Abstract
The subnuclear distribution of replication complex proteins is being recognized as an important factor for the control of DNA replication. Herpes simplex virus (HSV) single-strand (ss)DNA-binding protein, ICP8 (infected cell protein 8) accumulates in nuclear replication domains. ICP8 also serves as helper function for the replication of adeno-associated virus (AAV). Using quantitative 3D colocalization analysis we show that upon coinfection of AAV and HSV the AAV replication protein Rep and ICP8 co-reside in HSV replication domains. In contrast, Rep expressed by a recombinant HSV, in the absence of AAV DNA, displayed a nuclear distribution pattern distinct from that of ICP8. Colocal ization of Rep and ICP8 was restored by the reintroduction of single-stranded AAV vector genomes. In vitro, ICP8 displayed direct binding to Rep78. Single-stranded recombinant AAV DNA strongly stimulated this interaction, whereas double-stranded DNA was ineffective. Our findings suggest that ICP8 by its strong ssDNA-binding activity exploits the unique single-strandedness of the AAV genome to form a tripartite complex with Rep78 and AAV ssDNA. This novel mechanism for recruiting components of a functional replication complex directs AAV to subnuclear HSV replication compartments where the HSV replication complex can replicate the AAV genome.
INTRODUCTION
Adeno-associated virus (AAV)-based gene therapy vectors have become increasingly successful over recent years. Their widespread application requires highly efficient production methods. Since the development of herpes simplex virus (HSV)-based packaging systems for recombinant AAV vectors (1) the interaction of AAV with its helper virus HSV has raised increasing interest. The AAVs rely on either adenovirus or a herpes group virus for productive infection (2). In the absence of a helpervirus, AAV type 2 integrates into the host cell genome with a high preference for a specific site on human chromosome 19q13.3qter (3–5). AAV contains a 4.7 kb linear ssDNA genome with two open reading frames, rep and cap, which are flanked by 145 bp inverted terminal repeats (ITRs). These comprise the origins of replication, the packaging signal and serve as cis elements for chromosomal integration. AAV encodes three capsid proteins, and four overlapping, non-structural proteins. These comprise Rep78, a C-terminally spliced variant, called Rep68 and N-terminally truncated versions of either protein, called Rep52 and Rep40. The Reps are multifunctional regulatory proteins required for most steps of the AAV life cycle including DNA replication, gene expression, chromosomal integration and viral packaging (2).
We have previously identified a subset of six out of seven HSV replication genes as helper functions for productive AAV replication (6). Of these, the HSV ssDNA-binding protein (ssDBP) (ICP8) encoded by the UL29 gene, and a three-component helicase/primase complex encoded by the genes UL5, UL8 and UL52 are sufficient to support AAV replication. The two subunit HSV DNA polymerase (UL30/UL42) is not necessary but enhances AAV replication, both in vivo (6) and in vitro (7). The HSV origin-binding protein (UL9) is dispensable. This finding appears to reflect the lack of sequence similarity between the HSV origins and the AAV-ITRs that serve as AAV origins of replication. AAV Rep78/68 is required for AAV DNA replication initiated at the hairpin-structured AAV-ITRs. Rep78/68 binds to the Rep-binding site (RBS), unwinds the ITR due to its ATP-dependent helicase activity and introduces a single-strand nick at the adjacent terminal resolution site (trs) (8–10). Due to these activities Rep can be regarded as an AAV ori-binding protein and initiator of AAV DNA replication. The aim of this study was to test whether on AAV templates comprising the ITRs, Rep functionally replaces the HSV origin-binding protein (UL9) in the sense that Rep is able to recruit components of the HSV replication complex to the single-stranded AAV genome.
HSV ICP8, one of the key functions for HSV-induced AAV replication was considered as the prime candidate for a direct interaction partner of Rep (6). The biochemical activities of purified ICP8 as a key component of the HSV replication complex have been extensively studied [reviewed in (11)]. Binding of ICP8 to ssDNA is highly cooperative, but without obvious sequence specificity. Unwound ssDNA regions will be covered by ICP8, which keeps them in an extended conformation (12,13). In vitro studies demonstrated that ICP8 represents the first HSV replication protein to be recruited by the HSV ori-binding protein (UL9) to assist ATP-dependent ori unwinding (14). ICP8 enhances the assembly of the HSV replication complex by interaction with the UL8 subunit of the helicase–primase complex (15) and by stimulation of polymerase processivity (16). Conventional and confocal microscopy described the accumulation kinetics of ICP8 in nuclear HSV replication centers: at 2–3 h post-HSV infection ICP8 colocalizes to HSV DNA within punctate pre-replication sites. These are the precursors of active HSV replication compartments that fuse to larger globular structures by 4–6 h post-infection (p.i.) (17,18).
Here we present a recombinant HSV that expresses Rep from its cognate AAV promoters. It serves as a tool to study the kinetics and subcellular distribution of Rep and ICP8 at the single cell level by closely mimicking conditions found upon coinfection with HSV and AAV. 3D-immunofluorescence and quantitative colocalization analysis reveals that Rep and ICP8 colocalize in HSV replication compartments in dependence of single-stranded AAV genomes. Rep and ICP8 also interact in vitro. The interaction is strongly enhanced by the presence of single-stranded but not double-stranded AAV template DNAs.
MATERIALS AND METHODS
Cells and viruses
Mouse hybridoma cells producing mAb 76.3, 303.9 or 39S (HB8180, ATCC) were grown as outlined previously (19). BHK-21 cells and HSV strain 1802 were grown as described (20). AAV-2 stocks were grown on HeLa cells as described (6).
Plasmids
For construction of psub201lac the XbaI fragment of psub201 (21) comprising nucleotides 191–4485 of AAV-2 devoid of the ITRs (GenBank accession number J01901) was inserted into pFJ3 (20) downstream of the SV40-promoter-driven lacZ gene to give psub201lac. For construction of the N-terminal GST–ICP8 fusion construct, HSV UL29 gene was PCR-amplified from pCM-DBP (22) and assembled in pGEX-4T1 (Pharmacia) to yield pGEX-ICP8.
Generation of rHSVrep/cap
HSV virions were purified from supernatants of infected BHK cells and high molecular weight HSV DNA was extracted. The XbaI-flanked AAV-2 rep/cap cassette of psub201lac was inserted into the unique XbaI site of HSV-1 strain 1802 by DNA ligation and transfected into BHK cells, as outlined previously (20). HSV plaques were purified for several plaque rounds at limiting dilution when 100% of the resulting plaques tested positive for lacZ and AAV cap gene expression. One recombinant HSV plaque was selected at random to yield rHSVrep/cap virus stocks.
Western blot analysis
Protein extracts, gel electrophoresis and western blotting followed the protocols in Harlow and Lane (23). For Rep detection mAb 303.9 was used (24), for AAV Cap, mAb B1 (Progen, Heidelberg) with an anti-mouse peroxidase coupled secondary antibody detected by enhanced chemiluminescence according to the procedure given by the manufacturer (Amersham Life Science).
Double-label immunofluorescence
For AAV Rep and HSV ICP8 colocalization analysis, an established double-labeling immunofluorescence procedure for two mouse monoclonal primary antibodies was used (25–27). Discrimination was achieved by using an excess of labeled F(ab) fragments to detect the anti-Rep mAb of the first step and ensure complete blocking of free Fc tails. The amount of Cy3-conjugated goat F(ab) fragments of anti-mouse IgG (Dianova) required was titrated on Rep-expressing cells stained with the anti-Rep mAb and post-stained with the Alexa 350-conjugated goat anti-mouse IgG (Molecular Probes) used as secondary antibody in the second step. Rep staining was entirely blocked beginning at a dilution of 1:1000. A dilution of 1:500 was subsequently used.
First step. Cells were grown on cover slips, fixed 5 h p.i., and anti-Rep mAb 76.3 was applied as outlined above. After three washings in PBS (see above) 150 µl of PBS containing 1% BSA and a 1:500 dilution of Cy3-conjugated goat F(ab) fragments of anti-mouse IgG(H+L) (Dianova) were added for 15 min at room temperature followed by three washings in PBS.
Second step. A 1:10 dilution of the 39S anti-ICP8 hybridoma supernatant was prepared in PBS containing 1% BSA. Cover slips were incubated with 150 µl for 15 min at room temperature followed by three washes in PBS (see above) followed by a 15 min incubation with 150 µl of PBS containing 1% BSA and a 1:100 dilution of an Alexa 350-conjugated goat anti-mouse IgG (Molecular Probes).
Rep was visualized with a 575–640 nm filter (BP 546, FT 560). ICP8 was visualized using a LP 397 filter (BP 365 FT 395). The choice of fluorophores and filters allowed complete separation of fluorescence channels. The data displayed as Supplementary Material Figure 5 show that the double staining protocol allowed complete discrimination of the two antibody stainings. Colocalization of Rep and ICP8 was analyzed after 3D reconstitution of deconvolved two-channel images (see legends of Fig. 2, and Fig. 5 of Supplementary Material). For each data set, 10 individual cells were analyzed for colocalization.
Microscopy and imaging
Image acquisition was performed with a motorized Zeiss Axiophot 2 widefield microscope equipped with a Zeiss 63×/1.4 NA Oil DIC objective, a 1.0–2.5× Optovar, and a Princeton Instruments ‘Micromax’ cooled (–15°C) slow-scan CCD camera (Kodak KAF-1400 chip). For acquisition of 3D image stacks a PIFOC objective z-stepper was driven by the piezo-amplifier E662 LVPZT (Physik Instrumente, Germany). The microscopic set-up was integrated and controlled using the scripting feature of the IPLab for Macintosh software (version 3.2, Scanalytics, Fairfax). All 3D image stacks were acquired using the optimal sampling density derived from the optical set-up and the respective fluorophor (z-step size ≥15 × 300 nm). For each recording situation the point-spread function of the microscope was measured using fluorescent 0.17 µm microspheres (PS-Speck Microscope Point Source Kit, Molecular Probes). In this way the complete experimental set-up was calibrated, and chromatic aberrations or pixel shifts caused by the fluorophores and/or the microscope were amended. Following image acquisition, the raw data were exported to the Huygens System software (version 2.1.8, Scientific Volume Imaging B.V., Hilversum, The Netherlands) and digital deconvolution was performed using 50 iterations of the ‘iterative constrained Tikhonov-Miller regularized inversion method’ (ICTM) algorithm. The restored image data set was visualized and processed with the Imaris software package (versions 2.6.8 or 3.0 for IRIX, Bitplane AG, Zurich, Switzerland; http://www.bitplane.com). Restored 3D images were analyzed with the colocalization software package (version 1.0 for IRIX, Bitplane AG) or ImarisColoc (version 4.0 for Windows2000, Bitplane AG). For details see figure legends and Demandolx and Davoust (28–30) and Rovere et al. (31). The summed 3D image data were finally exported as TIFF files for assembly in Adobe Photoshop 5.02.
Purification of GST–ICP8 fusion protein
Escherichia coli strain BL21 was transformed with pGEX-ICP8, grown at 30°C to an OD600 nm of 0.6–0.8, induced with 0.2 mM IPTG for 3 h. Cells were harvested by centrifugation (2500 g, 10 min, 4°C) and sonified in lysis buffer [50 mM phosphate pH 7.8, 300 mM NaCl, 1% (v/v) Triton X-100, 0.1 mM PMSF]. Cell debris was removed by centrifugation (22 000 g, 10 min, 4°C) and the supernatant containing GST–ICP8 fusion protein was adsorbed to glutathione–Sepharose beads (Pharmacia) for 2 h at 4°C. The beads were collected by centrifugation (2500 g, 2 min, 4°C) and washed four times with lysis buffer.
GST pull-down assays
35S-Labeled Rep78 was generated by coupled in vitro transcription/translation (Promega) in rabbit reticulocyte lysates with T7 RNA polymerase in the presence of 35S-methionine and 35S-cysteine according to the supplier’s instructions. Equal proportions of labeled proteins were incubated with 5 µg of either GST alone or GST–ICP8 fusion protein bound to 20 µl glutathione–Sepharose beads for 2 h at 4°C in 1 ml binding buffer (25 mM HEPES KOH pH 7.5, 2% glycerol, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 12.5 µg/µl bovine serum albumin, 0.01% Nonidet P-40 and 0.1 mM PMSF). For binding studies in the presence of DNA, a 3466 bp PvuI/AseI restriction fragment from pTR-UF5 (32) was used, with and without heat denaturation. Beads were washed five times in 1 ml of binding buffer, boiled in SDS sample buffer and loaded on a 10% polyacrylamide gel. 35S-Labeled Rep78 was visualized by autoradiography and quantified by phosphoimager analysis (Molecular Dynamics).
RESULTS
Construction of a recombinant HSV expressing AAV genes
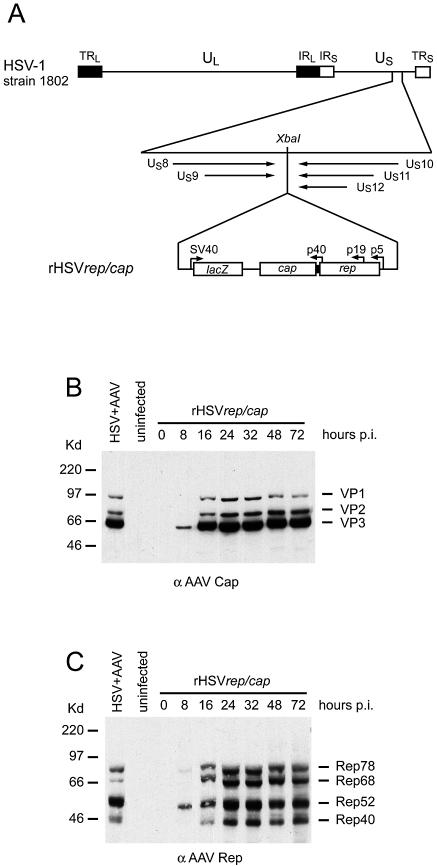
To study the interaction of HSV helper functions and AAV proteins at the single-cell level in the absence of replicating AAV DNA, a recombinant HSV was constructed that expressed the AAV promoter-driven rep/cap genes devoid of flanking ITRs, as outlined in Figure 1. To monitor AAV gene expression, extracts of rHSVrep/cap-infected BHK cells were analyzed on western blots with either anti-Rep mAb 303.9, or with anti-Cap mAb B1. Rep and Cap expression levels remained stable up to 72 h after rHSVrep/cap infection and are comparable with those of AAV replicating as free episome in the presence of HSV (Fig. 1).
Figure 1.
Construction of rHSVrep/cap. (A) The cloning strategy into the single XbaI site at nucleotide position 143.969 of HSV-1, strain 1802 flanked by non-essential HSV genes is depicted. The AAV-2 rep and cap gene expression cassette (nucleotides 191–4485) includes the AAV promoters and polyA+ signal, and is linked to an SV40 controlled lacZ gene. LacZ positive HSV clones were plaque purified and analyzed for AAV Rep and Cap expression. Lysates of rHSVrep/cap-infected cells were prepared at different times p.i. Samples equaling 1 × 104 cells were loaded per lane. Western blots were reacted with the indicated mAb, followed by peroxidase-coupled second antibody and enhanced chemiluminescence. (B) AAV capsid (VP) proteins analyzed with mAb B1. (C) AAV Rep detected with mAb 303.9. HSV + AAV infected cell extracts were prepared at 12 h p.i.
Subcellular localization of Rep in rHSVrep/cap-infected cells
The nuclear localization of Rep in rHSVrep/cap-infected cells and in cells infected with wild-type HSV and AAV was compared by immunofluorescence. Rep-positive nuclei became visible by 3 h p.i. The number of fluorescent cells rapidly increased up to 7 h p.i., when over 90% of the cells were positive (refer to Supplementary Material Fig. 4). The subnuclear distribution of Rep remained relatively homogeneous in rHSVrep/cap-infected cells, which is in agreement with previous reports (19,24). In contrast, cells coinfected with HSV and AAV displayed a more speckled staining pattern with foci of intense Rep staining that increased in size over time (data not shown). This pattern was reminiscent of developing HSV replication centers, where colocalization of ICP8 with other components of the HSV replication complex had been described previously (17,18,33,34). We therefore decided to analyze whether Rep and ICP8 colocalized in these nuclear domains by applying double-label 3D immunofluorescence microscopy and quantitative colocalization analysis.
Colocalization of AAV Rep and HSV ICP8 in HSV replication centers requires the presence of AAV genomes
Colocalization of Rep and ICP8 was analyzed at 5 h p.i. as outlined in the Materials and Methods. 3D-immunofluorescence with quantitative colocalization was performed by the aquisition of Z-axis stacks of the images in either fluorescence channel. After correction for bleaching, the 3D images were restored from the data sets, as described in the Materials and Methods. Thus, a quantitative and valid measure of the spatial distribution of fluorescence intensity in nuclei was obtained. The colocalization hypothesis was then tested under stringent criteria and the result of these calculations is graphically displayed in Figure 2 (colocalization). The fraction of green channel volume colocalized with red channel volume and the Pearson channel correlation in colocalized volume were calculated for a representative number of nuclei (Fig. 2). In cells coinfected with HSV and AAV a speckled immunofluorescence pattern indicative of HSV replication centers was displayed by ICP8, and Rep perfectly colocalized to ICP8 (Fig. 2, AAV + HSV). A similar ICP8 staining pattern was also seen in rHSVrep/cap-infected cells, but the Rep pattern was diffuse, with occasional small colocalizing spots. This difference in localization was surprising. The only obvious difference between an infection with rHSVrep/cap, which carries the entire AAV genome devoid of the ITRs, and a coinfection of HSV and of AAV was the presence of single-stranded AAV template DNA, flanked by the hairpin-structured AAV-ITRs. Therefore, we decided to test whether the introduction of DNAs flanked by AAV-ITRs into rHSVrep/cap-infected cells would change the nuclear localization pattern of Rep. Cells were transfected with a cloned AAV vector DNA (pTR-UF5), that carried a (green fluorescent protein) gfp/neo expression cassette flanked by AAV-ITRs, as the only AAV-derived sequence elements (32). DNA transfection of cloned vector DNA rigorously excluded the presence of AAV wild-type virus and is routinely used as a standard procedure to initiate AAV replication and Rep78/68-dependent ssDNA formation. Controls showed that Rep expressed from rHSVrep/cap led to pTR-UF5 DNA replication and ssDNA formation in transfected cells (data not shown). As anticipated, a change in the nuclear localization pattern of Rep expressed from rHSVrep/cap was seen in the presence of transfected pTR-UF5 DNA. Rep moved to the nuclear sites where ICP8 was accumulating and was found to colocalize with ICP8 (Fig. 2; panel: rHSVrep/cap and recombinant AAV). Controls with rHSVrep/cap-infected cells transfected with a GFP/neo construct devoid of AAV-ITRs did not show redistribution of Rep. Additional controls were performed to corroborate the data (refer to Supplementary Material Fig. 5). In summary, upon rHSVrep/cap infection the mere presence of AAV-ITR-flanked DNA led to a profound nuclear redistribution of Rep resulting in subnuclear colocalization with ICP8.
Figure 2.
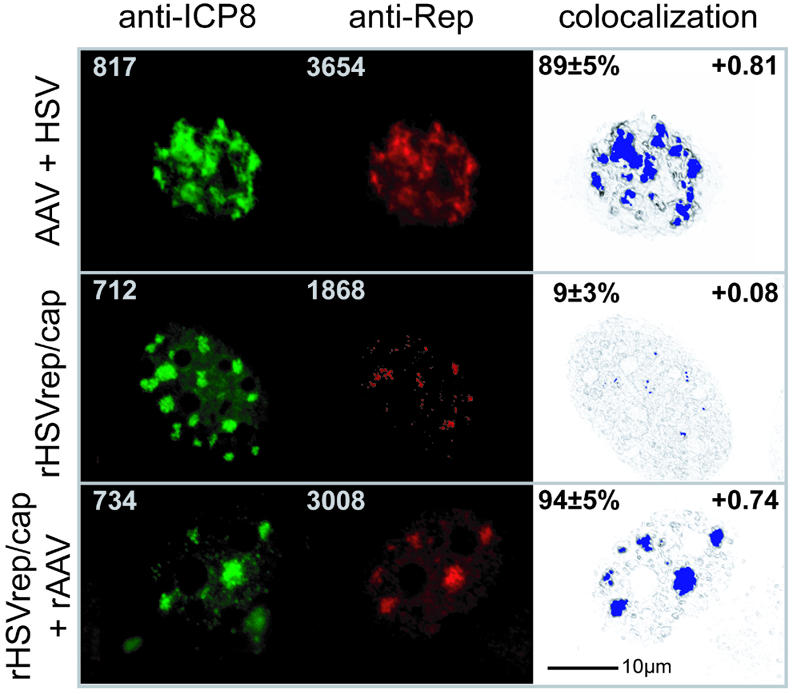
Subnuclear colocalization of AAV Rep and HSV ICP8. BHK cells were grown on coverslips, infected as indicated and fixed at 5 h p.i. Anti-Rep hybridoma 76.3 reactivity was visualized with Cy3-conjugated F(ab)-fragments of goat anti mouse IgG in the ‘red’ channel. The anti-ICP8 hybridoma 39S was applied and detected with Alexa 350-conjugated goat anti-mouse IgG in the ‘blue’ channel. Since blue color poorly reproduces on printed color panels, the Alexa 350 staining is represented as green false color (anti-ICP8 panel). For 3D-colocalization analysis the raw data set was restored using the ICTM algorithm of the Huygens 2 software (see Materials and Methods). The digitally deconvolved 2-channel 3D image was exported to the colocalization 1 software and to ImarisColoc for quantitative analysis, respectively. Following channel definition, a 2D histogram was calculated and a polygonal region was selected. This region was chosen by creating a selection proposal from the AAV + HSV data set. The threshold values and the histogram region were stored and applied to all data sets. After calculating the colocalization map, the colocalization voxels were displayed in blue color. The standard deviation of the intensities of all colocalization voxels in a map was found to be ≤8%. To facilitate navigation within the image the highlighted colocalization voxels were merged to a summed image (colocalization panel). Values given in the upper left edges of green and red panels are maximum fluorescence intensities measured within the raw 3D data set. The percent of green channel volume colocalized with red was calculated in ImarisColoc (Bitplane AG) for n = 6 nuclei and is given as mean ± SD in the upper left edges of the colocalization panel. The Pearson channel correlation in colocalized volume is given in the upper right edges of the colocalization panel (1 is perfect colocalization, 0 no correlation and –1 perfect inverse correlation). Recombinant AAV is transfected pTR-UF5 DNA.
Interaction of AAV Rep78 and HSV ICP8 in vitro and enhancement by ssDNA
The in vivo colocalization analysis was suggestive of an interaction of AAV Rep and HSV ICP8 in dependence of AAV-ITR-flanked, ssDNA genomes. To provide independent evidence for this assumption, the interaction of ICP8 and Rep was analyzed in vitro. GST-tagged ICP8 was purified and tested for binding of in vitro-translated Rep78 in GST pull-down assays. A weak but specific interaction of ICP8 and Rep78 could be seen repeatedly (Fig. 3A, lane 3). This interaction was not affected by the addition of increasing amounts of gel-purified double-stranded AAV vector DNA that comprised the 145 bp AAV-ITR sequences at either end. In contrast, AAV vector DNA in the ssDNA conformation enhanced ICP8–Rep78 interaction >10-fold (Fig. 3A, lanes 5 and 9; for quantification see B). ssDNA templates were generated by heat denaturation followed by snap cooling on ice. Under these conditions AAV ITR-flanked DNA adopts a ssDNA conformation flanked by 145 bp hairpin-shaped AAV-ITRs (35). The finding of a direct interaction of ICP8 and Rep in the absence of DNA (Fig. 3A, lane 3) in conjunction with ICP8’s known strong, cooperative and non-specific ssDNA-binding activity suggested the formation of a tripartite complex. This was supported by the 5-fold enhancement of the ICP8–Rep78 interaction in the presence of ssDNA devoid of AAV-ITRs and Rep-binding sites (Fig. 3C and D; +/–RBS). The even stronger interaction in the presence of ssDNA including the AAV-ITRs is likely due to Rep binding, since site-specific Rep78/68 binding to the double-stranded RBS in the hairpin-shaped AAV-ITRs has been recognized as an important step for AAV DNA replication in vivo (35,36). In summary, the AAV single-strand-dependent colocalization of Rep and ICP8 in vivo in infected cells is reflected by the demonstration of a direct in vitro interaction of purified ICP8 and Rep78 and its strong stimulation by AAV ssDNA.
Figure 3.
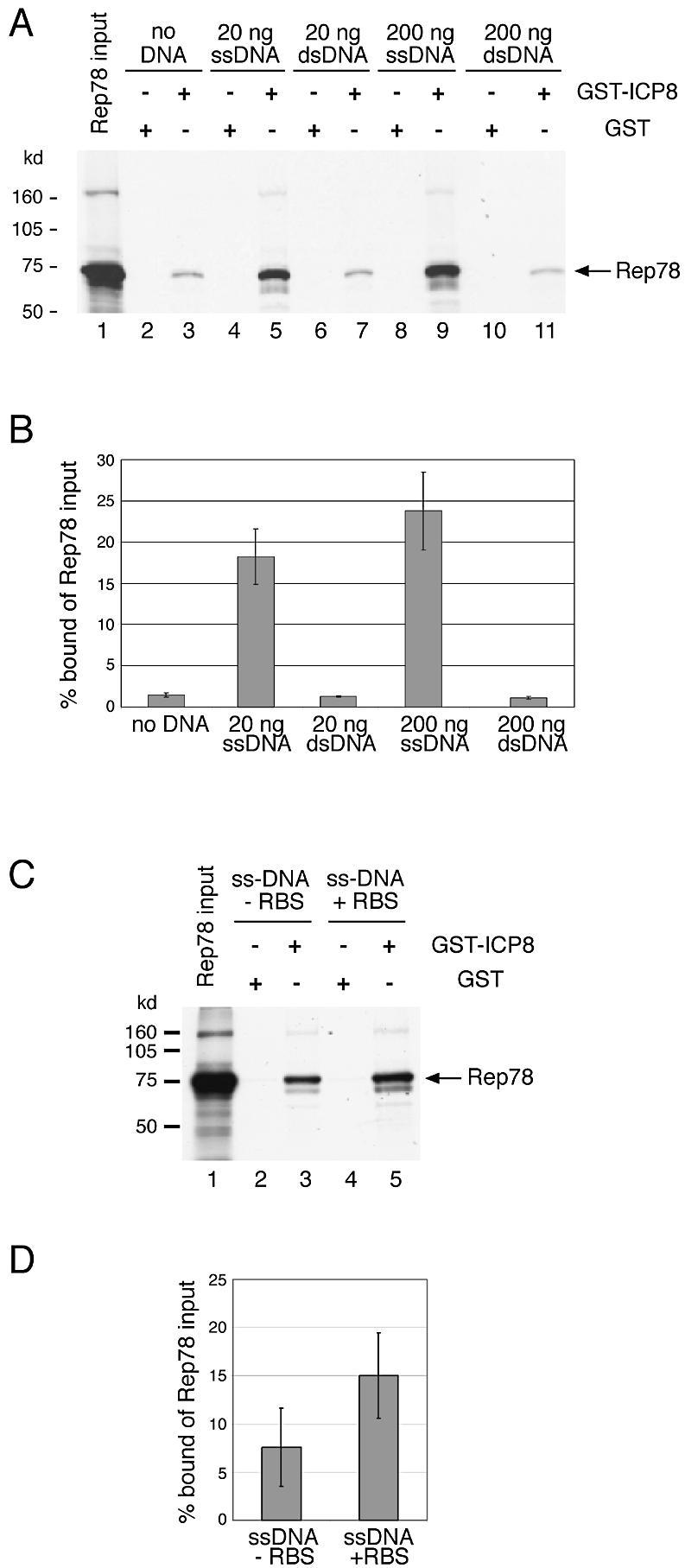
AAV ssDNA-dependent in vitro interaction of AAV Rep and HSV ICP8. In vitro-translated, 35S-labeled Rep78 was incubated with either GST–ICP8 or GST alone. Protein–protein interaction was studied either alone, or in the presence of varying amounts of gel-purified AAV template DNAs. The DNA was either in the double-stranded (ds) conformation, or was heat-denatured to adopt a single-stranded conformation, as indicated. (A and C) Autoradiographies of SDS–PAGE after separation of GST-pull-down products; +RBS, ssDNA flanked by AAV-ITRs with Rep-binding sites. –RBS, ssDNA without ITRs and deleted internal RBS homology sequences. Lanes 1 represent an aliquot of in vitro translated Rep78 alone. The faint high molecular weight band appears to represent multimerized Rep78. (B and D) Phosphoimager quantification of three separate experiments as displayed in (A) and (C).
DISCUSSION
In this report we use a novel recombinant HSV for coordinate expression of HSV and AAV gene products in the absence of single-stranded AAV DNA. This allowed us to analyze the kinetics and subnuclear distribution of HSV ICP8 and AAV Rep in the absence of replicating AAV DNA and to compare it with coinfections where both AAV and HSV DNA are replicating in parallel. The following observations were made. (i) Rep colocalizes perfectly with ICP8 in nuclear HSV replication compartments upon coinfection of AAV and HSV. (ii) Colocalization of Rep in replication compartments is dependent on the presence of single-stranded AAV DNA. (iii) Purified Rep78 and ICP8 interact in vitro. (iv) This interaction is strongly enhanced by single-stranded AAV vector DNA, whereas double-stranded DNA is ineffective. Taken together, these findings suggest that ICP8 and Rep form a tripartite complex with ssDNA. It is assumed that these interactions redirect Rep and the incoming single-stranded AAV genomes to the nuclear HSV replication domains, the assumed site for AAV DNA replication by the HSV replication machinery.
This is the first report to show that AAV Rep colocalizes and directly interacts with a component of the HSV replication complex that we identified before as being required for productive AAV replication (6). Although Rep has been shown to interact with a variety of cellular proteins (37,38) the direct interaction with the product of genetically identified HSV or adenovirus helper functions has as yet not been described. The strong enhancement of ICP8 and Rep interaction by ssDNA was unexpected for the following reasons: when expressed alone either of the two interacting proteins displayed a diffuse and uncharacteristic nuclear staining pattern (17–19,24,39,40). In addition, the diffuse staining pattern of Rep did not change, either during the accumulation of ICP8 in nuclear replication compartments upon HSV infection of Rep-expressing cells (unpublished observation), or upon infection with a Rep-expressing ICP27-defective HSV mutant (1). Here we show that the nuclear localization of Rep is highly dynamic in dependence of ICP8 and of single-stranded AAV template DNA.
Possible roles of HSV replication compartments for AAV replication
HSV DNA replication leads to a profound change in the nuclear architecture. HSV DNA accumulates adjacent to nuclear domains, called ND10 where HSV transcription and DNA replication initiate in close proximity to these domains (41,42). Upon HSV infection, ICP8 quickly accumulates in small, punctate nuclear pre-replicative sites adjacent to ND10 domains (33,39,43,44). These eventually fuse to HSV replication compartments where ICP8 was shown to colocalize to HSV DNA and to the other components of the HSV replication complex (18,33,34). Co-expression of the seven HSV replication genes together with an HSV origin was shown to be sufficient for the formation of mature HSV replication compartments (33,34). Furthermore, a subassembly of four proteins consisting of ICP8 and the helicase–primase complex was sufficient for the formation of pre-replication foci (33,45). We showed previously, that a complex of exactly these four proteins is sufficient for AAV replication (6). The recent finding that ICP8 in conjunction with the helicase–primase complex was able to catalyze DNA strand exchange and homologous DNA recombination in vitro (46), may help to explain the resolution of AAV DNA replication intermediates in HSV infected cells. AAV DNA replication is dependent on Rep78/68 binding to the AAV origin, similarly as is HSV DNA replication dependent on UL9-binding to the HSV origins. UL9 contacts ICP8, which then covers the unwound DNA strands of the nascent replication bubble (14). The direct interaction of ICP8 and Rep78 shown in this study provides evidence for the assumption that the AAV origins can serve as heterologous replicons. We envision, that template bound Rep, in analogy to UL9 can recruit ICP8 and thereby attract the required components of the HSV replication complex.
In adenovirus-infected cells single-stranded adenovirus-DNA was shown to colocalize with the adenovirus ssDBP. In contrast to HSV, the adenovirus nuclear replication centers were shown to be spatially separated from the sites of adenovirus transcription (47). ssDBP constitutes one of the adenovirus helper functions for AAV replication, whereas the other helper functions are transcription or translation factors (2,48). In cells coinfected with AAV and adenovirus, ssDBP was shown to display a nuclear immunofluorescence pattern similar to that of AAV Rep. Further analysis was however hampered by the fact that high levels of Rep78 severely inhibited the formation of mature adenovirus replication centers (49). This finding may reflect the recent description of a Rep78 domain that inhibits adenovirus replication by a protein kinase A-dependent mechanism (50). It will be interesting to see how comparable the roles of ssDBP of adenovirus and HSV ICP8 are during AAV replication.
Biochemical activities of ICP8 and Rep
The apparent binding constant (Kα) of ICP8 on ssDNA has been reported to lie in the range of 106 M–1 (51,52). ICP8 fully coats ssDNA templates, as demonstrated by EM analysis (16,53) with the C-terminal 60 amino acids shown to be essential for cooperativity (52). Both EM analysis and in vitro binding studies showed that Rep78/68 binds site-specifically to the double-stranded RBS of the hairpin-structured AAV-ITRs, and to the homologous sequence on human chromosome 19 (8,35,54,55). This opens the possibility that ICP8 covers ssAAV DNA without competing with Rep, which binds site-specifically to the double-stranded, hairpin- structured AAV-ITRs. Since quantitative Rep binding data on various DNA templates comparable with those of ICP8 are not available, a possible contribution of Rep ssDNA binding cannot be excluded. Alternatively, a tripartite complex on ssDNA devoid of ITRs may be formed via direct Rep–ICP8 interaction further enhanced by ITR-specific binding of Rep. The analysis of the interactions of AAV-Rep with components of the HSV replication complex will add to our understanding of how the heterologous AAV origins can be used as templates for the HSV replication complex. The detailed understanding of these mechanisms will also help to optimize packaging systems for recombinant AAV vectors based on engineered HSV-strains, one of the most promising tools for efficient AAV vector production.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. Theimer and E. Hammer for expert technical help, R. Joncker for secretarial assistance, J. Kleinschmidt for AAV antibodies, N. Mucyczka for plasmid pTR-UF5 and J. McLaughlin for HSV strains. We are grateful to T. Cremer for critical reading of the manuscript and helpful comments on the imaging experiments. The work was supported by grants from the Bundesministerium für Forschung und Technologie (BMBF), and the Deutsche Forschungsgemeinschaft (DFG-SFB 506) to R.H. and grants Bo1100, En305 and SFB190 from the Deutsche Forschungsgemeinschaft to M.B. and M.E.
REFERENCES
- 1.Conway J., Rhys,C., Zolotukhin,I., Zolotukhin,S., Muzyczka,N., Hayward,G. and Byrne,B. (1999) High-titer recombinant adeno-associated virus production utilizing a recombinant herpes simplex virus type I vector expressing AAV-2 rep and cap. Gene Ther., 6, 986–993. [DOI] [PubMed] [Google Scholar]
- 2.Muzyczka N. and Berns,K.I. (2001) Parvoviridae: the viruses and their replication. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott, Philadelphia, PA, Vol. 2, pp. 2327–2359. [Google Scholar]
- 3.Kotin R.M., Siniscalco,M., Samulski,R.J., Zhu,X.D., Hunter,L., Laughlin,C.A., McLaughlin,S., Muzyczka,N., Rocchi,M. and Berns,K.I. (1990) Site-specific integration by adeno-associated virus. Proc. Natl Acad. Sci. USA, 87, 2211–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samulski R.J., Zhu,X., Xiao,X., Brook,J.D., Housman,D.E., Epstein,N. and Hunter,L.A. (1991) Targeted integration of adeno-associated virus (AAV) into human chromosome 19 [published erratum appears in EMBO J. (1992), 11, 1228]. EMBO J., 10, 3941–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hüser D., Weger,S. and Heilbronn,R. (2002) Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR. J. Virol., 76, 7554–7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weindler F.W. and Heilbronn,R. (1991) A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol., 65, 2476–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward P., Falkenberg,M., Elias,P., Weitzman,M. and Linden,R.M. (2001) Rep-dependent initiation of adeno-associated virus type 2 DNA replication by a herpes simplex virus type 1 replication complex in a reconstituted system. J. Virol., 75, 10250–10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Im D.-S. and Muzyczka,N. (1990) The AAV origin-binding protein Rep68 is an ATP-dependent site-specific endonuclease with helicase activity. Cell, 61, 447–457. [DOI] [PubMed] [Google Scholar]
- 9.Snyder R.O., Im,D.-S., Ni,T., Xiao,X., Samulski,R.J. and Muzyczka,N. (1993) Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J. Virol., 67, 6096–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiorini J.A., Wiener,S.M., Owens,R.A., Kyöstiö,S.R.M., Kotin,R.M. and Safer,B. (1994) Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J. Virol., 68, 7448–7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehman I.R. and Boehmer,P.E. (1999) Replication of herpes simplex virus DNA. J. Biol. Chem., 274, 28059–28062. [DOI] [PubMed] [Google Scholar]
- 12.Ruyechan W.T. (1983) The major herpes simplex virus DNA-binding protein holds single-stranded DNA in an extended configuration. J. Virol., 46, 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C.K. and Knipe,D.M. (1985) An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J. Virol., 54, 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S.S. and Lehman,I.R. (1997) Unwinding of the box I element of a herpes simplex virus type 1 origin by a complex of the viral origin binding protein, single-strand DNA binding protein and single-stranded DNA. Proc. Natl Acad. Sci. USA, 94, 2838–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gac N.T.L., Villani,G., Hoffmann,J.S. and Boehmer,P.E. (1996) The UL8 subunit of the herpes simplex virus type-1 DNA helicase-primase optimizes utilization of DNA templates covered by the homologous single-strand DNA-binding protein ICP8. J. Biol. Chem., 271, 21645–21651. [DOI] [PubMed] [Google Scholar]
- 16.Ruyechan W.T. and Weir,A.C. (1984) Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J. Virol., 52, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Bruyn Kops A. and Knipe,D.M. (1988) Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA-binding protein. Cell, 55, 857–868. [DOI] [PubMed] [Google Scholar]
- 18.Liptak L.M., Uprichard,S.L. and Knipe,D.M. (1996) Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol., 70, 1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wistuba A., Kern,A., Weger,S., Grimm,D. and Kleinschmidt,J.A. (1997) Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol., 71, 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rixon F.J. and McLaughlin,J. (1993) Herpes simplex virus vectors. In Davidson,A.J. and Elliot,R.M. (eds), Molecular Virology: A Practical Approach. Oxford University Press, Oxford, pp. 285–307. [Google Scholar]
- 21.Samulski R.J., Chang,L.-S. and Shenk,T. (1987) A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol., 61, 3096–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilbronn R. and zur Hausen,H. (1989) A subset of herpes simplex replication genes induces DNA amplification within the host cell genome. J. Virol., 63, 3683–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E. and Lane,D. (1988) Antibodies, A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Kleinschmidt J.A., Möhler,M., Weindler,F. and Heilbronn,R. (1995) Sequence elements of the adeno-associated virus rep-gene required for suppression of herpes-simplex virus induced DNA amplification. Virology, 206, 254–262. [DOI] [PubMed] [Google Scholar]
- 25.Wessel G.M. and McClay,D.R. (1986) Two embryonic, tissue-specific molecules identified by a double-label immunofluorescence technique for monoclonal antibodies. J. Histochem. Cytochem., 34, 703–706. [DOI] [PubMed] [Google Scholar]
- 26.Lewis Carl S.A., Gillete-Ferguson,I. and Ferguson,D.G. (1993) An indirect immunofluorescence procedure for staining the same cryosection with two mouse monoclonal primary antibodies. J. Histochem. Cytochem., 41, 1273–1278. [DOI] [PubMed] [Google Scholar]
- 27.Negoescu A., Labat-Moleur,F., Lorimier,P., Lamarcq,L., Guillermet,C., Chambaz,E. and Brambilla,E. (1994) F(ab) secondary antibodies: a general method for double immunolabeling with primary antisera from the same species. Efficiency control by chemiluminescence. J. Histochem. Cytochem., 42, 433–437. [DOI] [PubMed] [Google Scholar]
- 28.Demandolx D. and Davoust,J. (1995) Multicolor analysis in confocal immunofluorescence microscopy. J. Trace Microprobe Tech., 13, 217–225. [Google Scholar]
- 29.Demandolx D. and Davoust,J. (1997) Multicolour analysis and local image correlation in confocal microscopy. J. Microsc., 185, 21–36. [Google Scholar]
- 30.Demandolx D. and Davoust,J. (1997) Multiparameter image cytometry: from confocal micrographs to subcellular fluorograms. Bioimaging, 5, 159–169. [Google Scholar]
- 31.Rovere P., Zimmermann,V.S., Forquet,F., Demandolx,D., Trucy,J., Ricciardi-Castagnoli,P. and Davoust,J. (1998) Dendritic cell maturation and antigen presentation in the absence of invariant chain. Proc. Natl Acad. Sci. USA, 95, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolotukhin S., Potter,M., Hauswirth,W.W., Guy,J. and Muzyczka,N. (1996) A ‘humanized’ green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol., 70, 4646–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukonis C.J. and Weller,S.K. (1997) Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol., 71, 2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong L. and Hayward,G.S. (1997) Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol., 71, 3146–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Im D.-S. and Muzyczka,N. (1989) Factors that bind to adeno-associated virus terminal repeats. J. Virol., 63, 3095–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder R.O., Samulski,R.J. and Muzyczka,N. (1990) In vitro resolution of covalently joined AAV chromosome ends. Cell, 60, 105–113. [DOI] [PubMed] [Google Scholar]
- 37.Weger S., Wendland,M., Kleinschmidt,J. and Heilbronn,R. (1999) The adeno-associated virus type 2 regulatory proteins Rep78/Rep68 interact with the transcriptional coactivator PC4. J. Virol., 73, 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiorini J.A., Zimmermann,B., Yang,L., Smith,R.H., Ahearn,A., Herberg,F. and Kotin,R.M. (1998) Inhibition of PrKX, a novel protein kinase and the cyclic AMP-dependent protein kinase PKA by the regulatory proteins of adeno-associated virus type 2. Mol. Cell. Biol., 18, 5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukonis C.J., Burkham,J. and Weller,S.K. (1997) Herpes simplex virus type 1 prereplicate sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol., 71, 4771–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Bruyn Kops A., Uprichard,S.L., Chen,M. and Knipe,D.M. (1998) Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology, 252, 162–178. [DOI] [PubMed] [Google Scholar]
- 41.Maul G.G., Ishov,A.M. and Everett,R.D. (1996) Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology, 217, 67–75. [DOI] [PubMed] [Google Scholar]
- 42.Ishov A.M. and Maul,G.G. (1996) The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol., 134, 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uprichard S.L. and Knipe,D.M. (1997) Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology, 229, 113–125. [DOI] [PubMed] [Google Scholar]
- 44.Burkham J., Coen,D.M. and Weller,S.K. (1998) ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol., 72, 10100–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrington-Lawrence S.D. and Weller,S.K. (2003) Recruitment of polymerase to herpes simplex virus type 1 replication foci in cells expressing mutant primase (UL52) proteins. J. Virol., 77, 4237–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nimonkar A.V. and Boehmer,P.E. (2002) In vitro strand exchange promoted by the herpes simplex virus type-1 single strand DNA-binding protein (ICP8) and DNA helicase-primase. J. Biol. Chem., 277, 15182–15189. [DOI] [PubMed] [Google Scholar]
- 47.Pombo A., Ferreira,J., Bridge,E. and Carmo-Fonseca,M. (1994) Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J., 13, 5075–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson W.D. and Westphal,H. (1981) A cascade of adenovirus early functions is required for expression of adeno-associated virus. Cell, 27, 133–141. [DOI] [PubMed] [Google Scholar]
- 49.Weitzman M.D., Fisher,K.J. and Wilson,J.M. (1996) Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol., 70, 1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Pasquale G. and Chiorini,J.A. (2003) PKA/PrKX activity is a modulator of AAV/adenovirus interaction. EMBO J., 22, 1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dudas K.C. and Ruyechan,W.T. (1998) Identification of a region of the herpes simplex virus single-stranded DNA-binding protein involved in cooperative binding. J. Virol., 72, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mapelli M., Mühleisen,M., Persico,G., van der Zandt,H. and Tucker,P.A. (2000) The 60-residue C-terminal region of the single-stranded DNA binding protein of herpes simplex virus type 1 is required for cooperative DNA binding. J. Virol., 74, 8812–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makhov A.M., Boehmer,P.E., Lehman,I.R. and Griffith,J.D. (1996) Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol., 258, 789–799. [DOI] [PubMed] [Google Scholar]
- 54.Weitzman M.D., Kyöstiö,S.R.M., Kotin,R.M. and Owens,R.A. (1994) Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl Acad. Sci. USA, 91, 5808–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young S.M. Jr, McCarty,D.M., Degtyareva,N. and Samulski,R.J. (2000) Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol., 74, 3953–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.