Abstract
Previously, the GRF-INTERACTING FACTOR1 (GIF1)/ANGUSTIFOLIA3 (AN3) transcription coactivator gene, a member of a small gene family comprising three genes, was characterized as a positive regulator of cell proliferation in lateral organs, such as leaves and flowers, of Arabidopsis (Arabidopsis thaliana). As yet, it remains unclear how GIF1/AN3 affects the cell proliferation process. In this study, we demonstrate that the other members of the GIF gene family, GIF2 and GIF3, are also required for cell proliferation and lateral organ growth, as gif1, gif2, and gif3 mutations cause a synergistic reduction in cell numbers, leading to small lateral organs. Furthermore, GIF1, GIF2, and GIF3 overexpression complemented a cell proliferation defect of the gif1 mutant and significantly increased lateral organ growth of wild-type plants as well, indicating that members of the GIF gene family are functionally redundant. Kinematic analysis on leaf growth revealed that the gif triple mutant as well as other strong gif mutants developed leaf primordia with fewer cells, which was due to the low rate of cell proliferation, eventually resulting in earlier exit from the proliferative phase of organ growth. The low proliferative activity of primordial leaves was accompanied by decreased expression of cell cycle-regulating genes, indicating that GIF genes may act upstream of cell cycle regulators. Analysis of gif double and triple mutants clarified a previously undescribed role of the GIF gene family: gif mutants had small vegetative shoot apical meristems, which was correlated with the development of small leaf primordia. gif triple mutants also displayed defective structures of floral organs. Taken together, our results suggest that the GIF gene family plays important roles in the control of cell proliferation via cell cycle regulation and in other developmental properties that are associated with shoot apical meristem function.
The size and shape of lateral organs, such as leaves and flowers, are under the control of developmental genetic programs exhibiting species-specific characteristics. In general, lateral organs from plant species of different size differ in cell number rather than cell size, as exemplified in the case where big petals of Brassica napus have the same size of cells as small petals of Arabidopsis (Arabidopsis thaliana) do (Mizukami and Fischer, 2000; Mizukami, 2001). Thus, cell number is a primary determinant of organ size of a species, although concomitant and/or subsequent cell expansion amplifies and modifies the effect of cell number, contributing to the final size and shape of lateral organs (Tsukaya, 2005; Cho et al., 2007).
Many genetic approaches have been employed to understand the control of cell proliferation and its role in organogenesis, and these have identified a number of genes that play important roles in the process. Some of these genes, such as AINTEGUMENTA (ANT), KLUH (KLU), and STRUWWELPETER (SWP), play positive roles (i.e. their loss-of-function mutants developed small lateral organs due to a reduction in cell numbers; Mizukami and Fischer, 2000; Autran et al., 2002; Anastasiou et al., 2007). On the other hand, overexpressors of those genes have bigger organs resulting from more cells, although the SWP overexpressors resulted in smaller organs despite there being more cells, because of smaller cell size. It has been proposed that those genes positively regulate cell proliferation and organ growth by controlling the duration of cell proliferation. ANT, KLU, and SWP encode the APETALA2 domain transcription factor, the P450 cytochrome oxidase CYP78A5, and the Med150/RGR1-like subunits of the Mediator transcriptional regulatory complex, respectively. There are also negative regulators of cell proliferation and organ growth, such as PEAPOD and BIG BROTHER genes, which encode putative DNA-binding proteins and an E3 ubiquitin ligase, respectively (Disch et al., 2006; White, 2006). Those negative regulators affect the duration of cell proliferation in lateral organs as well.
We have previously uncovered a novel class of transcriptional factors, GROWTH-REGULATING FACTOR (GRF), comprising nine members in Arabidopsis (Kim et al., 2003). The grf triple loss-of-function mutant of GRF1 through GRF3 developed narrow and small leaves, whereas overexpression of GRF1 and GRF2 resulted in large leaves. Reexamination of cellular parameters revealed that the grf triple mutant produced fewer palisade cells, whereas the GRF overexpressors contained more cells (Kim and Kende, 2004; Kim and Lee, 2006), indicating that GRF proteins act as positive regulators of cell proliferation. In search of partner proteins for GRF1, we found another gene family, GRF-INTERACTING FACTOR (GIF), which comprises three members (Kim and Kende, 2004). GIF proteins are thought to be functional homologs of the human SYT transcriptional coactivator. Like the grf triple mutant, the gif1 single mutant showed narrow and small leaves as well as flower petals, which were caused by a reduction in cell numbers, indicating that GRF and GIF1 proteins form a functional complex involved in regulating cell proliferation and, thus, growth and shape of lateral organs (Kim and Kende, 2004). Horiguchi et al. (2005) also confirmed the result by demonstrating that angustifolia3 (an3) mutations, which occurred at the GIF1 gene, and the grf5 mutation caused a reduction in cell numbers, leading to small and narrow lateral organs, while overexpression of GIF1/AN3 increased leaf size with normal shape.
Meanwhile, it still remains to be elucidated by what mechanism the GIF1/AN3 gene regulates cell proliferation and what biological roles the other two members, GIF2 and GIF3, play in Arabidopsis growth and development. In this article, we show that all of the GIF genes act as positive regulators of cell proliferation of lateral organs in a functionally redundant manner. Our results indicate that GIF genes may affect the duration of cell proliferation by modulating the expression level of cell cycle regulators. We also present experimental evidence that GIF genes are required for other developmental processes, such as plastochron and flower structure.
RESULTS
In Silico Expression Profiles of GIF Genes and Isolation of T-DNA Insertional Mutants
Using RNA gel-blot analysis, we have previously shown that the GIF family genes share an overlapping expression pattern and that GIF1/AN3 mRNAs are most abundant compared with GIF2 and GIF3 (Kim and Kende, 2004). To compare the tissue-specific expression pattern of GIF genes in more detail, we compiled the microarray data from the AtGenExpress expression atlas (Schmid et al., 2005), which contained expression profiles of all three GIF genes in 45 different organs or developmental stages (Supplemental Fig. S1). The overall expression patterns of GIF genes are similar, suggesting that those genes may be functionally redundant. It should be noted, however, that the maximum level of GIF1/AN3 mRNA was about three times higher than that of GIF2 and that of GIF2 was about two times higher than that of GIF3. All of the GIF genes show a distinctively high expression in the shoot apex containing lateral organ primordia and the apical meristem. The in silico expression pattern and strength are consistent with the previous RNA gel-blot results (Kim and Kende, 2004) and suggest that their biological function may be relevant especially to growth and development of the shoot apical meristem (SAM) and/or lateral organs developing from the flanking region of the SAM.
A phylogenetic analysis suggested that GIF2 and GIF3 genes might result from a duplication event of a common ancestral gene after diversification from GIF1/AN3 (Kim and Kende, 2004). This notion is also supported by the fact that GIF2 and GIF3 have the same gene structure, with four introns, whereas GIF1/AN3 lacks the last one (Fig. 1; Kim and Kende, 2004). To investigate whether or not GIF2 and GIF3 proteins have similar biological function, we isolated homozygous T-DNA insertional mutants, gif2 and gif3, from the Syngenta (SAIL_328_A03) and Salk (SALK_072950) lines, respectively (Sessions et al., 2002; Alonso et al., 2003). We found that the gif2 and gif3 mutations resulted from the T-DNA insertions at 69 bp from the start of the second exon and 45 bp upstream of the translational start site, respectively (Fig. 1). Reverse transcription (RT)-PCR analysis using cDNAs prepared from DNase-treated mRNAs revealed that the gif2 mutant completely lacked GIF2 mRNA, whereas the gif3 mutant showed a significant but much lower level of GIF3 mRNA than that of wild-type plants (Fig. 1, bottom). The T-DNA insertional sites and RT-PCR analyses indicate that gif2 and gif3 are likely to be a null and a hypomorphic mutant, respectively. Interestingly, wild-type plants exhibited unspliced species of GIF3 mRNA, whose level was further elevated in the gif3 mutant, suggesting that the primary transcripts of the GIF3 gene were not subject to a complete splicing and that the T-DNA insertion into the 5′ untranslated region of GIF3 interfered with normal splicing.
Figure 1.
Gene structure and representation of T-DNA insertional mutations. T-DNA integration sites are marked with inverse triangles. Arrows, Orientation of the T-DNA left border (LB); black boxes, exons; white boxes, introns; solid lines, 5′ untranslated region; arrowheads, primer sites for RT-PCR analysis at right. The ruler is a scale of bp. The gel images show RT-PCR determination of GIF mRNA content in wild-type Columbia (Col) and gif mutant plants. Amplification of ACTIN8 (ACT8) cDNA was used as a control. The single and double asterisks indicate DNA fragments derived from spliced and unspliced GIF3 mRNA species, respectively.
Growth and Developmental Phenotypes of Lateral Organs
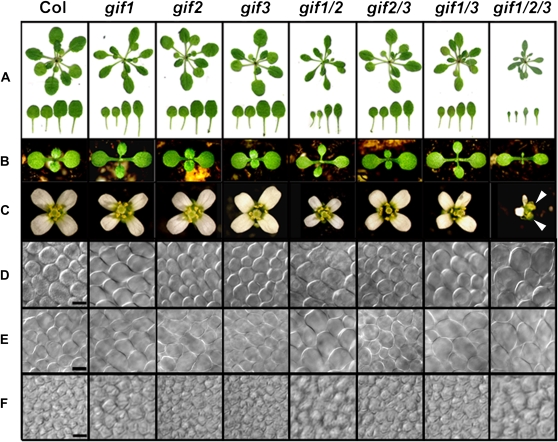
As described previously (Kim and Kende, 2004), the gif1 null mutant caused by T-DNA insertion into the third exon had small and narrow lateral organs, such as leaves, cotyledons, and flowers (Fig. 2, A–C). Quantification of the first pair of primary leaves at their maturity revealed that leaf-blade area of gif1 was decreased by 52% over the wild type, which was due to reduction in both blade length and width (Fig. 3A, left). Petiole length was also decreased as much as blade length was. The areas of cotyledons and petals were reduced as well, by approximately 22% and 40%, respectively (Fig. 3, B and C). Reduction in width was more prominent than in length, resulting in narrow morphology and, thus, increasing the index value (the ratio of length to width). To the contrary, dimensional and morphological phenotypes of gif2 and gif3 single mutants were not distinctive, although careful observation and measurement of leaf area revealed a minute change in the gif2 single mutant, which was statistically significant by Student's t test (P < 0.05; Figs. 2 and 3). To test synergistic interaction between gif mutations, we constructed double and triple mutants by genetic crossing and found that the gif1 gif2 gif3 (hereafter gif1/2/3) triple mutant developed tiny and narrow organs (Fig. 2, A–C, last column). Leaf-blade area of the triple mutant was only about 10% of the wild type; the areas of cotyledons and petals were 42% and 26%, respectively (Fig. 3, left panels). Phenotypic strength of the gif1/2 double mutant was intermediate between gif1 and the triple mutants. gif1/3 and gif2/3 double mutants, however, developed only slightly smaller organs than their stronger parental lines (Figs. 2 and 3). These results clearly demonstrate that gif mutations act synergistically to affect the growth and development of lateral organs, and the relative contribution of each mutation to the phenotype was gif1 > gif2 > gif3, indicating that GIF genes have an overlapping function in regulating the size and shape of lateral organs. The root growth of gif mutants was not affected at all (data not shown).
Figure 2.
Phenotypes of gif mutants. A, Wild-type Columbia (Col) and gif plants (top) and rosette leaves (bottom) from the first to fourth leaves at day 20. B, Seedlings at day 8. C, Mature flowers. Note that gif1/2/3 has only two petals and split carpels (arrowheads). D, Adaxial palisade cells of mature leaves. E, Adaxial palisade cells of mature cotyledons. F, Subepidermal cells at the abaxial side of mature petals. Scale bars = 25 μm.
Figure 3.
Quantification of mutant phenotypes at the organ and cell level. A, Phenotype of the first two leaves at day 25. Leaf size and shape are shown, including index values (left), number and size of adaxial palisade cells (middle), and number of trichomes (right). B, Phenotype of cotyledons at day 15. Cotyledon size and shape are shown, including index values (left) and number and size of adaxial palisade cells (right). C, Phenotype of mature petals. Petal size and shape are shown, including index values (left) and number and size of subepidermal cells at the abaxial side (right). The numerical marks on the x axis denote gif single and multiple mutants. Col, Columbia wild type. n = 10 for determination of organ-level parameters; n = 200 from 10 different organs for cellular parameters. Error bars indicate se.
Unexpectedly, we found that the triple mutant displayed aberrant structures of floral organs, including split carpels and reduction in numbers of petals and anthers, while the wild type, single mutants, and gif2/3 showed normal floral organization and organ numbers; gif1/2 and gif1/3 double mutants displayed such floral defects but only in the terminal two flowers (Fig. 2C, last column; details will be described elsewhere). This observation confirmed the contribution of single mutations once more. In addition, gif1/2 and gif1/2/3 mutants were completely sterile, whereas other mutants set seeds, the number of which was variable depending on their phenotypic severity (Table I). Because of their infertility, gif1/2 and gif1/2/3 mutant lines were maintained in the heterozygous gif1 mutation with other homozygous ones. The sterility problem of gif1/2 seemed to stem from an abnormal functionality of female reproductive organs rather than from pollen. In other words, the gif1/2 double mutant produced pollen that was fully functional on wild-type carpels, producing as many seeds as wild-type pollen did, whereas wild-type pollen did not produce even a single seed on gif1/2 carpels (Supplemental Table S1). The gif1/2/3 triple mutant, besides the structural abnormality of flower organs, produced no pollen at all. These results indicate that GIF genes are also required for the maintenance of structural integrity and reproductive function of flower organs.
Table I.
Plastochronic and other developmental phenotypes of gif mutants
Values shown are means ± se; n = 10.
| Leaf Order | Genotype
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Columbia | gif1 | gif2 | gif3 | gif1/2 | gif2/3 | gif1/3 | gif1/2/3 | |
| Day when leaf attains 1 mm in lengtha | ||||||||
| 1, 2 | 8.6 ± 0.1 | 7.8 ± 0.1 [0.8]b | 8.3 ± 0.1 [0.3] | 8.5 ± 0.1 [0.1] | 8.2 ± 0.1 [0.4] | 8.4 ± 0.1 [0.2] | 8.1 ± 0.1 [0.5] | 10.4 ± 0.2 [−1.9] |
| (3.9)c | (3.0) | (3.6) | (3.8) | (2.7) | (3.4) | (2.8) | (0.9) | |
| 3 | 12.5 ± 0.1 | 10.8 ± 0.1 [1.7] | 11.9 ± 0.1 [0.6] | 12.3 ± 0.1 [0.1] | 10.9 ± 0.1 [1.5] | 11.8 ± 0.1 [0.6] | 10.9 ± 0.2 [1.6] | 11.3 ± 0.3 [1.2] |
| (1.3) | (0.8) | (1.3) | (1.2) | (0.8) | (1.4) | (1.0) | (1.1) | |
| 4 | 13.8 ± 0.1 | 11.6 ± 0.2 [2.2] | 13.2 ± 0.1 [0.6] | 13.5 ± 0.1 [0.3] | 11.7 ± 0.1 [2.0] | 13.2 ± 0.2 [0.7] | 11.9 ± 0.1 [1.9] | 12.4 ± 0.2 [1.5] |
| Day when leaf attains 1 mm in lengthd | ||||||||
| 1, 2 | 15.8 ± 0.1 | 12.7 ± 0.0 [3.2] | 13.5 ± 0.0 [2.3] | 13.9 ± 0.3 [2.0] | 12.7 ± 0.0 [3.2] | 13.4 ± 0.2 [2.4] | 12.8 ± 0.1 [3.0] | 16.1 ± 0.1 [−0.2] |
| (4.6) | (3.9) | (4.4) | (4.6) | (4.0) | (4.7) | (4.1) | (1.5) | |
| 3 | 20.5 ± 0.2 | 16.6 ± 0.1 [3.9] | 17.9 ± 0.1 [2.5] | 18.5 ± 0.1 [2.0] | 16.6 ± 0.1 [3.8] | 18.1 ± 0.1 [2.3] | 16.9 ± 0.1 [3.6] | 17.6 ± 0.1 [2.9] |
| (1.4) | (1.4) | (1.5) | (1.7) | (1.6) | (1.5) | (1.5) | (0.9) | |
| 4 | 21.8 ± 0.1 | 18.0 ± 0.1 [3.8] | 9.5 ± 0.0 [2.4] | 20.2 ± 0.2 [1.6] | 18.2 ± 0.2 [3.6] | 19.7 ± 0.2 [2.2] | 18.4 ± 0.3 [3.5] | 18.5 ± 0.2 [3.4] |
| No. of rosette leaves when plants have the first open flowersa | ||||||||
| 10.7 ± 0.1 | 11.3 ± 0.4 | 10.3 ± 0.5 | 11.1 ± 0.5 | 10.3 ± 0.3 | 10.0 ± 0.3 | 11.0 ± 0.5 | 10.0 ± 0.3 | |
| Bolting time: day when the first flower bud develops to stage 12a | ||||||||
| 29.5 ± 1.6 | 27.3 ± 1.3 | 28.7 ± 1.0 | 29.5 ± 1.2 | 26.0 ± 0.9 | 24.0 ± 1.5 | 24.8 ± 1.5 | 21.4 ± 1.0 | |
| No. of seeds per siliquea | ||||||||
| 42.7 ± 1.4 | 19.6 ± 1.1 | 35.1 ± 1.0 | 40.5 ± 2.6 | 0.0 ± 0.0 | 28.8 ± 1.7 | 8.0 ± 1.4 | 0.0 ± 0.0 | |
| Primary stem length (cm)a | ||||||||
| 24.3 ± 2.5 | 28.9 ± 3.0 | 24.3 ± 3.2 | 22.9 ± 1.5 | 30.2 ± 2.3 | 24.1 ± 2.9 | 33.4 ± 2.5 | 14.5 ± 2.4 | |
Plants were grown under long days (18 h of light/6 h of darkness).
Numbers in brackets are differences in days between the wild type and the mutants.
Numbers in parentheses are plastochron lengths between successive leaves.
Plants were grown under short days (8 h of light/16 h of darkness).
We determined cell numbers and sizes of lateral organs by observing subepidermal cells under Nomarski optics. Numbers of adaxial palisade cells in the first two mature leaves of gif1/2/3 were only 27% in the longitudinal axis and 15% in the maximum-width axis over wild-type plants (Fig. 3A, middle). The total number of palisade cells of the triple mutant, calculated by dividing leaf area by cell area, was only 4% of the wild type. The number of trichomes on the first two leaves of gif1/2/3 was also reduced to 8% of wild-type plants, which is indicative of a great reduction in the number of epidermal cells as well (Fig. 3A, right). Reduction in those cell numbers of other single and double mutants corresponded to changes in the organ-level dimensional parameters (Fig. 3A, compare left and middle panels). To the contrary, cell area of the triple mutant increased by 2.6-fold over the wild type (Figs. 2D and 3A), which probably resulted from a “compensatory” effect due to the drastic reduction in cell number (Tsukaya, 2003). It should be noted, however, that the compensatory increase in cell area was only partially able to make up for the reduction in leaf size, and there was no change in cell shape (Fig. 2). The gif1/2 double mutant exerted its compensatory effect to an extent comparable with the triple mutant, and that of the gif1 single and gif1/3 double mutants was also apparent. Other single and double mutants, however, did not show a significant change in cell area. These results indicate that the compensation occurs in direct proportion to the change in cell numbers. This is also true for cotyledons and flowers: more gif mutations resulted in smaller organs with fewer but bigger cells (Fig. 3, B and C). Taken together, our data indicate that GIF2 and GIF3 as well as GIF1 are involved in the positive regulation of cell proliferation of lateral organs in a functionally redundant manner.
Kinematic Analysis of Leaf Area and Cellular Parameters during Leaf Growth
To obtain more information about the effect of gif mutations on cell proliferation, we performed kinematic analysis on leaf growth. The first pairs of leaves were harvested at the indicated days from 4-d-old seedlings. Because gif1/2 double and gif1/2/3 triple homozygous mutants showed distinctively small and narrow cotyledons (Fig. 2B), we were able to differentiate homozygous individuals in the segregating populations of gif1/+ gif2/gif2 and gif1/+ gif2/gif2 gif3/gif3 genotypes.
Leaf area continued to increase over time: slow during the early stage, then at an accelerated rate, and lastly, leaf expansion slowed toward the final size of mature leaves (Fig. 4A, left). Leaf expansion of wild-type plants was almost completed at day 20, whereas that of the gif1 single mutant and the gif1-harboring double mutants ceased at 17 and 13 d, respectively. The gif1/2/3 triple mutant showed only a minute leaf expansion during the whole growth period. Plotted on a natural log scale in a magnified version, the leaf area data from days 4 through 7 revealed that gif1/2/3 leaves were markedly smaller, by 5.5-fold, than wild-type leaves already at day 6 and afterward (Fig. 4A, right). A clear distinction between the gif1/2 double mutant and the wild type was also recognized at day 7. Intriguingly, however, leaf area of the gif1 single mutant was not smaller than that of the wild type in those early days; only later did it become smaller, eventually reaching half the size of wild-type leaves. This is consistent with what we have observed during plant growth: note the similar size of the first two leaves of the wild type and the gif1 single mutant even at day 8 in Figure 2B. We attribute this apparent discrepancy in size of young and mature gif1 leaves to the strong plastochronic properties of the gif1 mutant (see below).
Figure 4.
Kinematic analysis of leaf growth. Measurements were performed on the first two leaves of the wild-type Columbia (Col) and gif plants grown on soil for the indicated times. Adaxial palisade cells were analyzed. A, Leaf blade area. B, Numbers of cells in a line along the maximum width region. C, Relative cell proliferation rate derived from B. The rate was calculated by dividing the difference in cell numbers of two successive measurements by the cell numbers of the previous one. D, Area of adaxial palisade cells. The graphs at right show magnified versions to indicate differences in the early stages of leaf growth. Note that the right graph in A was plotted on the natural log scale. The same numbers of samples were analyzed as in Figure 3. Error bars indicate se.
To compare cell proliferation patterns of the wild type and mutants during leaf growth, we determined the number of adaxial palisade cells in a line along with a maximum-width axis of the first two leaves, and the resulting kinematic graph clearly showed that cell proliferation ceased at day 11 in the wild type but at day 7 in the strong mutants harboring gif1 (i.e. gif1, gif1/2, gif1/3, and gif1/2/3; Fig. 4B, left). Previously, Ferjani et al. (2007) also observed that an3-4, another mutant allele of GIF1, exhibited early cessation of the cell proliferation phase in the leaf-length axis. Moreover, leaves of gif1/2 double and gif1/2/3 triple mutants had only about half the cells of wild-type leaves already at days 6 and 4, respectively (Fig. 4B, right). Intriguingly again, however, gif1 leaves at the early stage between days 4 and 6 had more cells than wild-type leaves, at least partially explaining the reason why the leaf size of the gif1 mutant was similar to that of the wild type in those early days (Fig. 4B). This tendency in gif1 was consistently observed in an3-4 (Ferjani et al., 2007).
Calculation of cell proliferation rate (changes in cell numbers between consecutive days divided by the cell number of the previous day) showed that cell proliferation in leaf was most active during the interval between days 5 and 7, but maximum proliferation activity of most gif mutants was remarkably or slightly lower than that of the wild type; especially, that of triple mutants was close to zero (Fig. 4C). Furthermore, almost no cell proliferation activity of the strong mutant lines was detected around day 8 and thereafter, whereas wild-type activity remained at a substantial level until day 10. These data indicate that the gif mutations exert synergistic effects on both the rate and duration of cell proliferation.
Leaf cell expansion of the wild type and all of the gif mutants increased in a sigmoidal pattern throughout growth (Fig. 4D, left). The compensatory enhancement of cell expansion in the gif triple mutant became prominent from day 6, although it was not possible to make a clear distinction between the wild type and mutants before that day (Fig. 4D, right). It should be noted, however, that the enhanced cell expansion was not sufficient to mask the effect of the reduced cell proliferation on determining leaf size: already at day 6 and thereafter, leaf size of the triple mutant was at least five times smaller than that of the wild type (Fig. 4A).
Expression of Cell Cycle-Regulating Genes
The low rate of cell proliferation of gif mutants prompted us to examine the expression of cell cycle-regulating genes. Total RNA was isolated from the first two leaves of 6-d-old wild-type and gif1/2/3 triple mutant plants, in which their cell proliferation occurred at a maximum rate, and subjected to RT-PCR. The transcript levels of two cyclin genes, CycB1;1 and CycD3;1, as well as a cyclin-dependent kinase gene, Cdc2b, were significantly reduced in the triple mutant (Fig. 5). Both CycB1;1 and Cdc2b are involved in the G2/M transition (Mironov et al., 1999; Vandepoele et al., 2002), and CycD3;1 has been known to dominantly drive the G1/S transition (Dewitte et al., 2003; Menges et al., 2006). The S-phase-specific proliferating cell nuclear antigen PCNA1 (Kosugi and Ohashi, 2002; Anderson et al., 2008) was remarkably down-regulated as well. These data indicate that GIF genes may control cell proliferation by positively regulating cell-cycling gene expression in the primordial leaves.
Figure 5.
Determination of mRNA levels of various cell-cycling genes by RT-PCR. Total RNAs were isolated from the first two leaves of wild-type Columbia (Col) and the gif1/2/3 triple mutant (gif) at day 6 and used for RT-PCR analysis. The numbers at the top indicate dilution factors of cDNAs derived from the RT reaction. ACTIN8 (ACT8) was used as an amplification control.
Mutant Phenotypes in the SAM Tissues
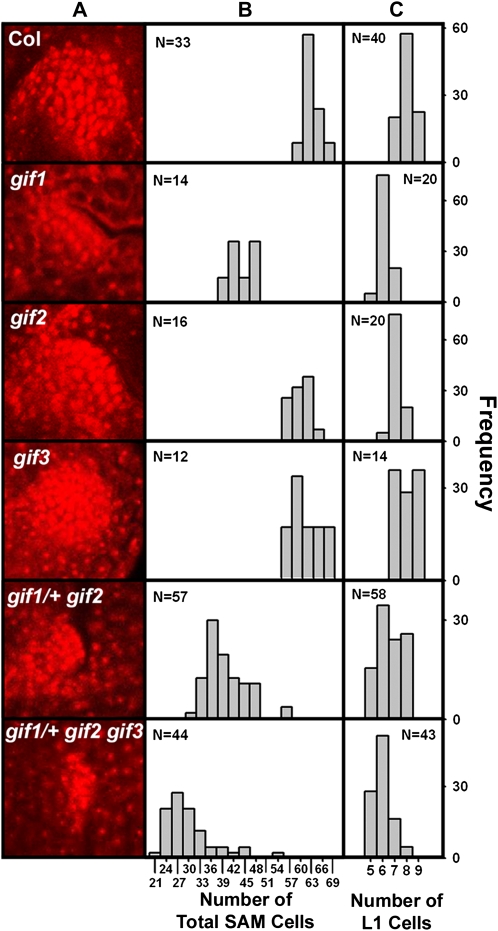
Since cells in the SAM continue to be recruited into developing leaf primordia (Telfer and Poethig, 1994) and since the gif mutations affect the cell number of primordial leaves, we investigated the effect of gif mutations in controlling the size of the SAM cell pool. Nuclei of mature embryos were stained with propidium iodide, and the SAM size and its cell number in a longitudinal median section were determined by confocal microscopy. We found that the SAM size of the gif1 mutant but not of gif2 and gif3 was smaller than that of the wild type (Fig. 6A). Counting the number of nuclei revealed that the gif1 SAM was made up of fewer cells in total and in its L1 layer and that a substantial portion of gif2 mutant individuals had the SAM with fewer cells (Fig. 6, B and C). We also determined the number of SAM cells in segregating individual embryos from the selfed siliques of gif1/+ gif2/gif2 and gif1/+ gif2/gif2 gif3/gif3 genotypes. A majority of the progeny of gif1/+ gif2/gif2 had the SAM comprising fewer cells than their parental mutant, resulting in a smaller SAM; most individuals of the F1 progeny of gif1/+ gif2/gif2 gif3/gif3 had even smaller SAM with fewer cells than the segregating population of gif1/+ gif2/gif2. These results indicate that the gif triple mutations synergistically reduced the size of the vegetative SAM cell pool.
Figure 6.
Phenotype of the SAM tissue. A, Size of the SAM. B, Total number of SAM cells. C, Number of L1 layer cells. Nuclei of mature embryos were stained with propidium iodide and visualized by confocal microscopy. Each rank at the x axis in B means a three-cell range (i.e. cell numbers in each rank are equal to the numerical marks ± 1). Col, Columbia wild type.
Plastochronic Properties of gif Mutants
Apart from the cell proliferation phenotype, we noticed that gif mutations seemed to accelerate various developmental processes. To estimate the developmental acceleration, we measured the time when leaves reach 1 mm in length with a stereoscope in long-day conditions. The first two leaves of the gif1 mutant attained that length at day 7.8, which was 0.8 d earlier than the wild type (Table I). The third and fourth gif1 leaves were much earlier than the wild-type leaves, by 1.7 and 2.2 d, respectively. The difference between the wild type and gif1 was profound in the short-day conditions (i.e. gif1 leaves attained the length more than 3 d earlier compared with the corresponding wild-type leaves, clearly indicating that the gif1 mutation accelerated the early developmental pace). The time intervals at which successive leaves reached that length (plastochron) were also significantly shortened in the gif1 mutant compared with the wild type: for instance, it took 3.9 and 4.6 d in the respective photoperiods for the wild-type third leaf to attain 1 mm of length after its first two leaves did so, but it took 3.0 and 3.9 d for the gif1 third leaf under the corresponding conditions. However, the final numbers of wild-type and gif1 leaves were not significantly different from each other, and neither was germination time (data not shown). Taken together, these results clearly indicate that the gif1 mutant displays short plastochronic length at the early developmental stage as well as the hasty emergence of the first two leaves; furthermore, they explain why the gif1 single mutant has more cells in the primordial leaves at the early stage than wild-type primordium does, despite its lower cell proliferation activity, as mentioned above.
In contrast, the time when the double mutants harboring the gif1 mutation, gif1/2 and gif1/3, attained 1-mm-long leaves was slightly delayed or similar, compared with their parental gif1 single mutant in both photoperiodic regimens, although they did so earlier than the wild type. Especially, the first two leaves of the gif1/2/3 triple mutant were more delayed than even the wild-type plant, by 1.9 and 0.2 d (compare the numbers in brackets in Table I for both photoperiodic regimens). It is important to realize, however, that this does not necessarily mean that no or antagonistic plastochronic synergism exists between those mutations. Rather, this observation means that leaves of gif double and triple mutants, unlike gif1 leaves, were so short that the time for the attainment of a 1-mm-long leaf could not be an appropriate parameter for plastochronic comparison with the wild type or gif1. In fact, the gif1/2/3 plastochron between the first two and third leaves was exceptionally shorter than that of gif1 (compare the numbers in parentheses in Table I), and synergistic acceleration of other developmental processes was evident: the gif1 mutant bolted 2 d earlier than the wild type did, and addition of the gif2 and gif3 mutations gradually shortened the bolting time, resulting in the gif triple mutant bolting 8 d earlier than the wild type (Table I). It is interesting that, in spite of their minute effect on leaf size, gif2/3 and gif1/3 double mutations shortened the bolting time a little more than the gif1/2 double mutation did.
We also found that mutants containing the gif1 mutation developed more axillary branches, as shown in Figure 7A, in which all of the primary shoots and leaves, except two cotyledons, were removed to expose axillary branches at day 30. The triple mutant showed a long primary branch (Fig. 7A, far right). During the vegetative stage or just before bolting, only the gif triple mutant was able to develop primary branches from cotyledons and rosette leaves (i.e. nine out of 10 gif1/2/3 mutants showed a pair of rapidly growing axillary leaves from both cotyledons and the first two rosette leaves at day 17.0 ± 0.7 and 19.1 ± 0.7, respectively, whereas none of the other mutants as well as the wild type did so during the same period, although gif1/2 double mutants occasionally did). The triple mutant produced about five more primary axillary branches from rosette leaves and even cotyledons than the wild type, and other mutants harboring gif1 mutations showed intermediate numbers (Fig. 7C). To the contrary, there was no significant difference in numbers of primary branches derived from cauline leaf axils (Fig. 7D). These data indicate that the loss of GIF genes accelerates various developmental processes, including leaf plastochron.
Figure 7.
Shoot branching phenotype. A, Branching phenotype of mature plants at 35 d. The main shoot and all of the rosette leaves were removed to expose rosette leaf branches and cotyledons. Arrowheads indicate rosette leaf branches with flower buds. B, Schematic representation of Arabidopsis branching structure. C, Number of rosette leaf primary branches. D, Number of cauline leaf primary branches. n = 10. Error bars indicate se. Col, Columbia wild type.
Finally, the gif triple mutant developed short inflorescence stems, although other mutant plants containing the gif1 mutation tend to have a little longer stem than the wild type (Table I), suggesting that all three GIF genes are necessary for normal growth of the inflorescence stem.
Overexpression of GIF1, GIF2, and GIF3 in gif1 Mutants and Wild-Type Plants
To test whether or not GIF2 and GIF3 genes will be able to complement the gif1 phenotype, gif1 mutant plants were transformed with GIF1, GIF2, and GIF3 cDNAs under the control of the cauliflower mosaic virus 35S promoter. Dozens of T1 generation plants were obtained for each construct, and most of them showed a wild-type-like phenotype (Fig. 8A; data not shown). Quantification data revealed that leaf dimension parameters of gif1 plants overexpressing each construct were very close to those of the wild type (Fig. 8B). Leaf indices were also recovered to the wild-type value, displaying leaves with round shape rather than the narrow one of the gif1 mutant. All of these dimensional changes corresponded exactly to those in cell numbers (Fig. 8C). We also established overexpressor lines whose gif1 mutation was segregated out after crossing the complementation lines to wild-type plants and found that overexpression of each gene in the wild-type genetic background increased both leaf area and cell numbers up to 14% (Fig. 8, D and E). These results indicate that the gain-of-function effects of GIFs on lateral organ growth are highly similar to each other, stimulating cell proliferation and organ growth.
Figure 8.
Overexpression phenotype of GIF genes in gif1 (A–C) and wild-type (D and E) plants. The first pairs of 23-d-old plants were used for the analysis of leaf dimensional and cellular parameters (n = 10). Error bars indicate se. Col, Columbia wild type. [See online article for color version of this figure.]
DISCUSSION
In this study, we presented, to our knowledge, the first loss- and gain-of-function evidence that GIF2 and GIF3 acted as positive regulators for lateral organ growth, as did GIF1. We were also able to determine contributions of individual GIF genes to the biological function through phenotypic analyses of a series of multiple gif mutants. gif triple mutants also revealed novel phenotypes that are relevant to functional activities of the shoot apical meristem, such as plastochronic and axillary branching properties, as well as reproductive organ function and structure.
GIF Genes Have Functional Redundancy in Determining Organ Size as Well as in Regulating Multiple Developmental Processes
We demonstrated that GIF genes have an overlapping expression pattern in many different tissues and that gif mutations act synergistically to induce dramatic reductions in cell numbers of lateral organs, such as leaves and flowers as well as cotyledons (Figs. 1–3), resulting in tiny plants. The mutational effect on cell proliferation was less severe in cotyledons than in other lateral organs, probably because the postembryonic cell division of cotyledons occurred for a narrow time window (Masubelele et al., 2005). The reduction in cell numbers was accompanied by a remarkable enlargement of cell size, which only partially compensated for the reduced organ size. The gif mutations also synergistically reduced the vegetative SAM size by decreasing cell numbers (Fig. 6). Furthermore, we have demonstrated that overexpression of GIF1, GIF2, and GIF3 promotes cell proliferation and leaf size and that GIF2 and GIF3 proteins are functional equivalents of GIF1 (Fig. 8). Horiguchi et al. (2005) have previously shown that overexpression of the GIF1/AN3 gene stimulates cell proliferation as well, leading to enlarged leaves by about 20%. These results led us to the conclusion that all of the GIF genes function redundantly as positive regulators of cell proliferation, thereby determining plant organ size. The overlapping expression pattern and functional redundancy are in line with our previous biochemical studies showing that GIF2 proteins interact with GRF1 proteins as does GIF1 (Kim and Kende, 2004), raising the possibility that all three GIF proteins may participate in the formation of the functional transcriptional complex with GRF proteins in a combinatorial manner. However, not all GRFs may be in the complex to the same extent, because GIF1 interacted strongly with GRF5 and GRF9 but weakly with GRF4 (Horiguchi et al., 2005).
Overexpression of GIF and GRF genes stimulated organ growth but to a limited extent (Fig. 8; Kim et al., 2003; Horiguchi et al., 2005). It may be due to other factors limited in the mechanisms of organ size determination, and GIFs and GRFs are very likely to be limiting factors to each other. To test this interesting possibility, we set out to construct double overexpressors of GIFs and GRFs. The resultant transgenic plants might not only give a detailed understanding of molecular mechanisms of organ growth with regard to the transcriptional complex but also might provide a basic knowledge for the development of agriculturally useful traits.
Judged from the abnormal structure of flower organs, as mentioned briefly, and the plastochronic phenotype (Figs. 2 and 7; Table I), it seems that GIF genes play an important role in regulating other aspects of shoot development. It remains unknown whether those phenotypes are in any causal relationship to the cell proliferation process. With respect to the pleiotropic phenotype of gif mutants, it is interesting that the human homolog of the GIF proteins, SYT transcription coactivators, also show diverse biological roles in human cells via interaction with a plethora of proteins (Eid et al., 2000; de Bruijn et al., 2001; Kato et al., 2002; Perani et al., 2003, 2005). For example, the human SYT proteins interact with the chromatin-remodeling proteins BRM and BRG1, which are known to act as positive or negative regulators of global expression (Thaete et al., 1999; Kato et al., 2002; Perani et al., 2003). SYT also interacts with the histone acetyltransferase p300/CBP to control cell adhesion (Eid et al., 2000). The Arabidopsis BRM gene has been reported to be involved in numerous aspects of plant growth and development (Hurtado et al., 2006; Kwon et al., 2006). Han et al. (2007) demonstrated that, in Arabidopsis, several genes encoding histone acetyltransferase play a role in regulating plant growth and flowering time. We have previously shown that the molecular behavior of GIF1 proteins is similar to that of SYT, including their existence as nuclear speckles and transcriptional activation activities (Kim and Kende, 2004). Therefore, apart from its known interacting partner GRF, GIF proteins may have other interacting proteins to affect multiple developmental processes.
Mechanisms by Which GIF Genes Regulate Cell Proliferation during Organogenesis
It is conceivable that cell numbers in a plant organ could be determined in several different ways: (1) the frequency of cell proliferation; (2) the duration of cell proliferation; and (3) the number of cells in the primordial cell pool (Autran et al., 2002). Studies on many genes that are involved in cell proliferation during organogenesis suggest that most of those genes seem to control cell numbers by changing the duration of cell proliferation rather than its rate, regardless of their role as positive or negative regulators. ANT and KLU affect cell proliferation by regulating the duration, rather than the rate, of cell proliferation (Mizukami and Fischer, 2000; Anastasiou et al., 2007). As positive regulators, GIF genes apparently influence the duration of cell proliferation as well, as the strong gif mutants ceased cell proliferation several days earlier than the wild type (Fig. 4). However, the mechanism by which GIF genes affect the duration seems to differ from that of ANT and KLU. First, ANT and KLU genes are involved in maintaining the meristematic competence of cells; thus, their mutational effect was manifested primarily in the later stage of the proliferation phase rather than in young organs (Mizukami and Fischer, 2000; Anastasiou et al., 2007). To the contrary, the effect of gif mutations was obvious in the primordial leaves, where most of the cells are actively dividing. This was clearly in line with the fact that GIF1/AN3 expression, assayed by GUS histochemical staining in promoter-GUS fusion transgenic lines, was highly active only in the very early stage of primordial leaves (Horiguchi et al., 2005). Second, while the frequency of cell proliferation in young organs of ant and klu mutants is very similar to that in the wild type (Mizukami and Fischer, 2000; Anastasiou et al., 2007), that of gif double and triple mutants is low, especially at the very early stage showing the maximum proliferating activity (Fig. 4). The low frequency of cell proliferation in gif mutants is supported by the finding that the transcript levels of cell cycle-regulating genes are reduced in the gif triple mutant at day 6 (Fig. 5). Like GIF genes that are highly expressed in the shoot apex (Fig. 1), those cell cycle genes have been shown to be actively expressed in both the SAM and developing primordia in Arabidopsis and tobacco (Nicotiana tabacum; Segers et al., 1996; Donnelly et al., 1999; Kosugi and Ohashi, 2002; Dewitte et al., 2003). This raises the possibility that the function of GIF genes may be mediated through the control of expression of cell cycle genes. For instance, the expression level of CycB1;1 was significantly reduced in the gif triple mutant (Fig. 5), and the GUS activity under the promoter of CycB1;1 was especially high at marginal positions of the wild-type leaf primordia that were rapidly expanding along the mediolateral axis (Donnelly et al., 1999). In addition, employing promoter∷GUS reporter lines and RT-PCR analysis, Horiguchi et al. (2005) also showed that the expression gradient of GIF1/AN paralleled that of the G2/M transition-specific cyclin gene along the longitudinal axis of developing leaves. Taken together, these results suggest that a decrease in the marginal proliferation activity would lead to a narrow leaf, which is in a good agreement with the gif phenotype.
The fact that the gif triple mutant produces leaf primordia with fewer cells raises the following possibility: besides low frequency of cell proliferation (Fig. 4), the leaf primordium of gif mutants may serve as a poor reservoir of progenitor cells available for further cell division. That, in turn, would lead to earlier exhaustion of meristematic cells, resulting in the earlier cessation and, thus, the short duration of cell proliferation. Autran et al. (2002) demonstrated that the swp mutant produced small leaf primordia with reduced cell numbers, suggesting that the primordial cell pool might be an important determinant of the final leaf size.
Relationship between the Sizes of the SAM Cell Pool, Leaf Primordium, and Plastochron
One of the key functions of the SAM is to form lateral organs, such as leaves and flowers, from its peripheral zone. It has been proposed that the growth of young developing primordium is preceded by active mitosis in a large part of the SAM L1 layer (Laufs et al., 1998b). Clonal and anatomical analyses suggested that 12 to 30 founder cells of the peripheral zone in a mature embryo of Arabidopsis contribute to each of the first two leaf primordia and that cells in the SAM continue to be recruited into the developing primordium even after imbibition of seeds (Furner and Pumfrey, 1992; Irish and Sussex, 1992). In the SAM of narrow sheath mutants of maize (Zea mays), fewer cells are recruited into leaf founder cells, producing extremely narrow leaves (Scanlon et al., 1996; Scanlon, 2000). Another maize mutant, rough sheath2, developed reduced numbers of leaf founder cells in the SAM, resulting in narrow or bladeless leaves (Schneeberger et al., 1998; Timmermans et al., 1999). Therefore, it is conceivable that the small size of leaf primordia in the gif mutants may be, in part, attributable to the small size of the SAM cell pool, probably because the SAM tissues do not have sufficient numbers of founder cells. This notion is in line with the fact that all of the cytokinin-signaling mutants and transgenic plants with reduced cytokinin activities have both the smaller SAM cell pool and leaf organs with fewer cells compared with the wild type (Werner et al., 2001, 2003; Higuchi et al., 2004; Nishimura et al., 2004; Miyawaki et al., 2006). However, precise data on the causal relationship between altered numbers of the SAM cells and leaf cells are scarce in Arabidopsis. A better understanding of the gif mutants may help clarify the relationship between the sizes of the SAM and the leaf primordium in the future.
Meanwhile, it has been well documented that, in general, plastochron length and SAM size are inversely correlated in Arabidopsis. The cytokinin mutants and transgenic plants mentioned above have much smaller SAM and fewer leaves compared with the wild type, which is indicative of a longer plastochron (Werner et al., 2001, 2003; Higuchi et al., 2004; Nishimura et al., 2004; Miyawaki et al., 2006). Up-regulation of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9 expression results in both a smaller size of the meristem and a longer plastochron (Wang et al., 2008). On the other hand, bigger SAM size of clavata1 and altered meristem program1 mutants was accompanied by shorter plastochron length (Chaudhury et al., 1993; Kwon et al., 2005; Vidaurre et al., 2007). The inverse correlation, however, does not appear to be robust, as gif mutants display a significant shortening of plastochron length, despite their reduced SAM size (Fig. 6; Table I). In addition, recessive mutations in MGOUN1 and MGOUN2 cause a reduction in leaf number and larger meristem (Laufs et al., 1998a). Therefore, it appears that alteration in the plastochronic property of mutant plants may depend not only on the size of the SAM cell pool but also on the regulatory nature of each mutation in the SAM tissue.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) seeds were sown on wet soil (Mix5; Sunshine), stratified at 4°C for 3 d, and transferred to a growth room at 23°C under a photoperiod of 16 h of light/8 h of darkness, which was marked as day 0 throughout the experiments. For measurement of leaf plastochron, plants were grown under the short-day conditions of 8 h of light/16 h of darkness. Wild-type plants and all of the T-DNA insertional mutants were in the Columbia ecotype. gif1 (SALK_150407; Kim and Kende, 2004), gif2 (SAIL_328_A03; Sessions et al., 2002), and gif3 (SALK_072950; Alonso et al., 2003) seeds were obtained from the Arabidopsis Biological Resource Center.
Identification of T-DNA Insertional Mutants and Construction of Multiple Mutants
Homozygous mutant plants were selected by PCR-assisted genotyping. Primers for amplification of wild-type GIF genes and T-DNAs are described in Supplemental Table S2. DNA fragments amplified with the gene-specific and left-border primers were sequenced to confirm the T-DNA insertion site. The double homozygous lines, gif1 gif2, gif2 gif3, and gif1 gif3, were established through crosses between each homozygous line. The gif1 gif2 double mutant was maintained in the gif1/+ gif2/gif2 genotype, as it was female sterile, although it produced fertile pollen. The gif1 gif2 gif3 triple mutant was obtained as the F2 progeny of the cross between gif1 gif2 (male) and gif2 gif3 (female) and maintained in gif1/+ gif2/gif2 gif3/gif3.
Construction of Transgenic Plants Overexpressing GIF Genes
GIF1, GIF2, and GIF3 cDNAs were amplified by PCR using primer pairs containing the Gateway partial recombination site at the 5′ end, attB1 or attB2 (for primer sequence information, see Supplemental Table S2). The amplified DNA fragments were used as template for a second round of PCR with adaptor primers. The final cDNA fragments were inserted into the entry vector pDONR221 by the BP recombinant reaction, according to the manufacturer's protocol (Invitrogen). The resulting plasmids were used in the LR reaction with the destination vector pB2GW7,0 (http://www.psb.ugent.be/gateway) to produce GIF overexpression constructs driven by the cauliflower mosaic virus 35S promoter. The recombinant binary plasmids were introduced into gif1 mutant plants by Agrobacterium tumefaciens-mediated transformation (Clough and Bent, 1998). Dozens of independent T1 plants overexpressing GIF1, GIF2, and GIF3 were selected on soil (Sunshine Mix1) soaked with the herbicide ammonium glufosinate (Bayer Cropscience). Single-insertion homozygous T3 lines of the GIF1, GIF2, and GIF3 overexpressors in the gif1 mutant background were selected and established. For the construction of GIF1, GIF2, and GIF3 overexpressors in the wild-type background, the established T3 lines were crossed to wild-type plants, after which plants homozygous for transgenes were established in the F3 progeny via PCR genotyping and herbicide resistance.
RT-PCR Analyses
For determination of GIF mRNA content in T-DNA insertional mutants, total RNAs were extracted with TRIzol reagent (Invitrogen) from 15-d-old wild-type and mutant plants, treated with DNase I (DNA-free; Ambion), and subjected to RT (SuperScript II; Invitrogen). The resulting cDNAs were used for PCR amplification, the conditions for which were as follows: denaturation at 95°C for 2 min, followed by 34 cycles of 95°C for 15 s, 52°C for 30 s, and 72°C for 1 min (for primer sequence information, see Supplemental Table S2).
For determination of transcript levels of cell-cycling regulators, cDNAs were prepared from the 6-d-old first two leaves as mentioned above, serially diluted to the indicated factors, and used for amplification. PCR conditions were as follows: denaturation at 95°C for 2 min, followed by 29 cycles of 95°C for 15 s, 52°C for 30 s, and 72°C for 1 min (for primer sequence information, see Supplemental Table S2).
Microarray Data Profiling
The microarray data of the AtGenExpress expression atlas were retrieved from The Arabidopsis Information Resource (Schmid et al., 2005; ftp://ftp.arabidopsis.org/home/tair/Microarrays/Datasets /AtGenExpress).
Measurement of Dimensional Parameters of Leaves, Cotyledons, and Petals
Digital images of detached leaves, cotyledons, and petals were acquired using a scanner. Area, length, and width of leaves, cotyledons, and mature petals as well as petioles were determined with the image-analyzing program SCIONIMAGE (Scion). For kinematic analysis of leaf area, young leaves up to day 9 were detached and mounted on slide glass for light-microscopic image analysis.
Number and Size of Leaf-Blade, Cotyledon, and Petal Cells
Leaf, cotyledon, and petal tissues were fixed with ethanol:acetic acid (6:1) for 4 h and were washed with 100% ethanol three times and then 70% ethanol once. Finally, all tissues were cleared in a chloral hydrate solution (8 g of chloral hydrate, 1 mL of glycerol, and 2 mL of distilled water) and mounted on slide glass. The microscopic images were obtained using a differential interference contrast microscope (Zeiss Axioplan). Subepidermal cells aligned along a longitudinal axis just beside the midvein or along a transverse axis in the maximum width region were counted. To determine cell area, 20 cells grouped halfway from the midvein to the leaf margin at the widest point were analyzed with SCIONIMAGE software.
Confocal Laser Scanning Microscopy for the SAM Cells
Mature embryos were prepared and stained with propidium iodide according to Running et al. (1995), after which the shoot apical region of mature embryos was analyzed and photographed by confocal laser scanning microscopy (Bio-Rad MRC-1024). Nuclei in the SAM region were counted to determine cell numbers.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AY102639 through AY102641 for GIF1 through GIF3, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression profile of GIF genes.
Supplemental Table S1. Numbers of seeds produced from crosses between Columbia and the gif1 gif2 double mutant.
Supplemental Table S2. Primer sequences for PCR amplification.
Supplementary Material
Acknowledgments
We thank Dr. Woo Taek Kim for technical support, critical reading of the manuscript, and helpful comments; Dr. Myeong Min Lee for technical support; and Drs. Soon Ki Park and Jong Tae Song for sharing their growth room. We also thank the Arabidopsis Biological Resource Center for the mutant seeds.
This work was supported by the Korea Research Foundation (grant nos. KRF–2006–331–C00264 and KRF–2008–314–C00345).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jeong Hoe Kim (kimjeon4@knu.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Anderson HJ, Vonarx EJ, Pastushok L, Nakagawa M, Katafuchi A, Gruz P, Di Rubbo A, Grice DM, Osmond MJ, Sakamoto AN, et al (2008) Arabidopsis thaliana Y-family DNA polymerase η catalyses translesion synthesis and interacts functionally with PCNA2. Plant J 55: 895–908 [DOI] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 21: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES (1993) amp1: a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4: 907–916 [Google Scholar]
- Cho KH, Jun SE, Lee YK, Jeong SJ, Kim GT (2007) Developmental processes of leaf morphogenesis in Arabidopsis. J Plant Biol 50: 282–290 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Bruijn DR, dos Santos NR, Thijssen J, Balemans M, Debernardi S, Linder B, Young BD, Geurts van Kessel A (2001) The synovial sarcoma associated protein SYT interacts with the acute leukemia associated protein AF10. Oncogene 20: 3281–3289 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M (2006) The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol 16: 272–279 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Eid JE, Kung AL, Scully R, Livingston DM (2000) p300 interacts with the nuclear proto-oncoprotein SYT as part of the active control of cell adhesion. Cell 102: 839–848 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H (2007) Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner IJ, Pumfrey JE (1992) Cell fate in the shoot apical meristem of Arabidopsis thaliana. Development 115: 755–764 [Google Scholar]
- Han SK, Song JD, Noh YS, Noh B (2007) Role of plant CBP/p300-like genes in the regulation of flowering time. Plant J 49: 103–114 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, Kobayashi M, Shinozaki K, Kato T, Tabata S, et al (2004) In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA 101: 8821–8826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68–78 [DOI] [PubMed] [Google Scholar]
- Hurtado L, Farrona S, Reyes JC (2006) The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol 62: 291–304 [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1992) A fate map of the Arabidopsis embryonic shoot apical meristem. Development 115: 745–753 [Google Scholar]
- Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, Takeuchi T, Ohtsuki Y, Sugano S, de Bruijn DR, Chait BT, et al (2002) SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem 277: 5498–5505 [DOI] [PubMed] [Google Scholar]
- Kim JH, Choi D, Kende H (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H (2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH (2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Kosugi S, Ohashi Y (2002) E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J 29: 45–59 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D (2005) WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev 19: 992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D (2006) A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133: 3223–3230 [DOI] [PubMed] [Google Scholar]
- Laufs P, Dockx J, Kronenberger J, Traas J (1998. a) MGOUN1 and MGOUN2: two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Laufs P, Grandjean O, Jonak C, Kiêu K, Traas J (1998. b) Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 10: 1375–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JA (2005) D-type cyclins activate division in the root apex to promote seed germination in Arabidopsis. Proc Natl Acad Sci USA 102: 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Samland AK, Planchais S, Murray JA (2006) The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov VV, De Veylder L, Van Montagu M, Inze D (1999) Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell 11: 509–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y (2001) A matter of size: developmental control of organ size in plants. Curr Opin Plant Biol 4: 533–539 [DOI] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C (2004) Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perani M, Antonson P, Hamoudi R, Ingram CJ, Cooper CS, Garrett MD, Goodwin GH (2005) The proto-oncoprotein SYT interacts with SYT-interacting protein/co-activator activator (SIP/CoAA), a human nuclear receptor co-activator with similarity to EWS and TLS/FUS family of proteins. J Biol Chem 280: 42863–42876 [DOI] [PubMed] [Google Scholar]
- Perani M, Ingram CJ, Cooper CS, Garrett MD, Goodwin GH (2003) Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene 22: 8156–8167 [DOI] [PubMed] [Google Scholar]
- Running MP, Clark SE, Meyerowitz EM (1995) Confocal microscopy of the shoot apex. Methods Cell Biol 49: 217–229 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ (2000) NARROW SHEATH1 functions from two meristematic foci during founder-cell recruitment in maize leaf development. Development 127: 4573–4585 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Schneeberger RG, Freeling M (1996) The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development 122: 1683–1691 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schneeberger R, Tsiantis M, Freeling M, Langdale JA (1998) The ROUGH SHEATH2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125: 2857–2865 [DOI] [PubMed] [Google Scholar]
- Segers G, Gadisseur I, Bergounioux C, de Almeida Engler J, Jacqmard A, Van Montagu M, Inzé D (1996) The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J 10: 601–612 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Poethig RS (1994) Leaf development in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 379–401
- Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner E, Cooper CS, Goodwin G (1999) Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet 8: 585–591 [DOI] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tsukaya H (2003) Organ shape and size: a lesson from studies of leaf morphogenesis. Curr Opin Plant Biol 6: 57–62 [DOI] [PubMed] [Google Scholar]
- Tsukaya H (2005) Leaf shape: genetic controls and environmental factors. Int J Dev Biol 49: 547–555 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T (2007) AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134: 2561–2567 [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmülling T (2001) Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA 98: 10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DW (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.