Abstract
Polychlorinated biphenyls (PCBs) are widely distributed persistent organic pollutants. In vitro research has shown that plant cell cultures might transform lower chlorinated congeners to hydroxylated PCBs, but there are few studies on in vivo metabolism of PCBs by intact whole plants. In this research, poplar plants (Populus deltoides × nigra, DN34) and switchgrass (Panicum vigratum, Alamo) were hydroponically exposed to 3,3′,4,4′-tetrachlorobiphenyl (CB77). Metabolism in plants occurred rapidly, and metabolites were detected after only a 24 h exposure. Rearrangement of chlorine atoms and dechlorination of CB77 by plants was unexpectedly observed. In addition, poplars were able to hydroxylate CB77 and the metabolite 6-hydroxy-3,3′,4,4′-tetrachlorobiphenyl (6-OH-CB77) was identified and quantified. Hybrid poplar was able to hydroxylate CB77, but switchgrass was not, suggesting that enzymatic transformations are plant specific. Sulfur-containing metabolites (from the action of sulfotransferases) were investigated in this study, but they were not detected in either poplar or switchgrass.
Short abstract
Whole poplars in vivo, candidate species for remediation at dredged material disposal sites, are able to hydroxylate and dechlorinate CB77 with chlorine substituent rearrangements.
Introduction
Polychlorinated biphenyls are persistent, bioaccumulating, toxic organic pollutants. Their general metabolic pathway in mammals involves cytochrome P-450 (CYP) enzyme-mediated oxidation to form hydroxylated metabolites and epoxides and further conjugation with glucuronide or glutathione to create corresponding conjugates, leading further to methylsulfonyl metabolites catalyzed by glutathione S-transferase (GST) (1). The formation of methyl sulfones is structure dependent. PCBs with vicinal hydrogens at the 3 and 4 positions of the aromatic ring as well as 2,5- or 2,3,6-substituted chlorines are most likely to be metabolized to methyl sulfones (2,3).
It is important to study the fate of individual PCB congeners because the properties, toxicities, and behavior of PCBs vary widely in the environment. The coplanar 3,3′,4,4′-tetrachlorobiphenyl (CB77, IUPAC number) is a dioxin-like congener, with a relatively high toxicity compared to most tetrachlorobiphenyls. Its metabolism in animals has been investigated (1,4). The main monohydroxy metabolites of CB77 in mammals includes 4-hydroxy-3,3′,4′,5-tetrachlorobiphenyl (4−OH-CB79), 5-hydroxy-3,3′,4,4′- tetrachlorobiphenyl (5−OH-CB77), a trace of 6-hydroxy-3,3′,4,4′-tetrachlorobiphenyl (6−OH-CB77), and 2-hydroxy-3,3′,4,4′-tetrachlorobiphenyl (2−OH-CB77). The methylsulfonyl metabolites of CB77 were documented in rats, mice, and fish, although CB77 is not one of the preferred structures for methyl sulfone metabolism (5−8).
According to the “green liver” model, metabolism in plants is similar to the detoxification mechanism in animal livers because of similar enzyme systems (9). To date, metabolism of PCBs in plants is generally reported in in vitro studies using cell or tissue cultures, which have the advantages of rapidly growing biomass and axenic conditions where microorganisms play no role in plant metabolism (10−12). Different plant species, including Paul’s Scarlet rose, tobacco, Solanum spp., horseradish, and alfalfa have been screened for their ability to biotransform PCBs using cell cultures. Mono- and dihydroxy-chlorobiphenyls were identified as the metabolites of mono- and dichlorobiphenyls (13). Other research showed only monohydroxy-PCBs as the metabolites of di-, tri-, tetra-, and penta-chlorobiphenyls (14). Metabolism depends on the plant species and specific PCB congeners to which the plant tissue cultures are exposed. In general, lower chlorinated congeners are metabolized more rapidly than those with higher chlorine substitutions. However the position of chlorine atoms and molecular structures are also important factors affecting PCB metabolism (10). In the study of Wilken et al., only three in 12 cell cultures, tomato (Lycopersicum esculentum), lettuce (Lactuca sativa), and Paul’s Scarlet rose (Rosa spp.), were able to hydroxylate CB77, and the main identified metabolites were 2-OH-CB77, 6-OH-CB77, and 5-OH-CB77 (15). More recently, methoxy- and hydroxy-methoxy-PCBs were reported as the metabolites of tobacco cells biodegrading dichlorobiphenyls (16).
Despite the advantage of cell and tissue cultures over whole plants as a fast screening tool, the extrapolation of metabolism from in vitro to in vivo is tenuous. To better understand the biodegradation of PCBs in ubiquitous plants in nature, we find in vivo studies are necessary. In our previous studies (17), it was found that whole hybrid poplars take up and bind CB77 strongly in root tissues, and metabolism was suggested but not confirmed. To study the biotransformation of CB77 in whole plants and the applicability of phytoremediation for such PCB congeners, we hydroponically exposed intact plants to CB77, and the presence of metabolites in hydroponic solution and plant tissues were tracked through time. To our knowledge, this is the first in vivo study on hydroxylation of PCB within intact plants.
Experimental Section
Chemicals
The standards of CB77, hydroxy metabolites (4-OH-CB79, 5-OH-CB77, 6-OH-CB77), and the derivatization reagent (diazomethane) were all synthesized by the National Institute of Environmental Health Sciences (NIEHS) Superfund Basic Research Program Center at the University of Iowa. CB77 was synthesized from 3,4-dichlorobenzene boronic acid and 3,4-dichlorobromobenzene using the Suzuki coupling reaction (18). The 4-OH-CB79 metabolite was synthesized using the Suzuki coupling of 3,4-dichlorobenzene boronic acid and 2,6-dichloro-4-bromoanisole, followed by demethylation of the methoxy PCB with boron tribromide (19). The 5-OH-CB77 and 6-OH-CB77 metabolites were prepared using the Cadogan coupling of 3,4-dichloroaniline with 2,3-dichloroanisole and subsequent demethylation with boron tribromide (20). All four compounds were >99% pure according to gas chromatographic analysis (based on relative peak area). A solution of diazomethane in diethylether was synthesized from N-methyl-N-nitroso-p-toluenesulfonamide (Diazald) using an Aldrich mini Diazald apparatus (Milwaukee, WI) following manufacturer instructions. The synthesization and use of diazomethane must be completed in a fume hood because of high toxicity.
Stock solutions of CB77 and hydroxy-PCBs at 1 mg mL−1 as compounds were prepared in acetone. Working solutions were prepared by gradual dilution of the stock solution. Because of no commercial methylsulfonyl-CB77 standards available, 3-methylsulfonyl-2,2′,5,5′-tetrachlorobiphenyl (3-MeSO2-CB52) and 4-methylsulfonyl-2,2′,5,5′-tetrachlorobiphenyl (4-MeSO2-CB52) with the same amounts of chlorine substitutions as CB77 were purchased from Accustandard as references. Internal standard 2,2′,3,4,4′,5,6,6′-octachlorobiphenyl (CB204, IUPAC number) was purchased from Cambridge Isotope Laboratories, Inc. All standards and solutions were stored hermetically in amber glass vials at room temperature in the dark.
Silica gel (70−230 mesh, Fisher Scientific, Inc.) was activated overnight at 450 °C. Two kinds of cleanup silica gel (concentrated and 90% sulfuric acid silica gel) were prepared by blending 100 g of activated silica gel with 50 g of concentrated H2SO4 and 50 g of 90% (w/w) H2SO4, respectively.
Acetone (HPLC grade), methyl tert-butyl ether (MTBE) (HPLC grade), and hexane (pesticide grade) were purchased from Fisher Scientific. All other chemicals and reagents used in this experiment were of analytical reagent grade or higher purity.
Hydroponic Exposure
Cuttings from male clones of the adult Imperial Carolina hybrid poplar tree (Populus deltoides × nigra, DN34) and switchgrass (Panicum vigratum, Alamo) were used in this study. Each cutting was fit snugly with a predrilled screw cap and predrilled PTFE-faced septum. The interface of the septum and the cutting was sealed with 100% silicon sealant. Then, the cuttings were grown hydroponically in half strength Hoagland nutrient solution (21). After 25 days, healthy whole poplar plants were selected to carry out the experiment.
Seeds of switchgrass were germinated on a bed of perlite with 1/10 strength Hoagland solution. The grasses were transferred into half strength Hoagland solution (without perlite) when they grew 3 cm high. After enough biomass developed, bunches of grass (five to six individual grasses in a bunch) were grown in 500 mL glass jars for 1 week and then used for exposure.
The exposure reactors were 500 mL glass screw top conical flasks for poplars, 500 mL glass screw top jars for switchgrass, which have a sampling port with a PTFE-faced silicon septum and predrilled screw cap. Hoagland nutrient solution was made by using autoclaved deionized water saturated with oxygen by bubbling with compressed air flowing through a sterilized 0.20 μm filter to prevent microbial contamination. After 2 h, the content of oxygen in the solution, 8.81 mg L−1, was measured by a HACH HQ10 oxygen meter. An oxygen-rich solution was used to eliminate any possible anaerobic dechlorinating microorganisms. Then, each autoclaved reactor was filled with 400 g of nutrient solution and a suitable amount of CB77. Two initial exposure concentrations were investigated at 1 and 10 μg kg−1. All these procedures were performed in a laminar flow hood.
Each reactor was planted with 5−6 switchgrass plants or a single 8 in. individual poplar plant. Three of nine poplar plants with shoots cutoff acted as the excised poplar controls, three of them were air-dried to act as dead poplar controls, and the remaining three were autoclaved before exposure to create autoclaved poplar controls. An untreated whole poplar control and an untreated switchgrass control, without CB77 in the hydroponic solutions, were used to detect any PCB contamination emanating from laboratory air during the experiment. Plants were not sterilized except for the autoclaved controls. All reactors were wrapped with aluminum foil, which supported roots in the dark environment and eliminated photolysis of CB77. The temperature during plant growth was maintained at 23 ± 1 °C, and the photoperiod was 16 h a day under fluorescent lighting with a light intensity between 120 and 180 μmol m−2 s−1. Transpiration of whole plants was determined daily by weighing the reactors. Oxygen-saturated, autoclaved deionized water was injected into the reactors to replace transpiration losses.
Roots and hydroponic solutions of whole plants and controls were sampled and analyzed after 1 and 5 day exposures. Roots were ground in liquid nitrogen with a ceramic mortar and pestle. All equipment was rinsed with reagent-grade acetone between samples.
Extraction, Separation, and Cleanup
The extraction, separation, and cleanup procedures for PCBs, hydroxy-PCBs, and methylsulfonyl-PCBs were modified from the method for plasma (22). In brief, the hydroponic solutions were amended with 10 drops of 37% HCl and 5 mL of 2-propanol, and then extracted with hexane/MTBE (1:1, v/v) for 30 min twice. Roots samples were mixed with 2 mL of 37% HCl and 5 mL of 2-propanol, homogenized, and extracted with 3 mL of hexane/MTBE (1:1, v/v) per gram of sample under vigorous shaking overnight. The organic extract was transferred after centrifugation. A second extraction was performed for half an hour.
Extracts were evaporated to dryness and redissolved in 1 mL of hexane. The hexane phase was partitioned with 500 μL of NaOH solution (0.5 M in 50% ethanol) and transferred to a clean vial after phase separation. The alkaline solution was re-extracted by another 1 mL of hexane. The combined hexane extract contained PCBs, methylsulfonyl-PCBs, and any other neutral metabolites (neutral fraction), while hydroxy-PCBs were left in alkaline solution. The alkaline solution was acidified with 125 μL HCl (2 M) and extracted with 1 mL of Hx/MTBE (9:1, v/v) twice. The extract was amended with a few drops of methanol and 0.5 mL of diazomethane to derivatize the OH-PCB to MeO-PCB. The derivatization reaction was performed in a refridgerator at 4−8 °C for 3 h. Excess diazomethane was evaporated under a gentle nitrogen stream. Then, the derivatization products, methyoxy PCB, were cleaned with acid silica gel column A (1 g of concentrated sulfuric acid silica gel with 0.1 g activated silica gel at the bottom) and eluted by 10 mL of dichloromethane.
The neutral fraction was partitioned with 0.5 mL of anhydrous dimethyl sulphoxide (DMSO) for 10 min. The hexane phase containing PCBs was transferred to a clean vial. The DMSO phase containing methylsulfonyl-PCBs was then mixed with 1 mL of water and extracted with 3 mL of hexane. The PCB fraction was cleaned by silica gel column A with 10 mL of dichloromethane for elution. The methylsulfonyl-PCBs were cleaned by silica gel column B (0.5 g anhydrous Na2SO4 on top, 0.5 g of 90% sulfuric acid silica gel in the middle, and 0.1 g of activated silica gel at the bottom) and eluted by 15 mL of dichloromethane. Eluates were evaporated to dryness and dissolved in hexane for gas chromatograph (GC) analysis.
Instruments
Qualitative and quantitative analysis of hydroxylated and methylsulfonyl metabolites was performed on an Agilent 6890 gas chromatograph equipped with split detector system, GC/MS/ECD. Analytes were separated in a HP-5 fused silica capillary column (5% phenyl methyl siloxane, 60.0 m × 250 μm × 0.25 μm), and then split to a mass spectrometry and electron capture detector. The GC was set as follows: injection port, 270 °C with splitless mode; high purity helium carrier gas at a constant flow rate of 1.9 mL min−1; ECD, 360 °C; 95% argon and 5% methane makeup gas flow at 30 mL min−1; electron impact (EI) ionization MS source, 250 °C, scan from m/z 50 to 500. The oven program started at 75 °C and was held for 3 min, then was increased at 5 °C min−1 to 150 °C and held for 1 min, and then heated at 2 °C min−1 to 300 °C and held for 5 min.
Identification and quantification of daughter PCBs derived from dechlorination were performed by an Agilent 6890N gas chromatograph coupled to a Waters Micromass Quattro micro GC mass spectrometer (GC/MS/MS) (Milford, MA) operating under electron impact (EI) positive mode at 70 eV and multiple reaction monitoring (MRM), and the trap current was 200 μA. The GC was equipped with an Agilent 7683 series autosampler (23). The retention windows were defined by PCB homologue parent and daughter ions. Analytes were separated in a Supelco SPB-Octyl capillary column (30 m × 250 μm × 0.25 μm) with helium at a constant flow rate of 0.8 mL min−1. The collision gas was ultra pure carrier grade Argon. The oven temperature was programmed from 75 °C and held for 5 min, heated to 150 at 15 °C min−1 and held for 1 min, and then heated to 280 at 2.5 °C min−1 and held for 3 min.
The mass recoveries and reproducibility of CB77 and 6-OH-CB77 were investigated by measuring the blank aqueous concentrations and root samples spiked with standards. Results in Table 1 show that the analytical method worked well without the occurrence of chemical or biological transformations during sample analysis.
Table 1. Mass Recoveries and Reproducibility for CB77 and 6-OH-CB77 Analysis in Aqueous and Root Samples.
| standards spiked (μg) |
recovery (%, n = 3) |
relative standard deviation (%, n = 3) |
||||
|---|---|---|---|---|---|---|
| samples | CB77 | 6−OH-CB77 | CB77 | 6−OH-CB77 | CB77 | 6−OH-CB77 |
| solution | 0.4 | 74−98 | n.d.a | 14 | ||
| 0.4 | 1 | 80−95 | 87−98 | 10 | 6 | |
| roots | 0.4 | 84−100 | n.d. | 9 | ||
| 0.4 | 1 | 68−83 | 92−102 | 13 | 6 | |
Nondetectable.
Results and Discussion
Hydroxylation in Plants
Identification of the hydroxy metabolites of CB77 in plant roots and nutrient solutions was based on a comparison of the retention times and mass spectra of derivatives of analytes (in samples) to those of the standards. The derivatives of hydroxy-CB77, 6-MeO-CB77, were coeluted with 4-MeO-CB79, but they were separated from 5-MeO-CB77 on the HP-5 capillary column. According to their mass spectra, they all have the molecular ion [M]+ at m/z 322 but have different fragmentation patterns due to the different positions of methyoxy substitutions. The 6-OH-CB77 metabolite was identified in roots and nutrient solutions of exposed whole poplar plants and excised poplar controls, but it was not found in autoclaved poplar controls, dead poplar controls, or untreated whole poplar controls (Table 2). The derivative of 6-OH-CB77, 6-MeO-CB77 (Figure 1), has a visible molecular ion and typical fragmentation clusters surrounding ions at m/z 267 [M-CH3-Cl]+ and 207 [M-COCH3-2Cl]+ in the mass spectrum, and the ions around 207 possess the highest abundance. Our study confirms that the metabolism of PCBs varies in different plant species. There were no hydroxy-PCB metabolites detected in whole switchgrass, indicating that switchgrass was not able to hydroxylate CB77 during the experimental period.
Table 2. Detection of Metabolites in Roots and Nutrient Solutions after Whole Plants were Hydroponically Exposed to CB77a.
| reactors | amount ofreactors | exposuretime | initial Cb(μg kg−1) | samples | 6-OH-CB77(μg kg−1) | CB73c(μg kg−1) | CB52(μg kg−1) | CB35(μg kg−1) | CB20/28d μg kg−1) | CB15(μg kg−1) | CB3(μg kg−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| exposed | 1 | 1 day | 10 | roots | 6.8 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| whole | solution | 0.0053 | n.d. | n.d. | 0.0083 | n.d. | n.d. | n.d. | |||
| poplars | 2 | 1 day | 1 | roots | 8.3e | 3.6 | 3.9 | 1.03 | 5.6 | 3.5 | 7.0 |
| solution | 0.011 | 0.074 | 0.082 | n.d. | 0.097 | 0.11 | 1.04 | ||||
| 2 | 5 days | 10 | roots | 8.9 | 0.030 | 0.017 | 3.0 | n.d. | n.d. | n.d. | |
| solution | 0.081 | 0.011 | 0.013 | 0.014 | 0.015 | 0.018 | 0.39 | ||||
| 5 | 5 days | 1 | roots | 10.7 ± 6.4f | n.d. | 0.74 ± 1.4 | 1.5 ± 0.98 | n.d. | 0.55 ± 0.71 | 5.0 ± 2.7 | |
| solution | 0.012 ±0.004 | 0.013 ±0.019 | 0.017 ±0.019 | n.d. | 0.021 ±0.026 | 0.016 ±0.026 | 0.024 ±0.017 | ||||
| excised | 3 | 5 days | 1 | roots | 10.4 ± 6.9 | n.d. | n.d. | 1.3 ± 0.14 | n.d. | n.d. | n.d. |
| poplars | solution | 0.0088 ±0.0056 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |||
| dead | 3 | 5 days | 1 | roots | n.d.g | n.d. | n.d. | 1.4 ± 0.76 | n.d. | n.d. | 0.77 ± 1.2 |
| poplars | solution | n.d. | n.d. | n.d. | 0.013 ±0.022 | n.d. | n.d. | 0.053 ±0.0012 | |||
| autoclaved | 3 | 5 days | 1 | roots | n.d. | n.d. | n.d. | 0.38 ± 0.38 | n.d. | n.d. | 0.88 ± 0.22 |
| poplars | solution | n.d. | n.d. | n.d. | 0.0033 ±0.0046 | n.d. | n.d. | 0.030 ±0.023 | |||
| untreated | 1 | 5 days | 0 | roots | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| poplars | solution | n.d. | −h | − | − | − | − | − | |||
| exposed | 3 | 5 days | 1 | roots | n.d. | 0.075 ±0.12 | 0.11 ±0.18 | 0.18 ±0.05 | 0.045 ±0.077 | 0.071 ±0.12 | 0.16 ±0.26 |
| switchgrass | solution | n.d. | n.d. | 0.0015 ±0.0015 | n.d. | 0.0053 ±0.0018 | n.d. | 0.0071 ±0.0091 | |||
| untreated | 1 | 5 days | 0 | roots | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| switchgrass | solution | n.d. | − | − | − | − | − | − |
All metabolites were identified and quantitatively analyzed by GC/MS/ECD and GC/MS/MS. Mechanism of chlorine rearrangement is unknown.
Initial concentration of CB77.
PCB congeners named as the IUPAC number.
Coeluted PCB congeners that were not able to be separated by the used GC column.
Mean value.
Mean value ± standard deviation (n≥3).
Nondetectable.
No samples were detected.
Figure 1.
Chromatogram of 6-MeO-CB77 after derivitization from 6-OH-CB77 in a root sample, detected by the split detector system (GC/MS/ECD). (a) Chromatogram of ECD. (b) Total ion chromatogram of MS. (c) Mass scan spectrum. Compound corresponds to the peaks highlighted in panel a and b.
The autoclaved poplar controls were dead plants, minimizing the action of microbes and eliminating the role of plant enzymes. In the unautoclaved dead poplar controls, microbes associated with the roots were considered to be active. The metabolite 6-OH-CB77 was not detected in either control, but it was detectable in the live poplars, suggesting that hydroxylation in poplars was caused by the plant itself, rather than by microorganisms in the root zone. Excised poplar controls had no significant evapotranspiration, but the plant tissue was viable during the exposure period. The 6-OH-CB77 metabolite was detected in the excised controls, and the concentration was similar to that in exposed whole poplars, indicating that transpiration was not required for metabolic transformation of CB77 in plant tissues.
Hydroxylation occurred very rapidly in poplars with 6-OH-CB77 detected in roots and solutions after 1 day exposure. The concentrations of 6-OH-CB77 increased slightly over time but without a significant difference between 1 and 5 day exposures, indicating that further transformation in poplars is possible if the exposure period was lengthened. There was no significant difference between the concentrations of hydroxy metabolites under different dosages, although the lower dosage of CB77 resulted in slightly higher concentrations of metabolites. Other researchers reported that CB77 at a low dose was metabolized more rapidly than that at high dose in scup (7). The mass and concentrations of hydroxylated metabolites in roots were far higher than those in nutrient solution. It is concluded that small amounts of 6-OH-CB77 exuded from roots into the rhizosphere compartment during the exposure period.
Sulfur-ContainingMetabolites
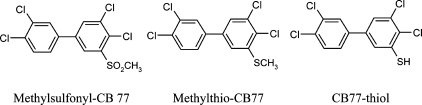
PCBs can be biotransformed to a multitude of sulfur-containing PCB metabolites, including thiol, methylthio-, and methylsulfonyl-PCBs under the function of CYP and GST enzymes in animals via forming glutathione conjugates of the PCBs (1,24−26). Most studies have focused on methylsulfonyl-PCBs (25−27), which have been found in various tissues of mammals and fish (28). Methylsulfonyl-CB77 and two possible sulfur-containing intermediates (Figure 2) are of concern in this study. Because the methylsulfonyl metabolites of CB77 are not commercially available, two standards of methylsulfonyl-CB52 with the same degree of chlorine substitutions as CB77 were selected as references. The retention time and mass spectra of 3- and 4-methylsulfonyl-CB52 were investigated. Similar mass spectra were detected in which typical molecular ion [M]+ at m/z 370 and fragmentation clusters surrounding at m/z 307 [M-CH3-CCl]+, 279 [M-CSO2CH3]+, and 184 [M-SO2CH3-3Cl]+ were displayed. Taking account of the possibility of various mass spectrum patterns of methylsulfonyl-CB77, molecular ions were powerful fragments to identify MeSO2-CB77. However, no similar mass spectra were found in the exposed poplar and switchgrass samples. There were also no molecular ions of CB77-thiol and methylthio-CB77 in any mass spectra of plant samples. It appears that neither poplar trees nor switchgrass are able to metabolize CB77 to methyl sulfones in 5 days. According to the literature (2,3,29), the structure of CB77, without not only vicinal 3,4 hydrogen substitutions but also 2,5-dichloro- or 2,3,6 trichloro-phenyl rings, makes it difficult to biotransform to methyl sulfones.
Figure 2.
Molecular structures of methylsulfonyl-CB77, methylthio-CB77, and CB77-thiol.
Dechlorination and Rearrangement in Plants
Various microbial dechlorination products of PCBs have been observed in anaerobic sediments (10). However, few reports are concerned with dechlorination of PCBs in animals and plants. Magee et al. found that the crude extract of nitrate reductase from leaves of the plant, Medicago sativa, was capable of dechlorination of CB153 by observing the decrease of CB153 after its incubation with the plant crude extract (30). Dechlorination metabolites were investigated to study the possible dechlorination of PCBs by whole plants. Anaerobic microbial dechlorination was avoided in the experiments of this study by autoclaving the reactors and solution, saturating the hydroponic solution with oxygen at the beginning of the exposure and replacing the transpiration loss with sterilized and oxygen saturated water. Thus, the hydroponic solution was aerobic during the short period of exposure (1 and 5 days). The dechlorination products in plant roots and nutrient solutions are shown in Table 2. A large proportion of the exposed whole plants were observed to contain significant concentrations of the metabolites of 2,3′,5′,6-CB (CB73), 2,2′,5,5′-CB (CB52), 3,3′,4-CB (CB35), 2,3,3′-CB/2,4,4′-CB (CB20/28), 4,4′-CB (CB15), and 4-CB (CB3) (CB20 and 28 are coeluted congeners on the Supelco SPB-octyl capillary column. Both of them have the possibility of occurrence.). For various controls, CB3 and 35 were the congeners detected most frequently. The concentrations of CB35 in autoclaved poplar control roots (with minimized roles of microbes and without the role of poplars) were far lower than that in dead poplar roots (with the presence of microbes but without viable poplar tissue) indicating that the symbiotic association of microbes in the root zone of the plant aid in the formation of CB35. The concentrations of CB35 in dead poplar roots were similar to that in excised poplar roots and exposed poplar roots (all with functioning root-associated microbes and poplar roots) at a lower dosage for 5 days, indicating that viable poplar tissue is not necessary for the formation of CB35. However, by comparison of the exposed whole poplars to the excised, dead, and autoclaved poplar controls, the formation of CB3, 15, 20/28, 52, and 73 were due to the role of the viable poplar (plant) tissues on dechlorination of the parent compound CB77.
According to the daughter PCBs detected in the reactors, the dechlorination of CB77 progressed from the tetra-chlorine (3,3′,4′4′-CB) congener to the di-(4,4′-CB) congener to the mono-chlorine (4-CB) congener. In addition, tetra-chlorine congeners, CB73, CB52, and trichlorine congeners CB20/CB28 were detected by GC/MS/MS, which were presumed to be the products of chlorine rearrangement of CB77 during metabolism by plants. The chlorine shift was suggested by our experiment in which a relatively high concentration of CB73 was detected after a whole poplar plant was exposed to CB52 for 5 days. The rearrangement of chlorines on the biphenyl rings was clearly observed in this study, although the mechanism is not understood. It is a surprising result that bears further confirmation and investigation.
The concentrations of the dechlorination products and chlorine-shift metabolites in switchgrass roots were lower than those in exposed poplar roots. This suggests that the enzymatic dechlorination and rearrangement capability of poplar is better than that of switchgrass.
Metabolism Pathways
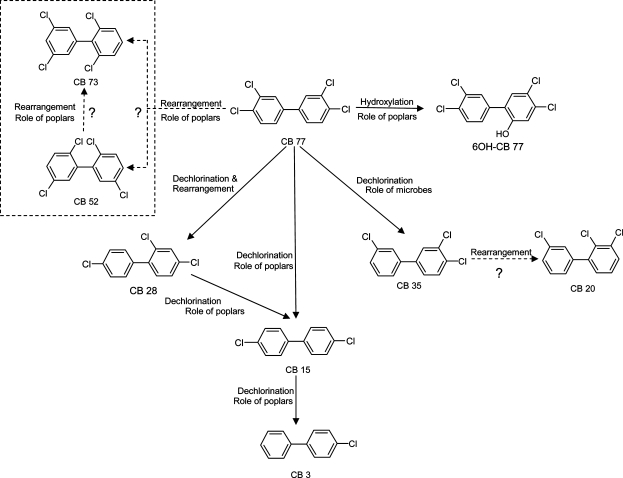
Generally, meta- and para-chlorines in PCB congeners are more susceptible to dechlorination (31). Dechlorination is usually a stepwise procedure. The 2,3,4,3′,4′-CB congener would biodegrade following the sequence as 2,3,4,3′,4′-CB, 2,4,3′,4′-CB, 2,4,3′-CB, 2,3′-CB, and 2-CB (32,33). Intermediates following dechlorination of CB77 by a palladium/iron nanoparticles system were determined to be (a) from 3,3′,4,4′-CB (CB77) to 3,3′,4-CB (CB35), 3,3′-CB (CB11), 3-CB (CB2), and biphenyl; (b) from 3,3′,4,4′-CB to 3,4,4′-CB (CB37), 4,4′-CB (CB15), 3,4-CB (CB12), 3,4′-CB (CB13), 4-CB (CB3), and biphenyl (34). Although the metabolism of pollutants in vivo is very complex and requires a wide range of enzymatic capability, a proposed mechanistic scheme (Figure 3) is shown for how poplars and associated microbes might interact to affect the observed transformation products from this study. All of the compounds shown in Figure 3 were confirmed by GC/MS/MS and GC/MS/ECD. The rearrangement of chlorines during metabolism of CB77 in poplar was suggested by the metabolites detected, CB52 and CB73 (Figure 3, compounds in the dotted square), during our experiments. The para/meta and meta/ortho chlorine shifts are hypothesized in the transformation from CB77 to CB52, from CB77 to CB73, from CB52 to CB73, and from CB35 to CB20. The meta/ortho shift was reported in the formation of PCDFs from PCB pyrolysis (35,36). However, these chlorine rearrangements have not been previously reported in microbial, mammal, or plant degradation studies of PCBs.
Figure 3.
Scheme of observed metabolism pathways of CB77 within poplars. Metabolites in the dotted square were also positively confirmed by GC/MS/MS in the poplar roots. Dotted arrows with question marks are the possible pathways which are not understood.
Our experiment showed that relatively high concentrations of CB15 and CB3 were detected after a whole poplar plant was exposed to CB28 for 5 days. Taking the daughter polychlorinated biphenyls in the reactors into account, we established the stepwise dechlorination pathway from CB28 to CB15 to CB3. However, the key intermediate, CB37, which should have occurred in the dechlorination from CB77 to CB15, was not detected. Possible hypotheses for the observed lack of CB37 are the following: (i) CB37 disappeared very quickly due to further transformation, including the formation of CB15 by dechlorination. (ii) Two meta chlorines on coplanar CB77 are of the same bond length, bond energy, and charge, which makes these two chlorines depart easily and form CB15 directly. (iii) CB77 was metabolized to CB28 directly under an enzymatic mechanism by poplar, which might involve dechlorination and rearrangement in one step.
There are two reported hydroxylation pathways for introducing hydroxyl substitution to the aromatic ring of PCBs (1). First is via an epoxide intermediate to form two hydroxylated metabolites. For example, CB77 could be metabolized to a 5,6-epoxide, which would form both 5-OH-CB77 and 6-OH-CB77. The second way is via an enzymatic hydroxyl substitution to a carbon on the aromatic ring directly to form the corresponding hydroxylated metabolite. These two hydroxylation pathways could be involved in poplars, but only the 6-OH-CB77 was detected in this paper.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) Superfund Basic Research Program (SBRP), Grant P42ES13661 and is a contribution from the W.M. Keck Phytotechnology Laboratory at the University of Iowa. We thank Hans-Joachim Lehmler, SBRP Center, University of Iowa, for providing PCB standards, and Anna Karin Norstrom and Craig Just, Civil Environmental Engineering, University of Iowa, for supporting the analytical methods.
References
- James M. O.Polychlorinated Biphenyls: Metabolism and Metabolites. In PCBs: Recent Advances in Environmental Toxicology and Health Effects; Robertson L. W., Hansen L. G., Eds.; The University Press of Kentucky: Lexington, KY, 2001; pp 34−46. [Google Scholar]
- Letcher R. J.; Norstrom R. J.; Bergman A. Geographical distribution and identification of methyl-sulphone PCB and DDE metabolites in pooled polar bear (Ursus maritimus) asipose tissue from western hemisphere Arctic and subarctic regions. Sci. Total Environ. 1995, 160/161, 409–420. [DOI] [PubMed] [Google Scholar]
- Letcher R. J.; Norstrom R. J.; Bergman A. An integrated analytical method for determination of polychlorinated aryl methyl sulfone metabolites and polychlorinated hydrocarbon contaminants in biologic matrices. Anal. Chem. 1995, 67, 4155–4163. [DOI] [PubMed] [Google Scholar]
- Letcher R. J.; Klasson-Wehler E.; Bergman A.. Methyl Sulfone and Hydroxylated Metabolites of Polychlorinated Biphenyls, In The Handbook of Environmental Chemistry; Paasivirta J., Ed.; Springer-Verlag; : Berlin, 2000; Volume 3, Part K, Chapter 11, New Types of Persistent Halogenated Compounds [Google Scholar]
- Haraguchi K.; Kato Y.; Masuda Y.; Kimura R. Metabolism of 3,4,3′,4′- tetrachlorobiphenyl via sulphur-containing pathway in rat: Liver-specific retention of methylsulphonyl metabolite. Xenobiotica 1997, 27, 831–842. [DOI] [PubMed] [Google Scholar]
- Yoshimura H.; Yonemoto Y.; Yamada H.; Koga N.; Oguri K.; Saeki S. Metabolism in vivo of 3,4,3′,4′-tetrachlorobiphenyl and toxicological assessment of the metabolites in rats. Xenobiotica 1987, 17, 897–910. [DOI] [PubMed] [Google Scholar]
- White R. D.; Shea D.; Stegeman J. J. Metablism of the aryl hydrocarbon receptor agonist 3,4,3′,4′-tetrachlorobiphenyl by the marine fish scup (Stenotomus chrysops) in vivo and in vitro. Drug Metab. Dispos. 1997, 25, 564–572. [PubMed] [Google Scholar]
- Wehler E. K.; Bergman A.; Brandt I.; Darnerud P. O.; Wachtmeister C. A. 3,4,3′,4′-Tetrachlorobiphenyl. Excretion and tissue retention of hydroxylated metabolites in the mouse. Drug Metab. Dispos. 1989, 17, 441–448. [PubMed] [Google Scholar]
- Sandermann H. Higher plant metabolism of xenobiotics: The “green liver” concept. Pharmacogenetics 1994, 4, 225–241. [DOI] [PubMed] [Google Scholar]
- Mackova M.; Barriault D.; Francova K.; Sylvester M.; Moder M.; Vrchotova B.; Lovecka P.; Najmanova J.; Demnerova K.; Novakova M.; Rezek J.; Macek T.. Phytoremediation of Polychlorinated Biphenyls. In Phytoremediation and Rhizoremediation; Mackova M., Dowling D., Macek T., Eds.; Springer: Dordrecht, The Netherlands, 2006; Chapter 11, pp 143−168. [Google Scholar]
- Harms H.; Bokern M.; Kolb M.; Bock C.. Transformation of Organic Contaminants by Different Plant System. In Phytoremediation Transformation and Control of Contaminants; McCutcheon S. C., Schnoor J. L., Eds.; John Wiley & Sons: Hoboken, NJ, 2003; Chapter 9, pp285−316. [Google Scholar]
- Mackova M.; Chroma L.; Kucerova P.; Burkhard J.; Demnerova K.; Macek T. Some aspects of PCBs metabolism by horseradish cells. Int. J. Phytorem. 2001, 7 (7), 401–414. [Google Scholar]
- Kucerova P.; Mackova M.; Chroma L.; Burkhard J.; Triska J.; Demnerova K.; Macek T. Metabolism of polychlorinated biphenyls by Solanum nigrum hairy root clone SNC-9O and analysis of transformation products. Plant Soil 2000, 225 (1−2), 109–115. [Google Scholar]
- Rezek J.; Macek T.; Mackova M.; Triska J. Plant metabolites of polychlorinated biphenyls in hairy root culture of black nightshade Solanum nigrum SNC-9O. Chemosphere 2007, 69 (8), 1221–1227. [DOI] [PubMed] [Google Scholar]
- Wilken A.; Bock C.; Bokern M.; Harms H. Metabolism of different PCB congeners by plant cell cultures. Environ. Toxicol. Chem. 1995, 14, 2017–2022. [Google Scholar]
- Rezek J.; Macek T.; Mackova M.; Triska J.; Ruzickova K. Hydroxy-PCBs, methoxy-PCBs, and hydroxy-methoxy-PCBs: Metabolites of polychlorinated biphenyls formed in vitro by tobacco cells. Environ. Sci. Technol. 2008, 42 (15), 5746–5751. [DOI] [PubMed] [Google Scholar]
- Liu J. Y.; Schnoor J. L. Uptake and translocation of lesser-chlorinated polychlorinated biphenyls (PCBs) in whole hybrid poplar plants after hydroponic exposure. Chemosphere 2008, 73, 1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmler H. J.; Robertson L. W. Synthesis of polychlorinated biphenyls using the Suzuki-coupling. Chemosphere 2001b, 45, 137–143. [DOI] [PubMed] [Google Scholar]
- Lehmler H. J.; Robertson L. W. Synthesis of hydroxylated PCB metabolites with the Suzuki-coupling. Chemosphere 2001a, 45, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Van den Hurk P.; Kubiczak G. A.; Lehmler H. J.; James M. O. Hydroxylated polychlorinated biphenyls as inhibitors of the sulfation and glucuronidation of 3-hydroxy-benzo[a]pyrene. Environ. Health Perspect. 2002, 110, 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E.Mineral Nutrition of Plants: Principles and Perspectives; John Wiley & Sons: New York, 1972. [Google Scholar]
- Bergman A.; Klasson-Wehler E.; Kuroki H. Selective retention of hydroxylated PCB metabolites in blood. Environ. Health Perspect. 1994, 102, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D.; Martinez A.; Hornbuckle K. C. Discovery of non-Aroclor PCB (3, 3′-dichlorobiphenyl) in Chicago air. Environ. Sci. Technol. 2008, 42 (21), 7873–7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke J.; Bergman A.; Larsen G. Methylsulfone formation and PCB metabolism. Organohalogen Compd. 1995, 25, 413–417. [Google Scholar]
- Bakke J.; Bergman A.; Larsen G. Catabolism of 2,4′,5-trichlorobiphenyl by the mercapturic acid pathway. Science 1982, 217 (13), 645–647. [DOI] [PubMed] [Google Scholar]
- Bergman A.; Norstrom R. J.; Haraguchi K.; Kuroki H.; Beland P. PCB and DDE methylsulfones in mammals from Canada and Sweden. Environ. Toxicol. Chem. 1994, 13, 121–128. [Google Scholar]
- Haraguchi K.; Kuroki H.; Masuda Y. Determination of PCB methylsulfone geners in Yusho and control patients. Chemosphere 1986, 15, 2027–2030. [Google Scholar]
- Karasek L.; Hajslova J.; Rosmus J.; Huhnerfuss H. Methylsulfonyl PCB and DDE metabolites and their enantioselective gas chromatographic separation in human adipose tissues, seal blubber and pelican muscle. Chemosphere 2007, 67, S22–S27. [DOI] [PubMed] [Google Scholar]
- Herman D. P.; Effler J. I.; Boyd D. T.; Krahn M. M. An efficient clean-up method for the GC-MS determination of methylsulfonyl-PCBs/DDEs extracted from various marine mammal tissues. Mar. Environ. Res. 2001, 52, 127–150. [DOI] [PubMed] [Google Scholar]
- Magee K. D.; Michael A.; Ullah H.; Dutta S. K. Dechlorination of PCB in the presence of plant nitrate reductase. Environ. Toxicol. Pharmacol. 2008, 25, 144–147. [DOI] [PubMed] [Google Scholar]
- Field J. A.; Sierra-Alvarez R. Microbial transformation and degradation of polychlorinated biphenyls. Environ. Pollut. 2008, 155 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- Abramowicz D. A. Aerobic and anaerobic biodegradation of PCBs: A review. Crit. Rev. Biotechnol. 1990, 10, 241–251. [Google Scholar]
- Abramowicz D. A.; Brennan M. J.; Van Dort H. M.; Gallagher E. L. Factors influencing the rate of polychlorinated biphenyl dechlorination in Hudson River sediments. Environ. Sci. Technol. 1993, 27, 1125–1131. [Google Scholar]
- Venkatachalam K.; Arzuaga X.; Chopra N.; Gavalas V. G.; Xu J.; Bhattacharyya D.; Hennig B.; Bachas L. G. Reductive dechlorination of 3,3′,4,4′- tetrachlorobiphenyl (PCB77) using palladium or palladium/iron nanoparticles and assessment of the reduction in toxic potency in vascular endothelial cells. J. Hazard. Mater. 2008, 159 (2−3), 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser H. R. Formation, occurrence and analysis of polychlorinated cibenzofurans, dioxins, and related compounds. Environ. Health Perspect. 1985, 60, 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser H. R. Formation of polychlorinated dibenzofurans (PCDFs) from pyrolysis of individual PCB isomers. Chemosphere 1979, 8 (3), 157–174. [Google Scholar]