogens are continually evolving to survive. Plants have developed a set of mechanisms to face the challenge of foreign pathogens through a long history of coevolution. Among these mechanisms, maintaining allele (or ortholog) variation or diversity, either at the gene structure level or the expression level, is an important way for plants to protect themselves from pathogen attack. Plant responses to pathogen infection are regulated by different types of genes. The disease resistance (R) genes mediate race-specific resistance by initiation of defense signaling. The allelic variation of most characterized R genes and their alleles is regulated at the gene structure level; different resistant alleles of an R gene and its susceptible allele frequently encode different proteins (Sun et al., 2004; Zhou et al., 2006). In a few cases, the variation of R genes and their susceptible alleles is regulated expressionally (Gu et al., 2005; Chu et al., 2006; Romer et al., 2007). Based on our understanding of R gene-mediated resistance, dominant R genes function as positive regulators, and their susceptible alleles have no function in host-pathogen interaction (Gu et al., 2005; Romer et al., 2007); in contrast, recessive R genes appear to have no function, and their susceptible (dominant) alleles function as negative regulators in defense responses (Chu et al., 2006; Jiang et al., 2006). There is no report that an R gene and its allele function as positive and negative regulators in defense responses, respectively.
A large number of other genes, which function in the defense signaling pathways initiated by R genes or the pathways leading to basal immunity, respond to pathogen attack by changing expression levels or by posttranslational modification of their encoding proteins. Thus, they are frequently called defense-responsive or defense-related genes. Although a large number of defense-responsive genes express differentially in host-pathogen interactions, the differential expression of most of these genes may be due to the activation of defense signaling in resistant reactions but not the variation of allelic expression, because gene expression was analyzed using near-isogenic lines for R genes (Zhou et al., 2002; Chu et al., 2004; Hulbert et al., 2007). Unlike the R genes, which are frequently subjected to positive selection, resulting in genetic diversity (Mondragon-Palomino et al., 2002; Sun et al., 2006), the defense-responsive genes have low levels of polymorphism and, in general, experience purifying selection (Bakker et al., 2008).
One important group of genes, which are also responsive to pathogen infection, is those encoding transcription factors that modulate the defense transcriptome. A number of WRKY-type transcription factors from different plant species have been identified to play important roles in host-pathogen interactions (Eulgem and Somssich, 2007; Pandey and Somssich, 2009). These WKRYs function either as positive or negative regulators in defense responses. Some WRKYs are both positive and negative regulators in different defense responses (Li et al., 2004, 2006; Wang et al., 2006; Xu et al., 2006). Some other WRKYs have partly redundant functions in defense signaling (Xu et al., 2006). However, there is no report that a WRKY gene or other type of defense-responsive gene and its allele function as a positive and negative regulators in defense responses, respectively.
Bacterial blight caused by Xanthomonas oryzae pv oryzae (Xoo), bacterial streak caused by Xanthomonas oryzae pv oryzicola (Xoc), and fungal blast caused by Magnaporthe grisea are devastating diseases of rice (Oryza sativa) worldwide. A numbers of R genes, but only a few resistance quantitative trait locus (QTL) genes for bacterial blight and blast resistance, have been isolated. A rice disease resistance QTL gene, OsWRKY13, which encodes a WRKY-type protein, is an important regulator of rice-Xoo and rice-M. grisea interactions (Qiu et al., 2007; Hu et al., 2008). Activation of OsWRKY13 can enhance rice resistance against Xoo and M. grisea (Qiu et al., 2007). In disease resistance, OsWRKY13's function is associated with activation of salicylic acid (SA)-dependent pathways and suppression of jasmonic acid (JA)-dependent pathways (Qiu et al., 2007, 2008). However, no R gene for Xoc resistance has been identified, and none of the resistance QTLs against Xoc has been characterized.
Our previous study showed that OsWRKY45 (locus identifier LOC_Os05g25770), according to the rice genome annotation of The Institute for Genomic Research (http://rice.tigr.org), functioned downstream of OsWRKY13 (Qiu et al., 2009). Activation of OsWRKY13 repressed OsWRKY45 expression, and suppression of OsWRKY13 enhanced OsWRKY45 expression; furthermore, Xoo infection influenced OsWRKY45 expression. These results suggest that OsWRKY45 may be involved in rice-Xoo interactions. In addition, one study reported that OsWRKY45 transcription factor plays a crucial role in benzothiadiazole-inducible blast resistance (Shimono et al., 2007); another study showed that overexpressing OsWRKY45 in Arabidopsis enhanced resistance to the bacterial pathogen Pseudomonas syringae tomato and enhanced tolerance to salt and drought stresses (Qiu and Yu, 2009). To study the role of OsWRKY45 in rice response to Xoo infection, we found that the two alleles of OsWRKY45 functioned differently in rice-pathogen interactions. We refer to the allele (Shimono et al., 2007) from japonica rice var Nipponbare as OsWRKY45-1 and to the allele from indica rice var Minghui 63 as OsWRKY45-2. OsWRKY45-1 acted as a negative regulator and OsWRKY45-2 as a positive regulator in both rice-Xoo and rice-Xoc interactions, although both alleles functioned as positive regulators in rice-M. grisea interactions. The opposite roles of this pair of alleles in bacterial resistance appear to be due to their regulation of different defense signaling pathways.
RESULTS
Two Alleles of the OsWRKY45 Gene in Different Rice Varieties
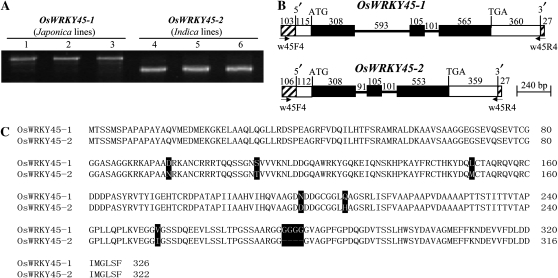
Asian cultivated rice consists of two major groups, which are known by the subspecies names indica and japonica. Amplification of OsWRKY45 from japonica rice var Nipponbare and indica rice var Minghui 63 using gene-specific PCR primers generated differently sized PCR products (Fig. 1A). Comparative analysis of the genomic and cDNA sequences of OsWRKY45 showed two homologous genes, defined as OsWRKY45-1 (GenBank accession no. GQ331932) from Nipponbare and OsWRKY45-2 (GQ331927) from Minghui 63 (Fig. 1B). The major differences between the two genes were a 502-nucleotide deletion in the first intron and a 12-nucleotide deletion in the third exon of OsWRKY45-2 compared with OsWRKY45-1. OsWRKY45-1 encodes a protein consisting of 326 amino acids, and OsWRKY45-2 encodes a protein of 322 amino acids; in addition, the two proteins have six amino acid substitutions (Fig. 1C). OsWRKY45 was also amplified from other rice varieties (Fig. 1A). Sequence comparison showed that another two japonica rice varieties, Mudanjiang 8 (GQ331930) and Dongjin (GQ331931), carried OsWRKY45-1, and another two indica rice varieties, Zhenshan 97 (GQ331928) and 93-11 (GQ331929), carried OsWRKY45-2.
Figure 1.
Structure and sequence comparison of OsWKRY45-1 and OsWRKY45-2. A, Amplification of OsWRKY45 using primers w45F4 and w45R4 showed differently sized PCR products in different rice varieties. The PCR products were 2,293 nucleotides in japonica var Nipponbare (1), Mudanjiang 8 (2), and Dongjin (3). The PCR products were 1,778 nucleotides for indica var Minghui 63 (4), Zhenshan 97 (5), and 93-11 (6). B, Gene structure. The coding regions (black boxes) of OsWRKY45-1 and OsWRKY45-2 are interrupted by introns (thick lines). The positions of 5′ and 3′ untranslated regions (white boxes), translation start codon (ATG), translation stop codon (TGA), and DNA fragments flanking the genes (hatched boxes) are also indicated. The numbers indicate the nucleotides of each substructure. C, Amino acid sequence alignment of OsWRKY45-1 and OsWRKY45-2 proteins. Dashes indicates a gap.
To determine whether the two genes are in the same locus of each genome or if there are alleles, an allelic test was performed using an F2 population segregating for OsWRKY45-1 and OsWRKY45-2. The numbers of F2 individuals carrying only OsWRKY45-1, both OsWRKY45-1 and OsWRKY45-2, and only OsWRKY45-2 were 42, 71, and 33, respectively, which fit the expected 1:2:1 ratio (χ2 = 1.22, P > 0.5). The two genes were mapped on chromosome 5 (Supplemental Fig. S1). In addition, BLAST analysis (Altschul et al., 1997) of the two gene sequences against Nipponbare and 93-11 whole genome sequences (http://rice.plantbiology.msu.edu/blast.shtml) identified only one gene with 100% coverage and 100% sequence identity to OsWRKY45-1 in the Nipponbare genome and only one gene with 100% coverage and 99.8% sequence identity to OsWRKY45-2 in the 93-11 genome. The homologs in Nipponbare and 93-11 genomes all localize on chromosome 5. These results suggest that OsWRKY45-1 and OsWRKY45-2 are alleles.
The promoter regions (approximately 1.5 kb upstream of the transcription initiation site) of different OsWRKY45 alleles from the six rice varieties were also compared. The OsWRKY45-1 promoters from japonica var Nipponbare, Mudanjiang 8, and Dongjin had identical sequences. The OsWRKY45-2 promoters from indica var Minghui 63, Zhenshan 97, and 93-11 also had identical sequences. There were 18 nucleotide substitutions and one insertion in the promoter region of OsWRKY45-2 compared with that of OsWRKY45-1 (Supplemental Fig. S2). These results suggest that there are at least two alleles of OsWRKY45, which appear to occur differentially in the two subspecies of Asian cultivated rice. Since OsWRKY45-1 is involved in benzothiadiazole-inducible blast resistance (Shimono et al., 2007), these results also suggest that the two alleles may function differently in rice-pathogen interactions.
OsWRKY45-1 and OsWRKY45-2 Play Opposite Roles in Rice-Xoo Interactions
To examine the above hypothesis, OsWRKY45-1 and OsWRKY45-2, driven by a constitutive (maize [Zea mays] ubiquitin gene) promoter (PUbi), were transferred into japonica rice var Mudanjiang 8, which is susceptible to Xoo strain PXO61 (Qiu et al., 2007). An RNA interference construct for OsWRKY45-2 was transferred into indica rice var Minghui 63, which is moderately resistant to PXO61 (Qiu et al., 2009). An OsWRKY45-1-knockout mutant, 2C-50229, which had the genetic background of japonica var Dongjin, was used for analysis.
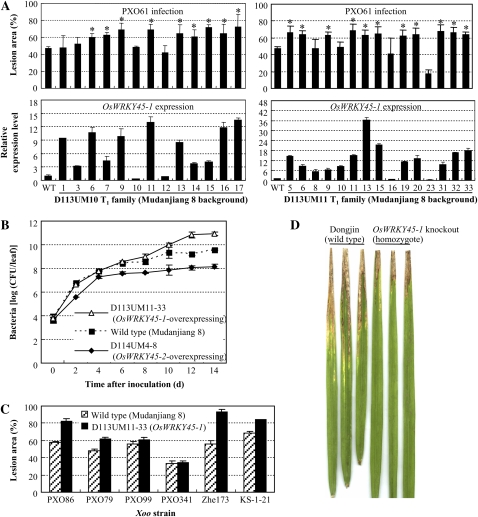
Ten independent transformants carrying PUbi:OsWRKY45-1, named D113UM3 to D113UM12, were obtained. Four of the 10 T0 plants carrying PUbi:OsWRKY45-1 were significantly more susceptible (P < 0.05) to Xoo strain PXO61, with lesion areas ranging from 66% to 69%, compared with 55% for wild-type Mudanjiang 8 (Supplemental Fig. S3). To verify that the increased susceptibility of the transgenic plants was due to overexpression of OsWRKY45-1, two T1 families from D113UM10 and D113UM11 were analyzed for susceptibility by inoculation with PXO61 and for OsWRKY45-1 expression level. The results showed that all of the susceptible plants overexpressed OsWRKY45-1 (Fig. 2A). The growth rate of bacteria on OsWRKY45-1-overexpressing plants was 2.5- to 43.8-fold higher than that on wild-type Mudanjiang 8 at 10 to 14 d after infection (Fig. 2B). T2 plants from the D113UM11-33 line were further examined for their responses to different Xoo strains. The transgenic plants were significantly more susceptible (P < 0.05) to Xoo strains PXO86, PXO79, PXO99, Zhe173, and KS-1-21 than the wild type (Fig. 2C). These results suggest that overexpression of OsWRKY45-1 resulted in a broad-spectrum susceptibility to Xoo strains.
Figure 2.
The role of OsWRKY45-1 in rice-Xoo interaction. Bars represent means (four to five replicates for the lesion area and three replicates for expression level) ± sd. A, Increased susceptibility to Xoo strain PXO61 was associated with overexpression of OsWRKY45-1 in OsWRKY45-1-overexpressing T1 families D113UM10 and D113UM11. The asterisks indicate that a significant difference (P < 0.05) in the lesion area was detected between transgenic plants and the wild type (WT). The expression levels of OsWRKY45-1 in transgenic plants are relative to that in the wild type. B, Growth of PXO61 in leaves of T2 plants of D113UM11-33 overexpressing OsWRKY45-1 and D114UM4-8 overexpressing OsWRKY45-2. Bacterial populations were determined from three leaves at each time point by counting colony-forming units (CFU). C, OsWRKY45-1-overexpressing transgenic line D113UM11-33 was also susceptible to other Xoo strains. D, Knockout of OsWRKY45-1 (2C-50229) enhanced rice resistance to Xoo strain PXO86. [See online article for color version of this figure.]
The response of the OsWRKY45-1-knockout mutant (2C-50229), which had a T-DNA inserted into its promoter (Supplemental Fig. S4A), to Xoo strain PXO86 was examined. The plants with homozygote T-DNA insertion, in which the expression level of OsWRKY45-1 was only approximately 2% to 5% of that in wild-type Dongjin, as detected by quantitative reverse transcription (qRT)-PCR, showed markedly enhanced resistance, with an average lesion area of 20% compared with 46% for the wild type (Fig. 2D; Supplemental Fig. S4, B and C). These results suggest that reducing OsWRKY45-1 transcripts can enhance rice resistance to Xoo. In conclusion, OsWRKY45-1 acts as a negative regulator in the rice response to Xoo infection.
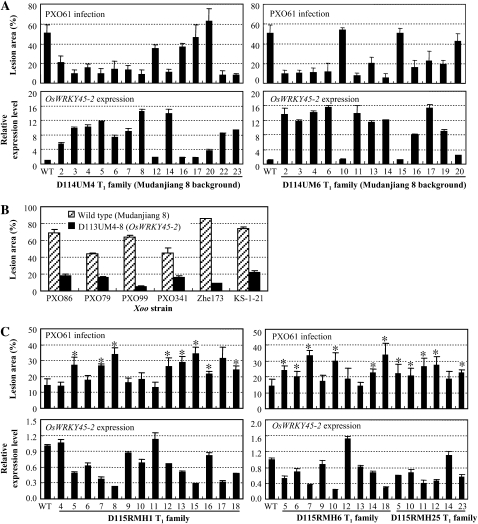
Seventeen independent transformants carrying PUbi:OsWRKY45-2, named D114UM1 to D114UM17, were obtained. Sixteen of the 17 T0 plants carrying PUbi:OsWRKY45-2 showed significantly enhanced resistance to Xoo strain PXO61, with lesion areas ranging from 5% to 28% compared with 55% for wild-type Mudanjiang 8 (Supplemental Fig. S5). Two T1 families from resistant D114UM4 and D114UM6 were further analyzed for resistance to PXO61 and for OsWRKY45-2 expression level. The results showed that the enhanced resistance was associated with overexpression of OsWRKY45-2 in the T1 families (Fig. 3A). The bacterial growth rate in OsWRKY45-2-overexpressing plants was 2- to 20-fold lower than that in the wild type at 2 to 14 d after infection (Fig. 2B). T2 plants from the D114UM4-8 line were further examined for their resistance spectrum to different Xoo strains. The transgenic plants showed markedly enhanced resistance to Xoo strains PXO86, PXO79, PXO99, PXO341, Zhe173, and KS-1-21 compared with the wild type (Fig. 3B). The lesion areas of the transgenic plants were reduced 51% to 94% compared with wild-type Mudanjiang 8. These results suggest that an increasing expression level of OsWRKY45-2 can promote a broad-spectrum resistance to Xoo strains.
Figure 3.
The role of OsWRKY45-2 in rice-Xoo interaction. The asterisks indicate that a significant difference (P < 0.05) in the lesion area was detected between transgenic plants and the wild type (WT). The expression levels of OsWRKY45-2 in transgenic plants are relative to that in the wild type. Bars represent means (four to five replicates for the lesion area and three replicates for expression level) ± sd. A, Enhanced resistance to Xoo strain PXO61 associated with overexpression of OsWRKY45-2 in T1 families D114UM4 and D114UM6. B, The OsWRKY45-2-overexpressing line D113UM4-8 was also resistant to other Xoo strains. C, Suppressing OsWRKY45-2 decreased rice resistance to Xoo strain PXO61. Minghui 63 is the wild type.
Thirty-one independent transformants carrying the OsWRKY45-2 RNA interference construct, named D115RMH1 to D115RMH31, were obtained. Minghui 63 is moderately resistant to Xoo strain PXO61 (Sun et al., 2004). Some of the T0 plants showed decreased resistance to PXO61 compared with wild-type Minghui 63 (data not shown). Three T1 families developed from three susceptible T0 plants, D115RMH1, -6, and -25, were further analyzed for their response to PXO61 and the OsWRKY45-2 transcript level. The results showed that the decreased expression levels of OsWRKY45-2 were associated with increased susceptibility in all of the T1 families (Fig. 3C). These results suggest that OsWRKY45-2 acts as a positive regulator in rice response to Xoo infection.
The Two Alleles Have Different Expression Responses to Xoo Infection
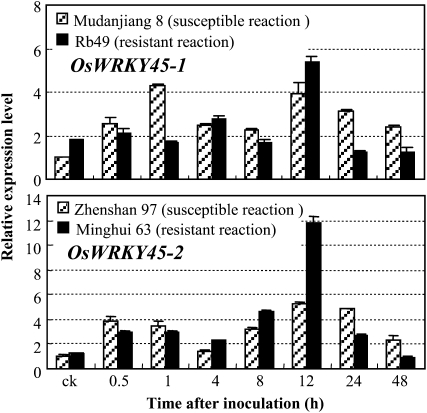
To evaluate whether the opposite functions of the two alleles in rice-Xoo interactions were due to their different expression patterns in pathogen infection, we examined their expression in different rice lines (Fig. 4). Rice var Mudanjiang 8, which is susceptible to Xoo strain PXO61, carries OsWRKY45-1. Transgenic line Rb49 carries an R gene, Xa3/Xa26, against PXO61 and has the genetic background of Mudanjiang 8 (Cao et al., 2007). Rice var Minghui 63 and Zhenshan 97 carry OsWRKY45-2, and the former, carrying Xa3/Xa26, is moderately resistant to PXO61, whereas the latter is susceptible to PXO61 (Cao et al., 2007). Although PXO61 infection induced OsWRKY45-1 and OsWRKY45-2 expression in both resistant and susceptible rice lines, the two alleles showed different expression patterns in resistant lines as compared with corresponding susceptible lines (Fig. 4). The expression level of OsWRKY45-1 in the resistant line was higher than that in the susceptible line without pathogen infection but was markedly lower than that in the susceptible line in early infection (1 h). In contrast, the expression of OsWRKY45-2 in the resistant line was markedly induced at 12 h after infection. Because OsWRKY45-1 and its promoter are identical in Mudanjiang 8 and Rb49 and OsWRKY45-2 and its promoter are identical in Zhenshan 97 and Minghui 63, the expression differences of the two alleles in susceptible and resistant lines may be due to the differential regulation by other transcriptional regulator(s) that is activated or suppressed during rice-Xoo interaction. Furthermore, the different expression patterns of OsWRKY45-1 and OsWRKY45-2 in resistant lines, as compared with those in their corresponding susceptible lines, suggest that the two alleles may be regulated by different transcriptional regulators due to promoter difference.
Figure 4.
OsWRKY45-1 and OsWRKY45-2 expression on pathogen infection. Plants were inoculated with Xoo strain PXO61 at the booting stage. ck, Before inoculation. Bars represent means (three replicates) ± sd.
OsWRKY13 Transcription Regulator Binds to the Promoters of OsWRKY45-1 and OsWRKY45-2 in Vivo
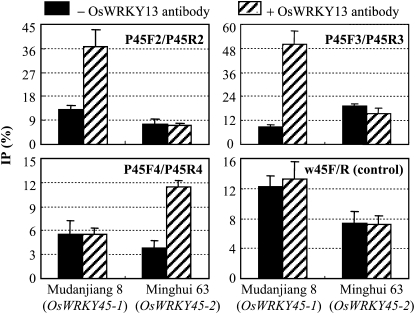
OsWRKY13 bound to the promoter of OsWRKY45 in vitro (Qiu et al., 2009). To evaluate whether OsWRKY13 was the direct regulator of OsWRKY45, we performed chromatin immunoprecipitation assay using anti-OsWRKY13 antibody (Supplemental Fig. S6). The samples were from rice var Mudanjiang 8, carrying OsWRKY45-1, and Minghui 63, carrying OsWRKY45-2. Three segments of OsWRKY45-1 and OsWRKY45-2 promoters, which putatively harbor W-boxes or W-box-like cis-elements for the binding of WRKY transcription factors or polymorphic sites, were analyzed (Supplemental Fig. S2). After immunoprecipitation with anti-OsWRKY13 antibody, enrichment of the first (P45F3/P45R3) and second (P45F2/P45R2) fragments of the OsWRKY45-1 promoter and the third (P45F4/P45R4) fragment of the OsWRKY45-2 promoter was detected by real-time PCR (Fig. 5). This result suggests that OsWRKY13 binds to the promoters of OsWRKY45-1 and OsWRKY45-2 in vivo, but it has partiality to bind to the different sites of the two promoters.
Figure 5.
OsWRKY13 protein binds to the promoters of OsWRKY45-1 and OsWRKY45-2. Chromatin immunoprecipitation assay was performed with the extracts from the leaves of Mudanjiang 8 and Minghui 63 seedlings. Real-time PCR was conducted before immunoprecipitation (input), after immunoprecipitation (IP) with anti-OsWRKY13 antibody (+), or after immunoprecipitation without anti-OsWRKY13 antibody (−). The amplification of the 3′ untranslated region of OsWRKY45-1 or OsWRKY45-2 using PCR primer pair w45F/R served as a sample quantity control. The amounts of target PCR products from IP are relative (IP/input %) to those from input. Bars represent means (three replicates) ± sd.
OsWRKY45-1 and OsWRKY45-2 Differentially Regulate the Expression of a Set of Defense-Responsive Genes
To ascertain which defense-responsive genes were influenced by OsWRKY45-1 or OsWRKY45-2, we analyzed the expression of 10 genes known to function in SA- or JA-dependent pathways in different rice plants after infection of Xoo strain PXO61 (Fig. 6). PAL1 (for Phe ammonia lyase 1; GenBank accession no. X16099) is involved in SA synthesis by the phenylpropanoid pathway. ICS1 (for isochorismate synthase 1; AK120689) and PAD4 (for phytoalexin-deficient 4; CX118864) are putatively involved in SA synthesis in rice by the isochorismate pathway (Qiu et al., 2007). Induced expression of PR1a (for acidic pathogenesis-related [PR] protein 1; AJ278436), NH1 (for Arabidopsis [Arabidopsis thaliana] NPR1 homolog 1; AY9123983), or OsWRKY13 (EF143611) was associated with activation of the SA-dependent pathway. LOX (for lipoxygenase; D14000) and AOS2 (for allene oxide synthase 2; AY062258) are involved in JA synthesis. PR1b (for basic PR protein 1; U89895) and PR10/PBZ1 (for ribonuclease; D38170) appear to function in both JA- and SA-dependent pathways (Qiu et al., 2007; Yuan et al., 2007; X. Shen and S. Wang, unpublished data).
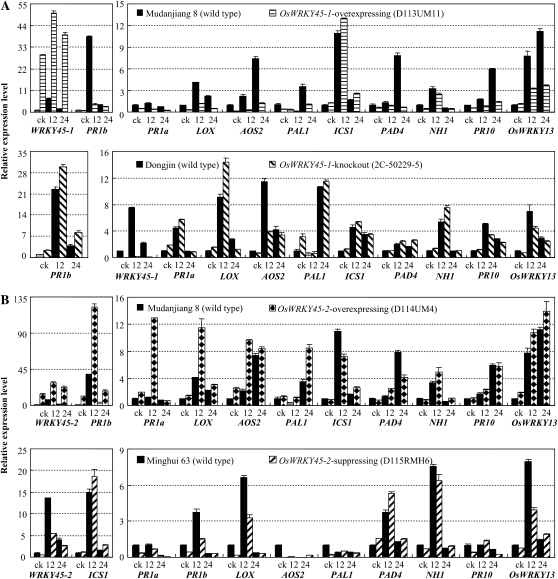
Figure 6.
Modulating OsWRKY45-1 (A) and OsWRKY45-2 (B) expression influenced the expression of other defense-responsive genes analyzed by qRT-PCR. AOS2, Allene oxide synthase 2; ICS1, isochorismate synthase 1; LOX, lipoxygenase; NH1, NPR1 homolog 1; PAD4, phytoalexin-deficient 4; PAL1, Phe ammonia lyase 1; PR1a, acidic pathogenesis-related protein 1; PR1b, basic pathogenesis-related protein 1; PR10/PBZ1, ribonuclease. Samples were collected before inoculation (ck) and at 12 and 24 h after inoculation with Xoo strain PXO61. Bars represent means (three replicates) ± sd.
In OsWRKY45-1-containing plants, the expression of PAL1, PAD4, PR1a, NH1, LOX, and PR1b was significantly increased (P < 0.01) in OsWRKY45-1-knockout plants (enhanced resistance) compared with wild-type Dongjin and was significantly suppressed (P < 0.01) in OsWRKY45-1-overexpressing plants (increased susceptibility) compared with wild-type Mudanjiang 8 in at least one time point examined (Fig. 6A). Both OsWRKY45-1-knockout and -overexpressing plants showed suppressed expression of AOS2, PR10/PBZ1, and OsWRKY13 and slightly increased expression of ICS1. These results suggest that OsWRKY45-1 may play an important role in regulating the expression of PAL1, PAD4, PR1a, NH1, LOX, and PR1b.
In OsWRKY45-2-containing plants, the expression of PAL1, PR1a, NH1, OsWRKY13, LOX, AOS2, and PR1b was significantly increased (P < 0.01) in OsWRKY45-2-overexpressing plants (enhanced resistance) compared with wild-type Mudanjiang 8 and was significantly suppressed (P < 0.01; NH1, P < 0.05) in OsWRKY45-2-suppressing plants (increased susceptibility) compared with wild-type Minghui 63 in at least one time point examined (Fig. 6B). The expression of ICS1 and PAD4 was suppressed in OsWRKY45-2-overexpressing plants and induced in OsWRKY45-2-suppressing plants compared with their corresponding wild-type plants in at least one time point examined. PR10/PBZ1 expression showed no obvious difference between OsWRKY45-2-overexpressing and wild-type plants but was significantly suppressed (P < 0.01) in OsWRKY45-2-suppressing plants compared with wild-type Minghui 63 (Fig. 6B). These results suggest that OsWRKY45-2 may play an important role in regulating the expression of PAL1, ICS1, PAD4, PR1a, NH1, OsWRKY13, LOX, AOS2, and PR1b. The different expression patterns of this set of defense-responsive genes in OsWRKY45-1- and OsWRKY45-2-containing plants suggest that this pair of alleles may regulate rice-Xoo interactions by different defense signaling.
OsWRKY45-1 and OsWRKY45-2 Influence the Levels of Endogenous SA and JA
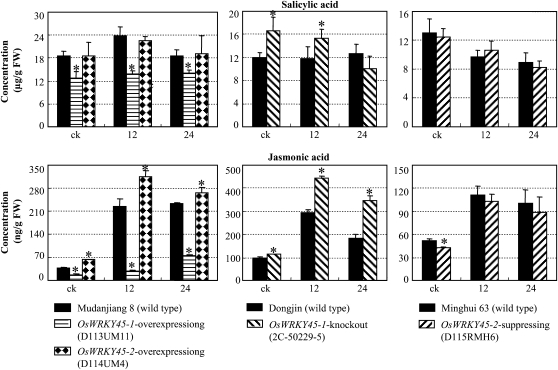
To examine whether the modified expression of defense-responsive genes caused by OsWRKY45 influences the endogenous levels of JA and SA, we quantified the concentrations of the two signal molecules in the leaves of the same plants used for analyzing the expression of defense-responsive genes after infection of Xoo strain PXO61. In OsWRKY45-1-containing wild-type plants, PXO61 infection markedly induced JA accumulation in both Mudanjiang 8 and Dongjin but only slightly induced SA accumulation in Mudanjiang 8 (Fig. 7). In OsWRKY45-2-containing wild-type Minghui 63, PXO61 infection induced JA accumulation and suppressed SA accumulation. The SA and JA levels were significantly reduced (P < 0.05) in OsWRKY45-1-overexpressing plants and were significantly increased (P < 0.05) in OsWRKY45-1-knockout plants compared with their corresponding wild types (Fig. 7). The JA level was significantly increased (P < 0.05) in OsWRKY45-2-overexpressing plants and decreased in OsWRKY45-2-suppressing plants compared with their corresponding wild types. The SA levels in both OsWRKY45-2-overexpressing and -suppressing plants showed no significant differences (P > 0.05) from their corresponding wild types. In conclusion, the Xoo resistance mediated by OsWRKY45-1-knockout plants was associated with increased accumulation of SA and JA, and the Xoo resistance mediated by OsWRKY45-2-overexpressing plants was associated with accumulation of JA but not SA. These results further support the hypothesis that the Xoo resistance negatively regulated by OsWRKY45-1 is different from that positively regulated by OsWRKY45-2.
Figure 7.
Modulating OsWRKY45-1 and OsWRKY45-2 expression influenced the accumulation of SA and JA. Samples were collected before inoculation (ck) and at 12 and 24 h after inoculation with Xoo strain PXO61. Bars represent means (three replicates) ± sd. Asterisks indicate that a significant difference (P < 0.05) was detected between transgenic plants and the corresponding wild type of the same treatment. FW, Fresh weight.
OsWRKY45-1 and OsWRKY45-2 Also Play Opposite Roles in Rice-Xoc Interactions
The OsWRKY45-1-overexpressing lines (D113UM10 and D113UM11) were more susceptible to Xoc strain RH3; the lesion lengths of these transgenic plants were increased 73% to 95% compared with wild-type Mudanjiang 8, which was moderately susceptible to Xoc (Table I). In contrast, the OsWRKY45-1-knockout plants (2C-50229-5) were more resistant to RH3; the lesion lengths of these plants were reduced 47% compared with wild-type Dongjin, which was moderately susceptible to Xoc. The OsWRKY45-2-overexpressing lines (U114UM4 and D114UM6) were more resistant to RH3 compared with wild-type Mudanjiang 8; the lesion lengths of these transgenic plants were reduced 52% to 56% (Table I). In contrast, the OsWRKY45-2-suppressing lines (D115RMH1 and D115RMH6) were more susceptible to Xoc; the lesion lengths of these transgenic plants were increased approximately 17% compared with wild-type Minghui 63, which was susceptible to Xoc. These results suggest that OsWRKY45-1 negatively regulates rice resistance to Xoc and that OsWRKY45-2 positively regulates rice resistance to Xoc.
Table I.
Performance of transgenic plants inoculated with Xoc strain RH3
| Rice Line | Genetic Background | Lesion Lengtha | P |
|---|---|---|---|
| cm | |||
| D113UM10 (OsWRKY45-1-overexpressing) | Mudanjiang 8 | 2.58 ± 0.07 | 0.0000 |
| D113UM11 (OsWRKY45-1-overexpressing) | Mudanjiang 8 | 2.29 ± 0.19 | 0.0019 |
| D114UM4 (OsWRKY45-2-overexpressing) | Mudanjiang 8 | 0.58 ± 0.04 | 0.0004 |
| D114UM6 (OsWRKY45-2-overexpressing) | Mudanjiang 8 | 0.63 ± 0.06 | 0.0001 |
| Mudanjiang 8 (wild type) | Mudanjiang 8 | 1.32 ± 0.08 | |
| 2C-50229-5 (OsWRKY45-1-knockout) | Dongjin | 0.73 ± 0.11 | 0.0009 |
| Dongjin (wild type) | Dongjin | 1.38 ± 0.11 | |
| D115RMH1 (OsWRKY45-2-suppressing) | Minghui 63 | 2.37 ± 0.10 | 0.0084 |
| D115RMH6 (OsWRKY45-2-suppressing) | Minghui 63 | 2.39 ± 0.18 | 0.0405 |
| Minghui 63 (wild type) | Minghui 63 | 2.03 ± 0.06 |
Data represents means (three plants each with four lesions per plant) ± sd.
Both OsWRKY45-1 and OsWRKY45-2 Positively Regulate Rice Resistance against M. grisea
A previous study reported that overexpression of OsWRKY45 (named OsWRKY45-1 in this study) enhanced rice resistance to blast disease (Shimono et al., 2007). To ascertain whether this pair of alleles also had different responses in rice-M. grisea interactions, we inoculated the transgenic plants with M. grisea isolate 91-17-2. Both OsWRKY45-1- and OsWRKY45-2-overexpressing plants showed enhanced resistance to M. grisea compared with wild-type Mudanjing 8, which was highly susceptible (Table II). However, the OsWRKY45-2-overexpressing plants were more resistant to 91-17-2 than OsWRKY45-1-overexpressing plants. In contrast, OsWRKY45-1-knockout and OsWRKY45-2-suppressing plants were more susceptible to 91-17-1 compared with wild-type Dongjin, which was resistant, and Minghui 63, which was moderately susceptible, respectively (Table II). These results suggest that both OsWRKY45-1 and OsWRKY45-2 act as positive regulators in rice-M. grisea interactions.
Table II.
Performance of transgenic plants inoculated with M. grisea isolate 91-17-2
| Rice Line | Genetic Background | Disease Indexa | Resistance/Susceptibility |
|---|---|---|---|
| D113UM11 (OsWRKY45-1-overexpressing) | Mudanjiang 8 | 38.4 | Moderately susceptible |
| D113UM10 (OsWRKY45-1-overexpressing) | Mudanjiang 8 | 39.3 | Moderately susceptible |
| D114UM4 (OsWRKY45-2-overexpressing) | Mudanjiang 8 | 17.9 | Moderately resistant |
| D114UM6 (OsWRKY45-2-overexpressing) | Mudanjiang 8 | 13.4 | Resistant |
| Mudanjiang 8 (wild type) | Mudanjiang 8 | 67.9 | Highly susceptible |
| 2C-50229-5 (OsWRKY45-1-knockout) | Dongjin | 23.4 | Moderately resistant |
| Dongjin (wild type) | Dongjin | 14 | Resistant |
| D115RMH1 (OsWRKY45-2-suppressing) | Minghui 63 | 42.2 | Moderately susceptible |
| D115RMH6 (OsWRKY45-2-suppressing) | Minghui 63 | 44.5 | Moderately susceptible |
| Minghui 63 (wild type) | Minghui 63 | 32.8 | Moderately susceptible |
Disease index was calculated with the individual leaf ratings using the following formula: disease index = [sum of numerical ratings from all leaves/(number of leaves assessed × maximum lesion rating)] × 100.
DISCUSSION
OsWRKY45-1 and OsWRKY45-2 Modulate Rice-Xoo Interactions via Different Mechanisms
The WRKY superfamily of rice consists of at least 98 members in japonica rice and 102 members in indica rice (Ross et al., 2007). Nine rice WRKY genes have been characterized to be involved in pathogen-induced defense responses. OsWRKY13 positively regulates rice defense responses against both Xoo and M. grisea (Qiu et al., 2007). OsWRKY71 positively regulates rice resistance to Xoo (Liu et al., 2007). OsWRKY53, OsWRKY45-1, OsWRKY89, and OsWRKY31 are positive regulators of rice resistance to M. grisea (Chujo et al., 2007; Shimono et al., 2007; Wang et al., 2007; Zhang et al., 2008). Although 15 Arabidopsis WRKY proteins have been identified to negatively regulate defense responses against different pathogens (Pandey and Somssich, 2009), only one rice WRKY protein, OsWRKY62, has been reported to be a negative regulator in rice pathogen defense (Peng et al., 2008). OsWRKY62 negatively regulates defense responses in basal and Xa21-mediated resistance against Xoo. One WRKY protein from pepper (Capsicum annuum) also functions as a negative regulator of pathogen defense (Oh et al., 2008). In adding to this list, our results suggest that both OsWRKY45-1 and OsWRKY45-2 are positive regulators in rice resistance against M. grisea, but the former is a negative regulator and the latter is a positive regulator in rice resistance against Xoo and Xoc.
The opposite functions of OsWRKY45-1 and OsWRKY45-2 in rice-Xoo interactions are controlled by different defense signaling pathways. This hypothesis is supported by the following evidence. First, the Xoo resistance, which was negatively regulated by OsWRKY45-1, was associated with increased accumulation of SA and JA, but the Xoo resistance, which was positively regulated by OsWRKY45-2, was only associated with increased accumulation of JA. Second, the expression patterns of a subset of defense-responsive genes were different in the OsWRKY45-1- and OsWRKY45-2-mediated disease resistance. The expression of ICS1 and PAD4, which are putatively involved in SA biosynthesis in rice via the isochorismate pathway (Qiu et al., 2007), was increased in OsWRKY45-1 negatively regulated resistance and appeared to be suppressed in OsWRKY45-2 positively regulated resistance. OsWRKY45-1-mediated resistance was accompanied by suppressed expression of OsWRKY13 and slightly induced expression of PR1a and PR1b, but OsWRKY45-2-mediated resistance was accompanied by increased expression of OsWRKY13 and markedly induced expression of PR1a and PR1b. Last, the OsWRKY45-2 protein has four amino acid deletions and six amino acid substitutions compared with OsWRKY45-1, which may result in different three-dimensional structures of the two proteins that preferentially regulate different sets of genes. However, further study is required to ascertain whether the two proteins have different DNA-binding abilities and preferences.
Rice Resistance against Xoo and M. grisea Is Regulated by Multiple Pathways
Resistance against biotrophic and hemibiotrophic pathogens is usually regulated by the SA-dependent pathway, whereas resistance against necrotrophic pathogens is usually controlled by the JA/ethylene-dependent pathway (Bari and Jones, 2009). SA- and JA/ethylene-dependent defense signals interact with each other either synergistically or antagonistically (Durrant and Dong, 2004). Although Xoo and Xoc belong to the same species and are both biotrophic pathogens, they have different pathogenic mechanisms. Xoo invades rice plants through hydathodes or wounds and lives in the vascular system. Xoc penetrates the leaves of rice plants through stomata and wounds and lives in the intercellular spaces of parenchyma and mesophyll cells (Nino-Liu et al., 2006). M. grisea is a hemibiotrophic pathogen, which involves initial proliferation inside living host cells before switching to a destructive necrotrophic mode. The infection of M. grisea follows a developmental process in the plant surface to leaf epidermal cells (Park et al., 2009).
Rice resistance against the biotrophic pathogen Xoo has been reported to be accompanied by increased accumulation of SA and suppressed accumulation of JA (Qiu et al., 2007, 2008; Yuan et al., 2007; Xiao et al., 2009) or by reduced accumulation of both SA and JA (Ding et al., 2008). Our results suggest that OsWRKY45-1 negatively regulated Xoo resistance is associated with increased accumulation of SA and JA and OsWRKY45-2 positively regulated Xoo resistance is associated with increased accumulation of JA but not SA. These results suggest that multiple mechanisms may be involved in rice resistance against Xoo, which may include the antagonistic or synergistic cross talk of SA- and JA-dependent signaling, SA- and JA-independent signaling, and JA-dependent signaling.
A previous study reported that OsWRKY45 (named OsWRKY45-1 in this study) acted in an SA signaling pathway that is independent of NH1, the rice ortholog of Arabidopsis NPR1, in response to the infection of the hemibiotrophic pathogen M. grisea (Shimono et al., 2007). Consistently, our results show that NH1 expression is suppressed in OsWRKY45-1-overexpressing plants, which showed enhanced resistance to M. grisea. In contrast, OsWRKY45-1-overexpressing plants showed significantly suppressed expression of SA- and JA-responsive genes PR1a, PR1b, and PR10/PBZ1 and reduced accumulation of SA and JA when without pathogen infection, suggesting that OsWRKY45-1 positively regulated blast resistance may be independent of SA and JA. The NH1 expression was also slightly suppressed in OsWRKY45-2-overexpressing plants, which showed enhanced resistance to M. grisea. However, OsWRKY45-2-overexpressing plants showed significantly induced expression of JA synthesis-related genes PR1a and PR1b and increased accumulation of JA when without pathogen infection, suggesting that OsWRKY45-2 positively regulated blast resistance may be dependent on JA. This hypothesis is also supported by the report that overexpressing AOS2 in rice accumulated higher levels of JA, induced expression of PR1a, PR1b, and PR10, and enhanced resistance to M. grisea (Mei et al., 2006). Thus, multiple signaling pathways, which are dependent on or independent of SA or JA, may be involved in rice resistance against M. grisea.
OsWRKY45 and OsWRKY13 May Regulate Each Other in a Feedback Loop
Activation of OsWRKY13 enhanced rice resistance to both Xoo and M. grisea (Qiu et al., 2007). The expression patterns of OsWRKY45 in OsWRKY13-overexpressing and -suppressing plants suggest that this pair of alleles function downstream of OsWRKY13 in rice-pathogen interactions (Qiu et al., 2009). In addition, OsWRKY13 bound to OsWRKY45-1 promoter in vitro (Qiu et al., 2009). Our results here further show that OsWRKY13 also binds to the promoters of OsWRKY45-1 and OsWRKY45-2 in vivo, suggesting that OsWRKY13 may directly regulate the expression of this pair of alleles. OsWRKY13 regulates OsWRKY45-1 and OsWRKY45-2 perhaps via the binding to W-box, W-box-like, or even other cis-acting elements, because OsWRKY13 also bound to the OsWRKY45-1 promoter region flanked by PCR primers P45F2 and P45R2, which did not contain W-box or W-box-like element (Fig. 5). Previous studies have reported that a barley (Hordeum vulgare) WRKY protein binds to a sugar-responsive cis-element specifically and a tobacco (Nicotiana tabacum) WRKY binds to a WK-box (Sun et al., 2003; van Verk et al., 2008). Thus, further study is required to determine whether OsWRKY13 could regulate OsWRKY45-1 via binding to a non-W-box or non-W-box-like element. Since overexpressing OsWRKY13 in japonica Mudanjiang 8 suppressed OsWRKY45-1 expression and suppressing OsWRKY13 in indica Minghui 63 increased OsWRKY45-2 expression (Qiu et al., 2009), OsWRKY13 may function as a transcriptional repressor of OsWRKY45-1 and OsWRKY45-2. The differential expression of OsWRKY45-1 and OsWRKY45-2 in rice-Xoo interactions may be at least partly due to the binding of OsWRKY13 to different sites of OsWRKY45-1 and OsWRKY45-2 promoters. It has been reported that WRKY proteins form homocomplexes or heterocomplexes for DNA binding in pathogen-induced defense responses and other physiological processes (Xie et al., 2006; Xu et al., 2006). OsWRKY13 and other proteins were also detected to bind to the same pathogen-responsive cis-element PRE4 by yeast one-hybrid assay and gel mobility shift assay (Cai et al., 2008). The different preferential binding sites of OsWRKY13 to the promoters of OsWRKY45-1 and OsWRKY45-2 may be due to the facts that other activated protein(s) are required for this binding and indica and japonica backgrounds may have different proteins interacting with OsWRKY13 for DNA binding. Furthermore, the nucleotide difference of the two promoters may result in different three-dimensional DNA structures, which also influence OsWRKY13 binding.
Modulating OsWRKY45-1 and OsWRKY45-2 expression also influenced OsWRKY13 transcript level, suggesting that OsWRKY45-1 and OsWRKY45-2 may also regulate OsWRKY13 expression at least in rice resistance to Xoo. Because OsWRKY13 expression was suppressed in both OsWRKY45-1-overexpressing and -suppressing plants in rice-Xoo interactions, other factors in addition to OsWRKY45-1 may contribute to the regulation of OsWRKY13. The expression patterns of OsWRKY13 in OsWRKY45-2-overexpressing and -suppressing plants are complementary both without and after Xoo infection, suggesting that OsWRKY45-2 may play an important role in the regulation of OsWRKY13 and that activation of OsWRKY45-2 induces OsWRKY13 expression. However, further studies are needed to ascertain whether OsWRKY45-1 and OsWRKY45-2 directly or indirectly regulate OsWRKY13 expression.
CONCLUSION
The OsWRKY45 alleles, encoding different proteins, play opposite roles in rice resistance against the bacterial pathogens Xoo and Xoc, but both alleles positively regulate rice resistance against the fungal pathogen M. grisea. This pair of alleles regulates rice resistance to the same pathogen via different signaling pathways. These results provide evidence that a pair of allelic defense-responsive genes function oppositely in disease resistance, which may lead us to pay more attention to the roles of this class of genes in host-pathogen interactions caused by allelic diversity. Our results also provide a candidate gene (OsWRKY45-2), which regulates a race-nonspecific disease resistance in rice, for breeding programs.
MATERIALS AND METHODS
Gene Isolation, Sequence Comparison, and Allelic Analysis
The genomic fragments of OsWRKY45-1 and OsWRKY45-2 genes were amplified from japonica rice (Oryza sativa) var Nipponbare and indica rice var Minghui 63 using primers w45F4 and w45R4 (Supplemental Table S1), respectively, for transformation. The genomic fragments of the genes were sequenced using primers w45F4, w45R4, w45F6, and w45R6 (Supplemental Table S1). Gene structures of OsWRKY45-1 and OsWRKY45-2 were determined by alignment of the genomic sequences with the full-length cDNAs AK066255 (http://cdna01.dna.affrc.go.jp/cDNA/) from Nipponbare and EI77K16 (GenBank accession no. CX102514) from Minghui 63 (Zhang et al., 2005). The OsWRKY45 genes from other indica and japonica rice lines were also sequenced using primers w45F4, w45R4, w45F6, and w45R6. These lines were indica lines Zhenshan 97 and 93-11 and japonica lines Mudanjiang 8 and Dongjin. The upstream regions (approximately 1.5 kb upstream of translation start codons) that included the promoters of different OsWRKY45 alleles were amplified using PCR primers w45F6 and w45R1, and the PCR products were sequenced using primers w45F1, w45F2, w45R1, and P45F2 (Supplemental Table S1).
To determine the allelic relationship of OsWRKY45-1 and OsWRKY45-2, an F2 population, consisting of 146 individuals from a cross between Mudanjiang 8 carrying OsWRKY45-1 and Minghui 63 carrying OsWRKY45-2, was used to map the two genes. A molecular linkage map consisting of 136 markers relatively evenly distributed on 12 rice chromosomes has been constructed using this population (Y. Zhou and S. Wang, unpublished data). A pair of PCR primers, P45F3 and w45R6 (Supplemental Table S1), which generated differently sized PCR products of the two genes, were use as markers. Mapmaker/Exp 3.0 (Lincoln et al., 1992) was used for linkage analysis.
Rice Transformation
The overexpression constructs of OsWRKY45-1 from japonica line Nipponbare and OsWRKY45-2 from indica line Minghui 63 were created by inserting a genomic fragment (2,277 nucleotides for OsWRKY45-1 and 1,762 nucleotides for OsWRKY45-2) containing the complete gene into vector pU1301, which contained a maize (Zea mays) ubiquitin gene promoter (Cao et al., 2007). To construct an RNA interference vector for OsWRKY45-2, a 724-nucleotide fragment digested with restriction enzyme PstI from cDNA clone EI77K16 (Zhang et al., 2005) of rice line Minghui 63 was inserted into the pDS1301 vector (Yuan et al., 2007). The recombinant plasmids were introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Agrobacterium-mediated transformation was performed using calli derived from mature embryos of Mudanjiang 8 or Minghui 63, according to a published protocol (Lin and Zhang, 2005).
Examination of a Knockout Mutant
Seeds of OsWRKY45-1 T-DNA insertion mutant 2C-50229 (POSTECH; http://signal.salk.edu/cgi-bin/RiceGE) were kindly provided by professor Gynheung An (Jeong et al., 2006). The background of this mutant is Dongjin (japonica). The T-DNA was inserted in the promoter of OsWRKY45-1. The insertion site was 401 nucleotides from the translation start codon. The genotype of this mutant was confirmed by PCR amplification using rice primer pair w45F2 and w45R1 (Supplemental Table S1) and T-DNA and rice primer pair RB1 (5′-TTGGGGTTTCTACAGGACGTAAC-3′) and w45R1 (Supplemental Fig. S3, A and B).
Pathogen Inoculation
To examine the resistance of rice plants to bacterial blight disease, plants were inoculated with Philippine Xanthomonas oryzae pv oryzae strains PXO61 (race 1), PXO86 (race 2), PXO79 (race 3), PXO99 (race 6), and PXO341 (race 10) and with Chinese Xoo strains Zhe173 and KS-1-21 at the booting stage by the leaf-clipping method (Chen et al., 2002). Disease was scored by measuring the percentage lesion area (lesion length/leaf length) at 2 to 3 weeks after inoculation. Xoo growth rate in rice leaves was determined by counting colony-forming units (Sun et al., 2004).
For blast disease evaluation, seedlings at the three- to four-leaf stage were inoculated with Magnaporthe grisea isolate 91-17-2 (kindly provided by Dr. Youliang Peng, China Agricultural University) by the spraying method (Chen et al., 2003). Disease was scored using the 0-to-9 scale rating system (International Rice Research Institute, 2002) at 7 d after inoculation. In this rating system, the disease index, obtained by calculating the number of infected leaves, the number of total screened leaves, and the ratings of infected leaves, was used to evaluate the disease. A disease index greater than or equal to 0 and less than or equal to 5 indicates high resistance, greater than 5 and less than or equal to 15 indicates resistance, greater than 15 and less than or equal to 30 indicates moderate resistance, greater than 30 and less than or equal to 45 indicates moderate susceptibility, greater than 45 and less than or equal to 60 indicates susceptibility, and greater than 60 indicates high susceptibility.
To evaluate the resistance of plants to bacterial streak disease, plants were inoculated with the Chinese Xanthomonas oryzae pv oryzicola strain RH3 by needle stab method at the booting stage (Chen et al., 2006). Disease was scored by measuring lesion length at 21 d after inoculation. For all the disease evaluations, mock-inoculated (control) plants were treated under the same conditions, except that the pathogen suspension was replaced with water.
Gene Expression Analysis
The qRT-PCR was conducted as described by Qiu et al. (2007). Supplemental Table S1 lists the PCR primers for the genes. To examine the influence of pathogen infection on gene expression, 3-cm leaf fragments next to bacterial infection sites were used for RNA isolation. The expression level of the rice actin gene detected with actin-specific primers was used to standardize the RNA sample for each qRT-PCR. The assays were repeated at least twice, with each repetition having three replicates; similar results were obtained in repeated experiments. sd was calculated for each datum.
Quantification of Hormones
To quantify free SA, the samples were prepared and quantified as described previously (Qiu et al., 2007). In brief, three replicates of each ground sample weighing 300 to 1,000 mg (exact weights were recorded) were used for extraction of SA. The organic extracts containing free SA were quantified using HPLC. SA was detected using a 230-nm wavelength.
To quantify JA, the samples were prepared and quantified as described by Ding et al. (2008). In brief, three replicates of each leaf sample (1 g) were used for JA purification. The purified sample was quantified using the HPLC-electrospray ionization-tandem mass spectrometry system. The quantitative data of JA and 10-dihydro-JA (internal standard; Olchemim) were obtained using the peaks of the precursor ions 209.1 and 211.2 and the product ions 59 and 59, respectively.
Chromatin Immunoprecipitation Assay
Immunoprecipitation was performed as described previously (Benhamed et al., 2006). In brief, 3 g of three- to four-leaf-stage rice seedlings grown on Murashige and Skoog medium were used for sample preparation. Sonicated chromatin fragments were immunoprecipitated with OsWRKY13-specific antibody, which was custom synthesized by NewEast Biosciences against peptide LEVPEPEPEQESEP (201 to 214 of OsWRKY13). The samples were reverse-cross-linked for 6 to 8 h at 65°C and then treated with proteinase K to remove all proteins. DNA was purified by phenol-chloroform extraction and recovered by ethanol precipitation in the presence of 1 μg of glycogen. The precipitated chromatin was resuspended in 50 μL of buffer containing 10 mm Tris-Cl, pH 7.5, and 1 mm EDTA and was used for real-time PCR analysis. An aliquot of nonimmunoprecipitated and sonicated chromatin was reverse-cross-linked for use as the total input DNA control. Immunoprecipitated DNA was analyzed by PCR analysis. PCR was performed in 20 μL with 1 μL of immunoprecipitated DNA and promoter-specific primers (Supplemental Table S1).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ331930 (Mudanjiang 8), GQ331931 (Dongjin), and GQ331932 (Nipponbare) for OsWRKY45-1 and GQ331927 (Minghui 63), GQ331928 (Zhenshan 97), and GQ331929 (93-11) for OsWRKY45-2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The location of OsWRKY45-1 and OsWRKY45-2 on the molecular linkage map.
Supplemental Figure S2. Alignment of the promoter regions of OsWRKY45-1 and OsWRKY45-2.
Supplemental Figure S3. Overexpressing OsWRKY45-1 increased susceptibility to Xoo strain PXO61.
Supplemental Figure S4. OsWRKY45-1 knockout mutant 2C-50229.
Supplemental Figure S5. Overexpressing OsWRKY45-2 enhanced rice resistance to Xoo strain PXO61.
Supplemental Figure S6. The quality of anti-OsWRKY13 antibody was examined using OsWRKY13-overexpressing and OsWRKY13-suppressing plants.
Supplemental Table S1. Primers used for PCR amplification.
Supplementary Material
This work was supported by the National Program on the Development of Basic Research in China (grant no. 2006CB101904), the National Program of High Technology Development of China (grant no. 2006AA10A103), and the National Natural Science Foundation of China (grant no. 30621065).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Shiping Wang (swang@mail.hzau.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker EG, Traw MB, Toomajian C, Kreitman M, Bergelson J (2008) Low levels of polymorphism in genes that control the activation of defense response in Arabidopsis thaliana. Genetics 178: 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Benhamed M, Bertrand C, Servet C, Zhou DX (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31: 86–96 [DOI] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S (2007) The expression pattern of a rice disease resistance gene Xa3/Xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zheng W, Huang X, Zhang D, Lin X (2006) Major QTL conferring resistance to rice bacterial leaf streak. Agric Sci China 5: 216–220 [Google Scholar]
- Chen H, Wang S, Xing Y, Xu C, Hayes PM, Zhang Q (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci USA 100: 2544–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q (2002) New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92: 750–754 [DOI] [PubMed] [Google Scholar]
- Chu Z, Ouyang Y, Zhang J, Yang H, Wang S (2004) Genome-wide analysis of defense-responsive genes in bacterial blight resistance of rice mediated by the recessive R gene xa13. Mol Genet Genomics 271: 111–120 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20: 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, et al (2007) Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta 1769: 497–505 [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al (2005) R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435: 1122–1125 [DOI] [PubMed] [Google Scholar]
- Hu K, Qiu D, Shen X, Li X, Wang S (2008) Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant 1: 786–793 [DOI] [PubMed] [Google Scholar]
- Hulbert SH, Bai J, Fellers JP, Pacheco MG, Bowden RL (2007) Gene expression patterns in near isogenic lines for wheat rust resistance gene lr34/yr18. Phytopathology 97: 1083–1093 [DOI] [PubMed] [Google Scholar]
- International Rice Research Institute (2002) Reference guide. In Standard Evaluation System for Rice (SES). International Rice Research Institute, Los Banos, Philippines, p 56
- Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al (2006) Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 45: 123–132 [DOI] [PubMed] [Google Scholar]
- Jiang GH, Xia ZH, Zhou YL, Wan J, Li DY, Chen RS, Zhai WX, Zhu LH (2006) Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAγ1. Mol Genet Genomics 275: 354–366 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Tapio Palva E (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46: 477–491 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zhang Q (2005) Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23: 540–547 [DOI] [PubMed] [Google Scholar]
- Lincoln S, Daly M, Lander E (1992) Constructing genetics maps with MAPMAKER/EXP 3.0. Technical Report. Whitehead Institute, Cambridge, MA
- Liu X, Bai X, Wang X, Chu C (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Mei C, Qi M, Sheng G, Yang Y (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant Microbe Interact 19: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino M, Meyers BC, Michelmore RW, Gaut BS (2002) Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res 12: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7: 303–324 [DOI] [PubMed] [Google Scholar]
- Oh S-K, Baek K-H, Park JM, Yi SY, Yu SH, Kamoun S, Choi D (2008) Capsicum annuum WRKY protein CaWRKY1 is a negative regulator of pathogen defense. New Phytol 177: 977–989 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Jin J, Lee YW, Kang S, Lee YH (2009) Rice blast fungus (Magnaporthe oryzae) infects Arabidopsis via a mechanism distinct from that required for the infection of rice. Plant Physiol 149: 474–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Laura EB, Chen XW, Dardick C, Chern MS, Ruan R, Patrick EC, Pamela CR (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1: 446–458 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Cheng H, Li X, Wang S (2009) Exploring transcriptional signalling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biol 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S (2008) Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant 1: 538–551 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Yu D (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65: 35–47 [Google Scholar]
- Romer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T (2007) Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318: 645–648 [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49: 827–842 [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19: 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15: 2076–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Wang S (2006) Point mutations with positive selection were a major force during the evolution of a receptor-kinase resistance gene family of rice. Plant Physiol 140: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encoding a LRR receptor kinase-like protein. Plant J 37: 517–527 [DOI] [PubMed] [Google Scholar]
- van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ (2008) A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol 146: 1983–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65: 799–815 [DOI] [PubMed] [Google Scholar]
- Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S (2009) A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 4: e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Yang G, Komatsu S, Shen QJ (2006) Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J 46: 231–242 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Shen X, Li X, Xu C, Wang S (2007) Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226: 953–960 [DOI] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S (2005) Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42: 772–780 [DOI] [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z (2008) Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res 18: 508–521 [DOI] [PubMed] [Google Scholar]
- Zhou B, Peng K, Chu Z, Wang S, Zhang Q (2002) The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice (Oryza sativa L.). Sci China C Life Sci 45: 449–467 [DOI] [PubMed] [Google Scholar]
- Zhou B, Qu SH, Liu GF, Dolan M, Sakai H, Lu GD, Bellizzi M, Wang GL (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19: 1216–1228 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.